Abstract
Duckweeds are the smallest free-floating flowering aquatic plants. Their biotechnological applications include their use as food, bioenergy, and environmental sustainability, as they can help clean polluted water. The high growth capacity and their chemical properties make them suitable for human health applications. Here we evaluated the ethanolic extracts from five species of duckweeds by HPLC-DAD/MS-MS for chemical characterization. Sixteen compounds were identified and quantified, in which three were chlorogenic acid derivatives and eleven apigenin and luteolin derivatives. We describe for the first time the presence in duckweeds of 5-O-(E)-caffeoylquinic acid (1), 3-O-(E)-coumaroylquinic acid (2), luteolin-7-O-glucoside-C-glucoside (3), 4-O-(E)-coumaroylquinic acid (4), luteolin-6-C-glucoside-8-C-rhamnoside (5), and luteolin-8-C-glucoside-6-C-rhamnoside (6). The flavonoids diversity showed a significant content of luteolin and its derivatives, except for Landoltia punctata that had significant apigenin content. Flavones identified in duckweeds were mostly C-glycosides, which can benefit human diets, and its abundance seems to be related to the higher antioxidant and anticancer capacities of Wolffiella caudata, Wolffia borealis, and Landoltia punctata. Our findings reinforce the idea that duckweeds could be valuable additives to the human diet, and their potential should be further explored.
Duckweeds are the smallest free-floating flowering aquatic plants.
Introduction
Duckweed is the given name for the smallest flowering aquatic plants that belong to Lemnaceae, which comprises 36 species grouped in five genera: Landoltia, Lemna, Spirodela, Wolffia, and Wolffiella.1,2 Furthermore, these plants are subdivided into the subfamilies Lemnoideae and Wolffioideae, which contain respectively the species of Landoltia, Lemna and Spirodela, and the rootless species of Wolffia and Wolffiella.3 These plants are capable of duplicating their biomass in 96 hours, conferring enormous applicability in biotechnology as water remediators, for environmental monitoring, food, biofuels, and cosmetic production as well as pharmaceutical intakes.4–10 The evolution trend in Lemnaceae has a relationship with secondary metabolites and cell wall composition despite its diversity.11,12
In plants, the secondary metabolites consist of a broad group of compounds produced to benefit the organism for different purposes: those related to the internal functioning of the plant and others selected by their applications for human health.13,14 Among the secondary metabolites, the flavonoids are a group of compounds synthesized from cinnamic acid derivatives coupled with three acetate units that display several biological roles.15 These polyphenol compounds are characterized by the C15 skeleton arranged in two phenyl rings and a heterocyclic ring (C6–C3–C6). The substitution pattern of the central ring along with the degree of unsaturation and the oxidation of the flavonoid structure resolve this group of compounds into six main classes: flavonol, flavone, flavanonol, flavanone, anthocyanin, and isoflavonoid.14,16,17 Despite their grossly similar chemical properties, the flavonoids in plants are involved in diverse processes such as growth, photosynthesis, and protective biological systems such as seed development, pollination, light screening, defence against pathogens, protection against UV-damage, temperature acclimation, and drought resistance. Also, some flavonoids can act as signaling molecules for allelopathic interactions, such as phytoalexins and detoxifying agents.13,14
Besides the natural bioactive properties in plants, flavonoids are phytochemicals with several medically-relevant biological activities such as antiviral, antifungal, antibacterial antihepatotoxic, anti-osteoporotic, antiulcer, anti-proliferative.16,18–21 Flavonoids display several other properties such as immunomodulation, apoptotic effects, and consolidation of applications on carcinogenesis, inflammation, atherosclerosis, and thrombosis.16,18,19 Flavonoids are also considered functional foods that benefit human health.16,18,19 Considering the applications for human food, the antioxidant capacity of flavonoids has an essential action in preventing the formation of reactive oxygen and nitrogen species, causing damage in DNA, proteins, lipids, and other biomolecules.22,23 Remarkable knowledge is found in the literature about the wine and tea phenolic compounds, which are responsible for their activity in the prevention of heart diseases and cancer.24–27 Thus, the intake of polyphenols present in plants is desirable for human health. Recently, flavonoids such as quercetin and rutin have been shown to interfere with the entrance of the SARS-Cov-2 in cells, highlighting their potential to treat the COVID-19 disease.28
Duckweeds can have high levels of flavonoids and together with its nutritional value of amino acids and proteins make it suitable for human consumption with health benefits.29–34 Duckweeds have been sources of human food in several Asian countries.35 Moreover, one duckweed species (Spirodela polyrhiza) is being used to treat urticaria, acute nephritis, influenza, and inflammation in Japan, Korea, and China.36,37 Several studies identified flavonoids mainly quercetin, apigenin, and luteolin derivatives as compounds in Spirodela polyrhiza extracts18,31,38 and these components are probably related to the pharmacological applications of this duckweed in traditional medicine.
