Abstract
We report on the structural diversity of mecA gene complexes carried by 38 methicillin-resistant Staphylococcus aureus and 91 methicillin-resistant coagulase-negative Staphylococcus strains of seven different species with a special reference to its correlation with phenotypic expression of methicillin resistance. The most prevalent and widely disseminated mec complex had the structure mecI-mecR1-mecA-IS431R (or IS431mec), designated the class A mecA gene complex. In contrast, in S. haemolyticus, mecA was bracketed by two copies of IS431, forming the structure IS431L-mecA-IS431R. Of the 38 S. haemolyticus strains, 5 had low-level methicillin resistance (MIC, 1 to 4 mg/liter) and characteristic heterogeneous methicillin resistance as judged by population analysis. In these five strains, IS431L was located to the left of an intact mecI gene, forming the structure IS431L-class A mecA-gene complex. In other S. haemolyticus strains, IS431L was associated with the deletion of mecI and mecR1, forming the structure IS431L-ΔmecR1-mecA-IS431mec, designated the class C mecA gene complex. Mutants with the class C mecA gene complex were obtained in vitro by selecting strain SH621, containing the IS431L-class A mecA gene complex with low concentrations of methicillin (1 and 3 mg/liter). The mutants had intermediate level of methicillin resistance (MIC, 16 to 64 mg/liter). The mecA gene transcription was shown to be derepressed in a representative mutant strain, SH621-37. Our study indicated that the mecI-encoded repressor function is responsible for the low-level methicillin resistance of some S. haemolyticus clinical strains and that the IS431-mediated mecI gene deletion causes the expression of methicillin resistance through the derepression of mecA gene transcription.
Despite the introduction of mecA into the chromosome, some clinical strains are found to remain susceptible to methicillin (pre-methicillin-resistant Staphylococcus aureus [MRSA] status). In those strains, mecA gene transcription is strongly repressed by the function of the accompanying regulator genes, mecI and mecR1 (9, 10). Transcription of mecA, encoding PBP2′ (or PBP2a), is essential for the phenotypic expression of methicillin resistance (23, 28). Since mecI encodes a strong repressor of mecA gene transcription, pre-MRSA strains have only a marginal level of resistance to methicillin (MIC < 8 mg/liter) (9, 12). Experimental inactivation of mecI causes derepressed production of PBP2′ and makes the cell express methicillin resistance (22, 27). This requirement of mecI inactivation for the phenotypic expression of methicillin resistance coincides well with the observation with clinical MRSA isolates in which the mecI gene is either mutated or completely deleted in the tested strains (19, 27, 32).
In pre-MRSA strain N315, mecA is flanked by the intact set of mec regulator genes on the left-hand side, and by a DNA region of about 3 kb, called the hypervariable region on the right-hand side (26). The right boundary of the latter region is demarcated by a copy of insertion sequence IS431mec (or IS431R) (3). The mecA locus of pre-MRSA strains thus forms a prototypic structure of mecI-mecR1-mecA-IS431R that is designated the class A mecA gene complex (mec complex). In other clinical MRSA strains, the entire mecI gene and the 3′ part of mecR1 are deleted from the class A mec complex and a fragment of the insertion sequence IS1272 (ψIS1272) has been association with the deletion point (1, 2). We designate this structure, ψIS1272-ΔmecR1-mecA-IS431R, the class B mec complex.
Recently, it has been recognized that some methicillin-resistant coagulase-negative Staphylococcus (MRC-NS) strains express a low-level methicillin resistance in spite of their carriage of mecA (15). We have previously shown that mecI is widely disseminated in C-NS species (29). Dickinson and Archer have demonstrated that intact mecI is found in some S. epidermidis clinical strains with low-level oxacillin resistance (6). They have shown that allelic replacement inactivation of mecI raises the oxacillin resistance of these strains by severalfold, indicating a role for mecI-mediated repression in the low-level beta-lactam resistance in certain S. epidermidis strains. On the other hand, Kobayashi et al. have found some S. epidermidis clinical strains in a Japanese hospital that carry intact mecI in spite of their overt beta-lactam resistance expression (18).
We have been studying the genetic organization of the chromosomal region surrounding mecA in clinical C-NS strains to trace the evolutionary makeup of the mec complex. During the course of this study, we found a few S. haemolyticus strains with low-level methicillin resistance and carrying a copy of IS431 (IS431L) inserted upstream of and close to the class A mec complex. Using a representative strain, SH621, in vitro experiments were performed to evaluate the role of IS431L in the expression of methicillin resistance in S. haemolyticus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Thirty-eight epidemic strains of MRSA isolated from patients in 20 countries in 1961 to 1985 have been described previously (12). Ninety-one MRC-NS strains of seven staphylococcal species (38 S. haemolyticus, 34 S. epidermidis, 9 S. sciuri, 5 S. caprae, 2 S. hominis, 2 S. capitis, and 1 S. warneri strains) were isolated from patients in five hospitals in Japan in 1981 to 1993. The species were determined using API Staph-Ident strips (Analytical Profile Index System S.A., Montaliew-Vercieu, France) and Staphyogram (Terumo Co., Tokyo, Japan). All bacterial strains were grown in Luria-Bertani broth with gentle shaking at 37°C (22). Pre-MRSA strain N315 has been described previously (9). S. haemolyticus strain SH621, used for the derivation of resistant mutants in this study carried a beta-lactamase plasmid (7) and had a low-level methicillin resistance (MIC = 2 mg/liter).
PCR for the analysis of mec complex structure.
The preparation of chromosomal DNA has been described previously (11, 23). PCR amplification was performed using 1 unit of AmpliTaq (Perkin-Elmer Cetus, Foster City, Calif.) in 50 μl of reaction mixture (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% [wt/vol] gelatin, 50% [vol/vol] glycerol, 1.5 mM MgCl2, 200 mM each deoxynucleoside triphosphate, 1.0 mM each primer, and template DNA). The reaction was carried out by using a Gene Amp PCR system 9600 (Perkin-Elmer). Thermal cycling was set at 30 cycles (30 s for denaturation at 94°C, 1 min for annealing at 50°C, and 2 min for elongation at 72°C).
Long-range PCR amplification was performed using 2.6 U of Expand high-fidelity PCR system enzyme mix as recommended by the manufacturer (Boehringer Mannheim Biochemica, Mannheim, Germany). A 5-μl portion of the reaction volume was subjected to electrophoresis in a 0.8% agarose gel containing 1 μl of ethidium bromide per ml to detect the amplified DNA fragment.
