Abstract
Pseudomonas aeruginosa exhibits high intrinsic resistance to penem antibiotics such as faropenem, ritipenem, AMA3176, sulopenem, Sch29482, and Sch34343. To investigate the mechanisms contributing to penem resistance, we used the laboratory strain PAO1 to construct a series of isogenic mutants with an impaired multidrug efflux system MexAB-OprM and/or impaired chromosomal AmpC β-lactamase. The outer membrane barrier of PAO1 was partially eliminated by inducing the expression of the plasmid-encoded Escherichia coli major porin OmpF. Susceptibility tests using the mutants and the OmpF expression plasmid showed that MexAB-OprM and the outer membrane barrier, but not AmpC β-lactamase, are the main mechanisms involved in the high intrinsic penem resistance of PAO1. However, reducing the high intrinsic penem resistance of PAO1 to the same level as that of penem-susceptible gram-negative bacteria such as E. coli required the loss of either both MexAB-OprM and AmpC β-lactamase or both MexAB-OprM and the outer membrane barrier. Competition experiments for penicillin-binding proteins (PBPs) revealed that the affinity of PBP 1b and PBP 2 for faropenem were about 1.8- and 1.5-fold lower, than the respective affinity for imipenem. Loss of the outer membrane barrier, MexAB, and AmpC β-lactamase increased the susceptibility of PAO1 to almost all penems tested compared to the susceptibility of the AmpC-deficient PAO1 mutants to imipenem. Thus, it is suggested that the high intrinsic penem resistance of P. aeruginosa is generated from the interplay among the outer membrane barrier, the active efflux system, and AmpC β-lactamase but not from the lower affinity of PBPs for penems.
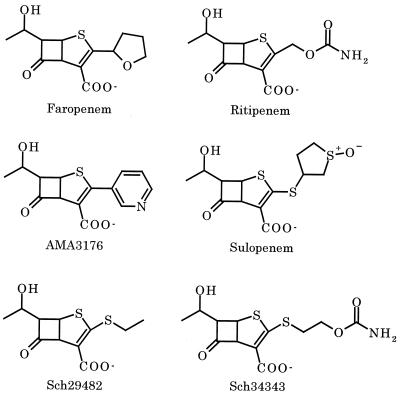
Penem antibiotics such as faropenem (formerly SUN5555 or WY-49605) (8, 16, 34), ritipenem (formerly FCE22101) (4, 31, 51), AMA3176 (K. Okonogi, T. Iwahi, M. Nakao, and Y. Noji, Program Abstr. 33rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 289, 1993), sulopenem (formerly CP-65,207) (9), Sch29482 (30, 36), and Sch34343 (27, 42), which were designed based on the structure-activity relationship of penicillins and cephalosporins (Fig. 1), display potent antibacterial activities toward a variety of gram-positive and gram-negative bacteria. Penems are stable to all classes of β-lactamases except for carbapenem-hydrolyzing enzymes such as class B metallo-β-lactamases (16, 27) and class A/group 2f β-lactamase (41). The stability of penems to β-lactamases can explain their potent antibacterial activities. However, several gram-negative pathogens such as Pseudomonas aeruginosa and Burkholderia cepacia demonstrate high intrinsic resistance to penem antibiotics, although neither of these bacteria innately produces penem-hydrolyzing β-lactamase.
FIG. 1.
Chemical structures showing the charge(s) in the penems used in this study.
P. aeruginosa is a clinically significant pathogen that exhibits intrinsic resistance to various antimicrobial agents including β-lactam antibiotics. Although this organism has an outer membrane with low permeability (3, 52), this alone does not adequately explain its intrinsic resistance (32, 33). As an additional mechanism interfering with the access of the agents to their targets in this bacterium, tripartite efflux systems such as MexAB-OprM (12, 37, 38), MexCD-OprJ (39), MexEF-OprN (18) and MexXY/OprM (1, 26, 28) contribute to the intrinsic and acquired resistance via proton-dependent extrusion of antimicrobial agents and other xenobiotics from the cell interior. MexAB-OprM, which is constitutively expressed in the wild-type strains, plays a role in the intrinsic resistance of P. aeruginosa to most β-lactams and many other structurally unrelated antimicrobial agents (12, 19). More recently, it has been demonstrated that MexXY/OprM is also involved in the intrinsic resistance of P. aeruginosa to several agents such as tetracycline, erythromycin, and gentamicin (26). Since the expression of MexCD-OprJ and MexEF-OprN is strictly suppressed by the respective regulator genes in wild-type P. aeruginosa cells, neither of these efflux systems is involved in the intrinsic antibiotic resistance (18, 39). AmpC β-lactamase encoded on the chromosome of P. aeruginosa may also contribute to the intrinsic resistance of this bacterium to β-lactams (22). Recent studies have demonstrated that the intrinsic resistance of P. aeruginosa to β-lactams is due to the interplay among multiple resistance mechanisms (21, 25, 29, 45).
Chelators such as EDTA and hexametaphosphate, as well as cationic compounds such as aminoglycosides, polymyxin B, and poly(l-lysine), remove divalent cations acting as bridges between neighboring lipopolysaccharide molecules in the outer membrane of gram-negative bacteria including P. aeruginosa, and disturb the outer membrane structure (14, 15, 21). Then, an enlarged pore that allows permeation of enzymes such as lysozyme and leakage of periplasmic enzymes such as β-lactamase is formed and drastically increases the permeability of the outer membrane (21). However, the growth of P. aeruginosa cells is exceptionally hypersusceptible to most outer membrane permeabilizers (14, 15). Furthermore, the addition of these agents may affect the activities of the antimicrobial agents, and excess disturbance of the outer membrane structure may generate physiological alterations in P. aeruginosa. The major porin OmpF of Escherichia coli forms a water-filled channel, the so-called porin pore, in the outer membrane and provides a diffusion pathway for hydrophilic and low-molecular-weight (Mr) molecules such as β-lactam antibiotics, quinolones, saccharides, and amino acids (33, 53). Accordingly, we can expect that induction of OmpF expression in P. aeruginosa cells would destroy the low permeability barrier of the P. aeruginosa outer membrane.