Based on the importance of the duckweed family for pharmacology and nutrition, this paper aimed at investigating the phenolic composition of ethanolic extracts and antioxidant activity from five species of duckweeds in order to characterize further the range of natural variations in the varieties of flavonoids produced by this plant family.
Experimental
Duckweed strains, cultivation method, and sample preparation
Five-species of duckweeds were investigated in this study. Three Lemnoideae species (Landoltia punctata, Lemna gibba, and Spirodela polyrhiza) and two Wolffioideae species (Wolffia borealis and Wolffiella caudata) were obtained from the Rutgers Duckweed Stock Cooperative (RDSC) collection. Landoltia punctata (7624), Lemna gibba (DWC128), Spirodela polyrhiza (9509), Wolffia borealis (9144) and Wolffiella caudata (9139) were cultivated on Schenk–Hildebrandt medium (1.6 g L−1) supplemented with 0.5% of sucrose, pH 6.5 at 25 °C with a photoperiod of 16 h of light in the intensity of 20 μmol m−2 s−1. After 21 days of cultivation, the plants were frozen in liquid nitrogen, freeze-dried and pulverized by Geno/Grinder®2010 SPEX SamplePrep.
Crude sample extraction
Twenty mg of each pulverized sample was extracted twelve times with 1.5 mL of 80% ethanol at 80 °C for 20 min. The extracted compounds were recovered, vacuum concentrated (ThermoScientific® Savant SC 250 EXP), and resuspended in 2 mg mL−1 in a hydroethanolic solution of 80% ethanol.
HPLC-DAD-ESI-MS/MS analysis of phenolic compounds in duckweeds
Samples diluted in ethanol 80% were filtered through a 0.45 μm Nylon syringe, and reversed-phase HPLC-DAD analyzed the extracts of duckweeds on an Agilent 1260. The phenolic compounds were separated using a Zorbax Eclipse Plus C18 column (4.6 × 150 mm, 3.5 μm) maintained at 45 °C. The injection volume of the sample was 5 μL. The solvents used in the mobile phase were 0.1% acetic acid in Milli-Q water (A) and acetonitrile (B), with the following concentration gradient of B: 0–3 min, 0%, 3–8 min, 0–10%, 8–8.1 min, 10–11%, 8.1–15 min, 11%, 15–15.1 min, 11–12%, 15.1–20 min, 12%, 20–20.1 min, 12–13%, 20.1–25 min, 13%, 25–30 min, 13–23%, 30–38 min, 23–30%, 38–43 min, 30–50%, 43–65 min, 50–100 min, 65–70 min. The phenolic acids and flavonoids were detected at 325 nm and 352 nm, respectively. Identification of compounds from duckweeds was carried out through HPLC-DAD-MS/MS (Agilent 1260 coupled to an Esquire 3000 Plus mass spectrometer) using the same chromatographic conditions and a negative ionization mode. Quantification of each phenolic compound was carried out by comparing the peak areas obtained from HPLC analysis with calibration curves of standards at the range of 1.5–150 μg mL−1, according to the phenolic class. Chlorogenic acid, vitexin, and orientin were used to quantify phenolic acids, apigenin, and luteolin derivatives.