PCR primers.
The primers used for the detection of mec complexes are listed in Table 1. The locations are direction of the primers are indicated in Fig. 1. Technical detection of the class A mec complex was based on the positive PCR test results for both sets of primers, mI-1 plus mI-2 and mcR2 plus mcR3 (Fig. 1). These sets of primers amplified DNA fragments of 0.5 and 1.6 kb, respectively. They included part of mecI and the penicillin-binding (PB) and membrane-spanning (MS) domains of mecRI (10). The class B mec complex was detected by long-range PCR using two sets of primers encompassing ψIS1272 and mecA genes: iS-3 plus mA5 and iS-4 plus mA5 (Fig. 1). The latter primer set (iS-4 plus mA5) was used to explore the possible presence of IS1272 inserted in the opposite orientation to the mecA gene.
TABLE 1.
Synthetic oligonucleotides primers used for the analysis of mec complex structure
| Primer designation | Sequencea | Localization | Positions | Reference(s) |
|---|---|---|---|---|
| mA2 | 5′-AACGTTGTAACCACCCCAAGA-3′ | mecA | 1461–1441 | 11,28 |
| mA4 | 5′-AGTGTATGATGAGCTATATGAGA-3′ | mecA | 1959–1981 | 28 |
| mA5 | 5′-CGCTCAGAAATTTGTTGTGC-3′ | mecA | 212–180 | 28 |
| mA6 | 5′-TATACCAAACCCGACAAC-3′ | mecA | 83–66 | 28 |
| mA7 | 5′-ATCGTTGACGATAATAGCAATACA-3′ | mecA | 907–930 | 28 |
| mA8 | 5′-ATGTTTGGATTATCTTTATCATAT-3′ | mecA | 1863–1886 | 28 |
| iS-1 | 5′-ACATTAGATATTTGGTTGCGT-3′ | IS431mec | 566–596 | 3 |
| iS-2 | 5′-TGAGGTTATTCAGATATTTCGATGT-3′ | IS431mec | 767–743 | 3 |
| iS-3 | 5′-TCGGATGCTATCATTAAGCAT-3′ | IS1272 | 1668–1688 | 2 |
| iS-4 | 5′-ACAATCTGTATTCTCAGGTCGT-3′ | IS1272 | 1722–1701 | 2 |
| mI-1 | 5′-AATGGCGAAAAAGCACAACA-3′ | mecI | 1923–1942 | 10, 30 |
| mI-2 | 5′-GACTTGATTGTTTCCTCTGTT-3′ | mecI | 2403–2283 | 10, 30 |
| mcR2 | 5′-CGCTCAGAAATTTGTTGTGC-3′ | mecR1 | 1954–1935 | 10, 30 |
| mcR3 | 5′-ATCTCCACGTTAATTCCATT-3′ | mecR1 | 357–376 | 10, 30 |
The location of each primer is shown in Fig. 1.
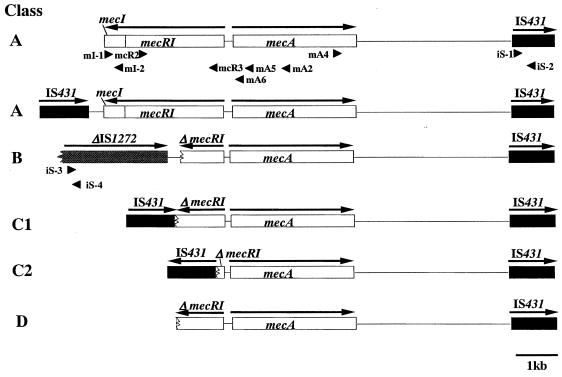
FIG. 1.
Genetic organization of the mecA gene complex in staphylococci and the PCR primers for the detection of IS431L and IS1272 upstream of mecA. The structures of five classes of mec complexes found in MRSA and MRC-NS are shown. In all five classes, IS431R was present downstream of mecA. The class A mec complex has the structure mecI-mecR1-mecA-IS431mec. In S. haemolyticus, the class A mec complex was accompanied by an upstream IS431 copy (IS431L). The class B mec complex has the structure ψIS1272-ΔmecR1-mecA-IS431. Class C1 and C2 mec complexes have a deletion in the left side of the class A mec complex associated with an insertion of IS431. In class C1, IS431L is inserted in the same orientation as mecA, and in class C2, IS431L is inserted in the opposite orientation relative to mecA. Class D has a deletion without any insertion sequence adjacent to the deletion point. Arrows above the genes indicate the direction of transcription. The arrowheads indicate the location and direction of primers (see Materials and Methods).
Nucleotide sequencing.
Nucleotide sequence determination was performed as described previously (22), using the Dye Terminator cycle-sequencing FS ready reaction kit (Perkin-Elmer). Description of primers synthesized specifically for the sequence determination is omitted from the text.
Computer analysis of nucleotide and protein sequences.
All the analyses were carried out using programs in the Wisconsin Package (version 9.0; Genetics Computer Group Madison, Wis.). A homology search was performed using BLAST and TFastA programs over the EMBL (release 55.0) and GenBank (release 107.0) databases and the FastA program over the SWISS-PROT database (release 35.0).
Cloning of the region upstream of mecA by IPCR.
Using inverse PCR (IPCR), DNA fragments corresponding to the region upstream (left-hand side) of mecA complex were amplified and then sequenced by primer walking. The experimental procedure for IPCR has been described previously (33). The chromosomal DNAs extracted from S. haemolyticus strains SH518, SH631, and JB16, S. epidermidis strain JK8, and S. caprae strain JA186 were digested with SphI. That of S. haemolyticus strain SH621 was digested with EaeI. The restriction enzymes were purchased from Takara Shuzo Co., Ltd., Shiga, Japan. Self-ligation of the SphI-digested DNA and the EaeI-digested DNA was performed at a DNA concentration of 1 to 2 μg/ml for 10 min using a ligation kit (Boehringer Mannheim Biochemica). The ligation mixtures were then used as template DNAs for PCR amplification. The primer sets were mI2 plus mA4 for the SphI self-ligates and mA4 plus mA6 for the EaeI self-ligates (Fig. 1).
Detection of PBP2′.
