Abstract
Background
Circular RNAs (circRNAs) are regarded as vital regulatory factors in various cancers. However, the biological functions of circDNER in the paclitaxel (PTX) resistance of lung cancer remain largely unexplored.
Methods
Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) was used to analyze circDNER, miR‐139‐5p, and ITGB8. Cell proliferation was assessed via colony formation and MTT assays. Cell apoptosis was evaluated by flow cytometry. Western blot was performed to assess protein expression. The targeted interaction among circDNER, miR‐139‐5p, and ITGB8 were validated using dual‐luciferase reporter or RNA immunoprecipitation assays.
Results
Inhibition of circDNER reduced IC50 of PTX, inhibited cell proliferation, invasion and migration, as well as promoted cell apoptosis in PTX‐resistant lung cancer cells. Mechanistically, circDNER sponged miR‐139‐5p to upregulate ITGB8 expression. Overexpression of miR‐139‐5p reversed the biological functions mediated by circDNER in PTX‐resistant lung cancer cells. MiR‐139‐5p overexpression suppressed PTX resistance and malignant behaviors of PTX‐resistant lung cancer cells, with ITGB8 elevation rescued the impacts. Moreover, we demonstrated that circDNER was upregulated in plasma exosomes from lung cancer patients. The plasma exosomes derived from these patients are the key factors enhancing the migration and invasion potential of lung cancer cells.
Conclusion
The circDNER mediated miR‐139‐5p/ITGB8 axis suppresses lung cancer progression. Our findings suggest that circDNER might act as a potential prognostic biomarker and therapeutic target for lung cancer treatment.
Keywords: circDNER, ITGB8, lung cancer, miR‐139‐5p, paclitaxel
The circDNER mediated miR‐139‐5p/ITGB8 axis suppresses lung cancer progression.
INTRODUCTION
Lung cancer, a major malignancy, is the leading cause of cancer death worldwide. 1 Although many therapies, such as surgery, radiotherapy and chemotherapy, have been applied to treat lung cancer, the 5‐year survival rate remains low due to tumor invasion and metastasis as well as lack of effective biomarkers and targets. 2 Hence, exploring novel approaches for the diagnosis and treatment of lung cancer are urgently required.
Circular RNAs (circRNAs) are a class of noncoding RNAs that possess a covalent closed loop. To date, it has been proven that circRNAs play a vital role and participate in proliferation, invasion, and metastasis in various malignancies, including bladder, pancreatic, and esophageal cancers. 3 Previous studies have shown that circRNAs can regulate tumor pathological progression and may serve as potential diagnostic biomarkers of cancer; 4 , 5 , 6 however, whether circDNER influences lung cancer remains unclear.
Exosomes are membrane vesicles produced by various cells and character with diameters ranging from 30–100 nm. The main functions of exosomes are transferring intercellular signal transduction to recipient cells. 7 , 8 It has been demonstrated that exosomes are involved in modulating drug response, as well as tumorigenesis, tumor invasion and metastasis by delivering micro (mi)RNA, long noncoding (lnc)RNA, circular (circ) RNA and proteins. 9 CircRNA are exceptionally stable in all cargos carried by exosomes. 10 However, the mechanism of circRNAs in the exosomes of lung cancer patients remains to be definitively elucidated.
In this study, circDNER was first identified by circRNA microarray analysis in exosomes from the plasma of lung cancer patients. Our data showed that circDNER was obviously upregulated in lung cancer tissues and plasma exosomes of lung cancer patients. Moreover, we found that circDNER competitively binds to miR‐139‐5p to regulate proliferation, invasion, metastasis and apoptosis via mediating ITGB8 expression in lung cancer. Our findings suggest that circDNER might act as a potential prognostic biomarker and therapeutic target for the treatment of lung cancer.
METHODS
CircRNA microarray analysis
A microarray assay was used to analyze the circRNA expression of lung adenocarcinoma and adjacent noncancerous tissues as well as plasma exosome samples collected from the Department of Thoracic Surgery, The Affiliated Hospital of Jiangnan University.
Clinical specimens
All human lung cancer tissue samples and plasma samples were obtained from the Department of Thoracic Surgery, The Affiliated Hospital of Jiangnan University. The tumors were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. The plasma samples were stored at −80°C. Written informed consent was obtained from all patients and this study was approved by the ethics committee of Affiliated Hospital of Jiangnan University.
Exosome isolation
All the samples were prepared by centrifuging for 5 min at 500 g. The supernatant was centrifuged at 10 000 g for 30 min to remove shed macrovesicles. The supernatant was collected and filtered through a 0.22 μm membrane filter. Finally, the supernatant was centrifuged at 110 000 g for 70 min.
Cell culture
The human lung cancer cell lines BEAS‐2B, HCC827, A549, H1975, H1299 and H460 were purchased from GeneChem. BEAS‐2B, HCC827, A549 and H1975 cells were cultured in RPMI‐1640 containing 10% FBS and 1% penicillin–streptomycin (Gibco). H1299 and H460 cells were cultured in DMEM (Gibco) containing 10% FBS and 1% penicillin–streptomycin. All cells were incubated at 37°C in an atmosphere of 5% CO2.
