Abstract
Endothelial cells differ from other cell types responsible for the formation of the vascular wall in their unusual reliance on glycolysis for most energy needs, which results in extensive production of lactate. We find that endothelium‐derived lactate is taken up by pericytes, and contributes substantially to pericyte metabolism including energy generation and amino acid biosynthesis. Endothelial–pericyte proximity is required to facilitate the transport of endothelium‐derived lactate into pericytes. Inhibition of lactate production in the endothelium by deletion of the glucose transporter‐1 (GLUT1) in mice results in loss of pericyte coverage in the retina and brain vasculatures, leading to the blood–brain barrier breakdown and increased permeability. These abnormalities can be largely restored by oral lactate administration. Our studies demonstrate an unexpected link between endothelial and pericyte metabolisms and the role of endothelial lactate production in the maintenance of the blood–brain barrier integrity. In addition, our observations indicate that lactate supplementation could be a useful therapeutic approach for GLUT1 deficiency metabolic syndrome patients.
Keywords: BBB permeability, endothelial metabolism, Glucose, lactate, pericyte metabolism
Subject Categories: Metabolism, Neuroscience, Vascular Biology & Angiogenesis
Metabolic shuttling of lactate between endothelial cells and neurovascular pericytes supports CNS vasculature integrity.

Introduction
Vascular integrity and the dynamic control of permeability are crucial to normal organ functions. Structurally, vascular integrity is maintained by formation of endothelial cell–cell junctions that can open and close in response to various signals. Another important element is the extent of pericyte coverage of blood vessels. Pericytes wrap around capillary and venular endothelial cells and share the basement membrane with the endothelium, thereby ensuring close physical and paracrine contacts between these two cell types. Pericytes are particularly important in the central nervous system (CNS) where they help to maintain the blood–brain barrier (BBB) (Armulik et al, 2010) and regulate microvascular blood flow (Yamanishi et al, 2006). A deficiency of pericytes in the CNS results in the BBB breakdown and death (Hellstrom et al, 2001; Armulik et al, 2010; Nikolakopoulou et al, 2019). While endothelial–pericyte communications are clearly important, their nature is not fully understood.
Endothelial cells (ECs) are unusual in their reliance on glycolysis as the primary means of energy generation (Krutzfeldt et al, 1990; De Bock et al, 2013a; Schoors et al, 2014; Kim et al, 2017; Yu et al, 2017a; Faulkner et al, 2020). The hallmark of glycolysis is the production of large amounts of lactate and, indeed, most of endothelial glucose (~90%) is catabolized to lactate and secreted as lactate extracellularly (Kim et al, 2017). With this much endothelial lactate entering the extracellular space, a significant amount of lactate would be expected to be present in the inner basement membrane space shared by ECs and pericytes, thereby making endothelial cells‐produced lactate available to pericytes. It was the purpose of this study to identify the metabolic fate of endothelium‐derived lactate in pericyte metabolism and the role of this metabolic crosstalk in maintenance of vascular homeostasis.
Several studies have shown that lactate can be used as fuel while quantitative analysis of lactate fluxes in mice suggests that it can be the primary source of carbons for the TCA cycle in a number of tissues (van Hall, 2010; Hui et al, 2017; Brooks, 2018; Jin et al, 2019). In the brain, lactate accounts for ~10–20% of energy generation under normal condition but that can rise to 60% when the lactate transporter is fully saturated (Boumezbeur et al, 2010; Hui et al, 2017). At the cellular level, neurons (Waagepetersen et al, 1998; Descalzi et al, 2019), fibroblasts (Shen et al, 2020), macrophages (Liu et al, 2020b; Zhang et al, 2020), and cancer cells (Bonuccelli et al, 2010) have all been reported to utilize lactate for energy generation albeit few details of this process are understood.
We found that lactate is an important source of carbons for pericyte metabolism and that it contributes directly to energy generation and amino acid biogenesis in these cells. Endothelial lactate export is mediated by MCT1 and MCT5 transporters while pericytes utilize MCT12 to import lactate. In the absence of endothelium‐derived lactate, pericytes in the CNS vasculature undergo apoptotic cell death resulting in impaired BBB and increased permeability while oral lactate supplementation can restore both pericyte coverage and the BBB integrity. These observations point to the crucial role of endothelial lactate in endothelium‐pericyte communications and maintenance of the BBB integrity.
Results
Deficiency of GLUT1 in ECs inhibits glucose uptake, glycolysis, and ATP production
The 14‐member family of glucose transporters (GLUTs) plays an important role in cellular glucose uptake and glucose homeostasis in mammals (Bertrand et al, 2020). To study the expression of glucose transporters in ECs, we took advantages of the publicly available endothelial scRNAseq atlas (EC atlas) (Kalucka et al, 2020). Analysis of the atlas data suggested that Glut1 is the predominant glucose transporter in brain ECs (Appendix Fig S1A). This was confirmed by immunostaining of the brain and retinal vasculature that showed high level and EC specificity of Glut1 expression (Appendix Fig S1B and C).
To explore the role of GLUT1 in the endothelium, we first examined glucose uptake after GLUT1 knockdown (siGLUT1) in BMECs (Appendix Fig S2A) using 2‐deoxy‐2‐[(7‐nitro‐2,1,3‐benzoxadiazol‐4‐yl)amino]‐d‐glucose (2‐NBDG), a fluorescence‐labeled glucose analog. As expected, there was a significant reduction in the number of 2‐NBDG positive cells following GLUT1 knockdown (Appendix Fig S2B–D) compared to control siRNA‐treated BMECs (siCON). Next, we evaluated the effect of GLUT1 knockdown on the extracellular acidification rate (ECAR) during sequential treatment with glucose, oligomycin, and 2‐DG. While control (siCON) BMECs were able to increase ECAR after glucose treatment, indicating glycolytic activity, it was significantly decreased in siGLUT1‐treated ECs (Appendix Fig S2E and F). The subsequent treatment with oligomycin triggered a further increase in ECAR in siCON‐treated ECs, revealing preserved glycolytic capacity, while no comparable increase was observed in siGLUT1‐treated BMECs (Appendix Fig S2E and G). With 2‐DG treatment, we found there was a strong reduction in glycolytic reserve in siGLUT1‐treated compared to siCON‐treated BMECs (Appendix Fig S2E and H). To investigate whether these changes in glycolytic activity following GLUT1 KD affected ATP production, we measured total ATP level and mitochondrial (mitoATP) and glycolytic (glycoATP) ATP production rates using a Seahorse analyzer. As expected, both total ATP and glycoATP production were decreased in siGLUT1‐treated ECs (Appendix Fig S2I). However, mitoATP production was increased, suggesting that an increase in oxidative phosphorylation is compensating for the decreased glycolytic ATP production (Appendix Fig S2I).
Deficiency of GLUT1 in ECs inhibits angiogenesis and vascular integrity
A previous study using a Glut1 chemical inhibitor (BAY‐876) reported reduced proliferation but not migration of endothelial cells in culture (Veys et al, 2020). In agreement with these data, siRNA‐mediated Glut1 knockdown also inhibited endothelial proliferation (Appendix Fig S3A and B). However, we observed a reduction in EC migration following GLUT1 KD (Appendix Fig S3C and D). As expected with the observed decrease in migration, there was a profound reduction in the number of filopodia in siGLUT1‐treated BMECs (Appendix Fig S3E and F) and Glut1‐deleted tipECs in retinal vasculature (Appendix Fig S3G and H).
To further investigate the role of GLUT1 in vivo, we generated endothelial‐specific inducible Glut1 knockout mice (Glut1 fx/fx Cdh5(PAC)CreERT2, hereafter denoted Glut1 iECKO) (Appendix Fig S4A–C). We first analyzed the effect of endothelial Glut1 deletion on angiogenesis using retinal vasculature. Neonatal littermates (WT, Glut1 iEC+/fx (Glut1 haplodeficient mice in EC) and Glut1 iECKO) were intraperitoneally injected with tamoxifen at P1 (postnatal day 1) and P2, and vasculature was observed at P6.5 using retinal whole mounts. Isolection‐B4 staining showed a gene dose‐dependent reduction in angiogenic outgrowth (Appendix Fig S5A and B). Consistent with a previous report using an inducible PDGFβ‐Cre‐driven Glut1 deletion mice (Veys et al, 2020), our Glut1 iECKO mice also displayed severe weight loss (Appendix Fig S5C) and lethality (Appendix Fig S5D) starting on P6 with all mice dying by P15. Endothelial Glut1 deletion in fully mature 8‐week‐old mice also resulted in universal lethality with similar kinetics (Appendix Fig S5E).
To further evaluate the effect of endothelial Glut1 deletion on blood vessels, we examined pericyte coverage in Glut1 iECKO and control mice. Immunostaining showed a striking reduction in pericyte coverage in the retinal vasculature of P6 Glut1 iECKO compared to wild‐type littermates (Fig 1A and B). The analysis of the retinal and brain vasculature in adult mice (8‐weeks old) 10 days after induction of endothelial GLUT1 deletion also showed a significant reduction in pericyte coverage (Appendix Fig S6A and B and Fig 1C and D) while SMC coverage was not altered (Fig 1C and E). In addition, a tight junction protein (claudin 5) was absent in the brain vasculature of Glut1iECKO mice (Fig 1F) suggesting that the pericyte loss is affecting the integrity of the BBB. To investigate the mechanism of the pericyte loss, we performed immunostaining for apoptotic cell death marker (cleaved caspase‐3) and found caspase‐3‐positive pericytes in Glut1iECKO mice showing that the loss of pericytes is mediated by apoptotic cell death (Fig 1G).
Figure 1. Loss of GLUT1 in EC decreases pericyte coverage on the vasculature and alters BBB permeability.