In this study, we determined the alterations in the susceptibility of P. aeruginosa to penem antibiotics in isogenic mutants from the laboratory strain PAO1 that have defects in the production of AmpC β-lactamase and the MexAB-OprM system and in which the expression of a major porin of E. coli, OmpF, was induced. We discuss the interplay among AmpC β-lactamase, the MexAB-OprM efflux system, and the outer membrane barrier in increasing the high intrinsic resistance of P. aeruginosa to penems.
MATERIALS AND METHODS
Organisms and media.
The P. aeruginosa strains used in this study are isogenic mutants of PAO1 and are listed in Table 1. Bacterial cells were grown in Luria (L) broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) or L agar (L broth plus 1.5% [wt/vol] agar) at 37°C. BM2 minimal medium (13) was used for selection of P. aeruginosa since E. coli cannot utilize citrate. Antibiotics were added to the media at the following concentrations: ampicillin, 100 μg/ml for E. coli; carbenicillin, 100 μg/ml for P. aeruginosa; streptomycin, 30 μg/ml for E. coli and 100 μg/ml for P. aeruginosa; tetracycline, 10 μg/ml for E. coli and for KG2225 and KG2505 of P. aeruginosa and 100 μg/ml for PAO1, KG2504, and OCR1 of P. aeruginosa; and chloramphenicol, 30 μg/ml for E. coli and 100 μg/ml for P. aeruginosa. L agar was supplemented with 5% (wt/vol) sucrose as required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Prototroph | |
| KG2225 | mexA::res-ΩSm of PAO1; Smr | This study |
| KG2504 | ampC::ΩSm of PAO1; Smr | This study |
| KG2505 | mexA::res-ΩSm of KG2504; Smr | This study |
| OCR1 | MexAB-OprM-overproducing nalB mutant of PAO1 | 24 |
| Plasmids | ||
| pTAK1900 | E. coli-P. aeruginosa shuttle cloning vector pAK1900 (38) carrying tet from pBR322 in bla; Tcr | This study |
| pKMF010 | pTAK1900 carrying the 3.5-kb SphI-PvuI fragment encompassing lacIq-Ptrc-ompF-rrnBT1T2; Tcr, inducible OmpF production | This study |
| pKMF012 | ompF-deficient pKMF010; Tcr | This study |
| pKMF014 | pTAK1900 carrying the 3.5-kb SphI-PvuI fragment encompassing lacIq-Ptrc-ompF-rrnBT1T2 and the 4.1-kb EcoRI-PstI fragment encompassing the IMP-1 gene; Tcr, inducible OmpF production and constitutive IMP-1 production | This study |
| pKMF016 | ompF-deficient pKMF014; Tcr, constitutive imipenemase production | This study |
Abbreviations: Smr, streptomycin resistant; Tcr, tetracycline resistant; rrnBT1T2, transcription terminator sequences; IMP-1, carbapenemase.
Insertion mutagenesis of ampC.
PCR was performed under conditions described previously (13) to amplify the 1.7-kb ampC gene (23) using the primer pair bla1 (5′-TGGACAGGCGCAGATCGATG-3′) in the ampR gene located ca.530 bp upstream of the ATG initiation codon and bla2 (5′-TCAGGCCTTCAGCGGCACC-3′) which included the TGA termination codon of the gene. The amplified gene was ligated between blunt-ended PstI and SmaI sites of pTAK1900 to yield pKMB001. The SmaI-ScaI fragment encompassing the ΩSm gene from pS1918 (40) was ligated into the NruI site in the ampC gene on pKMB001 to yield pKMB002. Then the EcoRI fragment including the ΩSm-inserted ampC gene from pKMB002 and the Mob cassette from pMT5071 (48) were cloned into the EcoRI and NotI sites, respectively, on pMT5059 (49) to yield pKMB004. The resulting plasmid was mobilized from E. coli strain S17-1 to the P. aeruginosa strain PAO1 to introduce the Smr determinant into the ampC gene on the recipient chromosomes to yield KG2504. Disruption of the ampC gene of KG2504 was confirmed by PCR (data not shown).
Insertion mutagenesis of mexA.
A 7.8-kb SacI-HindIII fragment encompassing mexAB-oprM of pPV20 (37) was cloned into the multiple-cloning site of pAK1900 (38). After blunting of the NotI site located in the flanking region of mexAB-oprM on the fragment, the EcoRI-HindIII fragment encompassing mexAB-oprM was subcloned into pMT5059 to yield pKMM026. The NruI-PstI region in the mexA gene was removed and a 10-bp XbaI linker was inserted to yield pKMM036. The res-ΩSm fragment from pMT5096 (48) and the Mob cassette from pMT5071 (48) were ligated into the XbaI and NotI sites, respectively, on pKMM036 to yield pKMM137. The resulting plasmid was mobilized from E. coli strain S17-1 to P. aeruginosa strains PAO1 and KG2504 to introduce the Smr determinant into the mexA gene on the recipient chromosomes by allelic exchange to yield KG2225 and KG2505, respectively. Disruption of the mexA gene was confirmed by PCR (data not shown).
Construction of an E. coli OmpF-expression plasmid.
PCR was performed to amplify the 1.0-kb ompF gene using E. coli K-12 chromosomal DNA as a template and the primer pair ompF1 (5′-CGCCCATGGGAATGAAGCGCAATATTCTGGCAG-3′) and ompF2 (5′-CGCAAGCTTAGAACTGGTAAACGATACCC-3′), which contain a newly added cutting site (underlined) for restriction nucleases. The PCR product was ligated into the NcoI-HindIII site in a multicloning site of pTrc99A (2) to yield pKMF001. The 3.5-kb SphI-PvuI region encompassing lacIq, Ptrc, ompF, and terminator sequences was blunt ended and ligated into the blunt-ended EcoRI site on pTAK1900, which had been constructed by insertion of the tet gene from pBR322 (5) into the bla gene on pAK1900, to yield pKMF010. To construct the ompF gene-deficient control vector, pKMF012, multicloning sites were removed from pTrc99A and the resulting 2.5-kb SphI-PvuI fragment encompassing lacIq, Ptrc, and terminator sequences was inserted into pTAK1900. Furthermore, for the permeability test, a 4.1-kb EcoRI-PstI region encompassing a carbapenemase gene (blaimp) on pMS363 (17) was ligated into the ScaI site on pKMF010 and pKMF012 to yield pKMF014 and pKMF016, respectively.