Cell transfection
Plasmids containing sh‐circDNER and sh‐NC, circDNER or ITGB8 overexpression vector, miR‐139‐5p mimics and miR‐NC, miR‐139‐5p inhibitor and anti‐miR‐NC were designed and transfected into lung cancer cells for up‐ or downregulating target genes according to the manufacturer's protocol. All plasmids were synthesized by GeneChem.
RNA isolation and qRT‐PCR
The total RNA of samples was isolated by RNAiso Plus reagent (Takara) according to the manufacturer's protocol. ReverTra Ace qPCR RT Kit (TaKaRa) was used to obtain cDNA. Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) was performed using ABI 7500 StepOnePlus system (Applied Biosystems). β‐actin were used as internal controls and each reaction was performed in triplicate. All primers are shown in Supplemental file 1.
Colony formation, invasion and migration assays
The transfected cells were seeded in a 6‐well plate for 14 days. The colonies were fixed with ethanol (75%, Beyotime), followed by staining with violet (0.1%, Beyotime). Finally, colonies (>50 cells/colony) were counted and photographed.
The transfected cells were resuspended with serum‐free medium and seeded in the transwell upper chamber. Then, 500 μl of DMEM medium supplemented with 10% FBS was added to the transwell lower chamber. After incubation for 48 h, the chambers were stained with 0.5% crystal violet and counted under a Nikon microscope (Minato).
Flow cytometry analysis
The transfected cells were collected and an apoptosis assay was conducted using the annexin V‐FITC/PI apoptosis detection kit (Sangon Biotech) after incubation in the dark for 20 min. Finally, flow cytometry was utilized to quantify cell apoptosis.
MTT assay
The transfected cells were seeded in a 96‐well plate with different concentrations of PTX (0, 200, 400, 600, 800, 1000, 1200 nM) for 24 h. The cell proliferation‐toxicity detection solution (CCK‐8, Tongren Chemical Institute) was added at 10 μl per well, and then incubated at 37°C for 1 h.
Dual‐luciferase reporter assay
The putative binding sequence of miR‐139‐5p and circDNER or ITGB8 was predicted by Circinteractome or TargetScan. Wild‐type (WT) sequencing of circDNER or ITGB8 3′UTR with binding sequence for miR‐139‐5p was cloned into the pmirGLO vector (YouBia) to construct WT‐circDNER or WT‐ ITGB8 3′UTR. At the same time, mutant reporter plasmids (MUT‐circDNER or MUT‐ITGB8 3′UTR) without binding sequence for miR‐139‐5p were generated in the same way. The indicated vector and miR‐NC/miR‐139‐5p were then cotransfected into A549/PTX cells. After 48 h of cotransfection, a dual‐luciferase reporter assay system (Promega) was used to examine luciferase activity.
RNA immunoprecipitation (RIP)
The Magna RIP RNA‐binding protein immunoprecipitation kit (Bersinbio) was used to perform RNA immunoprecipitation. The samples were lysed in lysis buffer followed by incubation with RIP immunoprecipitation buffer. Proteinase K buffer was then added to the samples. After purifying, all RNAs were analyzed by qRT‐PCR.
Western blot (WB)
The samples were lysed in RIPA buffer (Beyotime), separated using 10% SDS‐PAGE, and then transferred onto a nitrocellulose membrane. The membrane was blocked with 5% bovine serum albumin (BSA) for 1 h and then incubated with diluted primary antibodies to CD 63 (1:1000, ab118307), HSP70 (1:1000, ab2787), TSG101 (1:1000, ab125011), ITGB8 (1:2000, ab80673) and GAPDH (1:2000, ab371685) overnight at 4°C. After washing with TBST for three times, the membrane was incubated with secondary antibodies for 35 min at room temperature. The relative levels of proteins were analyzed by calculating the ratio of the gray value.
Tumor xenograft assay
BALB/c (nu/nu, N = 6, male, 5 weeks) mice were selected and randomly divided into two groups. After 1 week, cells (2 × 106) were subcutaneously injected into the right flank of mice. Two weeks later, one group of mice were injected with PBS and PTX, and another group of mice were injected with PTX‐induced exosome and PTX. Six weeks later, the mice were euthanized and the xenograft tumors were surgically resected. The animal experiments were approved by the Institutional Animal Care and Use Committees at Affiliated Hospital of Jiangnan University.
Statistical analysis
GraphPad 7.0 was used for data analysis. The data are shown as mean ± standard deviation (SD). All samples were analyzed by t‐test or one‐way ANOVA with a significance level of p < 0.05.
RESULTS
CircDNER is significantly upregulated in lung cancer tissues and cells, and increased circDNER expression predicts poor prognosis
To explore the effect of circDNER in the pathogenesis of lung cancer, GSE101684 microarray analysis was performed to evaluate numerous dysregulated circRNAs in tumor tissues. The analysis showed that circDNER was significantly upregulated in lung cancer tissue (Figure 1a,b), which indicated circDNER might play a crucial role in the development of lung cancer. At the same time, using qRT‐PCR, we found that circDNER was obviously increased in lung cancer tissue and A549 cells (Figure 1c,d). Higher expression of circDNER resulted in increased mortality (Figure 1e). These results indicated that upregulation of circ DNER may be vital in lung cancer progression.
FIGURE 1.