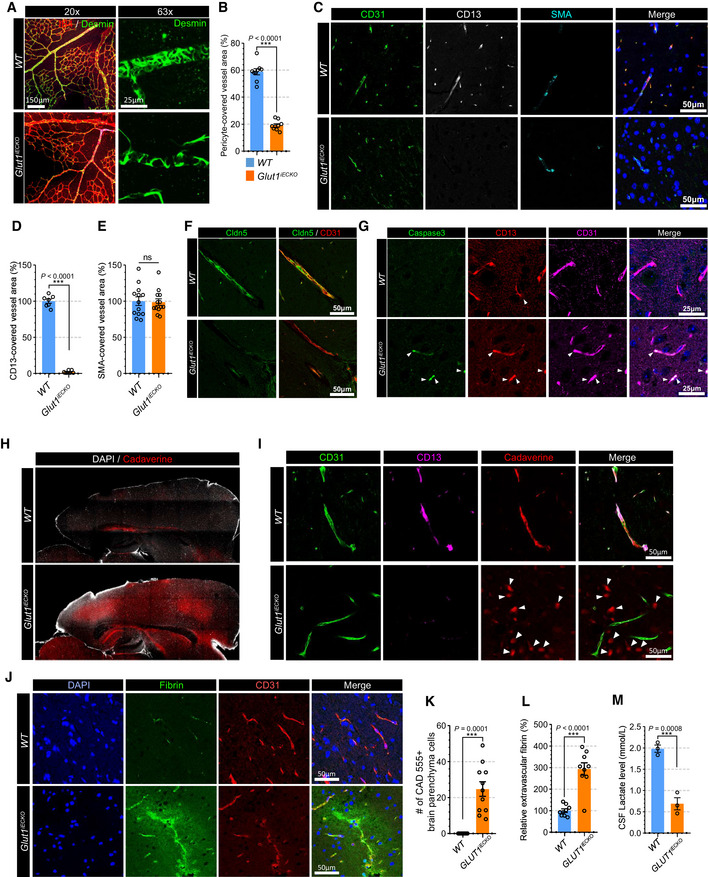
- Retinal whole mount immunostaining for Isolectin‐B4 (red) and Desmin (green) in WT (top panel) and Glut1 iECKO (bottom panel) mice showing pericyte coverage in the vasculature. Left and right panels show representative images with 20× or 63× magnification, respectively.
- Quantification of the pericytes coverage in the retinal vasculature of WT (blue) and Glut1 iECKO (orange) mice. (n = 9 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Immunostaining for CD31 (green), CD13 (white; pericyte marker) and αSMA (cyan) using paraffin section from WT (top panel) and Glut1 iECKO (bottom panel) mice.
- Quantification of the pericytes coverage using CD13 immunostaining in the brain vasculature of WT (blue) and Glut1 iECKO (orange) mice. (n = 7 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of the smooth muscle cell coverage using αSMA immunostaining in the brain vasculature of WT (blue) and Glut1 iECKO (orange) mice. (n = 13 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Immunostaining for Cldn5 (green) and CD31 (red) using a brain section of WT (top panel) and Glut1 iECKO (bottom panel) mice.
- Immunostaining for caspas‐3 (green), CD13 (red) and CD31 (cyan) using a brain section of WT (top panel) and Glut1 iECKO (bottom panel) mice.
- Permeability assay using vibratome section showing the leakage of retro‐orbital injected cadaverine dye (red) in the brain of WT (top panel) and Glut1 iECKO (bottom panel) mice. DAPI staining (white) shows the boundary of brain section.
- Permeability assay using vibratome section showing the leakage of retro‐orbital injected cadaverine dye (red) in the brain of WT (top panel) and Glut1 iECKO (bottom panel) mice. Note that cadaverine is retained in the intravascular area of WT mice (upper panel), but extravasated and accumulated in the brain parenchyma (white arrowheads) of Glut1 iECKO (bottom panel) mice.
- Representative confocal images of fibrin (green), CD31 (red) and DAPI (blue) immunostaining in the brain vasculature of WT (top panel) and Glut1 iECKO (bottom panel) mice.
- Quantification of the number of cadaverine 555 positive brain parenchyma cells in WT (blue) and Glut1 iECKO (orange) mice. (n = 13 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of extravascular fibrin deposits in WT (blue) and Glut1 iECKO (orange) mice. (n = 9 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of lactate in the cerebrospinal fluid of WT (blue) and Glut1 iECKO (orange) mice. (n = 3 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
Since the absence of pericytes in the vasculature results in increased permeability (Armulik et al, 2010), and given that the deletion of Glut1 in ECs causes pericyte depletion, we sought to determine whether there was an increase in permeability of the brain vasculature in Glut1 iECKO mice. To this end, we tested extravascular accumulation of an exogenous fluorescent dye (cadaverine) and endogenous plasma protein (fibrin). Retroorbital cadaverine injection demonstrated a significantly higher appearance of the dye in the brain of Glut1 iECKO compared to wild‐type littermates (Fig 1H). While cadaverine was retained almost exclusively intravascularly in WT mice, large amounts were observed in the brain parenchyma of Glut1 iECKO mice (Fig 1I and K). Similarly, there was a marked increase in extravascular fibrin in the brains of Glut1 iECKO mice compared with wild‐type littermates (Fig 1J and L). Finally, we examined whether Glut1 deficiency in ECs affects endothelial permeability. Staining of an endothelial monolayer for VE‐cadherin showed no changes in its localization or the appearance of adherence junctions in siGLUT1‐treated BMECs (Appendix Fig S7A and B) and in vitro permeability assay using FITC‐dextran also showed no differences in permeability between siCON and siGLUT1‐treated BMECs (Appendix Fig S7C). This indicates that the altered BBB permeability in Glut1 iECKO mice is not caused by endothelial autonomous phenotype, but by pericyte loss.
Reduced concentration of lactate in the cerebrospinal fluid (CSF) is one of the key abnormalities in patients with GLUT1 mutations (De Vivo et al, 1991). In agreement with these data, measurements of CSF lactate level demonstrated a significant reduction in Glut1 iECKO mice compared to wild‐type littermates (Fig 1M). The CSF lactate reduction and pericyte loss in Glut1iECKO mice imply that lactate plays an important role to maintain pericyte coverage.
Endothelium‐derived lactate feeds pericytes
Proper interaction with endothelial cells is essential for recruitment and migration of pericytes along blood vessels (Armulik et al, 2005; Yang et al, 2011). Since endothelial cells produce and release large amount of lactate into the vasculature (De Bock et al, 2013a; Kim et al, 2017) and given their close physical proximity to pericytes, we tested whether endothelial lactate plays an important role in pericyte biology. To check whether pericytes utilize endothelium‐derived lactate, we used Laconic, a Forster Resonance Energy Transfer (FRET)‐based quantitative intracellular lactate sensor (San Martin et al, 2013). HBVPs (human brain vascular pericytes) were infected with the Laconic‐expressing adenovirus and cultured in a presence of the glycolysis inhibitor (iodoacetic acid, 500 nM) to minimize basal glycolytic lactate production. As expected, increasing lactate concentration, from 2 to 100 mM, led to a dose‐dependent increase in the mTFP/Venus fluorescence ratio (Fig 2A and B). To study whether EC‐derived lactate is taken up by pericytes, we used a two‐well cell culture dish to coculture BMECs and HBVPs (Fig 2C). Both dish chambers were seeded with, respectively, BMECs and Laconic‐expressing HBVPs. Once the cells were attached (24 h after seeding), the culture insert was removed, allowing cell migration to take place (Fig 2D). When examined after another 24 h, we observed a striking increase in the mTFP/Venus fluorescence ratio in HBVPs in direct contact with ECs but not in HBVPs cells that were not in contact with the ECs (Fig 2E–G). Given that a knockdown of GLUT1 in ECs reduced their glycolysis, we further explored whether endothelial glycolysis affects lactate transport into pericytes. HBVPs in contact with siGLUT1‐treated BMECs showed markedly reduced mTFP/Venus fluorescence ratio than HBVPs in contact with siCON‐treated EC (Fig 2H and I). Taken together, those data indicate that ECs‐derived lactate can enter pericytes and that this process is facilitated by endothelial–pericyte proximity.
Figure 2. Endothelium‐derived lactate feed pericytes.
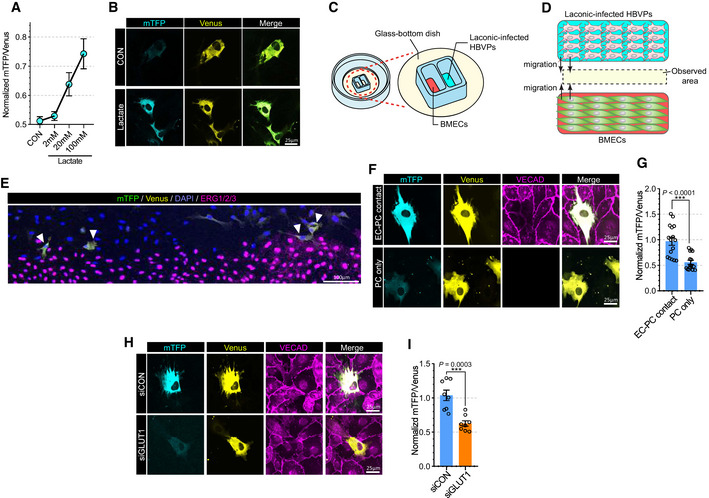
- Quantification of the mTFP/Venus fluorescence ratio of laconic (lactate sensor) showing concentration‐dependent lactate uptake in HBVPs. (n = 8 from 3 independent experiments. Error bars indicate the standard error of the mean (SEM)).
- Representative images showing mTFP (blue) and Venus (yellow) fluorescence in control and lactate (20 mM)‐treated HBVPs.
- Schematic illustration for the lactate uptake assay with 2 well chamber. Cells were seeded on a glass bottom dish. BMECs and adenoviral laconic‐infected HBVPs were separated by 2 well chamber. After attachment, the 2 well chamber was removed for cell migration.
- Schematic illustration for the lactate uptake assay after chamber removal. At 24 h after chamber removal, the fluorescence of mTFP and Venus were observed in the middle of the wells (yellow area).
- Representative stitched image showing mTFP (green), Venus (yellow), DAPI (blue) and ERG1/2/3 (purple) fluorescence from BMECs and laconic‐infected HBVPs on a 2 well chamber.
- Representative images (high magnification; 60×) showing mTFP (cyan), Venus (yellow) and VECAD (purple) fluorescence from BMECs and laconic‐infected HBVPs on a 2 well chamber. The upper panel shows HBVPs with endothelial contact and the lower panel shows HBVPs without endothelial contact.
- Quantification of the mTFP/Venus fluorescence ratio of laconic in HBVPs with endothelial contact (EC‐PC contact) or HBVPs alone (PC only). (n = 16 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Representative images showing mTFP (cyan) and Venus (yellow) fluorescence in HBVPs which have direct contact with control (upper panel) or GLUT1 (lower panel) siRNA‐treated BMECs. VECAD immunostaining (purple) was used for endothelial staining.
- Quantification of the mTFP/Venus fluorescence ratio of laconic in HBVPs which have direct contact with control (siCON) or GLUT1 (siGLUT1) siRNA‐treated BMECs (n = 12 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
Pericytes utilize lactate for oxygen consumption and pyruvate synthesis
We next explored the metabolic role of lactate in various vascular and nonvascular cell types. First, we evaluated the effect of lactate supplementation on oxygen consumption rate (OCR) to test whether cells use lactate for mitochondrial oxidation. Only HBVPs and mouse brain pericytes (mBPs), but not human aortic smooth‐muscle cells (HASMCs), human brain microvascular endothelial cells (BMECs), or fibroblasts (3T3L1) increased their OCR rate (Fig 3A). Importantly, pericyte OCR was increased in a lactate concentration‐dependent manner (Fig 3B). Furthermore, lactate treatment also increased OCR in HBVPs even in the presence of other nutrients such as glucose (5 mM), pyruvate (0.5 mM), and glutamine (0.5 mM) (Fig 3C). In agreement with these data, mass spectroscopy analysis demonstrated increase in ATP in HBVPs following lactate treatment (Fig 3D and Appendix Fig S8A and B).