A rapid slide latex agglutination assay kit was used to evaluate expression of PBP2′ by the procedure recommended by the manufacturer (8, 24) (Denka Seiken Co., Ltd., Niigata, Japan).
Selection of resistant mutants from a low-level methicillin-resistant S. haemolyticus strain, SH621.
S. haemolticus SH621 was cultivated in 5 ml of brain heart infusion (BHI) broth (Becton Dickinson Microbiology Systems). The overnight culture was diluted in the same medium to make a cell suspension with an optical density at 540 nm of 0.3. The 100-μl portions of the dilutions were spread onto BHI agar plates containing 1, 3, or 8 mg of methicillin per liter. For the enumeration of inoculum size (in CFU), the original cell suspension and its 10-fold serial dilutions were spread on BHI plates without antibiotic. The plates were incubated overnight at 37°C, and the mature colonies were enumerated. Six colonies were picked from the agar plate containing 1 mg of methicillin per liter, and 10 colonies each were picked from those containing 3 and 8 mg of methicillin per liter. Picked colonies were subjected to colony purification on plates containing the respective concentration of methicillin before being established as mutant strains.
MIC determination and population analysis.
The MIC of methicillin was determined by the plate dilution method as described previously (14). The plates were incubated for 24 h at 37°C before the inspection of cell growth.
Analysis of methicillin-resistant subpopulations of strain SH621 and its derivative strains was performed as described previously (14). Briefly, aliquots (100 μl) of an undiluted or appropriately diluted overnight culture were spread onto BHI agar plates containing various concentrations of methicillin. The plates were then incubated at 37°C for 48 h. The population curve was drawn by calculating and plotting the number of resistant cells contained in 100 μl of the undiluted overnight culture. Methicillin was obtained from Banyu Pharmaceutical Co., Ltd., Tokyo, Japan.
Total RNA isolation for RT-PCR from S. haemolyticus.
Total RNA extraction was performed as described previously (21). For methicillin induction, methicillin was added to the cultures to a final concentration of 8 mg/liter and cells were collected after a 30-min incubation at 37°C. Contaminating DNAs were degraded by digesting the RNA samples with RNase-free DNase (22). The yield and purity of total RNA were determined by measuring the absorbance of an aliquot at 260 and 280 nm using a DO 400 spectrophotometer (Beckman Instruments, Palo Alto, Calif.).
Preparation of the internal standard for comparative RT-PCR.
The internal standard DNA was synthesized by PCR amplification using pUC119 as a template. The primers used were 5′-AAAA-mA7-ATACGCCTATTTTTATAGGTTAATGTCAT-3′ and 5′-AAAA-mA8-AGTTGCTCTTGCCCGGCGTCAATACGGGAT-3′, where the underlined sequences corresponded to nucleotides 1179 to 1207 and 1598 to 1627 of pUC119, respectively (31). The set of primers amplified the DNA fragment of 0.5 kb. The amplified PCR product was subjected to agarose gel electrophoresis and was purified from the gel using the Suprec-01 purification system (Takara Shuzo co. Ltd., Shiga, Japan.). The integrity of the purified DNA was confirmed by nucleotide sequence determination. The concentration of DNA was determined by measuring the absorbance at 260 nm.
Quantitation of mecA gene transcripts by RT-PCR.
Reverse transcription-PCR (RT-PCR) was carried out using the Geneamp RT-PCR system (Perkin-Elmer Cetus). RT and PCR were carried out as described previously (22), except that serially diluted competitor DNAs were added to 0.0015 to 200 pM in 50 μl of PCR mixture. Preliminary experiments were performed to confirm the linear relationship between the amounts of input RNA and the amounts of PCR product for the entire set of experiments (25). The quantity of mecA mRNA in the RNA samples was calculated by the procedure described by Köhler (20).
RESULTS
Distribution of class A and class B mec complexes among C-NS strains.
Figure 1 illustrates the genomic structures around the mecA gene in 38 MRSA and 91 MRC-NS strains. The PCR amplification using primers mA4 plus iS-2 (shown in Fig. 1) demonstrated that all the strains possessed a copy of IS431R at about 2.5 kb downstream of mecA. The distance between mecA and IS431R varied slightly among the strains, ranging from 2.3 to 2.6 kb as judged from the differences in the electrophoretic mobility of PCR-amplified fragments in agarose gel. This minor difference in size was explained by the difference in the number of direct repeat units in the region between mecA and IS431R (reference 26 and data not shown). Another set of primers, mA4 plus iS-1, was used for the detection of an IS431 copy inserted in the opposite orientation relative to mecA gene. However, no DNA fragment was amplified, showing that IS431 and mecA were directed in the same orientation in all the strains.
Significant structural differences were present upstream of mecA gene. Four classes of mec complexes were distinguished (Fig. 1). The class A mec complex has an apparently intact structure mecI-mecR1-mecA-IS431R, and the class B mec complex has the structure ψIS1272-ΔmecR-mecA-IS431R. Both structures have been identified previously in MRSA strains (1, 29). The mec complexes of all S. aureus strains were classified into either class A (26 strains [68.4%]) or class B (12 strains [31.6%]) mec complexes (Table 2). Among the MRC-NS strains, 42 strains (46.2%) carried the class A mec complex, which was the most common mec complex distributed in all the seven species tested (Table 2). The class B mec complex was found in 16 strains of two C-NS species: 14 in 34 S. epidermidis strains (41.2%) and 2 in 38 S. haemolyticus strains (5.3%) (Table 2). The nucleotide sequences of the region between the right boundary of ψIS1272 and the deletion point of ΔmecR1 were determined for all the 28 strains of three staphylococcal species (S. aureus, S. haemolyticus, and S. epidermidis) (Fig. 2). All had an identical mecR1 deletion point (nucleotide 976 of mecR1), and the right boundary of IS 1272 was located 267 nucleotides away from the deletion point. The 267-nucleotide sequences were 100% identical among 28 strains. The nucleotide sequence of ψIS1272 revealed that the inverted repeat at the left end was missing and that there was a 23-base deletion at the 5′ end of orf1. This deletion was identical to that previously reported in MRSA strain COL (the GenBank accession number is U35635 [2]).
TABLE 2.