Permeability assay with intact cells.
The intact cell permeability assay was performed as described previously (46). An overnight culture of bacterial cells at 37°C was diluted 10-fold with L broth containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and this was further incubated. After 2 h, the bacterial cells were harvested by centrifugation (5,000 × g at 30°C for 10 min), suspended in 10 mM sodium phosphate buffer (pH 7.0)–5 mM MgCl2 and adjusted to an absorbance at 550 nm of 0.25 with the same buffer. The solution of 1,000 μl of 50 μM β-lactam prepared with 50 mM sodium phosphate buffer (pH 7.0)–5 mM MgCl2 was prewarmed to 37°C, and the reaction was started by adding 15 μl of this to the prewarmed cell suspension. The hydrolysis of imipenem, faropenem, and sulopenem was measured spectrophotometrically at 300, 305, and 328 nm, respectively. To determine the enzymatic parameters, plasmid-derived IMP-1 was purified from P. aeruginosa PAO2142Rp carrying pMS363 as described by Watanabe et al (50). The Km (micromolar) and Vmax (micromolar per minute) values of the purified enzyme for imipenem, faropenem, and sulopenem were 27.5 and 27.2, 18.0 and 101.0, and 4.1 and 13.6, respectively. The permeability parameters, Vi, Vt, and Vo, of a penem were examined using intact cells, sonicated cells, and supernatant, respectively. The hydrolysis of β-lactams by β-lactamase obeys Michaelis-Menten kinetics: V = Vmax × S/(Km + S), where V and S are the rate of hydrolysis and the concentration of β-lactam, respectively. Using this equation, we obtained Si, St, and So from Vi, Vt, and Vo, where Si and St are the periplasmic and external drug concentrations, respectively, and So is the concentration of drug that is hydrolyzed by the leaked IMP-1. Based on these parameters, the percentage permeability rate = 100 × (Si − So)/(St − So), was estimated.
Preparation of AmpC β-lactamase and hydrolysis activity assay.
An overnight culture of P. aeruginosa PAO1 was diluted 10-fold with 50 ml of prewarmed L broth, and this was further incubated at 37°C for 1 h. To induce AmpC β-lactamase expression, imipenem was added to a final concentration of 0.5 μg/ml and the incubation was continued. After 2 h, the cells were harvested by centrifugation at 10,000 × g at 4°C for 15 min, washed with 50 mM sodium phosphate buffer (pH 7.0), and resuspended in the same buffer. The cells were disrupted by oscillation from a W-225R sonicator (Heat Systems-Ultrasonic Inc., Plainview, N.Y.) (output, 3; 50% duty cycle; total, 3 min) in an ice-water bath. The residual cells and membranes were packed by centrifugation at 100,000 × g at 4°C for 30 min. The supernatant obtained was used as a crude preparation of AmpC β-lactamase. A reaction mixture containing 0.5 μM crude AmpC β-lactamase and 10 μM antibiotic in 50 mM sodium phosphate buffer (pH 7.0), was prepared and incubated at 37°C for up to 20 h. After the incubation, the reaction mixture was filtered using Ultrafree-MC centrifugal filter units (Japan Millipore Co., Tokyo, Japan) to remove AmpC β-lactamase, and the filtrate was used for measurement of the residual antibacterial activity by a bioassay using Bacillus subtilis ATCC 6633 as an indicator strain. The level of β-lactamase activity in the filtrate obtained from an AmpC β-lactamase preparation purified for another experiment was below the detection limit (data not shown). Thus, we confirmed effective removal of β-lactamase in the samples for the bioassay by the filtration.
Other procedures.
Preparation of plasmid DNA and related in vitro manipulation, agarose gel electrophoresis, transformation, restriction endonuclease digestion, ligation, and PCR were performed as described previously (13). Membrane preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblot analysis were performed as described previously (12). The MIC was determined by the twofold agar dilution technique with L agar containing 1 mM IPTG with an inoculum size of 104 cells. The competition assay for penicillin-binding proteins (PBPs) was performed using benzyl[14C]penicillin potassium (58 mCi/mmol; Amersham Pharmacia Biotech, Tokyo, Japan) as described previously (11).
RESULTS
Expression of E. coli OmpF in the P. aeruginosa outer membrane.
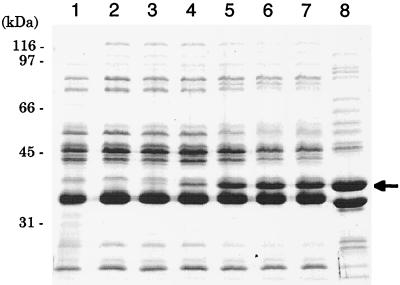
To examine whether the low permeability to penems results in intrinsic penem resistance in P. aeruginosa, we constructed a plasmid carrying the E. coli ompF gene downstream of lacIq-Ptrc regulation and introduced it into the P. aeruginosa cells. The expression of 37-kDa proteins corresponding to E. coli OmpF (Fig. 2, lane 8) depends on the amounts of IPTG added to the growth medium, and this was confirmed in experiments with the outer membranes of PAO1 cells transformed with pKMF010 (ompF) (lanes 3 to 7). PAO1 cells carrying pKMF012 (vector control) did not produce OmpF proteins even when grown in a medium containing 1 mM IPTG (lane 2). In PAO1 cells transformed with pKMF010, OmpF proteins were not found in the inner membrane or in the cytosolic fractions of PAO1 cells transformed with pKMF010 (data not shown). IPTG-dependent OmpF expression was also observed in the outer membranes of other PAO1 mutants, namely, KG2225, KG2504, KG2505, and OCR1, used in this study (data not shown). Furthermore, the growth of all P. aeruginosa strains tested was not affected even when induced with 1 mM IPTG (data not shown).
FIG. 2.