Figure 3. Lactate induces oxygen consumption in pericytes.
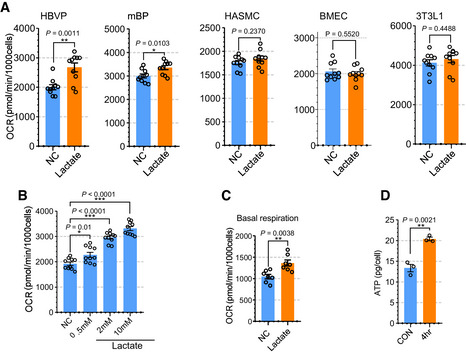
- Quantification of oxygen consumption rate (OCR) showing the effect of lactate treatment on the respiration (induced) in various cells including HBVP, mBP, HASMC, BMEC and 3T3L1. The media was changed into glucose‐, pyruvate‐ and glutamine‐free media before OCR measurement and lactate (2 mM) was added the media to measure induced OCR. (n = 10 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of OCR showing the effect of lactate dose dependent treatment (0.5, 2, and 10 mM) in HBVPs. (n = 9 from independent experiments. Error bars indicate the standard error of the mean (SEM)).
- Quantification of OCR (basal respiration) with full media (5 mM glucose, 0.5 mM pyruvate and 0.5 mM glutamine) showing the effect of lactate treatment (2 mM) in HBVPs. (n = 7 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of ATP using a LC‐MS/MS spectrometry in control and lactate treated (2 mM, 4 h) HBVPs (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
To understand the metabolic role of exogenous lactate in pericytes, we studied the fate of [13C3]‐lactate using tandem mass spectrometry (Appendix Fig S8C and D). First, we treated HBVPs with [13C3]‐lactate (2 mM) for 24 h and measured the concentration of [13C3]‐lactate in the media. As shown in Fig 4A, the concentration of [13C3]‐lactate was decreased after 24 h, likely due to its uptake by pericytes. To confirm this, we measured [13C3]‐lactate in HBVPs after 1‐hr treatment of [13C3]‐lactate (2 mM). As expected, [13C3]‐lactate was detectable in [13C3]‐lactate‐treated HBVPs, but not in control HBVPs (Fig 4B). Then we measured [13C3]‐pyruvate in HBVPs to confirm the cellular conversion of lactate into pyruvate (Appendix Fig S8E and F). While in control HBVPs, [13C3]‐pyruvate was not detectable, it was present in [13C3]‐lactate‐treated HBVPs (Fig 4C). To explore the dynamics of cellular uptake of lactate and conversion into pyruvate, HBVPs were exposed to [13C3]‐lactate (2 mM) for various times from 10 min to 4 h. There was a clear time dependence of 13C‐lactate to 13C3‐pyruvate conversion with concentration of [13C3]‐pyruvate in HBVPs increasing over time (Fig 4D). Similarly, the addition of increasing amounts of [13C3]‐lactate (0.2, 2, and 10 mM) to HBVP culture media showed a 13C‐lactate concentration‐dependent increase in cellular [13C3]‐pyruvate levels (Fig 4E). Interestingly, the lactate uptake/pyruvate conversion process did not reach saturation even at 10‐mM lactate concentration in the HBVP media (Fig 4E).
Figure 4. Pericytes utilize lactate for pyruvate synthesis.
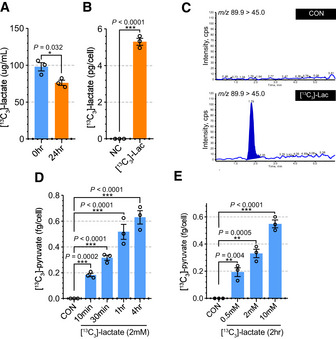
- Quantification of 13C3‐lactate in the media culturing HBVPs. [13C3]‐lactate (2 mM) was added to the media and measured at 0 and 24 h after incubation. (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Quantification of [13C3]‐lactate in the pellet of HBVPs showing HBVPs uptake [13C3]‐lactate from the media. (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Multiple reaction monitoring (MRM) chromatograms for [13C3]‐pyruvate in control (top) and [13C3]‐lactate treated (bottom) HBVPs. The molecular mass for M1 and M3 of [13C3]‐pyruvate were 89.9 and 45.0, respectively.
- Quantification of 13C3‐pyruvate in [13C3]‐lactate (2 mM) treated HBVPs at different time points. (0–4 h) (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from 1‐way ANOVA).
- Quantification of [13C3]‐pyruvate in HBVPs with different concentration of [13C3]‐lactate treatment. (0.5, 2, and 10 mM) (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from 1‐way ANOVA).
Metabolic fate of pericytes’ lactate
We next set out to elucidate the metabolic fate of exogenous lactate carbons in pericytes. Following isotopic labeling of HBVP with exogenous [13C3]‐lactate (2 mM), mass spectrometry was used to trace labeled carbon fraction of various metabolites (Fig 5A–K). A [13C6]‐glucose (5 mM)‐treated group was added for parallel comparison of lactate metabolism with glucose metabolism, and a high concentration of [13C3]‐lactate (20 mM)‐treated group (in glucose free media) was added to achieve higher labeling efficiency of lactate, respectively (Fig 5A and B).
Figure 5. Glucose and lactate metabolism in pericyte.
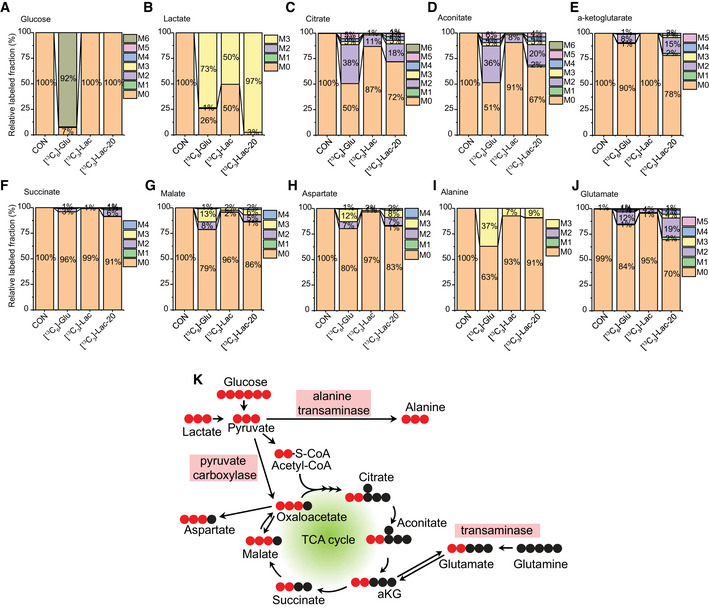
-
A–JIsotope‐labeled carbon tracing experiment showing glucose and lactate metabolism in HBVPs. HBVPs were treated with isotope‐labeled nutrients (CON: control, [13C6]‐glu: 5 mM [13C6]‐glucose, [13C3]‐Lac: 2 mM [13C3]‐Lactate, [13C3]‐Lac‐20: 20 mM [13C3]‐Lactate in glucose free media) for 12 h and analyzed by a Q‐exactive MS spectrometry. Barplots show relative fraction of unlabeled (M0) and labeled forms (M1–6) in various metabolites including A. Glucose, B. Lactate, C. Citrate, D. Aconitate, E. α‐ketoglutarate (αKG), F. Succinate, G. Malate, H. Aspartate, I. Alanine, J. Glutamate (n = 3 from independent experiments.)
-
KSchematic diagrams showing the glucose‐ or lactate‐derived carbon flow in pericytes (HBVPs) when [13C6]‐glucose or [13C3]‐lactate was used as labeling substrate(s). (Unlabeled carbon: black circle, labeled carbon: red circle).
When pyruvate enters the TCA cycle, it is oxidized to acetyl‐CoA, which contains 2 carbons (Fig 5K; Zhang et al, 2017). Therefore, the M2 fraction (the two carbon‐labeled fractions) in TCA components indicates that they were synthesized with the acetyl‐CoA bearing two labeled carbons from [13C6]‐glucose or [13C3]‐lactate. Citrate and aconitate displayed significant presence of the M2 fraction following HBVP treatment with [13C3]‐lactate (Fig 5C and D). With 50% of lactate labeled in HBVPs, the M2 fraction accounted for 11% of citrate and 8% of aconitate (Fig 5C). When lactate labeling increased to 97%, M2 contribution increased to 18% for citrate and 20% for aconitate (Fig 5C and D). The total fraction of labeled forms of aconitate (M1–M6) was 33% in [13C3]‐lactate (20 mM)‐treated HBVPs, indicating that ~30% of aconitate was synthesized from lactate. At the same time, 49% of aconitate contained labeled (M1–M6) fractions in [13C6]‐glucose‐treated HBVPs showing that glucose, as expected, is one of the major sources of carbon for biosynthesis of aconitate (Fig 5C).
Compared with aconitate and citrate, the total fraction of labeled forms in α‐ketoglutarate (αKG, Fig 5E) and succinate (Fig 5F) were decreased in both [13C6]‐glucose and [13C3]‐lactate‐treated HBVPs implying a strong efflux of citrate to the cytoplasm, where labeled carbons are removed for fatty acid biosynthesis or an influx of carbons from glutamine metabolism (Fig 5K). Interestingly, the major fraction of labeled carbon in malate was the M3 instead of M2 (Fig 5G), suggesting that this malate fraction was synthesized directly from pyruvate via pyruvate carboxylase (Merritt et al, 2011) (Fig 5K). Although oxaloacetate was not detected due to low sensitivity of the mass spectrometry, the same labeling pattern between malate (Fig 5G) and aspartate (Fig 5H) implies that the lactate‐derived M3 is the major fraction of oxaloacetate (Fig 5K).
The carbons from [13C3]‐lactate in HBVPs were also incorporated into amino acids including aspartate (Fig 5H), alanine (Fig 5I) and glutamate (Fig 5J). While M3 is the largest major fraction in alanine (Fig 5I), M2 was the largest fraction in glutamate (Fig 5J), indicating that alanine is synthesized from pyruvate by alanine transaminase (Liu et al, 2014) and glutamate from αKG by a transaminase (Suzuki & Knaff, 2005), respectively (Fig 5K). Two major fractions (M2 and M3) in malate (Fig 5G) and aspartate (Fig 5H) indicate that there are two different routes (M2 from TCA cycle‐derived oxaloacetate and M3 from pyruvate carboxylase‐derived oxaloacetate) for biosynthesis of these amino acids. Taken together, these data indicate that HBVPs have a distinct metabolic profile with significant contribution from lactate to the TCA cycle, acetyl‐CoA production, and biosynthesis of amino acids (Fig 5K).