Antioxidant activity
The antioxidant activity was evaluated by the 1,1-diphenyl-2-picryl-hydrazyl (DPPH) (Sigma-Aldrich®) method that is based upon the elimination of the free radical by the samples.39 For the assay, a 0.2 mM DPPH˙ solution in methanol was freshly prepared, and 200 μL of DPPH˙ solution was reacted with 20 μL of the plants' extracts (2 mg mL−1) diluted in 10, 20, 50, 100, and 200 μg mL−1. The reaction was incubated in the dark for 20 min, and the absorbance was measured at 515 nm with EPOCH equipment (Sellex Inc.). All samples were analyzed with three biological replicates and three technical replicates. The calibration curve was performed with a methanol Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) (Sigma-Aldrich®) solution in the concentration range of 5–175 μg mL−1. The 50% inhibitory capacity (IC50) was obtained from the equation of the straight line of the concentration graph by the percentage of the radical activity. The radical activity was determined, as shown in eqn (1).
 |
1 |
Statistical analysis
All data were expressed as means ± standard error for three replicates (n = 3). The interspecific analysis was performed by ANOVA one-way, followed by Tukey's test (p < 0.05) using the software R version 3.4.1. The Principal Component Analysis (PCA) was performed in Minitab-14.1 software to evaluate the phenolic compounds' distribution by the studied species.
Results
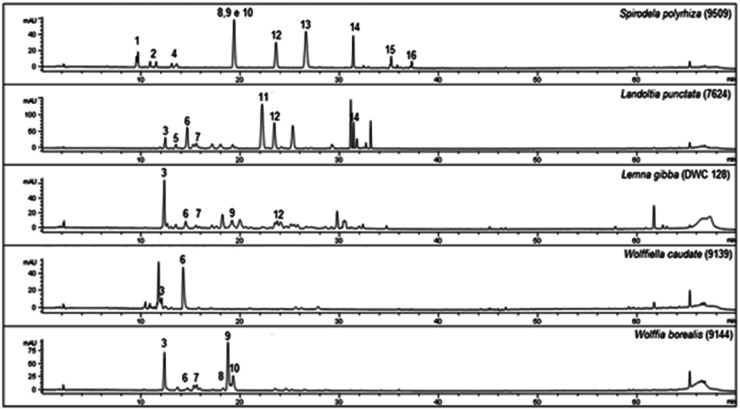
In the phenolic profile, we detected sixteen compounds (Fig. 1), of which 14 were identified by MS/MS through tandem MS patterns, along with retention time (Rt), and UV-Vis spectra using reference compounds for characterization (Table 1). Among the identified compounds, three were chlorogenic acid derivatives (1, 2, and 4) found only in S. polyrhiza and eleven were apigenin and luteolin derivatives (3, 5, 6, 7, 9, 11–16) found in all five-species (Table 1). The flavonoids were identified mostly as C-glycosides (5, 6, 7, 9, 11, 12, 15, and 16) than O-glycosides (3, 13, and 14) in the duckweed family (Table 1).
Fig. 1. Chromatographic profiles of Lemna gibba, Landoltia punctata, Spirodela polyrhiza, Wolffia borealis, and Wolffiella caudata obtained from ethanolic extracts. The numbers above peaks (1–16) refer to the identification of each compound. For the identified substances see 1–16 in Table 1 (n = 3).