Expression of OmpF in the outer membrane of P. aeruginosa and E. coli cells. Cells were grown at 37°C for 18 h on L agar containing various amounts of IPTG. The outer membrane proteins were prepared, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described previously (10). Lanes: 1, PAO1 (IPTG, 0 mM); 2, PAO1/pKMF012 (IPTG, 1 mM); 3, PAO1/pKMF010 (IPTG, 0 mM); 4, PAO1/pKMF010 (IPTG, 0.02 mM); 5, PAO1/pKMF010 (IPTG, 0.06 mM); 6, PAO1/pKMF010 (IPTG, 0.25 mM); 7, PAO1/pKMF010 (IPTG, 1.0 mM); 8, E. coli K-12 (IPTG, 0 mM). An arrow shows the position corresponding to E. coli OmpF (37 kDa).
We attempted to use IMP-1 (50), which has strong hydrolysis activity towards a broad range of β-lactams including penems, for measurement of the intact cell permeability of P. aeruginosa. We constructed a plasmid, pKMF014, in which the IMP-1 gene from pMS363 (17) was inserted into the ompF-carrying pKMF010. Furthermore, the IMP-1 gene-carrying pKMF016 was constructed from pKMF012 and used as a control. Intact cell permeability experiments (46) with P. aeruginosa strains transformed with these plasmids demonstrated that the faropenem and sulopenem rates of permeability through the outer membrane of P. aeruginosa PAO1 were 30 and 10%, respectively, of that of imipenem, which possesses strong antipseudomonal activity (Table 2), and that OmpF expression increased the permeability rate of imipenem, faropenem, and sulopenem by 2.9-, 1.8-, and 5.1-fold, respectively. However, the presence of pKMF016 alone in the tested strains did not change the permeability to any agents tested. The permeability of the outer membrane was only slightly affected by either deficiency or overexpression of the MexAB-OprM efflux system (refer to KG2225/pKMF014 and OCR1/pKMF014 in Table 2), indicating that the permeability in our experimental protocol was not affected by MexAB-OprM.
TABLE 2.
Alteration in the permeability rate of imipenem and penems through the outer membrane of P. aeruginosa PAO1 and its mutants by transformation with pKMF014, pKMF016a
| Strain/plasmid | Phenotypeb
|
Permeability ratec (%) of:
|
|||
|---|---|---|---|---|---|
| MexAB | OmpF | Imipenem | Faropenem | Sulopenem | |
| PAO1/pKMF016 | + | − | 4.98 ± 0.64 | 1.43 ± 0.40 | 0.48 ± 0.06 |
| PAO1/pKMF014 | + | + | 14.46 ± 2.17 | 2.58 ± 0.43 | 2.47 ± 0.39 |
| KG2225/pKMF016 | − | − | 4.26 ± 0.40 | 1.50 ± 0.32 | 0.58 ± 0.10 |
| KG2225/pKMF014 | − | + | 15.02 ± 1.45 | 2.52 ± 0.17 | 2.45 ± 0.02 |
| OCR1/pKMF016 | ++ | − | 4.50 ± 0.64 | 1.54 ± 0.35 | 0.55 ± 0.08 |
| OCR1/pKMF014 | ++ | + | 16.38 ± 2.17 | 2.56 ± 0.31 | 2.42 ± 0.14 |
As described in Results, for the permeability test, P. aeruginosa cells carrying the respective plasmids grown to exponential phase were further incubated for 2 h in L broth containing 1 mM IPTG.
+, wild-type level; −, nonexpression; ++, overexpression.
The results are presented as the mean ± standard deviation of three experiments.
The increased permeability by OmpF expression may be due to disturbance of the outer membrane structure caused by integration of heterogeneous protein rather than by the formation of a permeation route by OmpF. The MICs (in micrograms per milliliter) of antibiotics for PAO1 were as follows: erythromycin, 32; chloramphenicol, 4; crystal violet, 128; acridine orange, >1,024; vancomycin, 2,048; tobramycin, 2; and gentamicin, 8 (the last two are polycationic agents, and the others are hydrophobic agents). The MICs of the hydrophobic and polycationic agents did not change when they were induced with 1 mM IPTG (data not shown). These results indicated that OmpF expression did not affect the native barrier of the P. aeruginosa outer membrane to hydrophobic agents and polycationic agents. In fact, OmpF expression caused leakage of a small amount of IMP-1, i.e., leakage of less than 5% of the total IMP-1 activity in the P. aeruginosa cells tested (data not shown). However, treatment of P. aeruginosa cells with EDTA and hexametaphosphate induced the release of β-lactamase from intact cells (21). Thus, it is suggested that OmpF expression increased the permeability of PAO1 cells by inducing the formation of additional hydrophilic pathways in the P. aeruginosa outer membrane.
Identification of the mechanisms contributing to the intrinsic penem resistance of P. aeruginosa and their interplay.
To induce the loss of the MexAB-OprM efflux system or the AmpC β-lactamase, the mexA and/or ampC gene on the chromosome of P. aeruginosa PAO1 was disrupted by an allelic exchange technique using mexA::res-ΩSm or ampC::ΩSm, respectively. Western immunoblot analysis using the previously described mouse monoclonal antibodies (12) and rabbit polyclonal antibodies (13) showed that both mexA::res-ΩSm mutants, KG2225 and KG2505, produced a reduced level of OprM and no detectable amounts of MexA and MexB (data not shown). These results are consistent with a previous study (55) demonstrating the presence of a weak promoter upstream from oprM. The level of AmpC β-lactamase activity in crude cell extracts was examined by a spectrophotometric assay with 100 μM cephaloridine as a substrate as described previously (24). The ampC::ΩSm strains KG2504 and KG2505 did not express AmpC β-lactamase even when induced with 0.5 μg of imipenem per ml. However, the strains in which ampC expression was maintained, PAO1 and KG2225, produced the β-lactamase when induced with 0.5 μg of imipenem per ml (data not shown). Thus, loss of the MexAB-OprM efflux system and/or AmpC β-lactamase in the respective strains was confirmed.