Distribution of various classes of mec complex among C-NS species
| mec complex classa | No. (%) of isolates of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus(n = 38) | S. haemolyticus(n = 38) | S. epidermidis(n = 34) | S. sciuri(n = 9) | S. caprae(n = 5) | S. hominis(n = 2) | S. capitis(n = 2) | S. warneri(n = 1) | |
| A | 26 (68.4) | 5 (13.2) | 19 (55.9) | 9 | 4 (80.0) | 2 | 2 | 1 |
| B | 12 (31.6) | 2 (5.3) | 14 (41.2) | 0 | 0 | 0 | 0 | 0 |
| C1 | 0 | 1 (2.6) | 0 | 0 | 0 | 0 | 0 | 0 |
| C2 | 0 | 30 (78.9) | 1 (2.9) | 0 | 0 | 0 | 0 | 0 |
| D | 0 | 0 | 0 | 0 | 1 (20.0) | 0 | 0 | 0 |
Corresponds to the mec complex shown in Fig. 4.
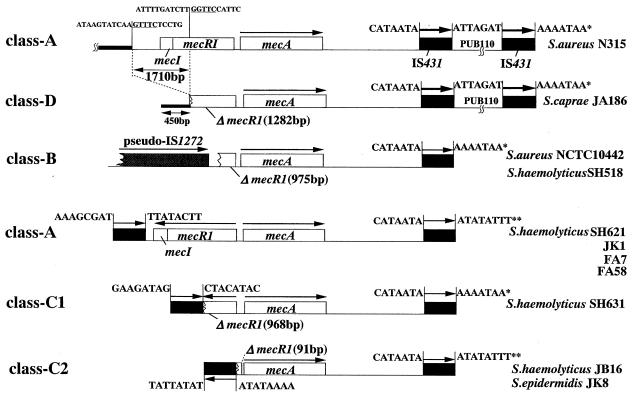
FIG. 2.
Nucleotide sequencing analysis of mec complexes and their surrounding regions. The arrows indicate the direction of transcription of genes. The region upstream of ΔmecR1 of S. caprae strain JA186 (class D) was compared with SCCmec of N315 (carrying the class A mec complex). Eight nucleotides flanking the IS431 copies are also shown to infer a possible relationship of individual IS copies. The single asterisk and the double asterisks indicate pairs of the same sequence. No conserved direct repeats indicating target duplication were found in the 8 bp flanking the IS431-mecA-IS431 structures (5).
The class C mec complex is prevalent in S. haemolyticus.
A total of 31 of 38 S. haemolyticus strains and 1 strain each of S. epidermidis and S. caprae strains had their mecI and mecR1 genes deleted, but there was no IS1272 copy associated with the deletion. Five of the strains (three S. haemolyticus strains [SH621, SH631, and JB16], one S. epidermidis strain [JK8], and one S. caprae strain [JA186]) were analyzed for the nucleotide sequences in the upstream region of mecA by using IPCR. A copy of IS431, designated IS431L, was found closely associated with the deletion point of ΔmecR1 in all strains except the S. caprae strain (see below). The structure of the mec complex, IS431L-ΔmecR1-mecA-IS431R, was designated the class C mec complex (Fig. 1 and 2; Table 2).
To find the prevalence of the class C mec complex, we proceeded to test all the other 126 staphylococcal strains for the presence of IS431 upstream of mec complex by using long-range PCR. Two primer sets were used: iS-1 plus mA5 and iS-2 plus mA5 (for the detection of an IS431L copy in possible reverse orientation) (Fig. 1). Positive PCR results were obtained with all 30 S. haemolyticus strains that did not carry either class A or class B mec complexes. However, no strain of other species except for one S. epidermidis strain JK8 was found to carry the class C mec complex.
Based on the extent of mecI and mecR1 deletion and orientation of IS431L, the class C mec complex was subgrouped into class C1 and class C2 as illustrated in Fig. 1. Curiously, it was found that all the five S. haemolyticus strains having the class A mec complex also possessed IS431L located 192 bp to the left of an intact mecI gene (Fig. 1). In the strains tested by us, IS431L was found only in species of S. haemolyticus and S. epidermidis. In other species, IS431L was not found within a ca 12-kb upstream region of the mecA gene, the distance that was covered by our long-range PCR.
Class D mec complex in S. caprae strain JA186.
The mecR1 gene of S. caprae strain JA186 was partially deleted at its 3′ portion from nucleotide 1283 to the end. Neither IS431 nor IS1272 was identified in the vicinity of the deletion point. Therefore, the structure was designated the class D mec complex. Nucleotide sequencing showed that the region flanking the 3′ deletion point of ΔmecR1 was identical to a stretch of DNA sequence in the type II SCC mec (carrying a class A mec complex [16]) found in pre-MRSA strain N315 (Fig. 2). When the two sequences were aligned, there was a deletion of 1,710 bp from JA186 chromosomal DNA, which involved the mecI gene and the 3′ one-third of the mecR1 gene. The nucleotide sequences of the deletion endpoints of JA186 had a modest homology to each other (4 bases identical in the 5 bases [GNTTC] [underlined in Fig. 2]).
Detection of IS431L movements in methicillin-resistant mutant strains derived from SH621.
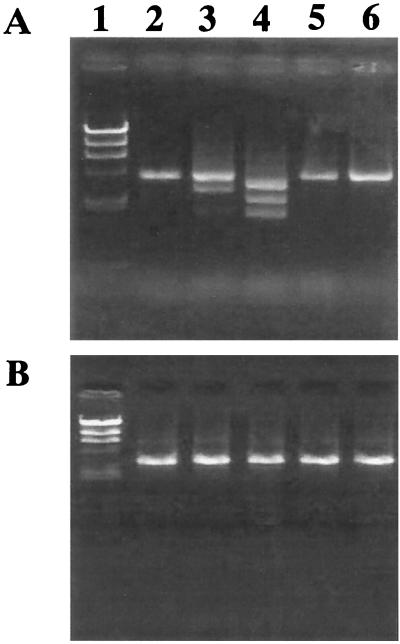
Methicillin MICs for 28 MRC-NS strains of seven species representing all classes of mec complexes (37 of 42 class A strains, 8 of 16 class B strains, class C1 strain SH631, 31 class C2 strains, and the unique class D strain JA186) were determined (Table 3). It was evident that the MICs for some of the class A strains were relatively low (<8 mg/liter). Strain SH621, for which the methicillin MIC was 2 mg/liter, was selected with various concentrations of methicillin to obtain mutants with raised methicillin resistance. A total of 6, 10, and 10 mutant strains were isolated from each of the three experiments using 1, 3, and 8 mg of methicillin, respectively, as selective concentrations. The 10 mutants obtained from the above three experiments were mixed, cultured overnight, and subjected to DNA extraction. Using three DNA templates thus prepared, PCR was performed to see possible movements of IS431L in any one of the 6 or 10 mutants, using primers iS-1 (detecting IS431L) plus mA2 (detecting the 5′ part of mecA). A single 4.1-kb DNA fragment was amplified by PCR performed on the template DNAs extracted from the mutants selected with 8 mg of methicillin per liter (Fig. 3A). On the other hand, several DNA fragments of different sizes were observed when PCR was performed on the template DNAs extracted from the mutants selected with 1 and 3 mg of methicillin per liter (Fig. 3A). The sizes ranged from 1.0 to 2.0 kb and were equal to or smaller than the 4.1-kb band amplified from the parent strain SH621. This indicated that IS431L was relocated to sites closer to mecA gene in some of the mutant strains. The control PCR experiment was performed using a set of primers, mA4 plus iS-2, intended for the detection of possible movements of IS431R relative to the mecA gene (Fig. 3B). This experiment did not show any DNA product with altered electrophoretic mobility; a single band of 2.8 kb was observed (Fig. 3B)
TABLE 3.