Lactate transporters involved in lactate shuttling between ECs and pericytes
Lactate is transported across cell membranes by monocarboxylate transporters (MCTs, also known as Slc16a family; Halestrap & Meredith, 2004). To determine the lactate transporter(s) in ECs, we screened the expression level of MCT family members using single‐cell RNAseq databases (EndoDB and Brain cell atlas; Fig 6A–D; Vanlandewijck et al, 2018; Kalucka et al, 2020). Among 14 MCT genes, MCT1, 4, 5, 6, 7, 12, and 13 were expressed in ECs, with MCT1, MCT5, and MCT8 showing the highest level of expression. Since MCT8 (Fig 6D) is widely expressed in brain cells, while MCT1 (Fig 6B) and MCT5 (Fig 6C) expression is mainly restricted to ECs, we have focused on the role of these two transporters (MCT1 and 5) in EC‐to‐pericyte lactate trafficking.
Figure 6. MCT5 in endothelial cells regulates lactate export into pericyte.
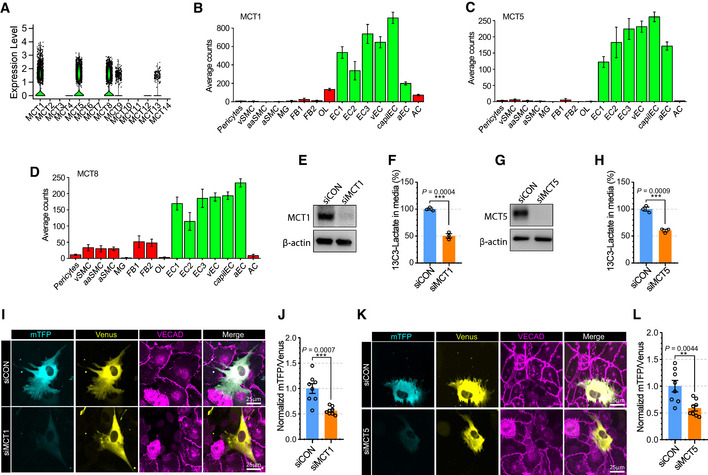
-
AViolin plot showing the expression of Slc16a family members (monocarboxylate transporters) in brain endothelial cells from scRNAseq database (EndoDB; 3,755 endothelial cells).
-
B–DBarplots showing the expression of B) MCT1 (Slc16a1), C) MCT5 (Slc16a4) and D) MCT8 (Slc16a2) across brain cells from the scRNAseq database (Brain cell atlas) (vSMC: venous smooth muscle cells, aaSMC: arteriolar SMCs, aSMC: arterial SMCs, MG: Microglia, FB1‐2: Fibroblast subtypes, OL: Oligodendrocytes, EC1‐3: EC subtypes, vEC: venous ECs, capiEC: capillary ECs, aEC: arterial ECs, AC: astrocytes) (1,100 endothelial cells, mean ± SD).
-
EWestern blot analysis for siRNA efficiency of MCT1.
-
FQuantification of 13C3‐Lactate using LC‐MS/MS in media of siCON or siSlc16a1 treated BMECs after 13C6‐glucose treatment (12 h). (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
-
GWestern blot analysis for siRNA efficiency of MCT12.
-
HQuantification of 13C3‐Lactate using LC‐MS/MS in media of siCON or siSlc16a4‐treated BMECs after 13C6‐glucose treatment (12 h). (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
-
IRepresentative images showing mTFP (cyan) and Venus (yellow) fluorescence in HBVPs which have direct contact with control (upper panel) or MCT1 (lower panel) siRNA‐treated BMECs. VECAD immunostaining was used for endothelial staining.
-
JQuantification of the mTFP/Venus fluorescence ratio of laconic in HBVPs which have direct contact with control (siCON) or MCT1 (siMCT1) siRNA‐treated BMECs. (n = 8 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
-
KRepresentative images showing mTFP (cyan) and Venus (yellow) fluorescence in HBVPs which have direct contact with control (upper panel) or MCT5 (lower panel) siRNA‐treated BMECs. VECAD immunostaining was used for endothelial staining.
-
LQuantification of the mTFP/Venus fluorescence ratio of laconic in HBVPs which have direct contact with control (siCON) or MCT5 (siMCT5) siRNA‐treated BMECs (n = 8 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
Measurements of [13C3]‐lactate in the media of [13C6]‐glucose (5 mM)‐treated BMECs revealed that secretion of lactate from ECs was decreased following MCT1 (Fig 6E and F) or MCT5 (Fig 6G and H) knockdown. Furthermore, BMEC‐HBVP coculture studies showed that pericytes in contact with ECs treated with either MCT1 (Fig 6I and J) and MCT5 (Fig 6K and L) siRNAs exhibited reduced mTFP/Venus ratios signifying decreased entry of endothelial lactate.
Given that MCT1 is predominantly expressed in ECs (Fig 6B), we explored whether MCT1 inhibition with a chemical inhibitor(Polanski et al, 2014) (AZD3965) (Appendix Fig S9A) would have a phenotype similar to that of Glut1 iECKO mice. The lactate level in the CSF fluid was significantly lower following AZD3965 treatment (Appendix Fig S9B) with the brain vasculature displaying the pericytes loss (Appendix Fig S9C and D) and BBB leakage (Appendix Fig S9E–G) compared to control group. At the same time, no lethality was observed following AZD3965 treatment.
We hypothesized that MCTs are involved not only in lactate transport out of ECs but also in its transport into pericytes. To identify the transporter(s) involved in lactate uptake in pericytes, we screened the expression of MCT family members in pericytes using an scRNAseq database (brain cell atlas; Vanlandewijck et al, 2018). Among the 14 transporters, MCT12 displayed the most abundant expression in pericytes (Fig 7A). Importantly, while MCT12 expression was high in vascular mural cells (smooth cells and pericytes), it was not abundantly expressed in various brain cell (Fig 7B). qPCR analysis and Western blotting confirmed high level of MCT12 expression in HBVPs but not BMECs (Fig 7C and D). To test whether MCT12 regulates lactate import in pericyte, we measured [13C3]‐lactate level in HBVPs pellet after [13C3]‐lactate treatment. [13C3]‐lactate was detectable in control siRNA‐treated HBVPs after 2‐h incubation with [13C3]‐lactate, but it was significantly decreased in siMCT12‐treated HBVPs (Fig 7E and F) indicating that MCT12 facilitates the cellular uptake of lactate in pericytes. Next, in order to explore whether MCT12 is involved in lactate transport from ECs to pericytes, we checked mTFP/Venus ratio in siCON or siMCT12‐treated HBVPs in coculture setting with BMECs. As expected, silencing of MCT12 reduced mTFP/Venus ratio in EC‐contacting HBVPs showing MCT12 expression in pericytes is required to transport lactate from ECs to pericytes (Fig 7G and H). Next, we traced isotope‐labeled carbon after [13C3]‐lactate treatment in siCON or siMCT12‐treated HBVPs. MCT12 silencing reduced lactate uptake and isocitrate level in HBVPs but had no effect on total α‐ketoglutarate (αKG) level due to the efflux of citrate into the cytoplasm and/or influx of carbons from glutamine metabolism (Fig 7I).
Figure 7. MCT12 in pericytes regulates lactate import from ECs.
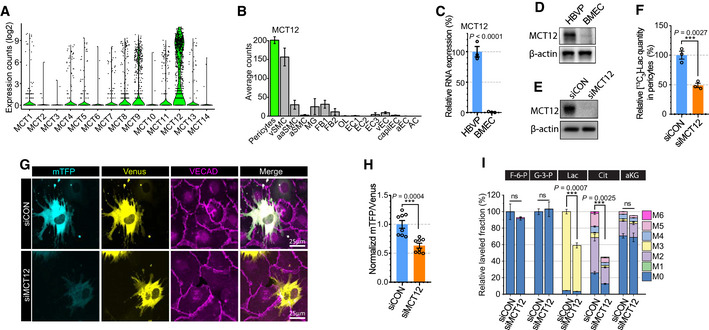
- Violin plot showing the expression of MCT family members (monocarboxylate transporters) in brain pericytes from scRNAseq database (Brain cell atlas; 1,100 endothelial cells).
- Barplot showing the expression of MCT12 across brain cells from the scRNAseq database (vSMC: venous smooth muscle cells, aaSMC: arteriolar SMCs, aSMC: arterial SMCs, MG: Microglia, FB1‐2: Fibroblast subtypes, OL: Oligodendrocytes, EC1‐3: EC subtypes, vEC: venous ECs, capiEC: capillary ECs, aEC: arterial ECs, AC: astrocytes) (1,100 endothelial cells, mean ± SD).
- Relative mRNA expression level of MCT12 in HBVPs (blue) and BMECs (orange). (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Western blot analysis showing expression level of MCT12 and β‐actin in HBVPs and BMECs.
- Western blot analysis for siRNA efficiency of MCT12.
- Quantification of [13C3]‐lactate using LC‐MS/MS in siCON (blue) or siMCT12 (orange) treated HBVPs after 20 mM [13C3]‐Lactate treatment (2 h). (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test).
- Representative images showing mTFP (cyan) and Venus (yellow) fluorescence in control (upper panel) or MCT12 (lower panel) siRNA treated HBVPs in a 2 well chamber assay system with BMECs. VECAD (purple) immunostaining was used for endothelial staining.
- Quantification of the mTFP/Venus fluorescence ratio of Laconic in control (blue) or MCT12 (orange) siRNA treated HBVPs. (n = 8 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test.
- Isotope carbon tracing with [13C3]‐lactate treatment (20 mM, 24 h) in siCON or siMCT12‐treated HBVPs showing the extent of downstream metabolites and incorporation rate of labeled carbons. (F‐6‐P: Fructose 6‐phosphate, G‐3‐P: Glycerol‐3‐phosphate, Lac: Lactate, Cit: Isocitrate and aKG: α‐ketoglutarate) (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from unpaired Student’s t test.
Lactate administration rescues pericyte coverage in Glut1 iECKO mice
To demonstrate functional importance of endothelial lactate to pericytes in vivo, we explored whether lactate supplementation would prevent pericyte loss and altered BBB permeability in the brain vasculature of Glut1 iECKO mice. Following induction of Glut1 deletion at 8 weeks of age, Glut1 iECKO mice were assigned to either sodium lactate (100 mM)‐supplemented or regular drinking water for 10 days. At that time, mice were injected with cadaverine and sacrificed. Immunocytochemical analysis of the brain vasculature demonstrated increased pericyte coverage in lactate‐supplemented compared to control mice (Fig 8A and B) while the amount of extravascular cadaverine was reduced (Fig 8A and C). The CSF lactate level was significantly increased with lactate supplementation (Fig 8B) indicating the loss of pericyte is associated with their access to lactate. There was, however, no rescue of lethality (Fig 8D). The lack of amelioration of lethality with lactate supplementation implies that the lethality is associated with lowered CSF glucose level in Glut1iECKO mice (Appendix Fig S10).