Table 3 shows the susceptibility of PAO1 and isogenic mutants lacking MexAB and/or AmpC β-lactamase and with or without the OmpF expression to various penems. The increases in outer membrane permeability due to OmpF expression in PAO1 caused a 16- to 128-fold increase in the susceptibility of PAO1 to the six penems tested (refer to PAO1/pKMF010). Loss of MexAB resulted in an 8- to 64-fold increase in the susceptibility of PAO1 to the penems tested except ritipenem. Although loss of MexAB seemed to barely affect susceptibility to ritipenem, overexpression of MexAB-OprM (refer to OCR1) caused an apparent reduction in the susceptibility of PAO1 to ritipenem as well as the other penems tested, indicating that MexAB-OprM does play a role in the intrinsic resistance of PAO1 to the six penems tested. However, the susceptibility of PAO1 to all penems tested was only slightly affected by the loss of AmpC β-lactamase, and there were only two- to fourfold increases in susceptibility (refer to KG2504). These results suggest that MexAB-OprM and the outer membrane barrier, but not AmpC β-lactamase, play a role in the intrinsic resistance of PAO1 to penem antibiotics. However, the MICs (4 to 32 μg/ml) of all penems tested for KG2225 or PAO1 carrying pKMF010 were still high even after the loss of MexAB-OprM or positive OmpF expression, respectively, compared with those for penem-susceptible gram-negative bacteria such as E. coli: the MIC for 90% of isolates (MIC90) of faropenem (34), ritipenem (31), AMA3176 (Okonogi et al., 33rd ICAAC), sulopenem (9), Sch29482 (30, 36), and Sch34343 (27) in E. coli are 1.56, 1, 0.39, 0.063, 1, and 0.2 μg/ml, respectively. It is suspected that the intrinsic penem resistance of P. aeruginosa results from the interplay among the identified mechanisms.
TABLE 3.
Susceptibilities of P. aeruginosa PAO1 and its isogenic mutants to penem antibioticsa
| Strain | Phenotypeb
|
MIC (μg/ml) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AmpC | MexAB | OmpF | Faropenem | Ritipenem | AMA3176 | Sulopenem | Sch29482 | Sch34343 | |
| PAO1 | + | + | − | 512 | 128 | 128 | 32 | 256 | 128 |
| PAO1/pKMF010 | + | + | + | 32 | 8 | 8 | 0.25 | 8 | 8 |
| KG2225 | + | − | − | 32 | 64 | 8 | 4 | 4 | 4 |
| KG2225/pKMF010 | + | − | + | 2 | 4 | 1 | 0.125 | 1 | 2 |
| KG2504 | − | + | − | 256 | 32 | 64 | 16 | 128 | 64 |
| KG2504/pKMF010 | − | + | + | 32 | 4 | 16 | 0.25 | 8 | 4 |
| KG2505 | − | − | − | 1 | 2 | 0.5 | 0.125 | 1 | 1 |
| KG2505/pKMF010 | − | − | + | 0.5 | 2 | 0.5 | 0.06 | 0.5 | 0.5 |
| OCR1 | + | ++ | − | 4,096 | 512 | 1,024 | 128 | 2,048 | 1,024 |
MICs were determined in experiments with L agar containing 1 mM IPTG. The presence of pKMF012 (vector control) alone in the tested strains did not change the susceptibility to any agent in this table.
+, wild-type level; −, nonexpression; ++, overexpression.
Loss of AmpC β-lactamase caused only an approximately twofold increase in the susceptibility of OmpF-expressing PAO1 to all penems tested (compared to the MICs for PAO1/pKMF010 and KG2504/pKMF010). In contrast, loss of either AmpC β-lactamase (refer to KG2505) or the outer membrane barrier (refer to KG2225/pKMF010) from the MexAB-OprM-deficient strain KG2225 caused 32- to 512-fold increases in the susceptibility of PAO1 and resulted the intrinsic resistance of PAO1 to all penems tested being reduced to the same as that of E. coli. The resistance of KG2505 was slightly reduced by further OmpF expression (refer to KG2505/pkMF010), and the resistance of KG2225/pKMF010 was slightly reduced by loss of AmpC β-lactamase (refer to KG2505/pKMF010).
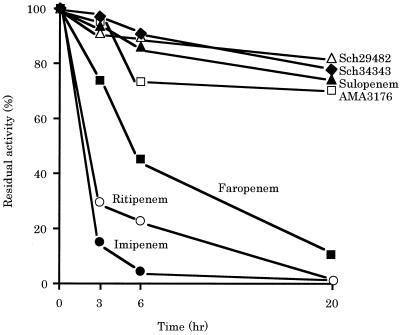
Loss of both AmpC β-lactamase and the MexAB-OprM system induced large increases in the susceptibility of PAO1 to all penems tested, suggesting that AmpC β-lactamase is among the mechanisms contributing to the intrinsic penem resistance of P. aeruginosa. However, several kinetic experiments (16, 27, 31, 34, 36) have demonstrated that all of the tested penems are highly stable to P. aeruginosa AmpC β-lactamase. To elucidate the discrepancy between the contribution of AmpC β-lactamase to intrinsic resistance and the stability of penems to AmpC β-lactamase, we examined the residual activities of various penem antibiotics after incubation of the agent with a crude AmpC β-lactamase preparation from PAO1. The antibacterial activity of all penems tested was reduced by incubation with AmpC β-lactamase, although AMA3176, sulopenem, Sch34343, and Sch29482 were more stable than ritipenem and faropenem (Fig. 3). Thus, AmpC β-lactamase is also one of the mechanisms contributing to the intrinsic penem resistance of P. aeruginosa.
FIG. 3.
Time course of the hydrolysis of various penems by the β-lactamase prepared from P. aeruginosa PAO1. The reaction mixture contained 0.5 μM β-lactamase and 10 μM penem in 50 mM sodium phosphate buffer (pH 7.0), and this was incubated for the indicated times. Each data point represents the level of residual antibacterial activity of the penem after incubation with β-lactamase for the indicated time.
Affinity of PBP.