Correlation between methicillin MIC and the class of mec complex carried by C-NS clinical strains
| Species | Total no. of strains tested | mec complex classa | No. of strains for which the MIC (mg/liter) is:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 8 | 16 | 32 | 64 | ≧128 | |||
| S. epidermidis | 16 | A | 1 | 4 | 5 | 6 | |||
| 5 | B | 1 | 1 | 3 | |||||
| 1 | C2 | 1 | |||||||
| S. haemolyticus | 5 | A | 3 | 2 | |||||
| 2 | B | 1 | 1 | ||||||
| 1 | C1 | 1 | |||||||
| 30 | C2 | 2 | 7 | 10 | 1 | 1 | 9 | ||
| S. sciuri | 7 | A | 2 | 4 | 1 | ||||
| S. caprae | 4 | A | 4 | ||||||
| 1 | D | 1 | |||||||
| S. hominis | 2 | A | 2 | ||||||
| S. capitis | 2 | A | 2 | ||||||
| S. warneri | 1 | A | 1 | ||||||
Corresponds to the mec complex shown in Fig. 4.
FIG. 3.
Detection of IS431L-mediated deletion in resistant mutants derived from S. haemolyticus SH621. Portions (3 μl) of the PCR mixture were applied to an agarose gel and subjected to electrophoresis. Lanes 1, λ-HindIII digest as a DNA molecular mass standard marker (the sizes were 23.1, 9.5, 6.6, 4.3, 2.3, and 2.2 kb from top to bottom); 2, SH621; 3 to 5, SH621-derived mutants with increased methicillin resistance. Mutants were selected with 1 (lane 3), 3 (lane 4), or 8 (lane 5) mg of methicillin per liter, respectively. (A) Primers iS-1 plus mA2 were used to detect the region between IS431L and mecA. (B) Primers mA4 plus iS-2 were used to detect the region between mecA and IS431R (IS431R). Note that there are varied band sizes only in lanes 3 and 4 in panel A.
Table 4 lists the methicillin MICs for the 26 mutant strains. It was noted that methicillin resistance was raised in all 26 mutants. Three different levels of resistance were noted among the strains. Thirteen mutants selected with 3 and 8 mg of methicillin per liter had high of methicillin resistance (MIC > 512 mg/liter). Eleven mutants selected with methicillin at 1 or 3 mg/liter expressed an intermediate level of methicillin resistance (MIC = 16 to 64 mg/liter). Two strains selected with 1 mg of methicillin per liter showed marginal or low-level methicillin resistance (MIC = 4 and 8 mg/liter for strains SH621-15 and SH621-16, respectively) (Table 4).
TABLE 4.
Structure of mec complex of and methicillin MIC for mutants derived from S. haemolyticus strain SH621
| Strain | PBP2 productiona | Selective methicillin concn (mg/liter)b | Class of mec complexc | PCRd structure of:
|
MIC of methicillin (mg/liter) | ||
|---|---|---|---|---|---|---|---|
| mecI |
mecR1
|
||||||
| PB | MS | ||||||
| SH621 | − | A | + | + | + | 2 | |
| SH621-15 | + | 1 | A | + | + | + | 8 |
| SH621-16 | + | 1 | A | + | + | + | 4 |
| SH621-32 | + | 3 | A | + | + | + | 512 |
| SH621-34 | + | 3 | A | + | + | + | <1,024 |
| SH621-310 | + | 3 | A | + | + | + | 512 |
| SH621-81 | + | 8 | A | + | + | + | <1,024 |
| SH621-82 | + | 8 | A | + | + | + | <1,024 |
| SH621-83 | + | 8 | A | + | + | + | 1,024 |
| SH621-84 | + | 8 | A | + | + | + | 1,024 |
| SH621-85 | + | 8 | A | + | + | + | <1,024 |
| SH621-86 | + | 8 | A | + | + | + | <1,024 |
| SH621-87 | + | 8 | A | + | + | + | 1,024 |
| SH621-88 | + | 8 | A | + | + | + | 1,024 |
| SH621-89 | + | 8 | A | + | + | + | <1,024 |
| SH621-810 | + | 8 | A | + | + | + | 512 |
| SH621-17 | + | 1 | C1a | Δ | + | + | 16 |
| SH621-37 | + | 3 | C1a | Δ | + | + | 16 |
| SH621-38 | + | 3 | C1a | Δ | + | + | 16 |
| SH621-39 | + | 3 | C1a | Δ | + | + | 16 |
| SH621-13 | + | 1 | C1b | − | Δ | + | 32 |
| SH621-35 | + | 3 | C1b | − | Δ | + | 64 |
| SH621-31 | + | 3 | C1b | − | Δ | + | 32 |
| SH621-33 | + | 3 | C1b | − | Δ | + | 16 |
| SH621-36 | + | 3 | C1b | − | Δ | + | 16 |
| SH621-11 | + | 1 | C1c | − | − | Δ | 16 |
| SH621-12 | + | 1 | C1c | − | − | Δ | 16 |
Judged by the rapid slide latex agglutination assay kit (24).