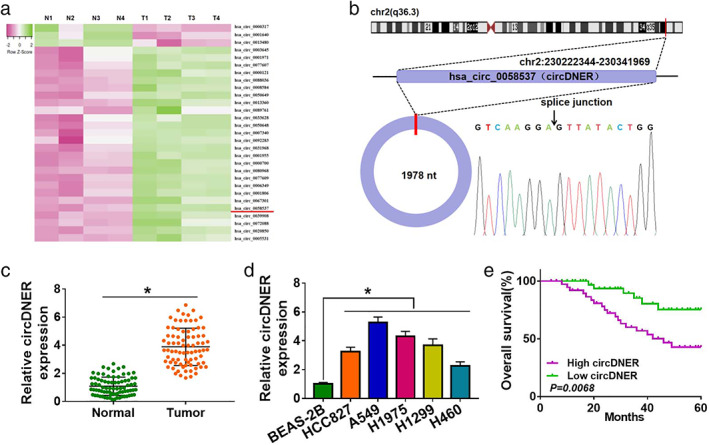
CircDNER expression is upregulated in lung cancer patients. (a) Thermography of GSE101684 microarray with differential circRNAs based on heatmap package. (b) Comparison between circ‐0058537 and circBase databases confirmed by Sanger sequencing. (c) qRT‐PCR was used to detect the circDNER expression in lung cancer tissue. (d) qRT‐PCR was used to test the circDNER expression in lung cancer cells. (e) Kaplan–Meier analysis was used to show the overall survival. Data are presented as the mean ± SD of at least three independent experiments. *p < 0.05
CircDNER could be delivered through exosomes
CircRNA has been widely reported in exosomes which play a significant role in tumorigenesis. 11 We isolated exosomes from PTX‐resistant cell culture media. First, exosomes of A549/PTX cells were observed using transmission electron microscopy (TEM) (Figure 2a). Nanoparticle tracking analysis (NTA) further confirmed that the dominant size of vesicles was 100 nm, and concentrations at 100 nm multiplied by dilution factors produced concentrations of 4.5 × 108/ml for A549 and 9.6 × 108/ml for A549/PTX, respectively (Figure 2b). We examined the exosomal markers CD63, HSP70 and Tsg101 by western blotting. The data showed that higher levels of CD63, HSP70 and Tsg101 were discovered confirming the purity of the exosomes (Figure 2c). Moreover, circDNER expression in A549 cells was strikingly lower than in A549/PTX cells (Figure 2d). In addition, markedly higher expression of circDNER and IC50 values were observed after treatment with exosomes (Figure 2e,f). To investigate the function of circDNER in vivo, a mice xenograft model was established. The data showed that A549 + PTX Exo + PTX group mice promoted tumor growth (Figures 2g–j).
FIGURE 2.
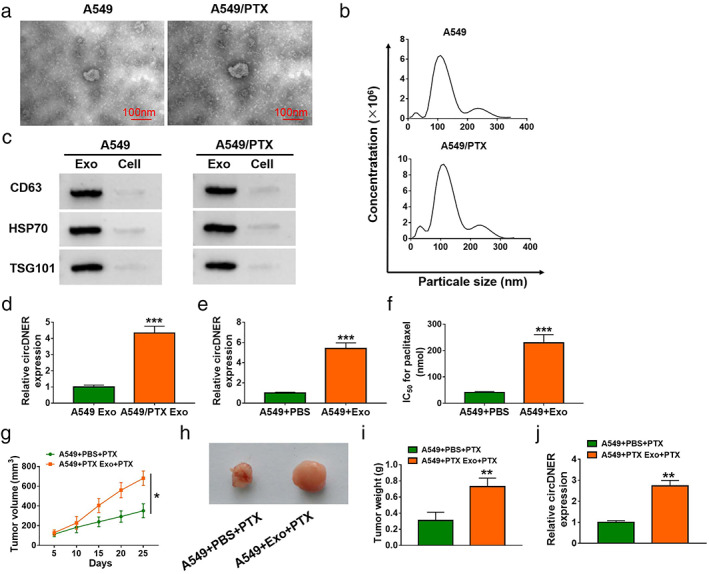
CircDNER could be transferred via exosomes. (a) Exosomes secreted by A549/PTX cells were detected by TEM. (b) Nanoparticle tracking analysis (NTA) showed dominant particle size. (c) WB assay was performed for determination of CD63, HSP70 and TSG101 protein levels in the supernatant or exosomes of A549/PTX cells. (d) CircDNER expression in A549 cells and A549/PTX cells was determined after treating with exosome. (e) CircDNER expression in A549 cells was detected after treating with PBS or exosome. (f) IC50 value was tested in A549 cells after treating with PBS or exosome. (g) Tumor volume was monitored every 3 days. (h) The tumor tissues were measured (six mice per group). (i) The weight of the tumor tissues was measured (six mice per group). (j) CircDNER expression in A549/PTX cells was detected after treating with PBS or exosome. Data are presented as the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
CircDNER plays a vital role in PTX‐resistant lung cancer cells
To further investigate the function of circDNER PTX‐resistant lung cancer, we established the PTX‐resistant A549 cells (A549/PTX) (Figure 3a). Meanwhile, we analyzed the circDNER expression in PTX‐senstive or PTX‐resistant tumor tissue, and found that circDNER expression was increased in PTX‐resistant tumor tissue and cells (Figures 3b,c). To further confirm the effect of circDNER in PTX resistance, we next examined whether knockdown of circDNER would resensitize these cells to PTX treatment. We silenced circDNER expression by transfection of sh‐circDNER. We found that IC50 value of PTX was decreased after knockdown of circDNER (Figure 3d,e).
