Graphical abstract
Keywords: Differential scanning fluorimetry, Thermal shift assay, ESP32, Protein melting point, Biophysical chemistry
Abstract
Differential scanning fluorimetry (DSF) is a widely used biophysical technique with applications to drug discovery and protein biochemistry. DSF experiments are commonly performed in commercial real-time polymerase chain reaction (qPCR) thermal cyclers or nanoDSF instruments. Here, we report the construction, validation, and example applications of an open-source DSF system for 176 €, which, in addition to protein-DSF experiments, also proved to be a versatile biophysical instrument for less conventional RNA-DSF experiments. Using 3D-printed parts made of polyoxymethylene, we were able to fabricate a thermostable machine chassis for protein-melting experiments. The combination of blue high-power LEDs as the light source and stage light foil as filter components was proven to be a reliable and affordable alternative to conventional optics equipment for the detection of SYPRO Orange or Sybr Gold fluorescence. The ESP32 microcontroller is the core piece of this openDSF instrument, while the in-built I2S interface was found to be a powerful analog-to-digital converter for fast acquisition of fluorescence and temperature data. Airflow heating and inline temperature control by thermistors enabled high-accuracy temperature management in PCR tubes (±0.1 °C) allowing us to perform high-resolution thermal shift assays (TSA) from exemplary biological applications.
Specifications table
| Hardware name | openDSF |
|---|---|
| Subject area | Chemistry and biochemistry |
| Hardware type | Measuring physical properties and in-lab sensors |
| Closest commercial analog | Real-time PCR thermocycler: e.g., Qiagen Rotor-Gene Q |
| Open source license | GNU General Public License (GPL) 3.0 |
| Cost of hardware | 176 € |
| Source file repository | openDSF (Mendeley data): https://doi.org/10.17632/73rt8s7pwd.2 |
Hardware in context
Differential scanning fluorimetry (DSF) is a method of biophysical protein analysis, which is utilized for the investigation of thermal protein unfolding [1], [2], [3]. In this context, protein-binding ligands may increase the protein stability and result in the protein unfolding being shifted to a higher melting temperature. The difference between the melting temperature of a protein in the presence or absence of a ligand is termed “thermal shift”. Thermal shift assays (TSA) are among the most common ligand characterization techniques with numerous applications for drug discovery and structural biology [4], [5]. TSAs are easy to establish and are typically performed in commercial real-time polymerase chain reaction thermal cycler (qPCR) instruments [6], [7]. To perform a typical DSF experiment, a sample containing a protein or protein complex is added with the SYPRO Orange dye. SYPRO Orange has a low intrinsic fluorescence with the natively folded protein present. Only upon heating, when hydrophobic patches of the protein are exposed, the merocyanine-type dye binds to them, resulting in a significant increase in fluorescence [3]. The apparent melting point (Tm) is usually determined as the inflection point of the fluorescence versus temperature curve and is considered a measure of protein stabilization [8]. Widely used qPCR machines used for this purpose utilize halogen lamps, laser diodes, or LEDs to monitor the fluorescence changes allowing the measurement of the SYPRO Orange fluorescence. Besides the high cost of commercial qPCR instruments, the factory filter sets frequently do not optimally fit the large Stokes shift of the SYPRO Orange dye (λex = 490 nm/λem = 624 nm) and cause reduced quantum yield and signal-to-noise (S/N)-ratios [9]. This might lead to the absence of a measurable melting curve for small and less hydrophobic proteins [3]. In contrast, fluorescence spectrometers with variable settings of wavelengths are limited in their temperature management, making them mostly impractical for DSF experiments.
To address this issue, Hoeser et al. developed a cheap and easy-to-build heating device for a 96-well plate reader that is coupled with a fluorescence spectrometer [10]. However, this requires the availability of a likewise expensive plate reader. Recently, open-source qPCR machines have been presented and described for DIY reproduction [11], [12]. However, some of these open-source qPCR devices (including some commercial instruments) were designed and proven for nucleic acid quantification and will likely be impractical for performing high-resolution DSF experiments because they show temperature homogeneity ranging up to ± 0.5 °C so that thermal shifts smaller than the temperature resolution limit might not be accurately detected [11], [13].
In addition to proteins, numerous RNA molecules also possess a native tertiary structure that can be characterized in the course of a thermal unfolding process. Similar to protein–ligand binding, small-molecule modulators of RNA often lead to stabilization and increased melting points [14], [15]. This can be obtained in the course of melting experiments from the intrinsic fluorescence properties of the nucleic bases or with an intercalating dye. For this purpose, Silvers et al. developed a DSF protocol using dyes from the Sybr series, which allow quantification with the same fluorescence setup as for SYPRO Orange when using the Sybr Gold dye (λex = 495 nm/λem = 537 nm) [16].
In this work, we describe the construction and performance of an optics-free 3D-printable low-cost differential scanning fluorometer (openDSF) with high sensitivity for SYPRO Orange (protein-DSF) or Sybr Gold (RNA-DSF) fluorescence detection and a melting temperature resolution of < 0.1 °C.
Hardware description
We designed an open-source differential scanning fluorometer as a low-cost alternative to existing qPCR thermal cyclers and commercial nanoDSF devices for biophysical characterization of protein–ligand complexes (Fig. 1). To support low-budget academic research campaigns, only inexpensive and easily available components (e.g., 3D-printed pieces, basic electronics) were used, so that the costs (176 €) are several magnitudes lower than those of a commercial counterpart (∼20,000 €) [17]. The fundamental principle of the system is that four DSF reaction mixtures, prepared in PCR tubes, are flowed with heated air, while both the temperature and the SYPRO Orange fluorescence of each PCR tube are measured and recorded. To achieve this functionality, the design of the device combines five modules to form the whole open-source differential scanning fluorometer: 1) the 3D-printed chassis, 2) the ESP32 microcontroller, 3) the air heating system, 4) the temperature probes, and 5) the fluorescence measurement path (Fig. 2B).
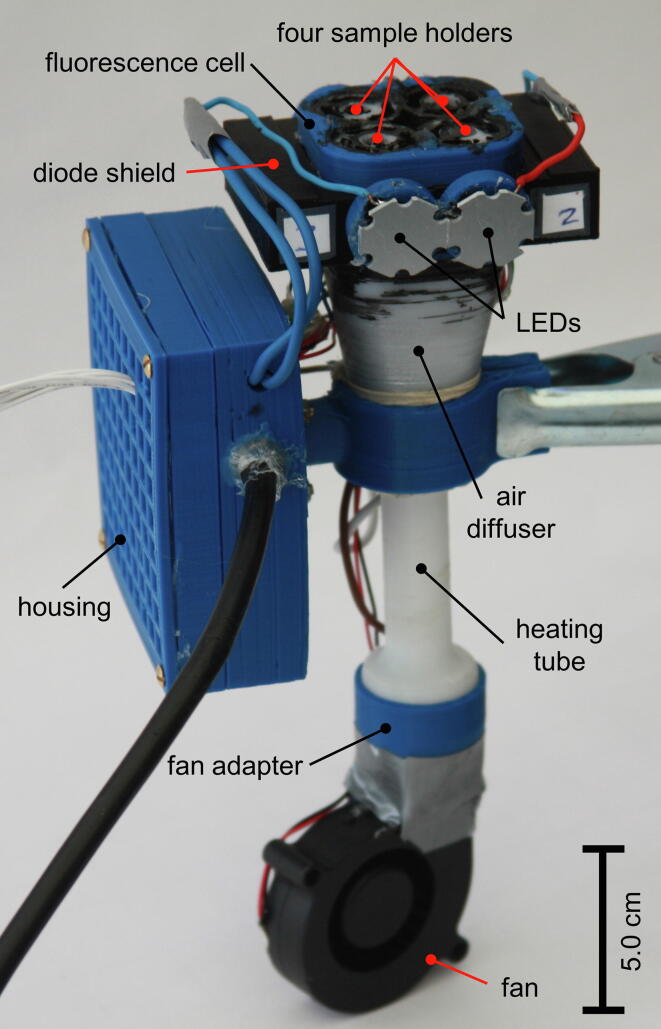
Fig. 1.
Device overview. Photograph of the openDSF instrument with opened sample holders and labeling of important features.
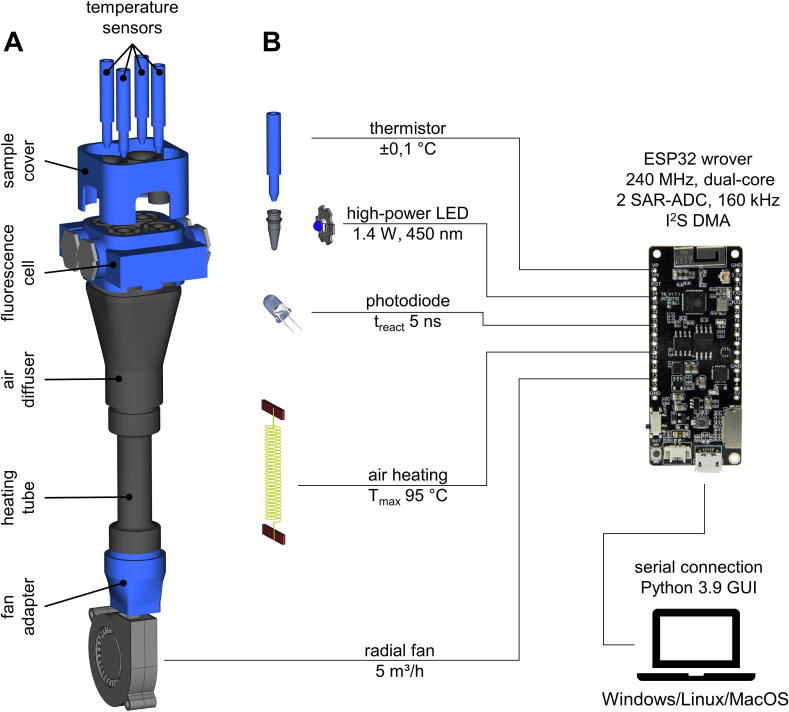
Fig. 2.