The concentration of methicillin used for the selection of mutants.
Corresponds to the mec complex shown in Fig. 4.
+ indicates that the size of the DNA fragment amplified by PCR was indistinguishable from that of SH621. Δ indicates that the amplified DNA fragment contained a deletion.
IS431L-mediated mecI gene deletion.
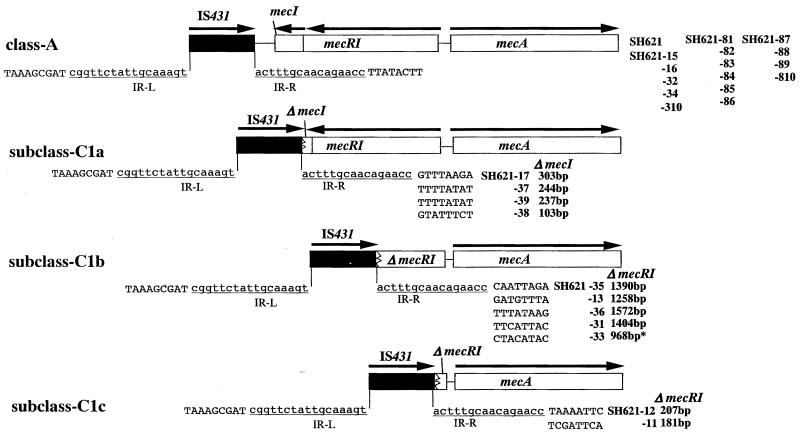
To correlate the movements of IS431L and the level of methicillin resistance expressed by each mutant strain, the 26 isolates were subjected to DNA extraction individually. Each chromosomal DNA was used as a template for PCR amplification with primers is1 plus mA5. Using the PCR product, nucleotide sequence of the region between IS431L and mecA gene was determined for each strain. The sequences are shown schematically in Fig. 4. The structure, IS431L-class A mec complex, remained intact in 15 mutants, and their mecI genes were intact as well. The rest of the mutants (11 strains), however, carried class C mec complex; i.e. their mec regulator genes were partially deleted. In these mutants, the right boundary of IS431L was found inside mecR1 or mecI (Fig. 4; Table 4). The boundary was in the 3′ part of mecI in four mutant strains (subclass C1a). In five mutant strains, the boundary was found in the 3′ part of mecR1 encoding the PB domain of mecR1 (subclass C1b). In two mutant strains, the boundary of IS431L was found in the central part of mecR1 encoding the MS domain (subclass C1c). On the other hand, all 10 mutant strains selected with 8 mg of methicillin per liter and 5 mutants selected with 1 and 3 mg of methicillin per liter retained the class A mec complex of parent strain SH621.
FIG. 4.
IS431L-mediated deletion of the class A mec complex. The structures of three subclasses of the class C1 deletion of the mec complex in the mutants are shown as follows: subclass C1a (IS431-ΔmecI-mecR1-mecA), subclass C1b (IS431-ΔmecR1 [retaining the MS domain]-mecA), and subclass C1c (IS431-ΔmecR1 [retaining only the 5′ end]-mecA). The flanking sequences of IS431L and inverted repeats (IR-L; underlined) are presented to show the deletion points in detail. The bold type indicates the names of the (mutant) strains and the sizes of the deletion. The arrows indicate the direction of transcription. A total of 15 mutants including all 10 mutants obtained by selection with 8 mg of methicillin per liter retained the class A mec complex. Others were classified as either one of the three class C subtypes. The asterisk in the mutant strain 33 shows that the position of IS431L was identical to that found in clinically isolated strain SH631.
The mecI deletion is correlated with heterogeneous methicillin resistance expression.
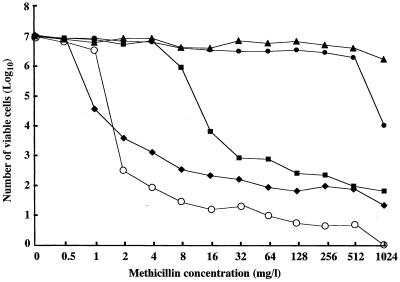
Comparison between methicillin resistance and the mec complex structure showed that all the mutants with class C mec complex had intermediate level of resistance (Table 2). In contrast, either high- or low-level resistance was associated with the class A mec complex. In this case, a selection concentration of 1 mg/liter was correlated with low-level methicillin resistance (Table 4). Figure 5 illustrates the patterns of methicillin-resistant subpopulations (population curves) of representative mutant strains in comparison with that of the parent strain, SH621. SH621-37 (carrying class-C1 mec complex) had a typical heterogeneous methicillin resistance. In contrast, two strains carrying the class A mec complex, SH621-34 and SH621-81, contained substantial sizes of cell subpopulations with high (>512 mg/liter) methicillin resistance. They expressed homogeneous methicillin resistance.
FIG. 5.
Population analysis of mutant strains SH621-37, SH621-34, and SH621-81 in comparison with the parent strains SH621 and N315. See Materials and Methods for the procedure of population analysis. Symbols: ⧫, SH621 (class A); ■, SH621-37 isolated with 3 mg of methicillin per liter (class C1); ●, SH621-34 isolated with 3 mg of methicillin per liter (class A); ▴, SH621-81 isolated with 8 mg of methicillin per liter (class A); ○, S. aureus N315 (class A).
Derepressed transcription of mecA in the mecI-deleted strain SH621-37.
We analyzed the quantitative expression of mecA in SH621 and its derivative strains SH621-31 and SH621-810 before and after methicillin induction (Table 5). The amount of mecA transcripts in SH621-810 was slightly larger than but almost comparable to that in SH621. The transcript increased after 30-min induction with 8 mg of methicillin per liter. This was in contrast to the parent strain SH621, in which methicillin could not induce increased mecA gene transcription. On the other hand, the amount of mecA gene transcripts in SH621-31 was increased by approximately 10-fold compared to the parent strain (Table 5), signifying that the mecA gene transcription was derepressed in the strain. Methicillin induction further increased the transcription level of mecA gene in SH621-31 (Table 5).
TABLE 5.
Quantitative analysis of mecA transcripts in SH621 and its derivative strains with and without methicillin induction
| Strains | MIC of methicillin (mg/liter) | Class of mec complex | Amt of mecA mRNA (amol)a
|
|
|---|---|---|---|---|
| Noninduced | Inducedb | |||
| SH621 | 2 | A | 1.48 | 2.66 |
| SH621-31 | 32 | C1b | 17.1 | 63.9 |
| SH621-810 | 512 | A | 1.79 | 39.8 |
Average of two experiments.