FIGURE 3.
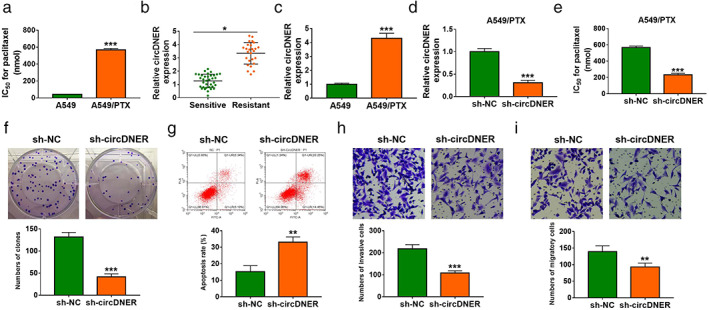
CircDNER knockdown increased PTX sensitivity in A549/PTX. (a) IC50 value of A549 or A549/PTX was examined via CCK‐8 analysis in A549 or A549/PTX cells treated with different concentrations of PTX (0, 200, 400, 600, 800, 1000, 1200 nM) for 24 h. (b and c) circDNER expression was determined in tumor and lung cell lines. (d) circDNER expression was detected after the introduction of sh‐NC or sh‐circDNER. (e) IC50 value of A549/PTX was examined after introducing sh‐NC or sh‐circDNER via CCK‐8 analysis. (f) The number of colonies was calculated after introducing sh‐NC or sh‐circDNER using clone formation assay. (g) Flow cytometry was used to assess cell apoptosis after introducing with sh‐NC or sh‐circDNER. (h and i) A transwell assay was utilized to evaluate cell migration and invasion after introducing with sh‐NC or sh‐circDNER. Data are presented as the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
To demonstrate the effect of circDNER on lung cancer cells, we examined cell proliferation using clone formation assays. Our findings suggested that silencing circDNER could obviously inhibit the proliferation of A549/PTX cells (Figure 3f). Flow cytometry data showed that silencing circDNER promoted cell apoptosis (Figure 3g). A transwell assay was utilized to detect cell invasion and motility. The results were consistent with proliferation, which indicated that downregulation of circDNER markedly restrained cell invasion and motility (Figure 3h,i). Together, these data implicated that circDNER downregulation could resensitize A549/PTX.
CircDNER serves as a miRNA sponge for miR‐139‐5p
It has previously been determined that circRNAs can serve as miRNA sponges to regulate miRNA targets. 12 We next explored the ability of circDNER to bind miRNAs. Our results indicated that the potential interactions between miR‐139‐5p and circDNER by online databases starBase, circNet, and RNA22 (Figure 4a). We observed that miR‐139‐5p upregulation obviously decreased the luciferase activity in the WT‐circDNER group but not in the MUT‐ circDNER group (Figure 4b). Moreover, lower expression of miR‐139‐5p was observed in resistant tissues and A549/PTX cells (Figure 4c,d). To assess the binding capacity of circDNER and miR‐139‐5p, RIP assays were performed which demonstrated that circDNER can bind with miR‐139‐5p (Figure 4e). As shown in Figure 4e, miR‐139‐5p expression was negatively correlated with circ DNER expression in lung cancer tissue (Figure 4f). Furthermore, miR‐139‐5p expression was increased when circDNER was silenced in PTX‐resistant cells. These data concluded that circDNER directly targeted miR‐139‐5p.
FIGURE 4.
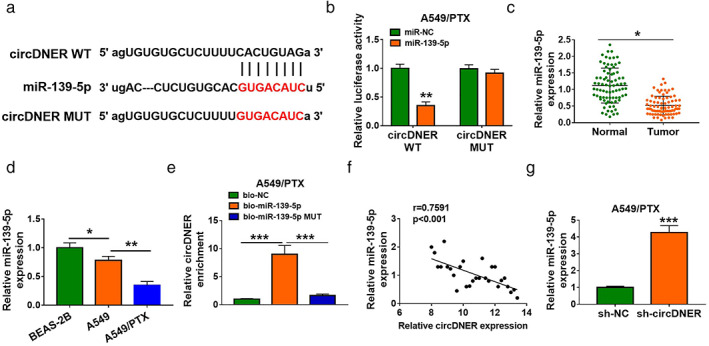
CircDNER functions as a sponge of miR‐139‐5p (a) Schematic of circDNER wild‐type (WT) and mutant (Mut) luciferase reporter vectors. (b) A luciferase reporter assay was performed to verify their targeting relationship in HEK‐293 T cells. The wild‐type binding site construct of circDNER and miR‐139‐5p. (c and d) The expression of miR‐139‐5p was determined in tumor and lung cell lines. (e) The RIP assay results demonstrated that circDNER could form silencing‐induced complexes with miRNA in A549 cells. (f) The association between the levels of miR‐139‐5p and circDNER in PTX‐resistant tissues was analyzed. (g) The abundance of miR‐139‐5p was detected in A549/PTX cells after transfection with sh‐NC, sh‐circDNER. Data are presented as the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Silencing CircDNER enhanced PTX sensitivity in A549/PTX cells by upregulating miR‐139‐5p
To investigate whether miR‐139‐5p mediated the function of sh‐circDNER, rescue experiments were performed in A549/PTX. Higher expression of miR‐139‐5p was observed by downregulating circDNER, which was attenuated by inhibiting miR‐139‐5p (Figure S1). Additionally, we found that miR‐139‐5p inhibition reduced the suppressive effects of knockdown circDNER colony‐forming ability (Figure 5a). Moreover, miR‐139‐5p silencing reversed sh‐circDNER‐triggered cell apoptosis (Figure 5b). Meanwhile, the inhibitory effects of circDNER knockdown on migration and invasion were also counteracted by interference of miR‐139‐5p (Figure 5c,d). In summary, circDNER mediated PTX sensitivity by targeting miR‐139‐5p.