β-Lactam antibiotics including penems display their antibacterial activity by binding to PBP, which are enzymes involved in the biosynthesis of bacterial peptidoglycan. Therefore, a low affinity of the PBPs of a bacterium for the β-lactam antibiotics would cause the bacterium to have intrinsic resistance to the agent. The affinities of each PBP of PAO1 for faropenem were examined by a competition assay with benzyl[14C]penicillin and compared with those for a potent antipseudomonal carbapenem, imipenem. The binding of faropenem to PBP1c and PBP3 was comparable to the respective binding of imipenem (Table 4). In contrast, the binding of faropenem to PBP1b and PBP2 was 1.5- to 1.8-fold lower than the respective binding of imipenem (Table 4). These low affinities cannot explain the high intrinsic resistance of P. aeruginosa to a penem, because triplicate loss of the AmpC β-lactamase, the MexAB-OprM efflux system, and the outer membrane barrier lowered the MICs of the penems tested except for ritipenem to approximately the MIC of imipenem for the AmpC-deficient PAO1 mutant (0.2 μg/ml) (25) (Table 3).
TABLE 4.
Binding of imipenem and faropenem to PBPs of P. aeruginosa PAO1
| PBP | Panem concn (μg/ml) required to inhibit the binding of [14C]benzylpenicillin by 50%
|
|
|---|---|---|
| Faropenem | Imipenem | |
| 1b | 0.23 | 0.13 |
| 1c | 0.15 | 0.15 |
| 2 | 0.19 | 0.13 |
| 3 | 0.20 | 0.18 |
| 4 | 0.13 | 0.61 |
| 5/6 | >1 | 0.16 |
DISCUSSION
We determined a mechanism contributing to the high intrinsic resistance of P. aeruginosa to penem antibiotics by using a series of isogenic mutants that had impaired MexAB-OprM and/or AmpC β-lactamase and E. coli OmpF expression plasmid.
MexAB-OprM expressed in wild-type strains of P. aeruginosa such as PAO1 plays a role in intrinsic penem resistance, because loss of MexAB-OprM (refer to KG2225 in Table 3) sensitized PAO1 to all penems tested and overexpression of MexAB-OprM had the opposite effect (refer to OCR1 in Table 3). Recent studies have led to the identification of a fourth operon, mexX-mexY (1, 28), which does not encode an outer membrane protein. Although expression of this operon is suppressed in wild-type strains of P. aeruginosa such as PAO1, MexXY expression is induced by antimicrobial agents such as tetracycline, gentamicin, and erythromycin and MexXY functionally associates with spontaneously expressed OprM (26). The production of OprM in mexA knockout mutants, KG2225 and KG2505 (reference 26 and data not shown), led to the assumption that MexXY-OprM is involved in the intrinsic resistance to penems. However, further loss of OprM from KG2505 did not change the susceptibilities to perens (K. Okamoto, N. Gotoh, and T. Nishino, unpublished data). This result suggested that MexXY is not involved in the intrinsic penem resistance of P. aeruginosa.
MexAB-OprM does not pump out imipenem (12, 24). The permeability rates of penem entry into the periplasm were not affected by either loss or overexpression of MexAB-OprM, similar to imipenem (Table 2). These results suggest that penem molecules that penetrate into the periplasm encounter a β-lactamase molecule in the periplasm prior to a substrate-trapping site of the MexAB-OprM efflux system and, furthermore, that the substrate-trapping site of the efflux system is located at a different site, probably in the inner membrane, rather than in the periplasm. This supports the substrate recognition model (20, 44) of MexAB-OprM for drug partitioning into the cytoplasmic membrane.
Loss of AmpC β-lactamase only slightly affected the susceptibility of PAO1 to penem antibiotics (refer to KG2504 in Table 3). This result may indicate that the slight contribution of AmpC β-lactamase to the intrinsic penem resistance of P. aeruginosa is due to the high stability of penems to β-lactamase as found in kinetic experiments (16, 27, 31, 34, 36). However, our study revealed that AmpC β-lactamase apparently contributes to penem resistance by its interplay with MexAB-OprM (refer to KG2505 in Table 3). Furthermore, incubation of various penems with a crude AmpC β-lactamase preparation showed that all of the penems tested were apparently hydrolyzed, although AMA3176, sulopenem, Sch29482, and Sch34343 were more slowly hydrolyzed than were ritipenem and faropenem (Fig. 3). Despite the nearly identical levels of the high stability of AMA3176, sulopenem, Sch29482, and Sch34343 to AmpC β-lactamase (Fig. 3), the differences in the susceptibility of KG2225 with or without the loss of AmpC to AMA3176 and sulopenem were much larger than those to Sch29482 and Sch34343 (refer to KG2225 and KG2505, respectively, in Table 3). This may be due to differences with which different penem antibiotics induce AmpC β-lactamase, although we do not have any results on the induction abilities. Taken together, although the removal of penem antibiotics by AmpC β-lactamase alone is very slight and is not important to penem intrinsic resistance, the interplay of AmpC β-lactamase with MexAB-OprM efflux system displays a potent ability to remove penems.
The contribution of the outer membrane barrier to penem intrinsic resistance was evaluated using the newly constructed E. coli OmpF expression plasmid instead of outer membrane permeabilizers such as chelators and polycationic agents. OmpF expression increased the permeability rate of faropenem and sulopenem entry into PAO1 cells by approximately two- and fivefold, respectively (Table 2), without inhibiting their growth. The difference between the increased permeability of faropenem, which has one negative charge in its molecular structure (Fig. 1), and that of sulopenem, which has two negative and one positive charges (Fig. 1), is in accordance with the earlier findings (53) that the OmpF pore in the E. coli outer membrane functions more selectively toward β-lactams with two negative and one positive charges than toward β-lactams with one positive charge. However, even when OmpF was expressed (refer to PAO1/pKMF014 in Table 2), the permeability rates of faropenem and sulopenem were about 50% of that of imipenem in PAO1. The higher permeability rate of imipenem than penems into PAO1 cells is due, in part, to the expression of the outer membrane protein OprD, which is permeable to positively charged carbapenems such as imipenem (10, 47). In fact, OprD deficiency caused decreases in the susceptibility of PAO1 to imipenem but not to any of the penems tested (Okamoto et al., unpublished). OmpF expression greatly increased the permeability rate of imipenem (refer to PAO1/pKMF014), and this appears to be due to higher selectivity of OmpF for zwitterionic β-lactams such as imipenem than others.