Characterization by HPLC-DAD-ESI-MS/MS and quantification of phenolic compounds (flavonoids) of S. polyrhiza, L. punctata, L. gibba, W. caudata, and W. borealis. Values are expressed as means (n = 3) ± standard deviationc.
| I.D. | RT (min) | Compounda | UV (nm) | [M − H]−, m/z → MS2 [M − H]−, m/zb | S. polyrhiza μg mg−1 DM | L. punctata μg mg−1 DM | L. gibba μg mg−1 DM | W. caudata μg mg−1 DM | W. borealis μg mg−1 DM |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 9.4 | 5-CQA | 220, 326 | 707, 353 (bp), 279, 191, 353 → 191 (bp), 179, 135 | 1.5 ± 0.0 | ||||
| 2 | 10.6 | 3-CoQA | 208, 228, 312 | 675, 337 (bp), 279, 163, 337 → 191, 163 (bp), 119 | 2.5 ± 1.2 | ||||
| 3 | 12.4 | Lut-7-O-gluc-C-gluc | 270, 350 | 609 (bp), 593, 463, 295, 609 → 591, 489, 447 (bp), 327 | 3.0 ± 0.4 | 0.3 ± 0.1 | 0.2 ± 0.1 | ||
| 4 | 13.5 | 4-CoQA | 242, 270, 336 | 337 (bp), 279, 173, 337 → 173 (bp), 163 | 0.4 ± 0.1 | ||||
| 5 | 13.6 | Lut-6-C-gluc-8-C-rham | 244, 334 | 593 (bp), 579, 279, 579 → 561, 531, 489 (bp), 459, 399, 369 | 1.3 ± 0.2 | 0.8 ± 0.0 | |||
| 6 | 14.9 | Lut-8-C-gluc-6-C-rham | 244, 272, 344 | 593 (bp), 579, 279, 579 → 561, 519, 489 (bp), 459, 429, 399 | 7.4 ± 0.8 | 0.5 ± 0.4 | 5.65 ± 0.29 | 0.8 ± 0.1 | |
| 7 | 16.0 | Lut-8-C-gluc-6-C-xyl | 270, 346 | 579 (bp), 447, 279, 579 → 561, 519, 489 (bp), 459, 429, 399 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.6 ± 0.1 | ||
| 8 | 18.4 | N. I. | 242, 326 | 563, 510, 447 (bp), 447 → 429, 387, 357, 327 (bp) | n.d. | n.d. | |||
| 9 | 19.2 | Lut-8-C-gluc (orientin) | 246, 272, 336 | 447 (bp) → 429, 393, 357, 327 (bp) | 6.8 ± 0.4 | 5.0 ± 0.5 | |||
| 10 | 19.7 | N. I. | 258, 350 | 563, 510, 447 (bp), 279, 447 → 429, 357, 327 (bp) | n.d. | n.d. | |||
| 11 | 22.0 | Apig-8-C-gluc-6-C-gluc | 232, 270, 338 | 593 (bp), 431, 281, 431 → 341, 311 (bp), 283 | 10.4 ± 1.0 | ||||
| 12 | 23.2 | Apig-8-C-gluc (Vitexin) | 236, 268, 338 | 431 → 341, 311 (bp), 283 | 2.3 ± 0.1 | 5.0 ± 0.5 | 0.3 ± 0.1 | ||
| 13 | 26.5 | Lut-7-O-gluc | 208, 256, 348 | 510, 447 (bp), 380, 279, 447 → 327, 285 (bp) | 7.1 ± 0.4 | ||||
| 14 | 31.5 | Apig-7-O-gluc | 240, 272, 330 | 494, 431 (bp), 279, 431 → 311, 269 (bp) | 1.5 ± 0.1 | ||||
| 15 | 35.0 | Lut-8-C-(2′-O-fer)-gluc | 208, 244, 336 | 686, 623 (bp), 329, 281, 623 → 447, 429 (bp), 309 | 0.1 ± 0.0 | ||||
| 16 | 37.1 | Apig-8-C-(2′′-O-fer)-gluc | 242, 268, 332 | 670, 607 (bp), 413, 279, 607 → 431, 413 (bp), 293 | 0.2 ± 0.0 |
5-CQA: 5-O-(E)-caffeoylquinic acid; 3-CoQA: 3-O-(E)-coumaroylquinic acid; 4-CoQA: 4-O-(E)-coumaroylquinic acid; Lut: luteolin; Apig: apigenin; gluc: glucoside; rham: rhamnoside; xyl: xyloside; fer: feruloyl.