FIGURE 5.
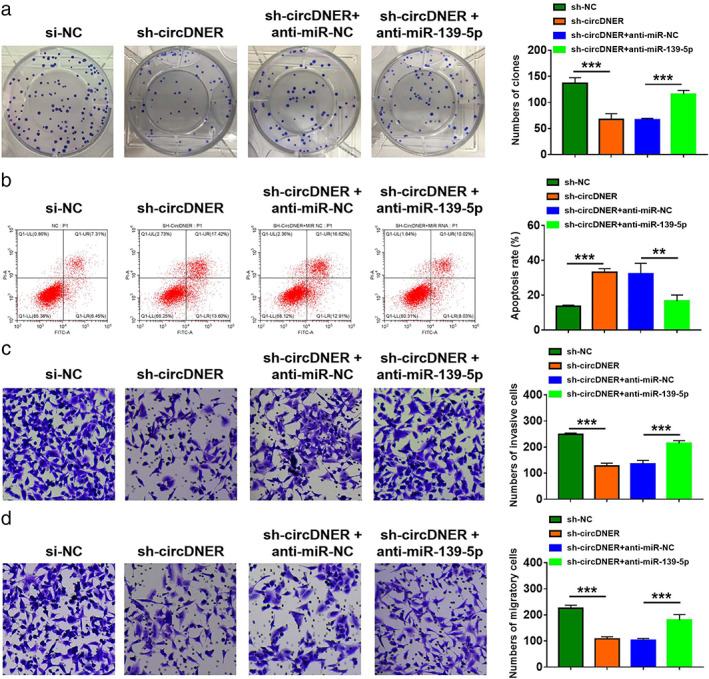
Downregulation of circDNER improved sensitivity in A549/PTX cells by sponging miR‐139‐5p. A549/PTX cells were transfected with sh‐NC, sh‐circDNER, sh‐circDNER + anti‐miR‐NC, or sh‐circDNER + anti‐miR‐139‐5p (a) A colony formation assay was used for assessing colony‐forming ability. (b) Flow cytometry was used to assess cell apoptosis. (c and d) A transwell assay was utilized to evaluate cell migration and invasion. Data are presented as the mean ± SD of at least three independent experiments. **p < 0.01, ***p < 0.001
CircDNER regulates expression of ITGB8 via miR‐139‐5p
It is well known that miRNAs mediate biological functions by directly targeting mRNAs. 13 , 14 We identified five genes that were screened in lung cancer cells transfected with mimic‐miR‐139‐5p and inhibitor‐miR‐139‐5p. The results showed that ITGB8 exhibited the most obvious change after treatment with mimic‐miR‐139‐5p and inhibitor‐miR‐139‐5p Figure S2). Complementary sites between miR‐139‐5p and ITGB8 were observed in Figure 6a, suggesting that miR‐139‐5p might directly target ITGB8. Moreover, we observed that miR‐139‐5p upregulation obviously decreased luciferase activity in the WT‐ITGB8 group but not in the MUT‐TGB8 group (Figure 6b). Meanwhile, the expression of ITGB8 including mRNA and protein was higher in tumor tissues compared with normal tissues (Figure 6c,d). RIP assays demonstrated that miR‐139‐5p could target ITGB8 (Figure 6e). In addition, the expression of ITGB8 was negatively modulated by miR‐139‐5p. There was a higher expression of ITGB8 in lung cancer than in corresponding paracancerous tissues (Figure 6f). Additionally, we found that overexpression of miR‐139‐5p inhibited the ITGB8 protein level in A549/PTX cells. In this study, we found that miR‐139‐5p directly targeted ITGB8 and negatively mediated its expression in A549/PTX cells.
FIGURE 6.