(A) Explosion drawing of the openDSF chassis. 3D-printed parts made of PETG are shown in blue. POM parts are shown in gray. A detailed explosion drawing and a cross-section of the fluorescence cell are shown in the figures below. (B) Schematic overview of all modules included in the system with information on characteristic specifications of the components. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3D-printed device chassis
Structural parts forming the chassis of the device were fabricated by fused deposition modeling with an Anycubic 4Max 3D-printer either from polyethylene terephthalate (PETG, diameter: 1.75 mm) or from polyoxymethylene (POM, diameter: 1.75 mm) filament. Functional tasks of the chassis are the assembly of the measuring, lighting, and heating modules as well as the directional flow control of the air heating system. DSF experiments are performed in the temperature range of 20–95 °C, which sets special requirements for the temperature stability of the 3D-printed parts. Hence, POM filament was used for parts with increased temperature demands (POM is dimensionally stable up to at least 130 °C [18]). To meet the known constructional limitations of POM 3D-printing, sharp corners and edges were avoided in the design of the POM parts, because these are particularly prone to warping [19]. The openDSF chassis consisted of 14 different 3D-printed parts (Fig. 2A). POM parts were designed double-walled and printed with an infill of 20% so that the high air content inside these parts makes them good thermal insulators. The low thermal losses allow for reproducible heating ramps and eliminate hot zones on the device surface so that it can be safely touched by the user at any time.
ESP32 microcontroller
The Espressif ESP32 wrover microcontroller (dual-core CPU, 8 MB PSRAM, 240 MHz clock) was chosen as the central control unit of the fluorometer. The ESP32 is a low-cost and relatively recent microcontroller family (released in 2016) [20]. The digital outputs of the chip were used to control the excitation LEDs, the air-supplying fan, and the heating wire. The peripheral I2S interface of the chip, which has been designed for transmitting audio data, has proven suitable for recording thermistor and photodiode analog input signals in the microsecond range (Fig. 2B). In this regard, two onboard multi-channel successive-approximation analog-to-digital converters (SAR-ADCs) were polled with a sampling rate of 60 kHz and the temperature resp. fluorescence values were recorded via direct memory access (DMA) protocol. Online data processing was handled by one of the two CPU cores, while the other core was tasked with general control and communication via the PC's serial interface. The firmware for the ESP32 was developed on the Espressif ESP-IDF 4.2 platform, while the serial interface was implemented with Python, Matplotlib, NumPy, SciPy, and Tkinter. A serial/USB driver must be installed on the connected PC.
Air heating system
Temperature control of the sample PCR tubes was performed by airflow heating, in which the air was tempered with a heating wire and conveyed by a radial fan (Fig. 3A). The heating power (heating wire current) was controlled by a 13-bit PWM MOSFET circuit, while the air volume (fan speed) was controlled by an 11-bit circuit. A Sunon Blower (MF50151V2-B00U-A99) was used as a radial fan, which was designed to convey approx. 5 m3/h of air along an SS430 awg30 heating wire (stainless steel 430, l = 70 cm, d = 0.25 mm, R = 12.2 Ω/m, X6Cr17). The power consumption of the heating module was designed to be max. 150 W to be able to run both slow and fast (0.1–9 °C/min) heating ramps for up to 4 PCR tube samples. A homogeneous temperature cross-section was ensured by turbulent airflow within the device’s mixing cell. The dimensions of the air-flowed round parts of the chassis were designed based on a calculated Reynolds number (Re ∼ 10,000). Additionally, to increase the velocity of the airflow and thus the heat transfer in the proximity of the sample vessels, the sample holders were shaped as Laval nozzles. The resulting pressure loss promotes uniform distribution of the airflow to the 4 individual sample holders.
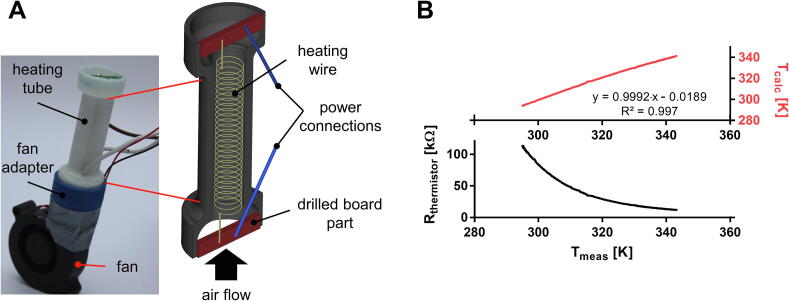
Fig. 3.
(A) Photograph and cross-section of the heating module. The air heating system works according to the principle that the fan conveys ambient air through the heating tube, in which the air is heated and turbulated on the coiled heating wire. The heating coil is self-supporting and is connected to the power supply at the ends on cut-to-size drilled board parts. (B) Bottom: The air heating system was calibrated using the thermistors of the sample holders. The resistance of the thermistors follows the logarithmic Steinhart-Hart relationship to the actual temperature. Top: Determination of the coefficients of the Steinhart-Hart equation () allows temperature adjustment in the PCR tubes with an absolute accuracy of 0.1 °C.
Temperature probes
The temperature of each sample can be determined separately using negative temperature coefficient (NTC) thermistors (NTC3950, 100 kΩ) within the airspace of the individual PCR tube. The temperature probes were dimensioned in a manner that they can be inserted into the PCR tube and thereby seal the upper opening. A thermistor circuit was designed in such a way that a voltage divider (Vref = 5.02 V) enabled the recording of relevant measurement temperatures (20–95 °C) within the ADC’s 12-bit resolution. The measured temperature-dependent voltages were recorded every second by the ESP32 chip and stabilized by oversampling (SAR-ADC at 60 kHz sampling rate). The conversion of voltages to sample temperatures was performed using the modified Steinhart-Hart equation () [21]. The apparent resistance was calculated from the measured voltages and the dimensions of our design: . The thermistors were calibrated by determining the coefficients (ai) using a precision contact thermometer (Pt100). With this setup, we were able to achieve linear heating ramps over the entire temperature range, with an absolute temperature inaccuracy of ± 0.1 °C (Fig. 3B).
Fluorescence measurement path
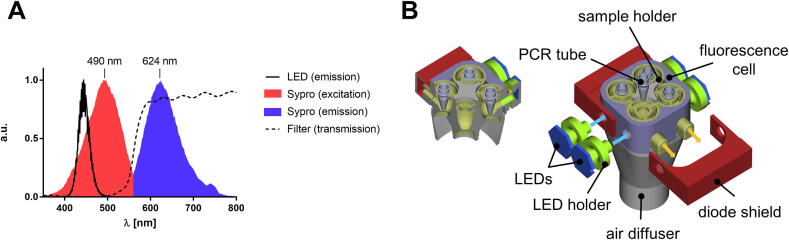
The conventional DSF dye SYPRO Orange is a merocyanine-type fluorescent dye with absorption and emission maxima of λex = 490 nm and λem = 624 nm (Fig. 4A). Commercial qPCR instruments often use costly lamps, optics, or filter equipment, which we have circumvented by using cheap high-power OSRAM Oslon SSL 80 royal-blue LEDs as the light source (on 20 mm stars, LD CQ7P). The fluorescent dye in the samples was excited with a 50 ms pulse of the high-power blue LED (λ = 450 nm, Wmax = 1400 mW, ϕE = 630 mW). Longer pulse widths resulted in significant photobleaching of the dye during long-term experiments, but since the fluorescence signal is recorded at a sampling rate of 60 kHz, a 50 ms pulse width was found to be sufficient to achieve low-noise recordings. Since the output fluorescence depends on the protein, the buffer, and the sample volume, the irradiation intensity can be modulated with the 11-bit PWM controller of a constant current source (LED BUCK V2, 1000 mA, 42 V, PWM < 5 kHz). Excitation of the sample solution was performed through the lateral outer wall of the PCR tube, with the emitted fluorescence captured at a 90° angle by OSRAM SFH 203 photodiodes (Fig. 4B). The SFH 203 photodiode was selected because of its exceptionally fast switching operational performance (rise/fall time: 5 ns). However, this photodiode exhibits sensitivity throughout the entire visible spectrum, so that the scattered and reflected excitation light must be eliminated with a filter. We have found that filter foils for stage spotlights are an excellent alternative to expensive optical filters. In this regard, the receiver photodiode was shielded with a piece of the LEE 158 Deep Orange filter foil, which efficiently prevented the passage of light with a wavelength of < 500 nm. The current of the receiver photodiode was evaluated with a transimpedance amplifier (Texas Instruments OPA380, RF = 12 MΩ) and the resulting output voltage was measured by the ESP32 SAR-ADC (sampling rate of 60 kHz, bit depth of 12-bit).