Induced with methicillin (8 mg/liter) for 30 min at 37°C.
DISCUSSION
Song et al. have proposed that the mecA gene was fused with mec regulator genes in the past to form the present operon structure of mec complex (28). Besides the prototypic class A mec complex thus formed, class B mec complexes have been found in MRSA (18, 23, 29, 30) as well as in MRC-NS strains (2, 6, 19, 29, 34). However, as this study and previous reports showed, at least a 5′ portion of mecR1 is identified adjacent to mecA in all the tested staphylococcal strains having various deletion patterns in mec complexes. Close linkage of mecA with IS431R was also demonstrated in C-NS strains, although there were minor size differences in the hypervariable region between the two genes (26). So far no mec complex without IS431R or a part of mec regulator genes has been found. Thus, it seems likely that the class A prototypic mec complex was formed in much early evolutionary times before the complex was transmitted into various staphylococcal species carried by a mobile genetic element, the staphylococcal cassette chromosome (SCC) (16, 17).
S. haemolyticus strains were characteristic in their possession of IS431L upstream of mec complex. The overall structure of IS431-mecA-IS431 reminded us of the structure of composite transposon (4, 5). However, analysis of nucleotide sequences flanking this structure revealed no conserved target duplication (Fig. 2). Thus, there is no evidence to support the possibility that the mec complex was transferred as a composite transposon (Fig. 2). It seems more likely that the structure (IS431L-mec complex) was constructed by the insertion of an IS431 copy upstream of the mec complex after the mec complex was integrated in SCC to form SCCmec. Although the structure was distributed preferentially in S. haemolyticus strains in our study, this structure is not confined to S. haemolyticus, since it has been identified recently in one S. sciuri strain as well (34).
The most common mec complex was class A, the prototype mec complex, which was found in practically all staphylococcal species including S. aureus (10). Therefore, it would be reasonable to assume that some S. haemolyticus strains had also acquired class A mec complex in the past. However, the observed high prevalence of the class C mec complex or IS431L-class A mec complex in S. haemolyticus requires explanation. The most plausible explanation is that an S. aureus clone, in which IS431L was inserted upstream of the class A mec complex, significantly expanded in the hospital environment. In this case, however, we have to assume some advantage for the strain with a class C mec complex to survive preferentially in the hospital environment (see below).
It has been postulated that the class B mec complex was transmitted from C-NS to S. aureus based on its carriage of ψIS1272; the insertion sequence (IS1272) is widely disseminated in S. haemolyticus strains but not in S. aureus strains (1). Extensive nucleotide sequencing of various mec complexes in this study provided further concrete evidence for the active transmission of mec complexes across staphylococcal species. The nucleotide sequence determination of the regions between IS1272 and ΔmecR1 unequivocally showed identity of the structures across three staphylococcal species. For example, the class C2 mec complex found in S. haemolyticus and S. epidermidis strains had an identical deletion point of the ΔmecR1, clearly showing that the two mec complexes have the same evolutionary origin. In S. haemolyticus strain FA83 carrying the class A mec complex, IS431L was located 190 bp to the left of the 3′ end of the intact mecI gene. The position coincided with the location of IS431 in S. sciuri strain SK8, which was previously identified by others (34). The data indicated that the structure, IS431L-class A mec complex, also has been transmitted across species barriers.
The ψIS1272 copies identified in three species, S. aureus, S. haemolyticus, and S. epidermidis, suffered an identical partial deletion, indicating that the insertion sequence was not capable of transposition (2). Therefore, neither of the insertion sequences IS431 (see above) or IS1272, found closely associated with mecA, seem to play a role in interspecies transfer of the mecA gene complex. On the other hand, nucleotide sequence analysis of four classes of mec complexes revealed that each complex was a component of SCCmec, a staphylococcal mobile genetic element (16, 17). For example, nucleotide sequencing of 450 bp of DNA upstream of ΔmecR1 of the class D mec complex in S. caprae JA186 revealed that it was identical to a part of type II SCCmec identified in S. aureus strain N315 (16, 17). Therefore, it is likely that the diversification of the mec complex occurred (presumably from the prototypic class A mec complex) after its establishment as a component of SCCmec in a C-NS strain. Diversified mec complexes then seem to have been transmitted to S. aureus and other staphylococcal species.
As demonstrated in pre-MRSA strain N315, the presence of an intact mecI gene causes repression of mecA gene transcription and makes the cell susceptible to methicillin (10, 12, 13, 22). S. haemolyticus strain SH621 had a class A mec complex whose mecI gene was intact, and only a small level of PBP2′ production was observed (Table 4). Population analysis of the strain showed a typical heterogeneous methicillin resistance, and the MIC of methicillin for this strain was 2 mg per liter. By selecting the strain with 8 mg of methicillin per liter, only the mutants with high-level methicillin resistance were obtained (MIC ≥ 512 mg/liter) (Table 4). On the other hand, mixture of mutants with intermediate-level (seven mutants) and high-level (three mutants) methicillin resistance were obtained by the selection with 3 mg of methicillin per liter. It was noted that in all seven mutants with intermediate resistance, mecI was partially or completely deleted by the IS431L-mediated deletion (Fig. 4). This coincides with the previous observation with N315 (10): when the strain is selected with 3 mg of methicillin per liter, heterogeneous methicillin-resistant mutants are obtained for which the methicillin-MICs are comparable to those for the seven mutants in this study. However, in N315, the inactivation of mecI function is caused by mutations incorporated either in the structural gene or at the mecA gene operator to which the MecI repressor is supposed to bind (10, 27). The observation presented in this study indicates that the IS431L-mediated mecI gene inactivation in SH621 occurs at least several times more frequently than the mutational inactivation of mecI. This coincides with the difference in the population curves between SH621 and N315 (Fig. 5): SH621 contains about 10-fold more subpopulations resistant to 2 and 4 mg of methicillin per liter than does N315 (Fig. 5). This high rate of generation of mutants with the mecI gene deleted may confer a survival advantage to the strain. This may explain the clonal dominance of IS431L-carrying S. haemolyticus strains in the hospital environment.