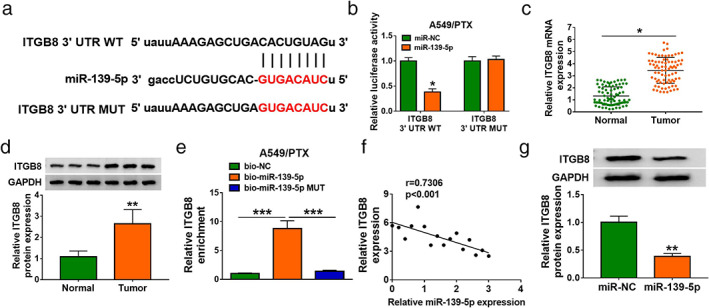
MiR‐139‐5p directly targeted ITGB8. (a) The binding sites between miR‐139‐5p and ITGB8 are shown. (b) The luciferase activity of A549/PTX cells cotransfected with WT‐ITGB8 3′UTR/MUT‐TPD52 3′UTR and miR‐139‐5p/miR‐NC was assessed. (c and d) ITGB8 mRNA and protein expression in PTX sensitive tissues were measured via WB and qRT‐PCR. (e) The RIP assay results demonstrated that ITGB8 could form silencing‐induced complexes with miRNA in A549 cells. (f) The correlation between miR‐139‐5p expression and ITGB8 mRNA expression in PTX‐resistant tissues was analyzed. (g) The abundance of ITGB8 was detected in A549/PTX cells after transfection with miR‐NC or miR‐139‐5p. Data are presented as the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
Overexpression of miR‐139‐5p improves PTX sensitivity in A549/PTX cells by targeting ITGB8
To confirm our previous results, gain‐of‐function and rescue experiments were performed. We found that downregulating miR‐139‐5p in A549/PTX cells clearly inhibited ITGB8 expression, while ITGB8 overexpression counteracted the inhibition effect of miR‐139‐5p on ITGB8 expression (Figure 7a). In addition, we observed that overexpression of miR‐139‐5p suppressed the colony‐forming ability and promoted the cell apoptosis, while addition of ITGB8 could reverse the effect (Figure 7b,c). Moreover, enforced expression miR‐139‐5p repressed migration and invasion of A549/PTX and the effect could be abated after overexpression of ITGB8 (Figure 7d,e).
FIGURE 7.
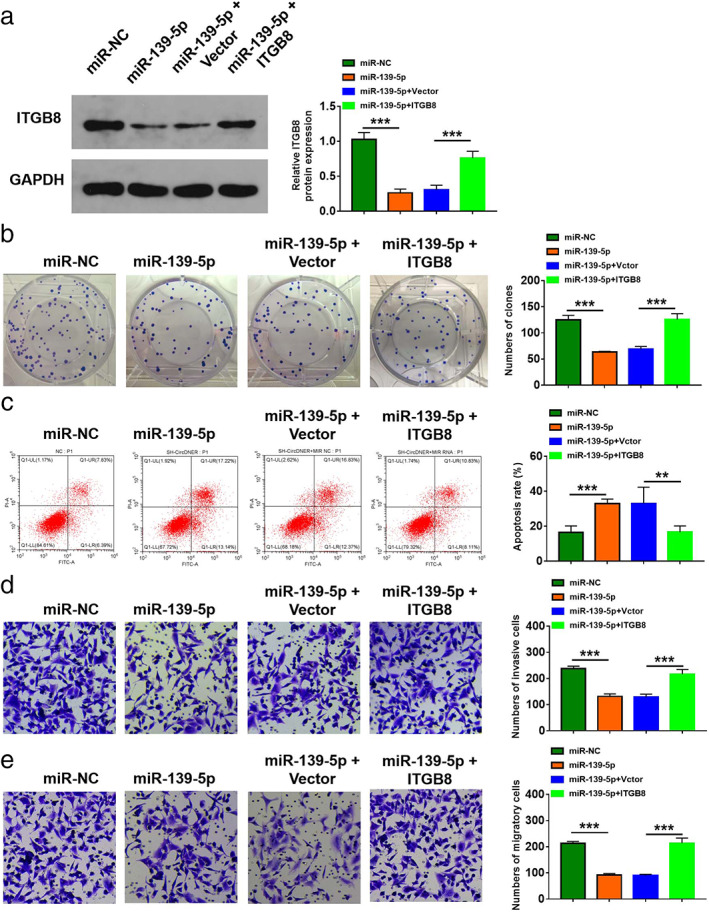
Upregulation of miR‐1182 enhanced DTX sensitivity in DTX‐resistant PCa cells by downregulating TPD52. A549/PTX cells were transfected with miR‐NC, miR‐139‐5p, miR‐139‐5p + vector, or miR‐139‐5p + ITGB8. (a) The protein level of ITGB8 was detected. (b) Colony‐forming ability was evaluated. (c) Flow cytometry was used to assess cell apoptosis. (d and e) A transwell assay was utilized to evaluate cell migration and invasion. Data are presented as the mean ± SD of at least three independent experiments. **p < 0.01, ***p < 0.001
CircDNER meditates ITGB8 expression by binding with miR‐139‐5p
Finally, we investigated whether circDNER could regulate ITGB8 expression by sponging miR‐139‐5p. Our data indicated that knockdown of circDNER decreased TPD52 protein expression, while silencing miR‐139‐5p could reverse the impact (Figure 8a), which indicated that circDNER could regulate ITGB88 expression by sponging miR‐139‐5p. A schematic diagram of the mechanism was created and is shown in Figure 8b.
FIGURE 8.