Figure 8. Lactate administration rescues pericyte coverage and vascular leakage but not lethality of Glut1iECKO mice.
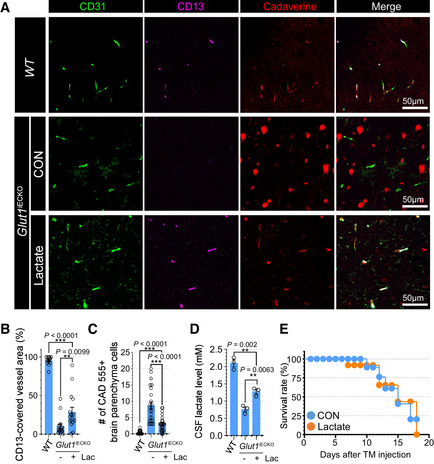
- Permeability assay using vibratome section showing the cadaverine 555 dye (red), CD31 (green) and CD13 (purple) in the brain vasculature of Glut1 iECKO mice with (bottom) or without (top) lactate administration. Note the extravascular accumulation of cadaverine dye in control Glut1 iECKO mice, but not in lactate supplemented Glut1 iECKO mice.
- Quantification of the CD13‐positive pericytes coverage in the brain vasculature of Glut1 iECKO mice with (orange) or without (blue) lactate administration. (n = 16 from 5 independent experiments. Error bars indicate the standard error of the mean (SEM) from 1‐way ANOVA).
- Quantification of the number of cadaverine 555 positive brain parenchyma cells in the brain vasculature of Glut1 iECKO mice with (orange) or without (blue) lactate administration. (n = 24 from 5 independent experiments. Error bars indicate the standard error of the mean (SEM) from 1‐way ANOVA).
- Quantification of CSF lactate level in WT and Glut1iECKO mice with/without lactate supplementation. (n = 3 from independent experiments. Error bars indicate the standard error of the mean (SEM) from 1‐way ANOVA).
- Kaplan‐Meier survival curve of Glut1 iECKO mice after tamoxifen injection at P60 with (orange) or without (blue) lactate administration (n = 8).
Discussion
The results of this study show that endothelium‐derived lactate is taken up and utilized by pericytes for energy generation and a number of biogenic processes. This is critical to normal pericyte function as blockade of endothelial lactate production leads to the loss of vascular pericyte coverage in the CNS and the loss of blood–brain barrier integrity.
While it has been widely accepted that glucose is the main nutrient used for ATP production, recent studies have shown that other nutrients, including lactate, can substantially contribute to energy generation in different tissues (Hui et al, 2020). Lactate in particular is emerging as an important source of energy in the brain although whether it contributes more to the TCA cycle than glucose is a matter of debate (Hui et al, 2020; Liu et al, 2020a). Furthermore, most studies of glucose and lactate metabolism have focused on analysis of whole organs to measure its contribution to the TCA cycle (Hui et al, 2017), leaving specific cell types that might be using lactate undefined. Two recent publication studies showed that neurons and macrophages use locally produced lactate for their energy needs. In particular, astrocytes‐derived lactate was shown to feed excitatory and inhibitory neurons to maintain long‐term memory (Descalzi et al, 2019) while endothelium‐generated lactate induces macrophages to promote muscle regeneration during ischemia (Zhang et al, 2020).
We and others have previously shown that ECs rely on glycolysis for ATP production, thereby generating large amounts of lactate (De Bock et al, 2013b; Kim et al, 2017; Yu et al, 2017b). The present study was designed to elucidate biological function of this lactate. We found that pericytes, but not smooth muscle cells, rely on endothelial‐produced lactate for energy generation. Critically, this is functionally important since the absence of endothelium‐derived lactate leads to pericyte loss from the vessel wall resulting in increased permeability of the BBB.
High local (perivascular) concentration of endothelium‐derived lactate makes it a readily available carbon source. Since pericytes share their basement membranes with endothelial cells, they are likely exposed to a particularly high lactate concentration. This is important as our data (Fig 4E) show that higher concentration of lactate facilitates its uptake into pericytes. Interestingly, pericyte lactate concentration (~pg/cell) is ~1,000‐fold higher than pyruvate (~fg/cell) (Fig 4B,D,E), which is much higher ratio that what is observed in the blood (~50‐fold) (Pianosi et al, 1995). This suggests that pericytes accumulate lactate and use it as a carbon reservoir. Since lactate–pyruvate conversion is a reversible process mediated by LDH (Vanderlinde, 1985), a high concentration of lactate would be expected to shift the equilibrium toward pyruvate synthesis as indeed was observed in this study. Alternatively, pericytes may normally exist in a highly reduced state given that high NADH/NAD ratio shifts the LDH equilibrium toward lactate (Silverstein & Boyer, 1964).
While endothelial cells and pericytes are in close contact, it is unlikely that there is a direct transporter‐mediated transfer of lactate between these cells. Our data show that EC transporters MCT1 and MCT5 are key to extracellular secretion of lactate, which then enters pericytes via the MCT12 transporter. The observation of higher amounts of lactate uptake in pericytes in close proximity to endothelial cells is consistent with this secretion/uptake paradigm and the concentration‐dependent nature of this uptake. In agreement with this hypothesis, high‐dose oral lactate supplementation partially rescued pericyte loss in endothelial Glut1iECKO mice. Although we showed that MCT12 plays a role in lactate uptake in the pericyte, its lactate binding affinity compared to other MCT members and in vivo phenotype of MCT12 mutant animal will require further studies.
Importantly, lactate supplementation, while improving the BBB integrity, did not prevent mortality in Glut1iECKO mice. This is not surprising given that CSF glucose level is lower in Glut1iECKO mice, and insufficient CSF glucose leads to neuronal loss in Glut1 deficient animals (Winkler et al, 2015). While lactate supplementation did raise CSF1 lactate levels in Glut1iECKO mice, it did not show any benefit in reducing lethality of Glut1iECKO mice caused by lowered CSF glucose level.
Prior studies reported that Glut1 is indispensable to normal BBB formation and function. Downregulation of GLUT1 in zebrafish results in impaired BBB permeability and brain edema (Zheng et al, 2010). Glut1 mutant mice also have altered BBB permeability in the brain vasculature as shown by extravascular accumulation of fibrin and reduced amounts of junction proteins in adult global Glut1 heterozygous (Glut1 +/−) mice and in endothelial‐specific Glut1 heterozygotes (Glut1 +/fx;Tie2‐Cre) (Winkler et al, 2015). However, a recent study reported no altered BBB permeability in inducible endothelial‐specific Glut1 knockout mice (Glut1 fx/fx;PDGFβCreERT2 ) using Evans Blue assay (Veys et al, 2020). The likely explanation is that impaired BBB permeability takes time to develop: it was present in 2‐week‐old Glut1 +/fx;Tie2‐Cre mice (Winkler et al, 2015) but not in one‐week‐old Glut1 fx/fx;PDGFβCreERT2 mice (Veys et al, 2020). While the latter strain did show electrocorticographic abnormalities, they did not have a full‐blown BBB breakdown. Our data are consistent with this interpretation; we could not find any significant differences in the BBB permeability using between WT and Glut1 iECKO mice 5 days after gene deletion, but observed a severe leakage in Glut1 iECKO mice 10 days after gene deletion (Fig 1H and I).
In patients, mutations in GLUT1 gene result in GLUT1‐deficiency syndrome (GLUT1DS), which causes a range of neurological abnormalities similar to those observed in Glut1 iECKO mice including movement disorders, stiffness, ataxia, and seizures (Wang et al, 2000, 2005; Brockmann, 2009). For decades, a ketogenic diet (high fat/low carbohydrate diet) has been the only therapeutic option available to GLUT1DS patients (Kass et al, 2016). However, ketogenic diet is less effective in older patients than in younger patients and has side effects including constipation, weight loss, or hunger. Thus, any alternatives are potentially appealing. In this study, we have shown that lactate supplementation in Glut1 iECKO mice improves pericyte coverage and the BBB breakdown although it had no effect on animal mortality. Given that lactate levels in the CSF of GLUT1DS patients are lower or at the low range of normal (De Vivo et al, 1991; Wang et al, 2005; Yang et al, 2011), while lactate supplementation alone might not be enough to prevent lethality, its addition to a ketogenic diet could be provide additional benefits in more severe cases.
In summary, we show that endothelial cell‐generated lactate is used by adjoining pericytes for energy generation and amino acid biosynthesis. In its absence, pericyte coverage of CNS blood vessels is impaired leading to increased BBB permeability (Movie EV1).
Materials and Methods
Mice
Cdh5(PAC)CreERT2 and Glut1 floxed animals were a generous gift from Dr. R. H. Adams (Wang et al, 2010) and Dr. E. D. Abel (Winkler et al, 2015), respectively. To induce Cre activity, fully adult mice (2‐months old) were injected with tamoxifen (2 mg/day) via intraperitoneal injection for 5 consecutive days. All experiments were performed using littermate on a C57BL/6 background. All mice required for this study were housed and maintained under specific pathogen‐free conditions in the animal facility of the Yale University School of Medicine, and all corresponding animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Yale University. This study was conducted in compliance with all relevant ethical regulations.
Cell culture
Human brain microvascular endothelial cells (BMECs) were purchased from ScienCell Research Laboratories and were cultured in EBM‐2 basal medium with EGM‐2 MV BulletKit (Lonza). Human Brain Vascular Pericytes (HBVPs) were purchased from ScienCell Research Laboratories and cultured in Pericyte Growth Medium‐2 (PromoCell) supplemented with 10% feral bovine serum (FBS), N‐2 Supplement, 1× glutaMAX and 100 IU/ml penicillin, and 100 mg/ml streptomycin. HASMCs were purchased from Thermo Fischer Scientific and cultured in Medium 231 (Thermo Fischer Scientific) supplemented with smooth muscle growth supplement including 5% FBS, 2 ng/ml hbFGF, 0.5 ng/ml hEGF, 5 ng/ml heparin, 0.01 µg/ml hIGF‐1, and 0.2 µg/ml BSA. 3T3L1 (fibroblast) cells were purchased from ATCC and cultured in DMEM supplemented with 10% FBS, 100 IU/ml penicillin, and 100 mg/ml streptomycin.
Primary mouse brain pericytes were isolated from brains of C57BL/6 (P5–7) mice. Briefly, dissected brains were chopped with sterile scissors and digested in a solution of 2 mg/ml collagenase type‐I (Sigma‐Aldrich) in DMEM media (Invitrogen) containing 5% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 mg/ml streptomycin for 1 h at 37°C. Thereafter, the cell suspension was immunomagnetically sorted in magnetic field using CD13 antibodies conjugated with dynabeads (Thermo Fischer Scientific). CD13+‐sorted cells were seeded in Pericyte Growth Medium‐2 (PromoCell) supplemented with 10% feral bovine serum (FBS), N‐2 Supplement, 1× glutaMAX and 100 IU/ml penicillin, and 100 mg/ml streptomycin.