Fig. 4.
(A) Normalized spectra of the SYPRO Orange fluorescence, LED emission, and filter foil transmission (all measured on a Tecan Spark 10 M spectrometer). The LED emission showed good spectral overlap with the excitation characteristics of SYPRO Orange, whereas the orange filter foil efficiently filtered out excitation light but allowed the SYPRO Orange emission to pass with a transmission rate of > 80%. (B) Exploded view drawing and cross-section of the fluorescence cell with labeling of the most important components. The blue arrows show the path of the excitation light while the orange arrows illustrate the emitted radiation path. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Data acquisition and processing
During a DSF run, both fluorescence and temperature data are acquired for each of the four holders individually (ADC serial inputs: 8 channels = 4 each for photodiodes and thermistors). In this regard, two values per data point, i. e. temperature and fluorescence, are recorded for regular intervals. The interval between two measurement points has been established with 1000 ms by default but can also be modified (def TEMP_SAMPLE_PERIOD). At the beginning of each interval, a sampling phase is performed in which the PCR tube is irradiated, and fluorescence resp. temperature values are recorded. The default duration for the sampling phase is determined by the LED pulse width (int led_on; default: 50 ms). The sampling frequency is 60 kHz on 8 channels (4 temperature, 4 fluorescence), hence, for each data point 375 raw values per channel () are recorded. Subsequently, these raw values are combined by arithmetic averaging and the mean fluorescence values are written to a log file for online plot and CSV export at specified temperatures (0.1–1 °C/datapoint). For post-acquisition processing, a smoothing function according to the Savitzky-Golay filtering algorithm was implemented (scipy.signal.savgol_filter). The signal acquisition and processing pipeline of the openDSF system is shown in SI Fig. 1.
-
•
The open-source differential scanning fluorometer (openDSF) allows fast, accurate, and reliable protein and RNA-DSF measurements.
-
•
The combination of high-power LEDs as the light source and stage light foil as filter components is a cost-effective alternative to laser lamps and optics equipment for fluorometric measuring cells.
-
•
Airflow heating and temperature control by thermistors allow high accuracy temperature management in PCR tubes (±0.1 °C).
-
•
The ESP32 microcontroller with the I2S interface is a powerful system for fast acquisition of analog measurement data (sampling rate 60 kHz).
-
•
A 3D-printable polyoxymethylene (POM) chassis allows biological experiments at up to 130 °C.
Design files summary
The repository containing the design data for reproduction and modification of an openDSF instrument can be found at https://doi.org/10.17632/73rt8s7pwd.2
AirDiffuser: The air diffuser (POM) divides the heated airflow among the four individual measuring cells and leads to turbulent air mixing and thus to a homogeneous temperature profile.
DiodeShield: The diode shield (PETG) is designed to shield the photodiodes from external light or daylight to reduce the noise of the recorded fluorescence signal.
FanAdapter: The fan adapter (PETG) converts the rectangular outlet of the Sunon radial fan to a circular fitting of the heating tube.
FluorescenceCell: The fluorescence cell (PETG) creates a framework into which the sample holders are mounted.
HeatingTube: The heating tube (POM) incorporates the coiled heating wire.
Housing: The housing of the circuit boards protects the electronic components during use.
LEDHolder: High-power LEDs are glued onto the LED holders and serve to stabilize them.
openDSF: Project folder (openDSF.zip) for insertion into an ESP_IDF development environment. Python GUI (openDSF.py) to control the device from the user’s PC.
PhotoDiodePCB: Material for the fabrication of printed circuit boards of the photodiode modules.
SampleCover: A light and temperature shielding cover (PETG + POM) is placed over the measuring cell, through which the temperature sensors are inserted into the PCR tubes. The heated airflow of the instrument exits through this cover, which is why the contact points to the hot air were protected with thermally robust POM inserts.
SampleHolder: The sample holders (POM) are shaped as Laval nozzles to ensure optimal airflow and heat transfer. Due to the pressure loss of the nozzle, homogeneous air distribution is achieved. The sample holders provide holes for the LEDs and photodiodes.
TemperatureSensor: The temperature sensors (PETG) hold the individual thermistors and shield the sensitive electrical contacts from moisture and physical contact.
WiringDiagram: Circuit diagram overview of the electronic components of the openDSF system created with KiCad Eeschema.
3D-printing profiles: List of 3D-printer settings (.ini) for the different filaments used (PETG and POM).
Bill of materials summary
| Designator | Component | Number | Cost per unit -currency | Total cost - currency | Source of materials | Material type |
|---|---|---|---|---|---|---|
| Constant current power supply | LED BUCK V2 constant current power supply (1000 mA, 42 V), PWM 2.5 V, < 5 kHz | 1 | 12.90 € | 12.90 € | https://bit.ly/31gELmI | Semi-conductor |
| ESP32 board | ESP32 wrover 8 MB Psram, development board | 1 | 7.21 € | 7.21 € | https://bit.ly/3l1rbda | Semi-conductor |
| Fan | Sunon Blower MF50151V2-B00U-A99 | 1 | 6.93 € | 6.93 € | https://bit.ly/3yV6btO | Composite |
| Heating wire | SS430 awg30 stainless steel wire 430, 0.25 mm, resistance 12.2 Ω/m, | 1 | 3.95 € | 3.95 € | https://bit.ly/3EbirKD | Metal |
| High-Power LED | OSRAM Oslon SSL 80 royal-blue on 20 mm star | 4 | 2.99 € | 11.96 € | https://bit.ly/38XAg0Y | Semi-conductor |
| PETG filament | Maertz 1.75 mm PETG,1.0 kg spool | 1 | 14.31 € | 14.31 € | https://bit.ly/2VqAYkk | Polymer |
| Photodiodes | OSRAM SFH 203 | 4 | 0.67 € | 2.56 € | https://bit.ly/2X7wqQc | Semi-conductor |
| Polyurethane varnish | OBI PU color varnish deep black semi-gloss, 125 mL | 1 | 6.99 € | 6.99 € | https://bit.ly/3Eh1luS | Organic |
| POM filament | Hobbyking 1.75 mm POM, 1.0 kg spool | 1 | 22.70 € | 22.70 € | https://bit.ly/3ngNkad | Polymer |
| Power supply | 36 V / 10 A | 1 | 18.15 € | 18.15 € | https://bit.ly/3jZoZDP | Composite |
| Power supply | 12 V / 3 A | 1 | 12.99 € | 12.99 € | Local electronics store | Composite |
| Power supply | 5 V / 3 A | 1 | 10.50 € | 10.50 € | Local electronics store | Composite |
| Stage light filter foil | LEE Filters: 158 Deep Orange | 1 | 5.50 € | 5.50 € | https://bit.ly/3yZGmbX | Other |
| Superglue | UHU Superflex Gel 3.0 g | 1 | 2.70 € | 2.70 € | Local store | Other |
| Thermistors | NTC3950, 100 kΩ | 4 | 0.95 € | 3.80 € | https://bit.ly/2X4BPrg | Semi-conductor |
| Transimpedance amplifier | TI OPA380AIDGKR | 4 | 4.49 € | 17.96 € | https://bit.ly/3l5jiUl | Semi-conductor |
| Accessories and electronic consumables | Resistors, capacitors, MOSFETs, diodes, … | var. | 15.00 € | 15.00 € | Local electronics store | Other |
| Sum | 176.11 € |
Build instructions
Safety hazards
Never heat or turn on the LED lights while the openDSF instrument is disassembled. Although the device uses low voltages (36 V), hot parts are inside the apparatus (heating wire). When assembled correctly, the high-power LEDs are directed into the interior of the apparatus, so they emit very little stray light to the environment. However, looking directly into the disassembled LEDs can irritate the user’s eyes. The SYPRO Orange resp. Sybr Gold DSF dye does not have a hazardous material classification, however, the biological samples and chemicals used in individual DSF experiments may harbor specific hazards. We recommend that the openDSF instrument, like all heating laboratory equipment, should not be left running unattended for several hours as they pose a potential fire risk in the event of a malfunction.
3D-printing:
We fabricated the 3D-printed parts with an Anycubic 4Max printer. Utilizing the open-source software FreeCAD 0.18 and Ultimaker Cura 15.04.6 39, we designed, meshed, and sliced individual components. We performed printing through a 0.4 mm nozzle with 1.75 ± 0.02 mm PETG filament at 225 °C. The layer thickness was set to 0.2 mm. POM was printed at 240–250 °C and a bed temperature of 70 °C. The infill percentage and infill style were 20% resp. the zig-zag pattern for both filaments. The complete printer settings were added as Cura profiles (.ini) to the repository of design files. An overview of all 3D-printed components can be seen in Fig. 2. For better adhesion to the printing table, POM was printed on a three-layer bed of PETG filament. A layer printing time of 6–8 s/layer was found to be most effective against warping effects for the POM filament.
Winding of the heating tube:
In the POM heating tube, a coil of the heating wire (8 mm diameter with 28 windings corresponding to a total length of approx. 0.7 m; pitch approx. 1.8 mm) was inserted (Fig. 3A). The ends of the coil were screwed onto cut-to-size drilled board parts (24 × 5 mm). To fix the coil vertically, the drilled board parts were clamped in the inlet and outlet of the heating tube, respectively. The power connections were soldered on the drilled boards and the connections were led out through 3 mm drilled holes in the heating tube.