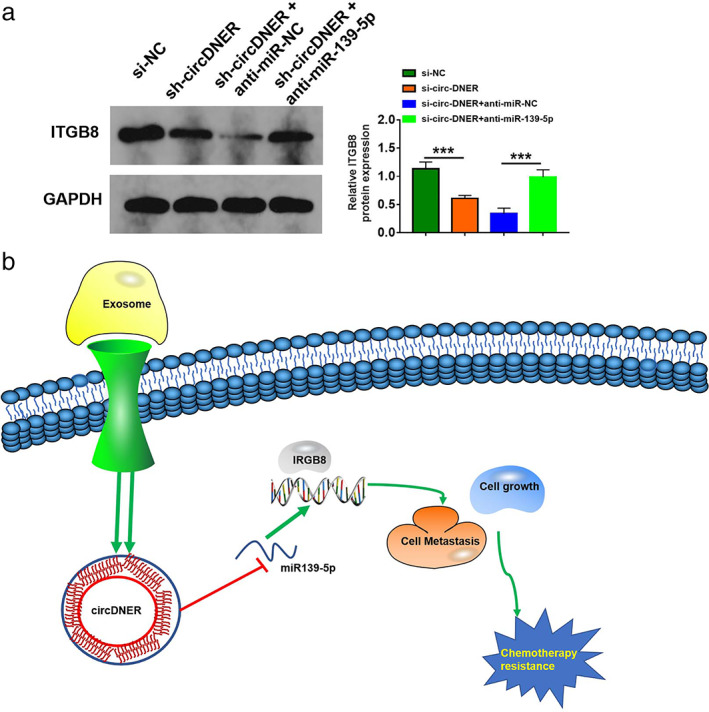
Circ‐DNER regulated ITGB8 expression by sponging miR‐139‐5p. (a) ITGB8 protein expression was examined by Western blot in A549/PTX cells after the introduction of sh‐NC, sh‐circDNER, sh‐circDNER + anti‐miR‐NC, or sh‐circDNER + anti‐miR‐139‐5P. Data are presented as the mean ± SD of at least three independent experiments. (b) Schematic diagram of the mechanism in which exosomal circ‐DNER modulated chemotherapy resistance by regulating the miR‐139‐5p/ITGB8 axis. ***p < 0.001
DISCUSSION
Lung cancer is a common cancer characterized by high recurrence and poor 5‐year survival rates. 15 A number of therapies which have been applied for the treatment of lung cancer have failed due to lack of specific molecular targets and effective interventions. 16 Therefore, searching for novel genetic targets and the pursuit of prognostic markers are critical for lung cancer.
In recent years, RNAs (circRNAs) have attracted great attention and have been shown to have a close relationship with various human malignant tumors. 17 , 18 Previous studies have found that circ_0002346 and circ_0001658 are downregulated in lung cancer, which are positively correlated with tumor size, lymphatic invasion, or lymph node metastasis in patients. In our study, we found upregulated expression of circDNER in lung cancer tissues and cells, suggesting circDNER could promote lung cancer proliferation, invasion and migration in vitro and in vivo, hence downregulating the circDNER had the opposite effect. It is well accepted that circRNAs can negatively regulate miRNA activity. 11 Our data indicated that circ DNER increased lung cancer invasion and metastasis by sponging miR‐139‐5p which had been reported was downregulated in human tumors. 19 Our finding is consistent with previous studies.
Integrin beta‐8 (ITGB8) has been shown to be essential for tumor cell survival and serves as a target gene of miR‐139‐5p according to our data. As a master regulator of angiogenesis, ITGB8 has previously been reported to correlate with poor prognosis in various malignancies. 20 In our study, we implicated for the first time that circ DNER promoted lung cancer proliferation, invasion and metastasis by modulating ITGB8 expression via sponging miR‐139‐5p, implying a novel regulatory axis formed by circ DNER /miR‐139‐5p/ITGB8 in lung cancer.
Exosomes are small endosomal‐derived membrane microvesicles with a diameter of approximately 30–100 nm, which can be actively secreted by a variety of cells. 21 A number of studies have demonstrated that exosomes are enriched in circRNAs, which may regulate tumor cell proliferation, migration and invasion. 22 , 23 Previous studies have shown that exosomal circ‐ciRS‐122 mediates PKM2 in colorectal cancer cell lines and that plasma exosomal circ‐PDE8A is associated with tumor invasion. 11 , 24 Plasma exosomal circular RNA circ‐RanGAP1 modulates VEGFA expression by targeting miR‐877‐3p to facilitate invasion and metastasis. 14 In this study, we demonstrated that plasma exosomal circDNER transferred from lung cancer cells increased lung cancer cell proliferation, invasion and migration, elucidating a novel mechanism of tumor metastasis in lung cancer. Moreover, our results demonstrated that plasma exosomal circ DNER can act as a tumor marker to distinguish between tumor patients and healthy people. Certainly, this conclusion needs further experimental and clinical studies to be confirmed.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Supplementary Figure 1. The expression of miR‐139‐5p was examined by qRT‐PCR. ***P<0.001.