Culture media was replaced every other day. Cells were regularly tested for mycoplasma contamination with PCR assay with primers for 16SrRNA of mycoplasma (F: 5’‐ACACCATGGGAGCTGGTAAT‐3’, R: 5’‐CTTCWTCGACTTYCAGACCCAAGGCAT‐3’) (Tabatabaei‐Qomi et al, 2014) and used between passages 1 and 5. Tissue culture plates were coated with 0.1% gelatin (Sigma) for 30 min at 37°C before cell plating. Lipofectamine RNAimax reagent (Invitrogen) was used for gene knockdown. For siRNA‐mediated gene knockdown, GLUT1 (siRNA ID: s12926), MCT1 (siRNA ID: s580), MCT5 (siRNA ID: s225086), MCT12 (siRNA ID: s533485) or control (siRNA ID: 4390843) Silencer™ Select Pre‐Designed siRNA was transfected by Lipofectamine RNAimax reagent (Life Technologies) according to the manufacturer’s protocol.
Drug administration
AZD3965 (Cayman chemical company) was prepared as a 25 mg/ml stock solution in 50% DMSO in normal saline and stored as single use aliquots at −20°C. 50% DMSO (in normal saline) solution was used as the vehicle control for control group mice. Eight‐weeks‐old mice were randomized into two groups, treated with 100 mg/kg BID (twice a day) AZD3965 by intraperitoneal (IP) injection for 7 days and then sacrificed for further analysis.
Metabolic assays with Seahorse analyzer
Metabolic analyses to measure oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were performed with a Seahorse XF96 analyzer and Seahorse XFe96 FluxPak mini (Agilent Seahorse) according to the manufacturer’s recommendations. Briefly, a day before the experiment, a 96 well sensor cartilage (Agilent Technologies) was hydrated with Agilent Seahorse XF Calibrant solution and incubated at 37°C without CO2. Cells were seeded at 30,000 cells per well in a fibronectin (Sigma‐Aldrich) coated Seahorse 96‐well plate (Agilent Technologies) and incubated overnight at 37°C in a CO2 incubator. On the day of the experiment, the Seahorse XF96 analyzer (Agilent Technologies) was calibrated using the hydrated cartilage and cells were maintained in FBS‐free Agilent Seahorse XF DMEM Medium (pH7.4) in a non‐CO2 incubator for 1 h before the assay. After measurement of OCR in basal level, lactate (final concentration: 2 mM or as indicated in the figure legend) were injected to the cells and the OCR (induced) was monitored. For ECAR measurements, glucose (5 mM), oligomycin (3 μM) and 2‐DG (100 mM) were sequentially added to the cells and the ECAR was monitored over time according to the manufacturer’s protocols.
Adenovirus
The construct for Laconic (Plasmid #44238; San Martin et al, 2013) was obtained from addgene. To generate the adenovirus, AdEasy™ Adenoviral Vector System (Agilent Technologies) was used. The expression construct was inserted into pShuttle‐CMV vector. After DNA purification, the vector was linearized by Pme‐I enzyme and electronically transformed into BJ5183 competent cells with pAd‐Easy vector for homologous recombination. After colony screening, the final adenoviral vector was purified with NucleoBond Xtra Midi kit (Takara) and verified by sequencing with CMV‐F and SV40pA‐Rev primers. Adenoviral vector was linearized with Pac‐I and transfected into HEK293A cells using lipofectamine 2000 (Invitrogen) for viral packaging. The amplified adenovirus was titered using an Adeno‐X Rapid Titer Kit (Clontech Laboratories) and used for the overexpression of Laconic in cells.
Edu staining
For detection of proliferating cells, cells were treated with 5‐ethynyl‐2‐deoxyuridine (EdU) (1 μg/ml) for 12 h and fixed with freshly made 4% paraformaldehyde for 15 min. The cells were subsequently incubated with EdU staining solution (Click‐iT EdU Imaging Kits, Invitrogen) for 20 min in the dark according to the manufacturer’s protocols. The EdU‐stained cells were visualized by Leica confocal laser scanning microscope SP8 (Leica, Wetzlar, Germany).
Proliferation assay
Proliferation was performed using xCELLigence RTCA System (Acea Biosciences). Briefly, 2000 HBVPs per well were seeded in gelatin‐coated E16 plates in EGM media with or without lactate supplementation. Cell index values at 48 h were used to calculate the effect of lactate supplementation in proliferation of HBVPs.
Tissue sections preparation
For frozen section, tissues were fixed in freshly made 4% paraformaldehyde (Sigma‐Aldrich) overnight at 4°C, incubated in 15% sucrose overnight at 4°C, and transferred to 30% sucrose at 4°C until the tissue sank. Fixed tissues were infiltrated with Tissue‐Tek O.C.T. embedding medium (Sakura) for 30 min at room temperature, transferred to an embedding mold filled with OCT, and frozen on dry ice. Frozen sections (10 μm thick) were cut at −20°C using Leica CM1510S‐3 Cryostat (Leica), and slides were kept at −70°C until use for immunostaining. For paraffin section, tissues were fixed in formalin overnight, dehydrated by titrating in ethanol (50%, 75%, and 95%), and submitted to the Pathology Tissue Services Core at the Yale university for paraffin embedding, sectioning, and staining. For vibratome section, brains were dissected and fixed in freshly made 4% paraformaldehyde overnight at 4°C and rehydrated with PBS. Rehydrated brains were cut using Leica VT1000 S vibratome (150‐μm thickness) and incubated with 0.1% Triton X‐100 (Sigma‐Aldrich) in PBS (PBS‐T) for 30 min at 37°C for permeabilization. Vibratome tissue sections were kept at 4°C until use for immunostaining.
Cerebrospinal fluid (CSF) collection
A straight, pulled, and sharpened glass micro‐capillary pipette was used for CSF sampling from mice. Glass capillary tubes (Drummond) were pulled with a P‐87 Flaming Brown Micropipette Puller (Sutter Instrument), beveled with a custom‐built micropipette beveler with a grit 600 sandpaper and used for CSF collection. Each mouse was anesthetized with intraperitoneal ketamine (100 mg/kg) plus xylazine (50 mg/kg) and placed under a Leica M125 dissection microscope. The surgical site was swabbed with 10% povidone, followed by 70% ethanol. Under a dissection microscope, a surgical skin incision approximately 1 cm long was made on the area posterior to the occipital crest, and the subcutaneous tissue and muscles were carefully removed by sterile scissors and forceps to expose the glistening surface of the dura covering the cisterna magna. The glass micro‐capillary pipette mounted on an MT1 single‐axis translation stage (Thorlab) was then advanced slowly to the dura membrane in an anterior direction by a micrometer‐driven micromanipulator. Once the dura membrane was punctured by the glass pipette, CSF was drawn into the micropipette by capillary action and assistance of custom‐built mouth pipette. Fifteen microliters of CSF per mice were used for mass spectrometry analysis.
Immunostaining
Before antibody staining, tissues were treated with blocking buffer (1% FBS, 3% BSA, 0.5% Triton X‐100, 0.01% Na deoxycholate, in TBS) for 1 h at room temperature. All antibody staining were performed in IHC buffer (1% FBS, 3% BSA, 0.1% Triton X‐100 in TBS). Primary antibodies were incubated overnight at 4°C. After three washes with TBS‐T (0.1% Triton X‐100 in TBS), secondary antibodies were added and incubated for 1 h with DAPI (Sigma Aldrich) at room temperature. Negative controls were generated by staining with the secondary antibodies and DAPI only. After staining, the tissues were covered with homemade mounting medium (0.2 mg/ml p‐phenylenediamine hydrochloride (PPD), 90% glycerol, 4 mM of sodium azide in 0.1 M of Tris buffer (pH9.0)) and analyzed with Nikon ECLIPSE 80i fluorescence microscope, Leica spinning disk confocal microscope, or a Leica SP8 confocal microscope. The following antibodies were used for immunostaining: Rhodamine Phalloidin (1:20, Invitrogen) CD31 (1:100, RND (AF3628)), Glut1 (1:100, Abcam (ab652)), ERG1/2/3 (1:100, Abcam (ab92513)), Desmin (1:100, SantaCruz (sc‐7559)), CD13 (1:100, BioLegend (301702)), SMA (1:100, Abcam (ab5694)), PDGFRb (1:100, Abcam (ab32570)), Fibrin (1:100, Millipore (MABS2155)), caspase‐3 (1:100, RND (AF835)), and isolectin‐B4 (1:100, Invitrogen). Alexa Fluor 488, 555 and 647 donkey secondary antibodies were from Invitrogen.
LC‐MS/MS
Sodium L‐lactate, sodium pyruvate, sodium adenosine 5'‐triphosphate (ATP), sodium adenosine‐13C10,15N5 5′‐triphosphate (13C10,15N5‐ATP), and 13C3‐pyruvic acid were purchased from Sigma‐Aldrich, and sodium 13C3‐lactate and 1,3‐13C2‐malonic acid were purchased from Cambridge Isotope Laboratories, respectively. Methanol and acetonitrile were obtained from J.T. Baker as HPLC grade. Water was purified using a Milli‐Q purification system (Millipore Corp). All other chemicals and solvents were of the highest analytical grade available. A stock solution of ATP was prepared in 10 mM of ammonium acetate (pH 7.0) at 1 mg/ml and working solutions were prepared from stock solution by serial dilution with 10 mM of ammonium acetate (pH 7.0) to concentrations of 10, 20, 50, 100, 200, 500, and 1,000 μg/ml. 15N5‐ATP was used as internal standard (IS) for ATP quantification. 15N5‐ATP was dissolved in 10 mM of ammonium acetate (pH 7.0) to prepare a stock solution (10 m/ml), and this solution was further diluted with 10 mM of ammonium acetate (pH 7.0) to 1 mg/ml to use as a working IS solution. A stock solution of 13C3‐pyruvic, 13C3‐lactate, and 1,3‐13C2‐malonic acid was prepared in DW at 1 mg/ml and working solutions were prepared from stock solution by serial dilution with DW to concentrations of 1, 2, 5, 10, 20, 50, 100, and 500 ng/ml for 13C3‐pyruvic, and 1, 2, 5, 10, 20, 50, 100, and 500 μg/ml for 13C3‐lactate, respectively. 1,3‐13C2‐malonic acid was used as IS for 13C3‐pyruvic and 13C3‐lactate quantification. 3‐13C2‐malonic acid stock solution was diluted with DW to 100 μg/ml to use as a working IS solution. Stock and working solutions were stored at −20°C. Calibration curves for each analyte were prepared by spiking standard working solution (75 µl) and IS working solution (10 µl) to DW (65 µl). After isotope labeled nutrient treatment, the number of cells was counted and cells were washed with ice‐cold water and immediately collected with 140 μl of 70% ice‐cold methanol. After mixing briefly, 10 μl of IS working solution were added to each sample, shaken for 10 min, and then centrifuged for 10 min at 15,000 g. Then supernatant was transferred to autosampler vials and 5 μl of aliquots were injected into the HPLC system.