Main observed fragments.
RT: retention time, N.I.: non-identified, n.d. not detected; DM: dry matter; bp: base peak.
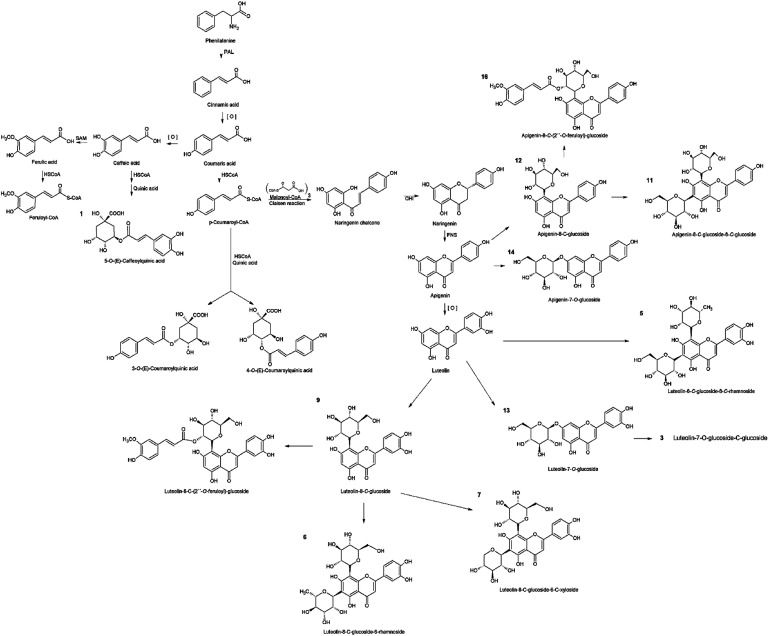
The chlorogenic acid derivatives (1, 2, and 4), found exclusively from S. polyrhiza in this study, accounted for 4.4 μg mg−1 of its dry mass (DM), representing 19.6% of the phenolic compounds. The other compounds of S. polyrhiza were categorized as apigenin derivatives (12, 14, and 16), 17.9% (4 μg mg−1 DM) and luteolin derivatives (9, 13, and 15), corresponding to 62.5% (14 μg mg−1 DM) (Table 1). S. polyrhiza was the only evaluated species in which feruloyl groups were identified as bound to the glycosyl unit (Table 1 and Fig. 2). L. punctata was distinguished from the other evaluated species due to the high content of apigenin, which corresponded to 81% (22.1 μg mg−1 DM – 3, 5, 6, and 11) of the phenolic content. L. gibba had a low level of apigenin (12–0.3 μg mg−1 DM) and 88% of luteolin derivatives (5, 6, and 9–2.2 μg mg−1 DM) (Table 1). The species within the family Wolffioideae displayed only luteolin derivatives (3, 5, 6, 7, and 9) that amount to 5.95 μg mg−1 DM for W. caudata, and 7.4 μg mg−1 DM for W. borealis (Table 1 and Fig. 2).
Fig. 2. The metabolic pathway involving the flavonoids and chlorogenic acid derivatives identified in the five species of duckweed. The network in black represents the building blocks for the biosynthesis of phenolic compounds identified that are represented in orange. The products from the shikimate pathway are the precursors for the chlorogenic acid and the flavonoids compounds synthesis. Chlorogenic acids are formed between cinnamic and quinic acids, while flavonoids are products from cinnamoyl-CoA where chalcones (naringenin-chalcone) act as precursors. The apigenin and luteolin backbones are synthesized from the naringenin-chalcone, which can be C– or O– branched with sugars and feruloyl radicals.