Mounting of 3D-printed parts:
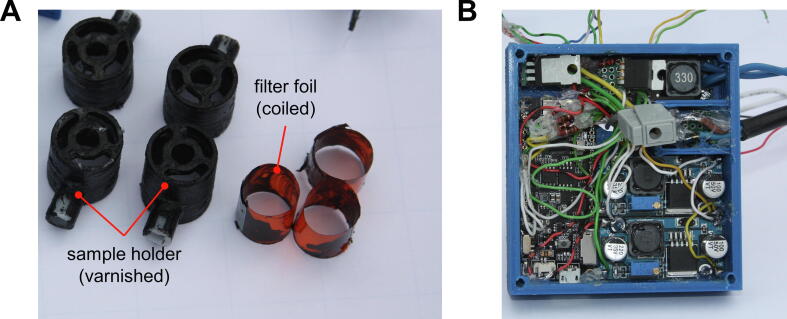
Before the 3D-printed parts, the heating tube, and the fan were assembled by their plug-in joints, the inner surfaces of the colorless POM parts (diffuser, sample holder, and LED holder) were coated with a black polyurethane varnish layer to absorb stray light. Alternatively, the corresponding parts might be printed with a non-transparent material, e.g., black filament. Previously, we also found that the mechanical durability of varnished 3D-printed parts is increased compared to the raw form [22]. Subsequently, a rolled piece of orange filter foil was inserted into the four individual sample holders and the foil was fixed with superglue (Fig. 5A). Caution: The filter foil is attacked by the contained solvents.
Fig. 5.
(A) Example of black painted POM parts: The four sample holders were varnished black because the colorless filament, in its untreated form, acts as an effective optical fiber to disperse stray light and causes significant tube-to-tube bleeding. Next to them, the rolled filter foil inserts, which shield the photodiodes from the blue excitation light. (B) Housing of the electronic components (ESP32 development board, MOSFET circuits, and constant current power supply). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The individual parts of the openDSF instrument were assembled according to the exploded view drawings (Fig. 2A and 4B). Note: Make sure that the joints are well sealed! Any printing imperfections must be corrected by filing. The critical plug connections (fan – fan connector – heating tube – diffuser) can optionally be sealed with Teflon insulating tape. To facilitate rebuilding, we have added a 3D model of the exploded view drawing from Fig. 2A to the repository of design files (https://data.mendeley.com/public-files/datasets/73rt8s7pwd/files/90f0a996-645d-4283-8ca8-ae1a65b3404b/file_downloaded), so that a rebuilder can examine the topology of the design while zooming in or moving parts around.
Assembly of the fluorescence cell:
For assembly of the fluorescence cell, two LED modules and two photodiode modules were fabricated, so that a sample holder is equipped with one LED and one photodiode each (Fig. 4B). For a single LED module, two high-power LEDs were soldered together with proper polarity and provided with connecting cables so that they could be glued onto their LED holder with hot glue and inserted into the cut-outs at the fluorescence cell. To assemble the photodiode module, two photodiodes were mounted (20 mm apart from each other) on a PCB board according to the PCB layout specifications (https://data.mendeley.com/public-files/datasets/73rt8s7pwd/files/81dcaa9e-9a6d-44c0-a272-a710958638ab/file_downloaded).
We manufactured the PCBs ourselves using the toner transfer method [23]. The electronic components were soldered onto the circuit board using a standard soldering iron. For the operational amplifier, assembly instructions from the manufacturer’s datasheet are to be followed. The photodiodes were inserted into the holes provided in the fluorescence cell and later covered by the two PETG diode shields (Fig. 4B).
Wiring of electronic components:
The thermistors were inserted into the 3D-printed temperature sensors and fixed so that the glass sphere of the thermistor just peeks out of the bottom hole (Fig. 6B). The MOSFET circuits (fan and heater) were assembled on drilled boards. A flyback diode was provided for the fan. The resistance values from the PWM supply were 15 Ω and 10 kΩ to the ground. Subsequently, the ESP32 development board, the constant current power supply, and the MOSFET circuits were placed in the housing (Fig. 5B). Finally, the individual electronic components were cabled according to the circuit diagram (https://data.mendeley.com/public-files/datasets/73rt8s7pwd/files/2dbbf304-9985-4c7c-988e-8e972766ae3e/file_downloaded). We noticed that using a USB cable without supplying voltage between the ESP32 and the computer leads to more stable ADC characteristics. The possible interference between simultaneous supply via the USB voltage common collector (VCC) and the 5 V pin of the ESP32 board has already been described for Vext < 5.2 V [24]. Therefore, we prepared a data-only USB cable from a standard micro-USB cable for data transfer to the computer according to literature instructions [25].
Fig. 6.
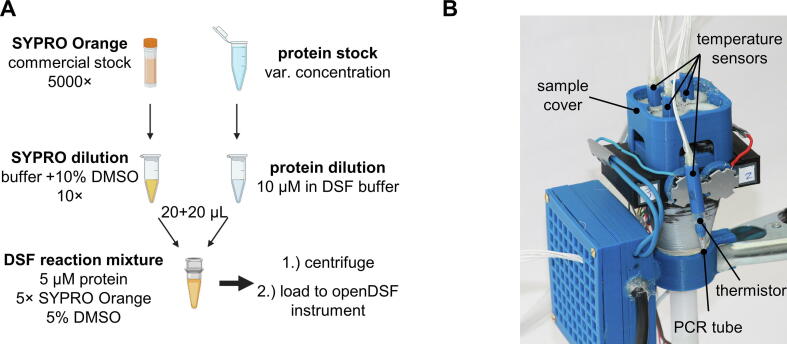
(A) Overview of sample preparation instructions (mix, centrifuge, and load) based on a commercially available SYPRO Orange solution (5000 × ) and a protein stock solution. (B) Close-up of the openDSF instrument with the sample holders closed by the sample cover highlighting the thermistors in the 3D-printed temperature sensors. Three temperature sensors are inserted ready-for-use in their respective sample holders, while one PCR tube equipped with a temperature sensor hangs in front of the instrument for illustration purposes. The thermistor can only be seen by its glass sphere, which extends a few millimeters from the temperature sensor. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Firmware compilation & openDSF GUI:
The firmware was built in the ESP-IDF development environment (v4.2.2). The steps for installing the development environment have been described in detail by the manufacturer: https://docs.espressif.com/projects/esp-idf/en/v4.2.2/esp32/get-started/index.html. After the development environment has been installed, proceed as follows (here for Windows users): The archive “openDSF.zip” (https://data.mendeley.com/public-files/datasets/73rt8s7pwd/files/ba2f88ef-547c-4ca7-8ef7-ed597cbf53b8/file_downloaded) has been unpacked. The unpacked openDSF folder was copied into the root directory of the development environment. Before the firmware can be compiled, the development environment was slightly modified. The file “…\components\driver\esp32\adc.c” was replaced by the custom file “…\openDSF\adc.c”. Subsequently, the firmware was compiled and transferred to the ESP32 board via USB cable.
During experimental usage, the ESP32 board is controlled by a GUI programmed in Python (version 3.9.2). Installation instructions for Python can be found here: https://docs.python.org/3/using/windows.html. The following libraries and modules must be installed in the Python environment: signal, time, datetime, serial, threading, queue, serial.tools.list_ports, scipy.optimize, numpy, tkinter, matplotlib, pyplot, backend_bases, backends.backend_tkagg, scipy.signal.savgol_filter.
Operation instructions
Sample preparation:
The following instructions are described as examples for the preparation of a single DSF reaction mixture but can be scaled up as desired. Prepare 30 µL of a protein solution (10 µM) in an appropriate buffer (e.g., 50 mM HEPES pH 7.5, 150 mM NaCl). In parallel, prepare 30 µL of a 10 × SYPRO Orange dilution (1:500 dilution of the commercial 5000 × stock solution) in the same buffer as used for the protein solution but supplemented with 10% DMSO. Mix 20 µL of the protein solution with 20 µL of the 10 × SYPRO Orange dilution to yield the DSF reaction mixture (final concentration: 5 µM protein and 5 × SYPRO Orange). Note: Other concentrations may also be possible, which could be determined for each protein in the course of an assay optimization. For the study of protein–ligand interactions, compounds or mock treatment additives can be added from DMSO or buffered solutions. Centrifuge (1 min at 300 rcf) and transfer 30 µL of the particle-free solution into a PCR tube (Fig. 6A). Insert 1–4 PCR tubes into the sample holders of the openDSF instrument, seal the reaction tubes with the thermistor caps, and close the instrument with the sample cover (Fig. 6B).