HPLC‐mass spectrometric (LC‐MS/MS) analysis was performed on an API 4000 QTrap® mass spectrometer (Applied Biosystems Sciex, Toronto, Canada) coupled with Agilent HP1200 HPLC system (Santa Clara, CA, USA). The Agilent HP1200 HPLC system (Santa Clara, CA, USA) consisted of a degasser (model G1322A), binary solvent delivery system (Model G1312A), ALS autosampler (model G1329A), and TCC column heater (model G1316A). The autosampler temperature was maintained at 4°C and chromatographic separations were achieved at 37°C using a SeQuant ZIC‐pHILIC 5 μm polymer PEEK coated HPLC column (50 × 2.1 mm I.D., 5 μm, Waters). The mobile phase used for analysis consisted of a mixture of acetonitrile and 10 mM of ammonium acetate buffer (30:70, v/v). The mobile phase was adjusted to pH 7.0 with ammonium hydroxide solution, filtered through a 0.45‐μm filter, and delivered at a rate of 0.4 ml/min. To optimize electrospray injection (ESI) conditions, quadrupole full scans were carried out in positive and negative ion detection modes. The ESI was carried out in positive ion detection mode for ATP and 15N5‐ATP, or negative ion mode for 13C3‐pyruvic, 13C3‐lactate, and 1,3‐13C2‐malonic acid. Two channels of multiple reaction monitoring (MRM) mode were used to detect analytes. The most abundant m/z pairs (Q1/Q3) for quantification were m/z 508.3/136.2 for ATP, m/z 523.4/146.3 for 13C10 15N5‐ATP, m/z 89.9/45.0 for 13C3‐pyruvate, m/z 91.8/45.0 for 13C3‐lactate, m/z 88.8/42.9 for lactate, and m/z 105.2/60.1 for 13C2‐malonic acid, respectively. After measurement of analytes with mass spectrometry, the total amount of analyte was adjusted to the total number of cells per group. Analyst 1.7 software was used for data acquisition and analysis (Applied Biosystems Sciex, Toronto, Canada).
Untargeted MS spectrometry
Cells grown in 6‐cm tissue culture dishes were washed with PBS twice and then preincubated with DMEM (Biological Industries) containing 10% dialyzed FBS, 5 mM of glucose, 0.5 mM of glutamine, and 1 mM of lactate. After 12 h of preincubation, media was replaced with fresh media containing isotope‐labeled nutrient (5 mM of [13C3]‐glucose, 2 mM of [13C3]‐lactate or 20 mM of [13C3]‐lactate (Cambridge Isotope Laboratories)). After 12‐ or 24‐hr incubation, cells were washed with ice‐cold water and metabolites were immediately extracted by aspirating water and adding 1 ml of 80% methanol. Precipitates were pelleted by centrifugation at 15,000 g in an Eppendorf 5424R‐refrigerated centrifuge (Eppendorf), and the supernatants were recovered and dried by DNA110 SpeedVac evaporator (Savant). After reconstitution with 100 μl of isopropanol:methanol (1:1; v/v) solution, the supernatant was subjected to MS analysis. The liquid chromatography‐MS (LC–MS) analysis was performed as previously described (Zhao et al, 2020). Separation was performed by reversed‐phase ion‐pairing chromatography on a C8 column coupled to negative‐ion mode, full‐scan LC–MS at 1‐Hz scan time, and 100,000 resolving power (stand‐alone orbitrap; Thermo Fischer Scientific). Data analysis with MAVEN software and natural isotope correction were performed as previously described (Su et al, 2017).
ibidi coculture
Ibidi 2‐well inserts (ibidi) were installed into a Nunc Lab‐Tek II one well chamber slide (Thermo Fischer Scientific) with sterile tweezers and gently pushed with a gloved fingertip. Pretreated BMECs or HBVPs with Laconic infection or siRNA treatment were seeded into opposite chambers at a density of 1 × 105 cells/cm2 and incubated overnight. The culture insert was then removed with sterile tweezers and the well topped up with 1 ml of EGM2 media. After 24‐hr incubation, cells were fixed with freshly prepared 4% PFA for 10 min at room temperature and treated with blocking buffer (1% FBS, 3% BSA, 0.5% Triton X‐100, 0.01% Na deoxycholate, in TBS ) for 1 h at room temperature. After immunostaining, cells were subjected to imaging using a Leica confocal laser scanning microscope SP5 (Leica, Wetzlar, Germany) and treated with blocking buffer (1% FBS, 3% BSA, 0.5% Triton X‐100, 0.01% Na deoxycholate, in TBS) for 1 h at room temperature. After immonostaining, cells were subjected to imaging using a Leica confocal laser scanning microscope SP8 (Leica, Wetzlar, Germany). Statistical analysis involved use of ImageJ and GraphPad Prism (GraphPad Software) followed by ANOVA multiple comparisons test.
NBDG uptake
2‐NBDG (Sigma‐Aldrich) stock solution was prepared by dissolving in DMSO at 5 mM concentration. Cells were treated with 2‐NBDG (5 mM; final concentration) for 30 min at 37°C. Free 2‐NBDG was washed with PBS from cultures and 2‐NBDG was measured by Nikon ECLIPSE 80i fluorescence microscope (Nikon) or flow cytometry (BD Biosciences) at excitation and emission 488 nm and 525 nm, respectively. For each sample, 20,000 events were acquired in the regions of the FSC × SSC plot. Data analysis involved flow cytometry software (BD Biosciences), and the normalized median fluorescence intensities were calculated as the ratios in median fluorescence intensities between treated and untreated cells. Statistical analysis involved the use of GraphPad Instat (GraphPad Software Inc., La Jolla, CA, USA) by ANOVA followed by Tukey‐Kramer multiple comparisons test.
Vessel permeability assay
Cadaverine Alexa fluorescence 555 (Thermo Fischer Scientific) was used for the in vivo vessel permeability test. 0.2 ml of cadaverine (0.1 mg/ml) was injected through retro‐orbital vein using insulin needles. Tissues were harvested 30 min later and fixed by 4% PFA overnight at 4°C. Brains were sectioned by Leica VT1000 S vibratome or Leica CM1510S‐3 Cryostat and stained as previously described. All images were captured by Leica SP8 and analysis by ImageJ.
In vitro permeability assay
Two days after siRNA transfection, siCON‐ or siGLUT1‐treated BMECs were seeded into the luminal side of filters (0.4–μm pore size; Corning, NY) in gelatin‐coated (1% gelatin) 12‐well plates (1 × 105 cells/well). After twenty‐four hours, cells were then cultured in EBM medium (serum‐starved) for 2 h. Then, FITC‐dextran (1 mg/ml; Sigma, St. Louis, MO) and VEGF‐A (50 ng/ml) were added to the upper compartments of the transwell cultures. After 20‐min incubation, a Synergy HTX Multi‐Mode Microplate Reader was used to measure the values for absorbance of the lower chamber solution at 492‐nm (excitation) and 520‐nm (emission) wavelengths.
Analysis of publicly available scRNAseq atlas
To check the expression of GLUT1 in various tissues, we used the publicly available processed data and corresponding metadata of BrainEC and EC atlas to avoid any discrepancies between processing and annotation of the data. For BrainEC atlas, preprocessed and normalized data as well as the accompanying metadata were used from the Vanlandewijck et al (2018). For EC atlas, normalized dataset was downloaded from EndoDB (https://endotheliomics.shinyapps.io/ec_atlas/; Khan et al, 2019; Kalucka et al, 2020). The R (R‐3.6.3) implementation of the ggplot2 and Seurat 3.0 software was used for t‐SNE, UMAP, and bar graph visualization (Macosko et al, 2015; Satija et al, 2015).
Statistics
Data are presented as the mean ± SD or SEM. Multiple group means were compared by 1‐way ANOVA, followed by a 2‐tailed Student’s t test for pair‐wise comparisons. Statistical significance was set at a P‐value < 0.05.
Author contributions
Heon‐Woo Lee: Conceptualization; Resources; Data curation; Software; Formal analysis; Validation; Investigation; Visualization; Methodology; Writing—original draft; Writing—review and editing. Yanying Xu: Conceptualization; Data curation; Software; Formal analysis; Validation; Investigation; Visualization; Methodology. Xiaolong Zhu: Investigation; Visualization. Cholsoon Jang: Software; Formal analysis; Investigation. Hosung Bae: Software; Formal analysis; Investigation. Woosoung Choi: Data curation; Software; Investigation; Visualization. Weiwei Wang: Data curation; Software; Formal analysis; Validation. Liqun He: Software; Formal analysis; Validation. Suk‐Won Jin: Supervision; Investigation; Visualization. Zoltan Arany: Supervision. Michael Simons: Conceptualization; Supervision; Funding acquisition; Writing—original draft; Project administration; Writing—review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
H‐WL, YX, and MS conceived the study and designed experiments. H‐WL, YX, XZ, CJ, HB, and WW performed experiments. H‐WL, YX, CJ, WC, HB, WW, and LH analyzed experiments. H‐WL, S‐WJ, and MS wrote the manuscript with support from all authors. ZA performed data analysis and manuscript preparation.
Supporting information
Appendix
Movie EV1
Acknowledgments
We thank R. Adams for Cdh5(PAC)CreERT2 and E. Dale Abel for Glut1 flox/flox mice, respectively. We also thank C. Betsholtz for useful comments on scRNAseq data analysis and R. Cardone for instruction and assistance on the Seahorse metabolic analyzer. Supported, in part, by NIH grants HL135582, HL062289, and HL107205.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
The EMBO Journal (2022) 41: e109890.
See also: M Castro & M Potente (May 2022)
Data availability
This study does not contain data amenable to public repository deposition.