The susceptibility of PAO1 to hydrophobic and bulky compounds such as erythromycin and polycationic compounds such as tobramycin was not affected by OmpF expression, as described in Results. These results confirm that OmpF expression does not induce the formation of a diffusion pathway for hydrophobic or polycationic compounds. We have also observed that OmpF expression induces increases in the permeability rates of other hydrophilic compounds such as cephems and fluoroquinolones through the P. aeruginosa outer membrane (Okamoto, et al., unpublished). These results show that destruction of the outer membrane barrier using the OmpF expression plasmid can be used for studying the resistance to other hydrophilic agents, although the diffusion pore maintains selectivity with regard to the molecular size, charge, and hydrophobicity (53) of the test solute. To solve these problems, we attempted to express Neisseria gonorrhoeae PIB (6) (GenBank accession no. X52823), which forms a much larger pore than OmpF. We were unsuccessful in expressing this gene in P. aeruginosa cells, because its expression is probably toxic to those cells. Although OmpF expression caused only partial destruction of the outer membrane barrier, OmpF expression in PAO1 (PAO1 carrying pKMF010 in Table 3) reduced the penem resistance level of PAO1 to that of MexAB-OprM-deficient PAO1 (refer to KG2225 in Table 3). This suggests that the outer membrane barrier plays a larger role in penem resistance than the MexAB-OprM efflux system does.
In this study, we demonstrated that the high intrinsic resistance of P. aeruginosa to penems could be reduced to the same level as that of penem-susceptible gram-negative bacteria such as E. coli, by loss of both MexAB-OprM and the outer membrane barrier or by loss of both AmpC β-lactamase and MexAB-OprM. Wild-type laboratory strains of E. coli also express the multidrug efflux system AcrAB/TolC (7, 35). However, comparison of the susceptibility of the AcrAB-deficient mutant and its parent E. coli strain demonstrated that no penems used in this study were pumped out by the major efflux system AcrAB of E. coli (Okamoto et al., unpublished). This supports the susceptibility tests demonstrating that the resistance mechanism in P. aeruginosa was not stronger than that in E. coli after loss of either the outer membrane barrier or AmpC β-lactamase from the MexAB-OprM-deficient mutant. Thus, the high intrinsic penem resistance in P. aeruginosa is caused by the interplay among at least these three resistance mechanisms.
The inhibition activity of faropenem for PBPs was slightly lower than that of imipenem in the competition assay (Table 4). These results may indicate that the low affinity of PBPs for penems is also one of the mechanisms contributing to intrinsic penem resistance. However, the MICs of almost all of the penems tested for KG2505/pKMF010, which lacked all three of the MexAB-OprM, AmpC β-lactamase, and outer membrane barrier, were identical to or lower than the MIC of imipenem for AmpC-deficient P. aeruginosa (Table 3).
Thus, we concluded that the interplay among the MexAB-OprM, AmpC β-lactamase, and outer membrane barrier, rather than the slightly low affinity of PBPs for penems, endows P. aeruginosa with high intrinsic resistance to penems. Accordingly, one feature of a new penem antibiotic exhibiting antipseudomonal activity should be an ability to escape from at least one of these resistance mechanisms, especially the outer membrane barrier and the efflux system. Moreover, clinical usage of outer membrane permeabilizers including antimicrobial cationic polypeptides (54) and efflux system inhibitors (43), which are still in development, may expand the clinical application of penem antibiotics to P. aeruginosa. Another important finding of this study is that the OmpF expression plasmid and the efflux system-deficient or AmpC-deficient strains are valuable tools for screening candidate compounds possessing antipseudomonal activity.
ACKNOWLEDGMENTS
We thank H. Tsujimoto for his help with the DNA recombination experiments.
This research was supported by grants for Scientific Research to N.G. from the Ministry of Education, Science, Sports, and Culture of Japan and from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Aires J R, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann E, Ochs B, Abel K J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 3.Angus B L, Carey A M, Caron D A, Kropinski A M B, Hancock R E W. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982;21:299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry A L, Aldrige K E, Allen S D, Fuchs P C, Gerlach E H, Jones R N, Pfaller M A. In vitro activity of FCE22101, imipenem, and ceftazidime against over 6,000 bacterial isolates and MIC quality control limits of FCE22101. Eur J Clin Microbiol Infect Dis. 1988;7:794–798. doi: 10.1007/BF01975053. [DOI] [PubMed] [Google Scholar]
- 5.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 6.Douglas J T, Lee M D, Nikaido H. Protein I of Neisseria gonorrhoeae outer membrane is a porin. FEMS Microbiol Lett. 1981;12:305–309. [Google Scholar]
- 7.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs P C, Barry A L, Sewell D L. Antibacterial activity of WY-49605 compared with those of six other oral agents and selection of disk content for disk diffusion susceptibility testing. Antimicrob Agents Chemother. 1995;39:1472–1479. doi: 10.1128/aac.39.7.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gootz T, Retsema J, Girard A, Hamanaka E, Anderson M, Sokolowski S. In vitro activity of CP-65,207, a new penem antimicrobial agent, in comparison with those of other agents. Antimicrob Agents Chemother. 1989;33:1160–1166. doi: 10.1128/aac.33.8.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotoh N, Nishino T. Decreases of the susceptibility to low molecular weight β-lactam antibiotics in imipenem-resistant Pseudomonas aeruginosa mutants: role of outer membrane protein D2 in their diffusion. J Antimicrob Chemother. 1990;25:191–198. doi: 10.1093/jac/25.2.191. [DOI] [PubMed] [Google Scholar]
- 11.Gotoh N, Nunomura K, Nishino T. Resistance of Pseudomonas aeruginosa to cefsulodin: modification of penicillin-binding protein 3 and mapping of its chromosomal gene. J Antimicrob Chemother. 1990;25:513–523. doi: 10.1093/jac/25.4.513. [DOI] [PubMed] [Google Scholar]