Run a DSF experiment:
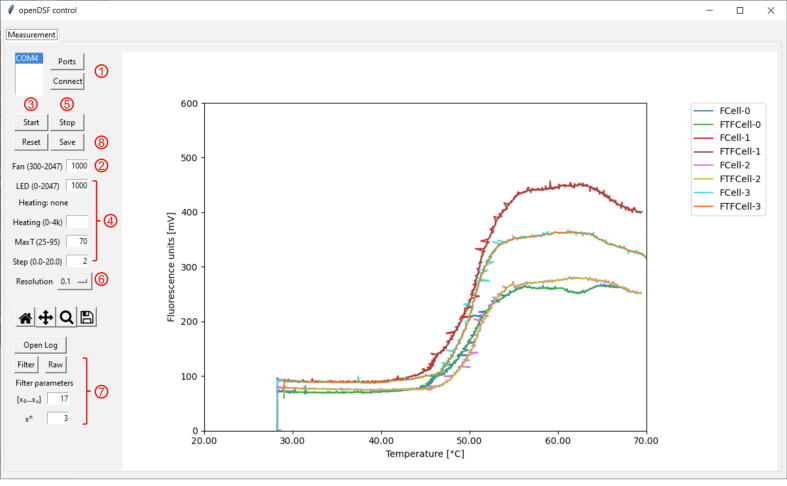
0.) Switch on the openDSF instrument by connecting the power supply. 1.) To establish proper communication between the computer and instrument, start the Python script (“openDSF.py”) from a console and press the “Ports”-button to select the serial port to the openDSF instrument (Fig. 7). Then click “Connect” to establish the connection. 2.) Initialize the heating module by entering the default parameter “1000” in the “Fan” input box. Other fan speeds can be selected (300–2047) and varied for assay optimization. 3.) Click on “Start” to initiate the acquisition of temperature data to check the function of the thermistors and to verify the temperature homogeneity between the different holders. 4.) Subsequently, the physical parameters of fluorescence detection and heating ramp are defined: Start the LED illumination by entering the default parameter “1000” in the “LED” input field. Check if the initial protein-dependent fluorescence value is between 20 and 300 mV and, if necessary, adjust the value to this range by varying the LED strength (0–2047). If a starting temperature above the prevailing room temperature is desired, a heating intensity parameter (0–4096) can be chosen in the “Heating” input field. The terminal temperature of the melting experiment is entered in the “MaxT” input field. The DSF experiment is started by launching the heating ramp, which can be run in varying gradients (0.1–20.0) via the “Step” input field. 5.) Once the melting process is completed and the specified maximum temperature has been reached, the data acquisition can be terminated by clicking the “Stop” button. The device cools back to room temperature automatically.
Fig. 7.
Graphical user interface of the openDSF.py control program. In the left control bar, the elementary functions of the openDSF instrument can be controlled (e.g., fan rpm, heating power, and LED power). The display panel on the right shows the raw and filtered data of the fluorescence vs. temperature plot (melting curves). The functionalities in the GUI are labeled according to the operating instructions (1–8) in the section “Run a DSF experiment”.
6.) For post-run processing of the melting curve data, the resolution of the temperature steps (typically: 0.1–1 °C) in the output file is defined. 7.) Since the acquired raw data is subject to ADC-related noise, it may be reasonable to smooth the raw data. A Savitzky-Golay filter function can be applied to the raw data by clicking on “Filter” and specifying the window size (x0…xn) and polynomial order (xn) of the smoothing function. The smoothing can be reversed by clicking on “Raw”. 8.) Both raw or filtered melting curves can be exported (“Save” button) as a CSV file for analysis with an external program. In this regard, various mathematical evaluation procedures for analyzing DSF datasets have already been described in the literature [3], [6], [8], [26]. The data sets of the following model applications were analyzed with the open-source DSF analysis software DSFworld (https://github.com/gestwicki-lab/dsfworld) and simpleDSFviewer (https://github.com/hscsun/SimpleDSFviewer-5.0) which were downloaded from their GitHub repositories for desktop usage [3], [8]. An example openDSF CSV data set can be found in the repository (https://data.mendeley.com/public-files/datasets/73rt8s7pwd/files/60ecd57d-24a1-4234-b7a9-4cbdbefd6794/file_downloaded).
Validation and characterization
Characterization of the instrument
To assess the capabilities of the openDSF system, various relevant benchmarks (absolute temperature accuracy, cross-holder temperature homogeneity, maximum heating/cooling rates, linear dynamic range, detector saturation, S/N-ratios, and tube-to-tube bleeding) were examined.
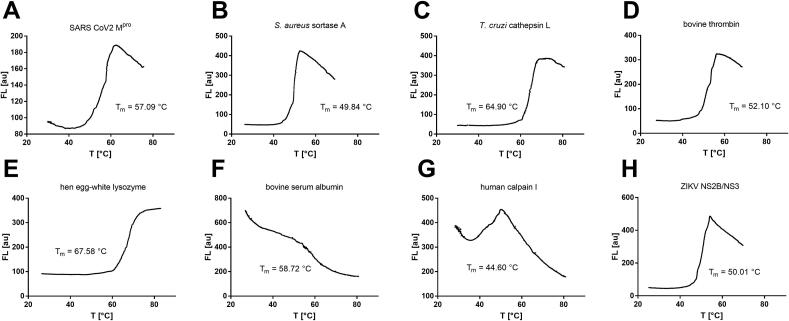
By online temperature control of each sample holder, we were able to obtain linear temperature ramps (R2 = 0.997) over the entire heating range we investigated (25–80 °C) (Fig. 3B). Calibration of the thermistors as temperature sensors resulted in an absolute temperature error of < 0.1 °C. This error was below the fluctuations caused by sample preparation (e.g., pipetting errors and material variations of the PCR tubes), which is why we qualified the temperature control by air heating and thermistor measurement as valid. However, due to the differential nature of DSF experiments, the reproducibility of the same temperature has to be evaluated prior to the absolute temperature accuracy. We did this by the reproduction of protein melting points in different holders during one experiment or during separate experiments (see below). The absolute deviation of protein melting points was in the range of 0.1–0.2 °C (Table 1). This error includes the statistical and systematic inaccuracy due to temperature inhomogeneities between different holders but also the variations caused during sample preparation. The standard error of the determined protein melting points is thus in the same range as commercial qPCR solutions [3], [10], [27], [28].
Table 1.
Overview of various protein melting points and S/N-ratios as recorded with the openDSF instrument in comparison to respective literature values. DSF reaction mixtures contained 5 µM protein and 5 × SYPRO Orange in 50 mM HEPES buffer at pH 7.5.
| Protein | Melting point [°C] | N (replicates) | Lit. melting point [°C] | S/N [dB] |
|---|---|---|---|---|
| Mpro | 57.09 ± 0.08 | 12 | 55.74[31] | 27.1 ± 0.32 |
| SrtA | 49.84 ± 0.13 | 16 | 50.50[32] | 31.1 ± 0.32 |
| Cruzain | 64.90 ± 0.14 | 8 | 66.40[33] | 30.5 ± 0.29 |
| Thrombin | 52.10 ± 0.08 | 4 | 58.30[30] | 28.8 ± 0.29 |
| Lysozyme | 67.58 ± 0.10 | 4 | 71.90[29] | 27.3 ± 0.27 |
| BSA | 58.72 ± 0.13 | 4 | 56.00[3] | 21.6 ± 0.53 |
| Calpain I | 44.60 ± 0.11 | 4 | 47.00[34] | 20.5 ± 0.16 |
| NS2B/NS3 | 50.01 ± 0.07 | 4 | 49.00[35] | 27.9 ± 0.26 |
Typical heating rates for DSF experiments are in the range of 0.5–4 °C/min [3]. For the openDSF system, these depend on both the power of the heating tube and the flow rate of the fan. We have performed the following example DSF experiments at a heating rate of 2 °C/min (GUI: Fan speed = 1000 and step = 2.0). However, even at very high heating rates of 9 °C/min (step = 9.0), we were still able to achieve good temperature homogeneity and melting point reproducibility (SI Fig. 3C). When operating the fan without heating, the openDSF system is also able to cool the sample holders uniformly with air. The cooling curve (with fan speed = 1000) follows an exponential relationship between the temperature reached (Tmax) and the ambient temperature (TR): . This feature can be utilized for a sequence of oscillating heating and cooling events, for example, to study protein unfolding and subsequent refolding (SI Fig. 3D) [29].
Using blue high-power LEDs and orange filter foil, we were able to remodel the fluorescence characteristics of the DSF reaction mixture (fluorescence transmission rate > 80%), while avoiding the need for expensive optics, lamps, or filter units (Fig. 4A). The practical fluorescence measurement range of the fluorometer is characterized by its linear dynamic range, which was determined by a series of different SYPRO Orange dilutions (SI Fig. 2A). The linear dynamic range of the openDSF was found to be between 0 and 3000 mV, which should be aimed at during DSF measurements. Above 3000 mV, the detector saturates and finally reaches its upper maximum at 3500 mV. Besides detector linear responsivity, S/N-ratios were determined as quality parameters for the fluorescence measurements and can be used as benchmarks for reproducing the system. The S/N-ratio was calculated as the absolute fluorescence increase during unfolding divided by the dark noise: . Where the dark noise corresponds to the standard deviation of 100 fluorescence data points before the actual heating ramp was started. In addition to the intrinsic protein properties, the S/N-ratio of the fluorescence measurement depends on the intensity of the LED illumination which was therefore optimized at the beginning of each run. An initial fluorescence value of 20–300 mV has been empirically found to be optimal for obtaining low-noise traces. The protein-specific S/N-ratios are shown in Table 1.