References
- Armulik A, Abramsson A, Betsholtz C (2005) Endothelial/pericyte interactions. Circ Res 97: 512–523 [DOI] [PubMed] [Google Scholar]
- Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, Niaudet C, He LQ, Norlin J, Lindblom P, Strittmatter K et al (2010) Pericytes regulate the blood‐brain barrier. Nature 468: 557–561 [DOI] [PubMed] [Google Scholar]
- Bertrand L, Auquier J, Renguet E, Ange M, Cumps J, Horman S, Beauloye C (2020) Glucose transporters in cardiovascular system in health and disease. Pflugers Arch 472: 1385–1399 [DOI] [PubMed] [Google Scholar]
- Bonuccelli G, Tsirigos A, Whitaker‐Menezes D, Pavlides S, Pestell RG, Chiavarina B, Frank PG, Flomenberg N, Howell A, Martinez‐Outschoorn UE et al (2010) Ketones and lactate "fuel" tumor growth and metastasis evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle 9: 3506–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumezbeur F, Petersen KF, Cline GW, Mason GF, Behar KL, Shulman GI, Rothman DL (2010) The contribution of blood lactate to brain energy metabolism in humans measured by dynamic 13C nuclear magnetic resonance spectroscopy. J Neurosci 30: 13983–13991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann K (2009) The expanding phenotype of GLUT1‐deficiency syndrome. Brain Dev 31: 545–552 [DOI] [PubMed] [Google Scholar]
- Brooks GA (2018) The Science and translation of lactate shuttle theory. Cell Metab 27: 757–785 [DOI] [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G et al (2013a) Role of PFKFB3‐driven glycolysis in vessel sprouting. Cell 154: 651–663 [DOI] [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G et al (2013b) Role of PFKFB3‐driven glycolysis in vessel sprouting. Cell 154: 651–663 [DOI] [PubMed] [Google Scholar]
- De Vivo DC, Trifiletti RR, Jacobson RI, Ronen GM, Behmand RA, Harik SI (1991) Defective glucose transport across the blood‐brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med 325: 703–709 [DOI] [PubMed] [Google Scholar]
- Descalzi G, Gao V, Steinman MQ, Suzuki A, Alberini CM (2019) Lactate from astrocytes fuels learning‐induced mRNA translation in excitatory and inhibitory neurons. Commun Biol 2: 247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner A, Lynam E, Purcell R, Jones C, Lopez C, Board M, Wagner KD, Wagner N, Carr C, Wheeler‐Jones C (2020) Context‐dependent regulation of endothelial cell metabolism: differential effects of the PPARbeta/delta agonist GW0742 and VEGF‐A. Sci Rep 10: 7849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP, Meredith D (2004) The SLC16 gene family‐from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch 447: 619–628 [DOI] [PubMed] [Google Scholar]
- van Hall G (2010) Lactate kinetics in human tissues at rest and during exercise. Acta Physiol (Oxf) 199: 499–508 [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Gerhardt H, Kalen M, Li X, Eriksson U, Wolburg H, Betsholtz C (2001) Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J Cell Biol 153: 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, Bartman C, Zhang Z, Jang C, Wang L et al (2020) Quantitative Fluxomics of Circulating Metabolites. Cell Metab 32: 676–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J et al (2017) Glucose feeds the TCA cycle via circulating lactate. Nature 551: 115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Bi A, Lan X, Xu J, Wang X, Liu Y, Wang T, Tang S, Zeng H, Chen Z et al (2019) Identification of metabolic vulnerabilities of receptor tyrosine kinases‐driven cancer. Nat Commun 10: 2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K et al (2020) Single‐cell transcriptome atlas of murine endothelial cells. Cell 180: 764–779 [DOI] [PubMed] [Google Scholar]
- Kass HR, Winesett SP, Bessone SK, Turner Z, Kossoff EH (2016) Use of dietary therapies amongst patients with GLUT1 deficiency syndrome. Seizure 35: 83–87 [DOI] [PubMed] [Google Scholar]
- Khan S, Taverna F, Rohlenova K, Treps L, Geldhof V, de Rooij L, Sokol L, Pircher A, Conradi LC, Kalucka J et al (2019) EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res 47: D736–D744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Li J, Jang C, Arany Z (2017) Glutamine fuels proliferation but not migration of endothelial cells. EMBO J 36: 2321–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt A, Spahr R, Mertens S, Siegmund B, Piper HM (1990) Metabolism of exogenous substrates by coronary endothelial cells in culture. J Mol Cell Cardiol 22: 1393–1404 [DOI] [PubMed] [Google Scholar]
- Liu S, Dai Z, Cooper DE, Kirsch DG, Locasale JW (2020a) Quantitative analysis of the physiological contributions of glucose to the TCA cycle. Cell Metab 32: 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Que S, Xu J, Peng T (2014) Alanine aminotransferase‐old biomarker and new concept: a review. Int J Med Sci 11: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xu J, Ma Q, Zhang X, Yang Q, Wang L, Cao Y, Xu Z, Tawfik A, Sun Y et al (2020b) Glycolysis links reciprocal activation of myeloid cells and endothelial cells in the retinal angiogenic niche. Sci Transl Med 12: eaay1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM et al (2015) Highly parallel genome‐wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt ME, Harrison C, Sherry AD, Malloy CR, Burgess SC (2011) Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc Natl Acad Sci USA 108: 19084–19089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakopoulou AM, Montagne A, Kisler K, Dai Z, Wang Y, Huuskonen MT, Sagare AP, Lazic D, Sweeney MD, Kong P et al (2019) Pericyte loss leads to circulatory failure and pleiotrophin depletion causing neuron loss. Nat Neurosci 22: 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianosi P, Seargeant L, Haworth JC (1995) Blood lactate and pyruvate concentrations, and their ratio during exercise in healthy children: developmental perspective. Eur J Appl Physiol Occup Physiol 71: 518–522 [DOI] [PubMed] [Google Scholar]
- Polanski R, Hodgkinson CL, Fusi A, Nonaka D, Priest L, Kelly P, Trapani F, Bishop PW, White A, Critchlow SE et al (2014) Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin Cancer Res 20: 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB, Barros LF (2013) A genetically encoded FRET lactate sensor and its use to detect the warburg effect in single cancer cells. PLoS One 8: e57712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single‐cell gene expression data. Nat Biotechnol 33: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors S, De Bock K, Cantelmo AR, Georgiadou M, Ghesquiere B, Cauwenberghs S, Kuchnio A, Wong BW, Quaegebeur A, Goveia J et al (2014) Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab 19: 37–48 [DOI] [PubMed] [Google Scholar]
- Shen Y, Jiang L, Wen P, Ye Y, Zhang Y, Ding H, Luo J, Xu L, Zen K, Zhou Y et al (2020) Tubule‐derived lactate is required for fibroblast activation in acute kidney injury. Am J Physiol Renal Physiol 318: F689–F701 [DOI] [PubMed] [Google Scholar]
- Silverstein E, Boyer PD (1964) Equilibrium reaction rates and the mechanisms of bovine heart and rabbit muscle lactate dehydrogenases. J Biol Chem 239: 3901–3907 [PubMed] [Google Scholar]
- Su XY, Lu WY, Rabinowitz JD (2017) Metabolite spectral accuracy on orbitraps. Anal Chem 89: 5941–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Knaff DB (2005) Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth Res 83: 191–217 [DOI] [PubMed] [Google Scholar]
- Tabatabaei‐Qomi R, Sheykh‐Hasan M, Fazaely H, Kalhor N, Ghiasi M (2014) Development of a PCR assay to detect mycoplasma contamination in cord blood hematopoietic stem cells. Iran J Microbiol 6: 281–284 [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde RE (1985) Measurement of total lactate dehydrogenase activity. Ann Clin Lab Sci 15: 13–31 [PubMed] [Google Scholar]
- Vanlandewijck M, He LQ, Mae MAA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L et al (2018) A molecular atlas of cell types and zonation in the brain vasculature. Nature 554: 475–480 [DOI] [PubMed] [Google Scholar]
- Veys K, Fan Z, Ghobrial M, Bouche A, Garcia‐Caballero M, Vriens K, Conchinha NV, Seuwen A, Schlegel F, Gorski T et al (2020) Role of the GLUT1 glucose transporter in postnatal CNS angiogenesis and blood‐brain barrier integrity. Circ Res 127: 466–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waagepetersen HS, Bakken IJ, Larsson OM, Sonnewald U, Schousboe A (1998) Comparison of lactate and glucose metabolism in cultured neocortical neurons and astrocytes using C‐13‐NMR spectroscopy. Dev Neurosci 20: 310–320 [DOI] [PubMed] [Google Scholar]
- Wang D, Kranz‐Eble P, De Vivo DC (2000) Mutational analysis of GLUT1 (SLC2A1) in Glut‐1 deficiency syndrome. Hum Mutat 16: 224–231 [DOI] [PubMed] [Google Scholar]
- Wang D, Pascual JM, Yang H, Engelstad K, Jhung S, Sun RP, De Vivo DC (2005) Glut‐1 deficiency syndrome: clinical, genetic, and therapeutic aspects. Ann Neurol 57: 111–118 [DOI] [PubMed] [Google Scholar]
- Wang YD, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U et al (2010) Ephrin‐B2 controls VEGF‐induced angiogenesis and lymphangiogenesis. Nature 465: 483–486 [DOI] [PubMed] [Google Scholar]
- Winkler EA, Nishida Y, Sagare AP, Rege SV, Bell RD, Perlmutter D, Sengillo JD, Hillman S, Kong P, Nelson AR et al (2015) GLUT1 reductions exacerbate Alzheimer's disease vasculo‐neuronal dysfunction and degeneration. Nat Neurosci 18: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi S, Katsumura K, Kobayashi T, Puro DG (2006) Extracellular lactate as a dynamic vasoactive signal in the rat retinal microvasculature. Am J Physiol Heart Circ Physiol 290: H925–934 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang D, Engelstad K, Bagay L, Wei Y, Rotstein M, Aggarwal V, Levy B, Ma L, Chung WK et al (2011) Glut1 deficiency syndrome and erythrocyte glucose uptake assay. Ann Neurol 70: 996–1005 [DOI] [PubMed] [Google Scholar]
- Yu PC, Wilhelm K, Dubrac A, Tung JK, Alves TC, Fang JS, Xie Y, Zhu J, Chen ZH, De Smet F et al (2017a) FGF‐dependent metabolic control of vascular development. Nature 545: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZX, Ruter DL, Kushner EJ, Bautch VL (2017b) Excess centrosomes induce p53‐dependent senescence without DNA damage in endothelial cells. Faseb Journal 31: 4295–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Muri J, Fitzgerald G, Gorski T, Gianni‐Barrera R, Masschelein E, D'Hulst G, Gilardoni P, Turiel G, Fan Z et al (2020) Endothelial lactate controls muscle regeneration from ischemia by inducing M2‐like macrophage polarization. Cell Metab 31: 1136–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Guo C, Jiang K, Ying M, Hu X (2017) Quantification of lactate from various metabolic pathways and quantification issues of lactate isotopologues and isotopomers. Sci Rep 7: 8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Jang C, Liu J, Uehara K, Gilbert M, Izzo L, Zeng X, Trefely S, Fernandez S, Carrer A et al (2020) Dietary fructose feeds hepatic lipogenesis via microbiota‐derived acetate. Nature 579: 586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng PP, Romme E, van der Spek PJ, Dirven CM, Willemsen R, Kros JM (2010) Glut1/SLC2A1 is crucial for the development of the blood‐brain barrier in vivo . Ann Neurol 68: 835–844 [DOI] [PubMed] [Google Scholar]