- 12.Gotoh N, Itoh N, Tsujimoto H, Yamagishi J, Oyamada Y, Nishino T. Isolation of OprM-deficient mutants of Pseudomonas aeruginosa by transposon insertion mutagenesis: evidence of involvement in multiple antibiotic resistance. FEMS Microbiol Lett. 1994;122:267–274. doi: 10.1111/j.1574-6968.1994.tb07179.x. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in ΔmexA-mexB-oprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock R E W, Wong P G W. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob Agents Chemother. 1984;26:48–52. doi: 10.1128/aac.26.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock R E W. Antibacterial peptides and the outer membranes of gram-negative bacilli. J Med Microbiol. 1997;46:1–3. doi: 10.1099/00222615-46-1-1. [DOI] [PubMed] [Google Scholar]
- 16.Inoue E, Mitsuhashi S. In vitro antibacterial activity and β-lactamase stability of SY5555, a new oral penem antibiotic. Antimicrob Agents Chemother. 1994;38:1974–1979. doi: 10.1128/aac.38.9.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyobe S, Tsunoda M, Mitsuhashi S. Cloning and expression in Enterobacteriaceae of the extended-spectrum β-lactamase gene from a Pseudomonas aeruginosa plasmid. FEMS Microbiol Lett. 1994;121:175–180. doi: 10.1111/j.1574-6968.1994.tb07096.x. [DOI] [PubMed] [Google Scholar]
- 18.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 19.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X-Z, Zhang L, Srikumar R, Poole K. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X-Z, Zhang L, Poole K. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 22.Livermore D M, Yang Y-J. β-Lactamase lability and inducer power of newer β-lactam antibiotics in relation to their activity against β-lactamase-inducibility mutants of Pseudomonas aeruginosa. J Infect Dis. 1987;155:775–782. doi: 10.1093/infdis/155.4.775. [DOI] [PubMed] [Google Scholar]
- 23.Lodge J M, Minchin S D, Piddock L J V, Busby S J W. Cloning, sequencing and analysis of the structural gene and regulatory resion of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem J. 1990;272:627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:400–402. doi: 10.1128/aac.43.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:2242–2246. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuda K, Sasaki K, Inoue K, Kondo H, Inoue M, Mitsuhashi S. In vitro antibacterial activity of Sch34343 and its stability to β-lactamases and renal dehydropeptidase 1. Antimicrob Agents Chemother. 1985;28:684–688. doi: 10.1128/aac.28.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakae T, Nakajima A, Ono T, Saito K, Yoneyama H. Resistance to β-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob Agents Chemother. 1999;43:1301–1303. doi: 10.1128/aac.43.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neu H C, Labthavikul P. Antibacterial activity of an oral penem, Sch29482. J Antimicrob Chemother. 1982;9(Suppl. C):49–57. doi: 10.1093/jac/9.suppl_c.49. [DOI] [PubMed] [Google Scholar]
- 31.Neu H C, Chin N X, Labthavikul P. The in-vitro activity of a novel penem FCE22101 compared to other β-lactam antibiotics. J Antimicrob Chemother. 1985;16:305–313. doi: 10.1093/jac/16.3.305. [DOI] [PubMed] [Google Scholar]
- 32.Nikaido H. Role of permeability barriers in resistance to β-lactam antibiotics. Pharmacol Ther. 1985;27:197–231. doi: 10.1016/0163-7258(85)90069-5. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishino T, Maeda Y, Ohtsu E, Koizuka S, Nishihara T, Adachi H, Okamoto K, Ishiguro M. Studies on penem antibiotics II. In vitro activity of SUN5555, a new oral penem. J Antibiot. 1989;42:977–988. doi: 10.7164/antibiotics.42.977. [DOI] [PubMed] [Google Scholar]
- 35.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips I, King A, Shannon K, Warren C. Sch29482: in-vitro antibacterial activity and susceptibility to β-lactamases. J Antimicrob Chemother. 1982;9(Suppl. C):25–30. doi: 10.1093/jac/9.suppl_c.25. [DOI] [PubMed] [Google Scholar]
- 37.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 38.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 40.Prentki P, Binda A, Epstein A. Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene. 1991;103:17–23. doi: 10.1016/0378-1119(91)90385-o. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen B A, Bush K, Keeney D, Yang Y, Hare R, O'Gara C, Medeiros A A. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob Agents Chemother. 1996;40:2080–2086. doi: 10.1128/aac.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves D S, Holt H A, Bywater M J. Comparative in-vitro activity of Sch34343, a new penem antibiotic. J Antimicrob Chemother. 1985;15(Suppl. C):57–66. doi: 10.1093/jac/15.suppl_c.57. [DOI] [PubMed] [Google Scholar]
- 43.Renau T E, Léger R, Flamme E M, Sangalang J, She M W, Yen R, Gannon C L, Griffith D, Chamberland S, Lomovskaya O, Hecker S J, Lee V J, Ohta T, Nakayama K. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem. 1999;42:4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 44.Srikumar R, Li X-Z, Poole K. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikumar R, Tsang E, Poole K. Contribution of the MexAB-OprM multidrug efflux system to the β-lactam resistance of penicillin-binding protein and β-lactamase-derepressed mutants of Pseudomonas aeruginosa. J Antimicrob Chemother. 1999;44:537–540. doi: 10.1093/jac/44.4.537. [DOI] [PubMed] [Google Scholar]
- 46.Trias J, Dufresne J, Levesque R C, Nikaido H. Decreased outer membrane permeability in imipenem-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33:1201–1206. doi: 10.1128/aac.33.8.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 48.Tsuda M. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutations in bacterial chromosome. Gene. 1998;207:33–41. doi: 10.1016/s0378-1119(97)00601-x. [DOI] [PubMed] [Google Scholar]
- 49.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wise R, Andrews J M, Danks G. Comparison of in vitro activity of FCE22101, a new penem, with those of other β-lactam antibiotics. Antimicrob Agents Chemother. 1983;24:909–914. doi: 10.1128/aac.24.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982;152:636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Falla T, Wu M, Fidai S, Burian J, Kay W, Hancock R E W. Determinants of recombinant production of antimicrobial cationic peptides and creation of peptide variants in bacteria. Biochem Biophys Res Commun. 1998;247:647–680. doi: 10.1006/bbrc.1998.8848. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Q, Li X-Z, Srikumar R, Poole K. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother. 1998;42:1682–1688. doi: 10.1128/aac.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]