As described during the build instructions, it is necessary to varnish or print the POM parts black, otherwise, they will act as an effective optical fiber and transport stray light from one sample to the neighboring PCR tube thereby falsifying the measurements. This so-called tube-to-tube bleeding has been quantified and must be requalified when building a comparable design. Using our setup, within the linear dynamic range, no tube-to-tube bleeding could be detected with varnished POM parts (SI Fig. 2B). In order to detect tube-to-tube bleeding, an overload of the detector far beyond the saturation limits (>3500 mV) was required, which in our case was correlated with optically perceptible fluorescence despite a closed device (250 × SYPRO Orange in 9:1 glycerol/water).
DSF experiments in the openDSF system were established for a sample volume of 30 µL in PCR tubes with the thermistor measuring temperature just above the liquid level (Fig. 6). The minimum sample volume was tested with additional sample sizes using 20, 10, and 5 µL of a DSF reaction mixture. The signal intensity decreased with lower sample volumes, leading to worse S/N ratios (SI Fig. 3A). The lowest practical volume that could be measured was 10 µL. On the other hand, when loading sample volumes>40 µL, these resulted in increased noise from the temperature sensors, as the thermistors were too close to the liquid level with these volumes.
To test the gas tightness of the openDSF system, the evaporation loss of the samples during the heating phases was investigated gravimetrically. A temperature ramp of 25–70 °C resulted in a loss of about 0.3 µL for a 30 µL sample (SI Fig. 3B). However, at higher temperatures (25–95 °C), the evaporation loss was increased to approx. 2 µL. Prolonged heating (10 min) at 95 °C also resulted in a similar loss of approx. 2 µL, leading to the hypothesis that the gas space in the temperature sensor becomes saturated with humidity and the liquid does not continuously evaporate at a leaky point. This hypothesis could be confirmed by repeated temperature ramps or heating intervals at 95 °C, as the evaporation loss was much lower in the following heating cycles (SI Fig. 3B).
We cannot yet conclusively judge the longevity of our design, but after about 100 heating ramps (25–80 °C), we did not notice any signs of wear or benchmark drops. Since the system is modular and open-source, broken parts can be replaced easily.
Exemplary DSF applications
To challenge the capabilities of the openDSF system eight different proteins were analyzed by their melting profiles: SARS-CoV2 main protease (Mpro), S. aureus sortase A (srtA), T. cruzi cathepsin L (cruzain), bovine thrombin, hen egg-white lysozyme, bovine serum albumin (BSA), human calpain I, and Zika virus (ZIKV) NS2B/NS3 protease. DSF reaction mixtures (5 µM protein, 5 × SYPRO Orange, 50 mM HEPES pH 7.5, 150 mM NaCl, 5% DMSO) were heated at temperature ramps of 2 °C/min from 25 to 80 °C, and subsequently, the melting points (Tm) were determined with the open-source analysis tool of DSFworld from the sigmoid RFU vs. temperature curves [3]. The melting curves of a representative replicate and the corresponding melting points are shown in Fig. 8.
Fig. 8.
Representative melting curve examples of various proteins (5 µM) and 5 × SYPRO Orange in 50 mM HEPES buffer at pH 7.5. (A) SARS-CoV2 main protease (Mpro), (B) S. aureus sortase A (srtA), (C) T. cruzi cathepsin L (cruzain), (D) bovine thrombin, (E) hen egg-white lysozyme, (F) bovine serum albumin (BSA), (G) human calpain I, (H) Zika virus (ZIKV) NS2B/NS3 protease.
A comprehensive overview of melting points and S/N-ratios is given in Table 1. Protein melting points determined by the openDSF instrument showed good agreement with the melting points determined by DSF or differential scanning calorimetry (DSC) as reported in the literature (Table 1). Only for thrombin (Fig. 8D), our measured Tm value deviates significantly from the literature melting point (52.10 °C vs. 58.30 °C), but this might be attributed to a different buffer composition in the literature experiment [30]. For BSA, we obtained a negative DSF curve (Fig. 8F). This is also in agreement with the literature since BSA binds SYPRO Orange already at room temperature due to numerous hydrophobic cavities and at higher temperatures, this binding is entropically reduced [3]. The melting point variance between different holders (intra-assay variation) and between different DSF runs (inter-assay variation) was 0.1–0.2 °C for all protein samples studied, which is low enough to investigate the influence of ligands on the thermal shift of the apparent melting point [2], [6], [7].
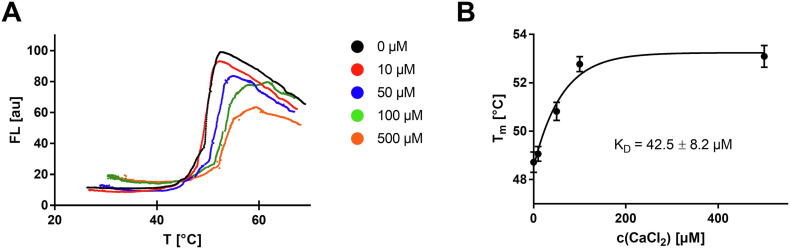
To demonstrate the practical applicability of the openDSF system to biological problems, selected protein–ligand interactions were investigated by DSF. The bacterial transpeptidase srtA utilizes calcium as a stabilizing cofactor. The protein melting point of srtA in the absence and presence of calcium has already been determined by DSC and differs by ΔTm = 3.4 °C [36]. Using ITC, the dissociation constant of this
srtA-Ca2+ complex was determined to be KD = 55 µM [37]. By studying a solution of 5 µM srtA supplemented with different concentrations of calcium chloride, we were able to reproduce the thermodynamic parameters determined with the other two biophysical methods using the openDSF instrument (Fig. 9A). In detail, we titrated 0, 10, 50, 100, and 500 µM calcium chloride to the DSF reaction mixtures and determined the dissociation constant KD(srtA-Ca2+) = 42.5 µM from the binding isotherm of the melting points versus the calcium concentration (Fig. 9B) [6].
Fig. 9.
(A) Representative srtA melting curves in the presence of different CaCl2 concentrations. At higher calcium concentrations, the srtA protein is stabilized towards higher melting points. (B) Determination of the thermodynamic dissociation constant KD for the srtA-Ca2+ complex from the melting point vs. CaCl2-concentration relationship by regression with a binding isotherm with . Shown are means ± standard deviation (n = 3).
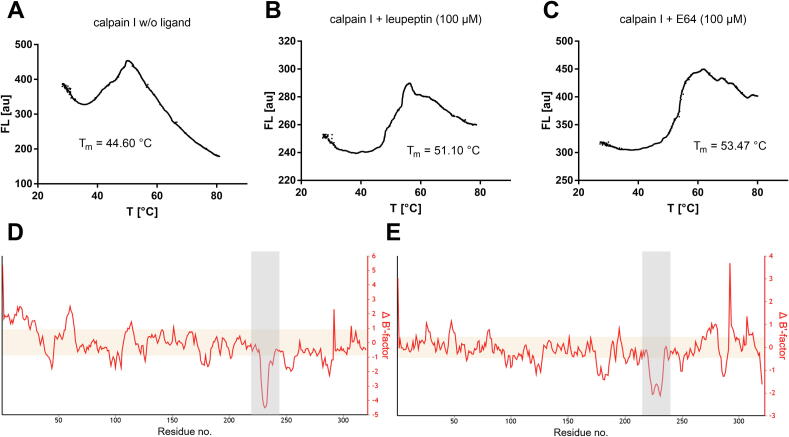
DSF is also useful for the identification and study of small-molecule drug substances on their protein targets. If a ligand binds to a protein, the free energy of ligand binding mostly results in protein stabilization and increased Tm-values [7]. Here, we studied the cysteine protease calpain I with or without treatment of the protease inhibitors leupeptin and E64 (structures in SI Fig. 4). The melting point of the untreated protease (5 µM) was determined with Tm = 44.60 °C (Fig. 10A). After treatment with 100 µM of the inhibitors leupeptin or E64, the melting point increased to 51.10 °C and 53.47 °C, respectively (Fig. 10B,C). The molecular background of this stabilization becomes evident from the crystal structures of the apo structure and the inhibitor-bound complexes [38], [39]. The structural rigidity of a protein can be determined by B'-factor analysis of the corresponding crystallographic dataset [40]. In the regions of the crystal structure where the inhibitor binds, a significant stabilization of the protein backbone takes place, which explains the increase in melting points (Fig. 10D,E). Here, it is shown that the combination of thermodynamic parameters from openDSF experiments with spatially resolved methods such as XRAY or NMR complement each other well.
Fig. 10.
DSF experiments on calpain I ligand complexes. (A) Melting curve of native calpain I (5 µM) in absence of a ligand, (B) Melting curve of calpain I (5 µM) in presence of the inhibitor leupeptin (100 µM), (C) Melting curve of calpain I (5 µM) in presence of the inhibitor E64 (100 µM), (D) B’-factor analysis with the open-source BANΔIT-web server [40]: The calpain-leupeptin complex (pdb: 1TL9) reveals rigidization (negative ΔB’) of the ligand-binding site (Asp259–Leu268) compared to the corresponding apo structure (pdb: 1KXR), (E) The calpain-E64 complex (pdb: 1TLO) reveals rigidization (negative ΔB’) of the ligand-binding site (Ile252–Lys266) compared to the corresponding apo structure (pdb: 1KXR).