The mutants listed in Table 4 possessed three different levels of methicillin resistance. Intermediate methicillin resistance seems to be caused by the inactivation of the mecI gene. On the other hand, except for the two strains with a marginal level of resistance, the mutant strains retaining intact mecI showed homogeneous methicillin resistance (Fig. 5). In the representative strain SH621-810, mecA transcription was inducible with methicillin despite the presence of intact mecI (Table 5). Considering the frequency of occurrence of the mutants with this property (appearance rate, 9.8 × 10−6 [Fig. 5]), a mutation would be postulated in a regulatory gene involved in mecA gene transcription. However, such a gene explaining this phenomenon is totally unknown at this moment. It is also unknown why a rapid slide latex agglutination assay for PBP2′ could discriminate between SH621 and its derivatives represented by SH621-810 even though the amount of mecA gene transcripts was almost comparable between the two (Table 4). This result was reproducible in repeated experiments, suggesting that some factors other than the level of transcript, such as mRNA stability, are involved with the phenomenon. Further studies are required to solve these questions and elucidate the regulatory system for methicillin resistance in S. haemolyticus.
ACKNOWLEDGMENTS
We thank T. Watanabe for her excellent technical assistance.
This work was supported by a Core University Program under Japan Society for the Promotion of Science (JSPS), coordinated by the University of Tokyo Graduate School of Medicine and University Sains Malaysia School of Medical Sciences; by a Specially Designated Research Promotion of Monbusho, Japan; and by a Grant for International Health Cooperation Research (11C-4) from the Ministry of Health and Welfare.
REFERENCES
- 1.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer G L, Thanassi J A, Niemeyer D M, Pucci M J. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1996;40:924–929. doi: 10.1128/aac.40.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberis-Maino L, Berger-Bächi B, Weber H, Beck W D, Kayser F H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59:107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- 4.Byrne M E, Gillespie M T, Skurray R A. 4′,4"-Adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid. 1991;25:70–75. doi: 10.1016/0147-619x(91)90008-k. [DOI] [PubMed] [Google Scholar]
- 5.Byrne M E, Littlejohn T G, Skurray R A. Transposons and insertion sequences in the evolution of multiresistant Staphylococcus aureus. In: Novick R P, editor. Molecular biology of staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 1303–1311. [Google Scholar]
- 6.Dickinson T M, Archer G L. Phenotypic expression of oxacillin resistance in Staphylococcus epidermidis: role of mecA transcriptional regulation and resistant-subpopulation selection. Antimicrob Agents Chemother. 2000;44:1616–1623. doi: 10.1128/aac.44.6.1616-1623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackbarth C J, Chambers H F. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanaki H, Hiramatsu K. Clinical evaluation of latex slide agglutination kit for penicillin-binding protein 2′ detection. Igaku Yakugaku. 1999;41:749–754. [Google Scholar]
- 9.Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995;39:531–543. doi: 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA) FEBS Lett. 1991;298:133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu K, Kihara H, Yokota T. Analysis of borderline-resistant strains of methicillin-resistant Staphylococcus aureus using polymerase chain reaction. Microbiol Immunol. 1992;36:445–453. doi: 10.1111/j.1348-0421.1992.tb02043.x. [DOI] [PubMed] [Google Scholar]
- 12.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu K, Ito T, Hanaki H. Mechanisms of methicillin and vancomycin resistance in Staphylococcus aureus. Clin Infect Dis. 1999;5:221–242. [Google Scholar]
- 14.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 15.Hussain Z, Stoakes L, Massey V, Diagre D, Fitzgerald V, El Sayed S, Lannigan R. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J Clin Microbiol. 2000;38:752–754. doi: 10.1128/jcm.38.2.752-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi N, Urasawa S, Uehara N, Watanabe N. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:717–720. doi: 10.1128/aac.42.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi N, Urasawa S, Uehara N, Watanabe N. Distribution of insertion sequence-like element IS1272 and its position relative to methicillin resistance genes in clinically important staphylococci. Antimicrob Agents Chemother. 1999;43:2780–2782. doi: 10.1128/aac.43.11.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhler T. General aspect and changes of nucleic acid quantitation by PCR. In: Köhler B T, Thamm B, Labner D, Rost A-K, Pustowoft B, Remake H, editors. Quantitation of mRNA by polymerase chain reaction. Vol. 1. Berlin, Germany: Springer-Verlag KG; 1995. pp. 1–14. [Google Scholar]
- 21.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 22.Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob Agents Chemother. 1996;40:2680–2685. doi: 10.1128/aac.40.12.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuhashi M, Song M D, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167:975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakatomi Y, Sugiyama J. A rapid latex agglutination assay for the detection of penicillin-binding protein 2′. Microbiol Immunol. 1998;42:739–743. doi: 10.1111/j.1348-0421.1998.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 25.Pannetier C, Delassus S, Darche S, Saucier C, Kourilsky P. Quantitative titration of nucleic acids by enzymatic amplification reactions run to saturation. Nucleic Acids Res. 1993;21:577–583. doi: 10.1093/nar/21.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryffel C, Bucher R, Kayser F H, Berger-Bächi B. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991;173:7416–7422. doi: 10.1128/jb.173.23.7416-7422.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma V K, Hackbarth C J, Dickinson T M, Archer G L. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J Bacteriol. 1998;180:2160–2166. doi: 10.1128/jb.180.8.2160-2166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song M D, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987;221:167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki E, Kuwahara-Arai K, Richardson J F, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicob Agents Chemother. 1993;37:1219–1226. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob Agents Chemother. 1992;36:429–434. doi: 10.1128/aac.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 32.Weller T M. The distribution of mecA, mecR1 and mecI and sequence analysis of mecI and the mec promoter region in staphylococci expressing resistance to methicillin. J Antimicrob Chemother. 1999;43:15–22. doi: 10.1093/jac/43.1.15. [DOI] [PubMed] [Google Scholar]
- 33.Wu S W, de Lencastre H, Tomasz A. The Staphylococcus aureus transposon Tn551: complete nucleotide sequence and transcriptional analysis of the expression of the erythromycin resistance gene. Microb Drug Resist. 1999;5:1–7. doi: 10.1089/mdr.1999.5.1. [DOI] [PubMed] [Google Scholar]
- 34.Wu S W, de Lencastre H, Tomasz A. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J Bacteriol. 1998;180:236–242. doi: 10.1128/jb.180.2.236-242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]