The phenolic compounds identified within the duckweed species studied in this work were mostly flavonoids. We investigated the antioxidant capacity for the species with significant contents of apigenin and luteolin. The IC50 value was higher in the species of the Lemnoideae subfamily, excluding L. punctata, than the Wolffioideae subfamily (Table 2).
Antiradical capacity of hydroalcoholic extracts of five duckweed species expressed as IC50 (μg mg−1).
| Species | IC50 (μg mg−1) |
|---|---|
| Spirodela polyrhiza | 41.45 |
| Landoltia punctata | 16.45 |
| Lemna gibba | 28.91 |
| Wolffiella caudata | 15.27 |
| Wolffia borealis | 11.17 |
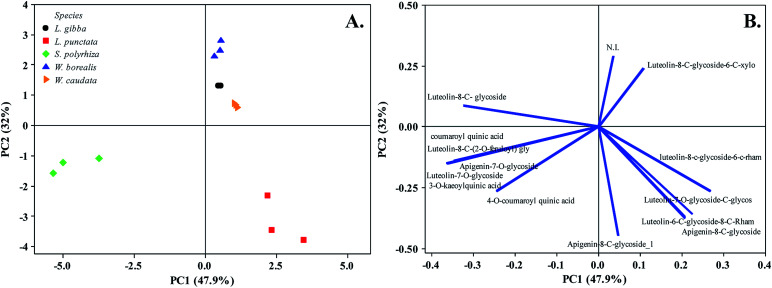
As previously mentioned, secondary metabolites are related to the evolutionary trend of duckweeds. We identified segregation between S. polyrhiza and L. punctata from L. gibba, W. borealis, and W. caudata probably due to the abundance phenolic compounds along with the presence of the 5-O-caffeoylquinic acid (1), 3-O-coumaroylquinic acid (2), and 4-O-coumaroylquinic acid (4) in S. polyrhiza and luteolin-6-C-glucoside-8-C-rhamnoside (5) and apigenin-8-C-glucoside-6-C-glucoside (11) in L. punctata (Fig. 1, 3, and Table 1). The more derived species (W. caudata and W. borealis – Wolffioideae) and L. gibba grouped in the PCA analysis due to the presence of compounds identified as luteolin-7-O-glucoside-C-glucoside (3), luteolin-8-C-glucoside-6-C-rhamnoside (6), and luteolin-8-C-glucoside (9) (Fig. 3).
Fig. 3. Principal Component Analysis (PC1 and PC2) of five duckweed species. (A) Duckweed species distribution in the plane defined by the first and second main components. (B) The relationship among variables of secondary metabolites from the plot of the PC1 and PC2 loading vectors describes the relationship among variables of compounds. Percentage values in parentheses (x and y axes) show the proportion of the variance explained by each axis. (n = 3). N. I. not identified. The vector values and statistical analyses are in ESI Table 1.†.
Discussion
Phenolic substances are the major bioactive compounds of some plants, acting on the defense against biotic and abiotic stresses.13,16,40 These biomolecules' chemical structures and properties have been reported to be essential for human health, being considered supplementary food and medicine.41 We identified chlorogenic acid derivatives and flavonoids as the phenolic compounds of five duckweed species.