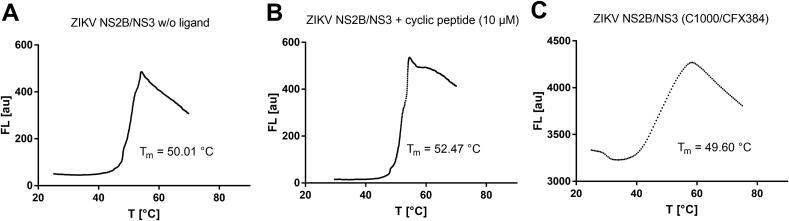
Furthermore, to compare a melting curve example with a commercial qPCR system, DSF reaction mixtures of the ZIKV NS2B/NS3 protease were analyzed using both the Bio-Rad C1000/CFX384 (Fig. 11C) and our openDSF instrument (Fig. 11A). The protein melting point was 49.60 °C and 50.01 °C, respectively, for both instruments, very close to the literature value (Table 1) [35]. The ZIKV protease is known to adopt multiple conformations due to the dynamic interaction of both NS2B and NS3 proteins. A catalytically active (mostly substrate-bound) form is referred to as the “closed” conformation, whereas the “open” conformation describes the predominate species of the protein resting state [41]. However, the addition of substrate mimetic inhibitors was reported to cause to the stabilization of the closed conformation and an increased melting point [42], [43]. In a DSF experiment, the presence of a cyclic peptide inhibitor (10 µM, structure in SI Fig. 4 [42]) led to an increase of the melting point by ΔTm = 2.46 °C (Fig. 11B).
Fig. 11.
DSF experiments on the ZIKV NS2B/NS3 protease. (A) ZIKV NS2B/NS3 fluorescence vs. temperature curve recorded with the openDSF instrument. (B) Fluorescence vs. temperature curve of ZIKV NS2B/NS3 in presence of the cyclic peptide ligand (10 µM, pdb: 6Y3B). (C) NS2B/NS3 fluorescence vs. temperature curve recorded with the C1000/CFX384.
To investigate the ability of our instrument to perform DSF analysis of RNA molecules, we examined two examples of literature-known tertiary-structured RNA aptamers. For this purpose, RNA (500 nM) was mixed with a suitable intercalating dye (1 × Sybr Gold) in a reaction buffer and, analogously to the protein DSF experiments, heated at a temperature ramp of 2 °C/min (25–70 °C). Melting points (Tm) were calculated using the differentiation methodology of Silvers et al. and the simpleDSFviewer software [8], [16].
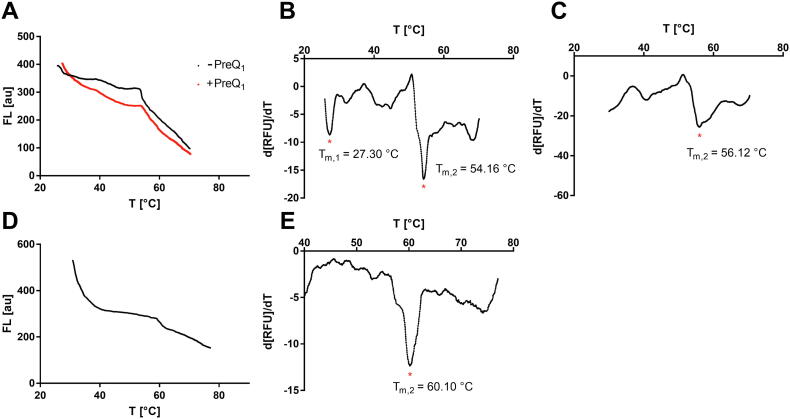
The aptamer domain of the preQ1 riboswitch from B. subtilis folds into a pseudoknot structure upon binding of its natural ligand preQ1. When preQ1 (10 µM, structure in SI Fig. 4) was added to the riboswitch (500 nM in 1 × phosphate-buffered saline, pH 7.5), the unliganded melting curve changed significantly (Fig. 12A). Examination of the ligand-free reaction mixture by the derivative analysis method yielded two melting points in agreement with the literature (Tm,1 = 27.30 °C, Tm,2 = 54.16 °C, Fig. 12B) [44]. In summary, the addition of the ligand preQ1 (10 µM) resulted in a shift of the second melting point towards higher temperatures (Tm,2 = 56.12 °C, Fig. 12C).
Fig. 12.
RNA-aptamer DSF experiments. (A) RNA-DSF trace for the preQ1 riboswitch aptamer domain (500 nM) with and without its natural ligand preQ1 (10 µM). (B) Derivative plot of the preQ1 aptamer in absence of a ligand. (C) Derivative plot of the preQ1 aptamer supplemented with 10 µM of preQ1. (D) RNA-DSF trace for a trypanosome-specific RNA aptamer. (E) Derivative plot of the trypanosome-specific RNA aptamer (temperature range: 40–75 °C).
Secondly, a trypanosome-specific RNA aptamer was investigated by DSF in 50 mM cacodylate buffer (pH 6.5) supplemented with 0.5 mM MgCl2 [45]. This parasite-specific aptamer was previously characterized with a melting point of 58 °C. Here, we could confirm the identity of our RNA construct by recording a DSF trace matching the melting point (Tm,2 = 60.10 °C, Fig. 12E). In contrast, the strong decrease in fluorescence at temperatures between 30 and 40 °C has not been observed in the literature, suggesting that some of the aptamer molecules are not in their native fold. In this respect, RNA-DSF is also suitable for an assessment of sample quality [16].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Research in the laboratories of Mark Helm and Tanja Schirmeister was funded by DFG grants (TRR 319 “RMaP” projects A01, C01, and A05). Cruzain was a gift from the group of Dr. Avninder Bhambra (University of Leicester, UK). We thank Prof. Torsten Steinmetzer (University of Marburg, Germany) for providing the cyclic peptide ligand (1-((8R,15S,18S)-15,18-bis(4-aminobutyl)-4,7,14,17,20-pentaoxo-3,6,13,16,19-pentaaza-1(1,3)-benzenacyclohenicosaphane-8-yl)guanidine).
Biography

Fabian Barthels received his M.Sc. in Biochemistry from the University of Tübingen (Germany) and after graduation, he joined the research group of Prof. Dr. Tanja Schirmeister for a Ph.D. in the field of Pharmaceutical Chemistry at the University of Mainz (Germany). During his doctorate, he discovered his interest in open-source hard- and software for drug discovery applications. Previously, he has developed and published an open-source liquid handling workstation (FINDUS) and a crystallographic B-factor analysis toolkit (BANΔIT). His current research interests involve designing biophysical instrumentations for the study of protein & RNA ligand interactions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ohx.2022.e00256.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lo M.-C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 2004;332(1):153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 2.Pantoliano M.W., Petrella E.C., Kwasnoski J.D., Lobanov V.S., Myslik J., Graf E., Carver T., Asel E., Springer B.A., Lane P., Salemme F.R. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screening. 2001;6(6):429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 3.T. Wu, J. Yu, Z. Gale-Day, A. Woo, A. Suresh, M. Hornsby, J.E. Gestwicki, Three Essential Resources to Improve Differential Scanning Fluorimetry (DSF) Experiments, preprint (BioRxiv), 2020. https://doi.org/10.1101/2020.03.22.002543 (accessed November 24, 2020).
- 4.D.E. Scott, C. Spry, C. Abell, Differential Scanning Fluorimetry as Part of a Biophysical Screening Cascade, in: D.A. Erlanson, W. Jahnke (Eds.), Methods and Principles in Medicinal Chemistry, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2016: pp. 139–172. http://doi.wiley.com/10.1002/9783527683604.ch07.
- 5.Gao K., Oerlemans R., Groves M.R. Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 2020;12(1):85–104. doi: 10.1007/s12551-020-00619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai N., Roder H., Dickson A., Karanicolas J. Isothermal analysis of thermofluor data can readily provide quantitative binding affinities. Sci. Rep. 2019;9:2650. doi: 10.1038/s41598-018-37072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 8.Sun C., Li Y., Yates E.A., Fernig D.G. SimpleDSFviewer: A tool to analyze and view differential scanning fluorimetry data for characterizing protein thermal stability and interactions. Protein Sci. 2020;29(1):19–27. doi: 10.1002/pro.v29.110.1002/pro.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg T.H., White H.M., Singer V.L. Optimal filter combinations for photographing SYPRO orange or SYPRO red dye-stained gels. Anal. Biochem. 1997;248(1):168–172. doi: 10.1006/abio.1997.2117. [DOI] [PubMed] [Google Scholar]
- 10.Hoeser J., Gnandt E., Friedrich T. Low cost, microcontroller based heating device for multi-wavelength differential scanning fluorimetry. Sci Rep. 2018;8:1457. doi: 10.1038/s41598-018-19702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza-Gallegos R.A., Rios A., Garcia-Cordero J.L. An affordable and portable thermocycler for real-time PCR made of 3D-printed parts and off-the-shelf electronics. Anal. Chem. 2018;90(9):5563–5568. doi: 10.1021/acs.analchem.7b04843. [DOI] [PubMed] [Google Scholar]
- 12.G. Mulberry, K.A. White, M. Vaidya, K. Sugaya, B.N. Kim, 3D printing and milling a real-time PCR device for infectious disease diagnostics, PLoS ONE. 12 (2017) e0179133. https://doi.org/10.1371/journal.pone.0179133. [DOI] [PMC free article] [PubMed]
- 13.J.T. Keer, Chapter 7:Quantitative Real-time PCR Analysis, in: Essentials of Nucleic Acid Analysis, 2008: pp. 132–166. https://doi.org/10.1039/9781847558213-00132.