Supplementary Figure 2. The effect of miR‐139‐5p on the expression of potential target mRNAs. The expression levels of ITGB8, FUS, IGF1R, GTF2E2, and ROCK1 were determined by qRT‐PCR in A549/PTX cells transfected with miR‐NC or miR‐139‐5p. **P<0.01, ***P<0.001
ACKNOWLEDGMENTS
This work was supported by the Wuxi Taihu Lake Talent Plan, Supports for Leading Talents in Medical and Health Profession and General project of Wuxi Municipal Health Commission (M202103).
Li J, Zhu T, Weng Y, Cheng F, Sun Q, Yang K, et al. Exosomal circDNER enhances paclitaxel resistance and tumorigenicity of lung cancer via targeting miR‐139‐5p/ITGB8 . Thorac Cancer. 2022;13:1381–1390. 10.1111/1759-7714.14402
Funding information Wuxi Taihu Lake Talent Plan Supports for Leading Talents in Medical and Health Profession and General project of Wuxi Municipal Health Commission, Grant/Award Number: M202103
REFERENCES
- 1. Yamaguchi K, Chikumi H, Shimizu A, et al. Diagnostic and prognostic impact of serum‐soluble UL16‐binding protein 2 in lung cancer patients. Cancer Sci. 2012;103:1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang S, Liu N, Ma M, et al. Methionine enkephalin (MENK) suppresses lung cancer by regulating the Bcl‐2/Bax/caspase‐3 signaling pathway and enhancing natural killer cell‐driven tumor immunity. Int Immunopharmacol. 2021;98:107837. [DOI] [PubMed] [Google Scholar]
- 3. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–8. [DOI] [PubMed] [Google Scholar]
- 4. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 5. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geng HH, Li R, Su YM, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR‐7a on its target genes expression. PLoS One. 2016;11:e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z, Wang Y, Xiao K, Xiang S, Li Z, Weng X. Emerging role of exosomes in the joint diseases. Cell Physiol Biochem. 2018;47:2008–17. [DOI] [PubMed] [Google Scholar]
- 8. Samanta S, Rajasingh S, Drosos N, Zhou Z, Dawn B, Rajasingh J. Exosomes: new molecular targets of diseases. Acta Pharmacol Sin. 2018;39:501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu R, Gao W, Yao K, Ge J. Roles of exosomes derived from immune cells in cardiovascular diseases. Front Immunol. 2019;10:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. [DOI] [PubMed] [Google Scholar]
- 11. Wang X, Zhang H, Yang H, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- 13. Pan Z, Lin J, Wu D, et al. Hsa_circ_0006948 enhances cancer progression and epithelial‐mesenchymal transition through the miR‐490‐3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY). 2019;11:11937–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu J, Wang YH, Yoon C, et al. Circular RNA circ‐RanGAP1 regulates VEGFA expression by targeting miR‐877‐3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. [DOI] [PubMed] [Google Scholar]
- 15. Fujimoto M, Kamiyama M, Fuse K, et al. ASK1 suppresses NK cell‐mediated intravascular tumor cell clearance in lung metastasis. Cancer Sci. 2021;112:1633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okimoto T, Kotani H, Iida Y, et al. Pemetrexed sensitizes human lung cancer cells to cytotoxic immune cells. Cancer Sci. 2020;111:1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Latowska J, Grabowska A, Zarebska Z, Kuczynski K, Kuczynska B, Rolle K. Non‐coding RNAs in brain tumors, the contribution of lncRNAs, circRNAs, and snoRNAs to cancer development‐their diagnostic and therapeutic potential. Int J Mol Sci. 2020;21:7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu Y, Kong S, Qin S, Shen X, Ju S. Exosomal circRNAs: sorting mechanisms, roles and clinical applications in tumors. Front Cell Dev Biol. 2020;8:581558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh AK, Rooge SB, Varshney A, et al. Global microRNA expression profiling in the liver biopsies of hepatitis B virus‐infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695–709. [DOI] [PubMed] [Google Scholar]
- 20. Zou W, Cao Y, Cheng K, et al. Downregulation of circ_0037655 impedes glioma formation and metastasis via the regulation of miR‐1229‐3p/ITGB8 axis. Open Life Sci. 2021;16:442–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bai S, Xiong X, Tang B, et al. Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR‐143 and targeting ERBB3/NF‐kappaB/MMP‐2 axis. Cell Death Dis. 2020;11:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu X, Ding J, Wang B, Wang J, Xu M. Circular RNA DLGAP4 is down‐regulated and negatively correlates with severity, inflammatory cytokine expression and pro‐inflammatory gene miR‐143 expression in acute ischemic stroke patients. Int J Clin Exp Pathol. 2019;12:941–8. [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR‐133/PRDM16 pathway. Int J Cancer. 2019;144:2501–15. [DOI] [PubMed] [Google Scholar]
- 24. Blanco FC, Soria MA, Klepp LI, Bigi F. ERAP1 and PDE8A are downregulated in cattle protected against bovine tuberculosis. J Mol Microbiol Biotechnol. 2017;27:237–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The expression of miR‐139‐5p was examined by qRT‐PCR. ***P<0.001.
Supplementary Figure 2. The effect of miR‐139‐5p on the expression of potential target mRNAs. The expression levels of ITGB8, FUS, IGF1R, GTF2E2, and ROCK1 were determined by qRT‐PCR in A549/PTX cells transfected with miR‐NC or miR‐139‐5p. **P<0.01, ***P<0.001