- 14.Draper D.E., Gluick T.C. In: Methods in Enzymology. Abelson J., Simon M., Verdine G., Pyle A., editors. Elsevier; 1995. Melting studies of RNA unfolding and RNA—Ligand interactions; pp. 281–305. [DOI] [PubMed] [Google Scholar]
- 15.Zamani F., Suzuki T. Synthetic RNA modulators in drug discovery. J. Med. Chem. 2021;64(11):7110–7155. doi: 10.1021/acs.jmedchem.1c00154. [DOI] [PubMed] [Google Scholar]
- 16.Silvers R., Keller H., Schwalbe H., Hengesbach M. Differential scanning fluorimetry for monitoring RNA stability. ChemBioChem. 2015;16(7):1109–1114. doi: 10.1002/cbic.v16.710.1002/cbic.201500046. [DOI] [PubMed] [Google Scholar]
- 17.QIAGEN Rotor-Gene Q - Average price, Bimedis. (2021). https://bimedis.com/qiagen-rotor-gene-q-m21547 (accessed November 18, 2021).
- 18.Brinkmann S., Oberbach K., Baur E., Schmachtenberg E., Osswald T.A. 29th ed. Carl Hanser Verlag GmbH; Munich: 2006. International Plastics Handbook. https://www.hanser-elibrary.com/isbn/9783446229051. [Google Scholar]
- 19.Muro‐Fraguas I., Sainz‐García E., Pernía‐Espinoza A., Alba‐Elías F. Atmospheric pressure air plasma treatment to improve the 3D printing of polyoxymethylene. Plasma Process Polym. 2019;16(7) doi: 10.1002/ppap.v16.710.1002/ppap.201900020. [DOI] [Google Scholar]
- 20.M. Babiuch, P. Foltýnek, P. Smutný, Using the ESP32 Microcontroller for Data Processing, in: 2019 20th International Carpathian Control Conference (ICCC), 2019: pp. 1–6. https://doi.org/10.1109/CarpathianCC.2019.8765944.
- 21.Lee W.Y., Kim T.M., Kim M.J., Ko Y.W., Kim J.D. User-friendly calibration tool for temperature measurements of PCR devices with NTC thermistors. Int. J. Control Autom. 2015;8(7):13–24. [Google Scholar]
- 22.Barthels F., Barthels U., Schwickert M., Schirmeister T. FINDUS: An open-source 3D printable liquid-handling workstation for laboratory automation in life sciences. SLAS Technol. 2020;25(2):190–199. doi: 10.1177/2472630319877374. [DOI] [PubMed] [Google Scholar]
- 23.simpletronic, Heatless (cold) Toner Transfer for PCB Making, Instructables. (2021). https://www.instructables.com/Heatless-cold-Toner-Transfer-for-PCB-Making/ (accessed September 11, 2021).
- 24.J. Kim, Analysis and Optimization of DC Supply Range for the ESP32 Development Board, preprint (TechRxiv), 2020. https://doi.org/10.36227/techrxiv.12798410.v3 (accessed September 12, 2021).
- 25.DavisT2, DIY Data Only USB Type B, Instructables. (2021). https://www.instructables.com/DIY-Data-Only-USB-Type-B/ (accessed September 12, 2021).
- 26.Wright T.A., Stewart J.M., Page R.C., Konkolewicz D. Extraction of thermodynamic parameters of protein unfolding using parallelized differential scanning fluorimetry. J. Phys. Chem. Lett. 2017;8(3):553–558. doi: 10.1021/acs.jpclett.6b02894. [DOI] [PubMed] [Google Scholar]
- 27.Vivoli M., Novak H.R., Littlechild J.A., Harmer N.J. Determination of protein-ligand interactions using differential scanning fluorimetry. J Vis Exp. 2014:51809. doi: 10.3791/51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosa N., Ristic M., Seabrook S.A., Lovell D., Lucent D., Newman J. Meltdown: A tool to help in the interpretation of thermal melt curves acquired by differential scanning fluorimetry. J Biomol Screen. 2015;20(7):898–905. doi: 10.1177/1087057115584059. [DOI] [PubMed] [Google Scholar]
- 29.Svilenov H.L., Menzen T., Richter K., Winter G. Modulated scanning fluorimetry can quickly assess thermal protein unfolding reversibility in microvolume samples. Mol. Pharmaceutics. 2020;17(7):2638–2647. doi: 10.1021/acs.molpharmaceut.0c00330. [DOI] [PubMed] [Google Scholar]
- 30.Crossen J., Diamond S.L. Thermal shift assay to probe melting of thrombin, fibrinogen, fibrin monomer, and fibrin: Gly-Pro-Arg-Pro induces a fibrin monomer-like state in fibrinogen. Biochim Biophys Acta Gen Subj. 2021;1865(2):129805. doi: 10.1016/j.bbagen.2020.129805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y.u., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barthels F., Marincola G., Marciniak T., Konhäuser M., Hammerschmidt S., Bierlmeier J., Distler U., Wich P.R., Tenzer S., Schwarzer D., Ziebuhr W., Schirmeister T. Asymmetric disulfanylbenzamides as irreversible and selective inhibitors of Staphylococcus aureus sortase A. ChemMedChem. 2020;15(10):839–850. doi: 10.1002/cmdc.201900687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cianni L., Rocho F.D.R., Rosini F., Bonatto V., Ribeiro J.F.R., Lameira J., Leitão A., Shamim A., Montanari C.A. Optimization strategy of single-digit nanomolar cross-class inhibitors of mammalian and protozoa cysteine proteases. Bioorg. Chem. 2020;101:104039. doi: 10.1016/j.bioorg.2020.104039. [DOI] [PubMed] [Google Scholar]
- 34.Moldoveanu T., Jia Z., Davies P.L. Calpain activation by cooperative Ca2+ binding at two non-EF-hand sites. J. Biol. Chem. 2004;279(7):6106–6114. doi: 10.1074/jbc.M310460200. [DOI] [PubMed] [Google Scholar]
- 35.Hammerstein F., Lauth L.M., Hammerschmidt S., Wagner A., Schirmeister T., Hellmich U.A. Cis autocatalytic cleavage of glycine-linked Zika virus NS2B-NS3 protease constructs. FEBS Lett. 2019;593(16):2204–2213. doi: 10.1002/feb2.v593.1610.1002/1873-3468.13507. [DOI] [PubMed] [Google Scholar]
- 36.I. Ugur, M. Schatte, A. Marion, M. Glaser, M. Boenitz-Dulat, I. Antes, Ca2+ binding induced sequential allosteric activation of sortase A: An example for ion-triggered conformational selection, PLoS One. 13 (2018) e0205057. https://doi.org/10.1371/journal.pone.0205057. [DOI] [PMC free article] [PubMed]
- 37.Wang X., Chen J.-L., Otting G., Su X.-C. Conversion of an amide to a high-energy thioester by Staphylococcus aureus sortase A is powered by variable binding affinity for calcium. Sci. Rep. 2018;8:16371. doi: 10.1038/s41598-018-34752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moldoveanu T., Hosfield C.M., Lim D., Elce J.S., Jia Z., Davies P.L. A Ca2+ switch aligns the active site of calpain. Cell. 2002;108(5):649–660. doi: 10.1016/S0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 39.Moldoveanu T., Campbell R.L., Cuerrier D., Davies P.L. Crystal structures of Calpain–E64 and – leupeptin inhibitor complexes reveal mobile loops gating the active site. J. Mol. Biol. 2004;343(5):1313–1326. doi: 10.1016/j.jmb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Barthels F., Schirmeister T., Kersten C. BANΔIT: B’-factor analysis for drug design and structural biology. Mol. Inf. 2021;40(1):2000144. doi: 10.1002/minf.v40.110.1002/minf.202000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahawaththa M.C., Pearce B.J.G., Szabo M., Graham B., Klein C.D., Nitsche C., Otting G. Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antivir. Res. 2017;142:141–147. doi: 10.1016/j.antiviral.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Braun N.J., Quek J.P., Huber S., Kouretova J., Rogge D., Lang‐Henkel H., Cheong E.Z.K., Chew B.L.A., Heine A., Luo D., Steinmetzer T. Structure-based macrocyclization of substrate analogue NS2B-NS3 protease inhibitors of Zika, West Nile and Dengue viruses. ChemMedChem. 2020;15(15):1439–1452. doi: 10.1002/cmdc.v15.1510.1002/cmdc.202000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y., Zhang Z., Phoo W.W., Loh Y.R., Wang W., Liu S., Chen M.W., Hung A.W., Keller T.H., Luo D., Kang CongBao. Structural dynamics of zika virus NS2B-NS3 protease binding to dipeptide inhibitors. Structure. 2017;25(8):1242–1250.e3. doi: 10.1016/j.str.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Hoeher J.E., Veirs M.A., Widom J.R. Impacts of fluorescent base analogue substitution on the folding of a riboswitch. preprint (BioRxiv) 2021 doi: 10.1101/2021.07.22.453416. [DOI] [Google Scholar]
- 45.Adler A., Forster N., Homann M., Göringer H.U. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen. 2008;11:16–23. doi: 10.2174/138620708783398331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.