Chlorogenic acids are a class of esters originated between cinnamic and quinic acids, synthesized through shikimate and phenylpropanoid pathway via p-coumaroyl CoA (Fig. 2), which can undertake chemical substitutions of the hydroxyl groups both axial (carbons 1 and 3) and equatorial (carbons 4 and 5).16 In the present study, the chlorogenic acid derivatives were identified only in S. polyrhiza (Table 1 and Fig. 1). The identified compounds caffeoylquinic acid (1) and coumaroylquinic acid (2 and 4) (Table 1 and Fig. 2) together with feruloylquinic acid that is one of the most common chlorogenic acids found in land plants.42
The flavone class consists of a 2-phenyl-1,4-benzopyrone structure common to apigenin, luteolin, chrysin, nobiletin, tangeretin, wogonin, and baicalin.17 The other 11 compounds (3, 5–16) isolated from duckweed extracts were identified as luteolin and apigenin-glycosides (Table 1). The MS/MS cleavage pattern was characterized by the range for mono-glycosylflavonoids, and di-glycosylflavonoids glycosyl-C-glycosylflavonoids (533–609 Da).43
The glycosylflavonoids identified were further characterized as 8-C- and 7-O-glycosides. The main distinction between the O- and C-glycosides is related to the glycosyl moiety attached via the anomeric carbon linked to the flavonoid backbone at the positions C-6 and C-8 of the ring A in the flavonoid structure.43 The glycosidic-O-linkages are cleaved with the rearrangement of H– leading to the loss of the monosaccharide residue with a mass of 162 Da for hexoses and 146 Da for pentoses.44 The cleavage pattern found for compounds 13 and 14 denotes a 7-O-glucoside, being therefore identified as luteolin-7-O-glucoside and apigenin-7-O-glucoside, respectively (Table 1). On the other hand, the flavonoids containing C-glycosides are directly linked to the sugar. Therefore, the fragmentation pattern is related to saccharide residue cleavages and the loss of a water molecule.44 These are more pronounced as 6-C groups of compounds.45,46
Usually, flavones are found in plants linked to sugars as O-glycosides, but C-glycosides can also occur.16,17,47 As mentioned above, the evaluated species had a predominance of C-glycosylated flavonoids (5, 6, 7, 9, 11, 12, 15, and 16) compared to O-glycosylated (3, 13, and 14) (Table 1 and Fig. 2). This corroborates the findings of Bai et al. (2018),48 Kim et al. (2010),36 McClure & Alston (1966),11 Qiao et al. (2011),18 Wallace & Alston (1966),49 and Wang et al. (2014).50 The glycosylation occurs after the synthesis of the flavonoid scaffold.51,52 The carbon–carbon bond in plant C-glycosylated flavonoids promotes protection from acid and enzymatic hydrolyses, which leads to divergence in degradation, pharmacokinetics, and bioactivity when compared with O-glycosides.53,54 The glycosylation is associated with the compound's hydrophobicity and promotes alterations in their antioxidant activity.55 As pointed out in Table 2, the higher accumulation of C-glycosylated flavonoids in L. punctata, W. caudata, and W. borealis may explain their higher antioxidant activity (Tables 1 and 2). Therefore, the catechol group and the phenolic hydroxyl groups were maintained free to scavenge radical species.
The dominant presence of C-glycosylated flavonoids in duckweeds hypothetically protect these aquatic plants from oxidative stress and predators due to the lack of lignin and high metabolic flux towards phenylalanine and flavonoids.34 Lignin is a phenolic compound responsible for plant mechanical support and pathogen protection in plants.56–59 Since duckweeds have trace amounts of lignin in all species60–63 with the absence of vessels, other biomolecules such as flavonoids are produced in higher quantities and possibly helps to protect duckweed tissues from pathogens.
The C-glycosylated flavonoids are thought to be suitable for human health applications due to their bioactivity.64 Flavonoids are pointed out as anti-diabetics, anti-inflammatory, anxiolytics, anti-spasmodic, anti-mutagenic, and hepatoprotection.54 Moreover, C-glycosylated flavonoids benefit the human diet, since the small intestine absorbs them more rapidly than the corresponding O-glycosides because it does not require de-glycosylation.54 This enrichment in C-glycosylated flavonoids highlights duckweed's potential as an attractive food supplement with health benefits.
Members of the Wolffioideae subfamily are being widely consumed in Asian countries as salad, mixed in soups, curries, and omelets.29 The presence of amino acids and fatty acid in the free forms make Wolffioideae more suitable for human consumption than the Lemnoideae species.29 We now show a further advantage of the consumption of Wolffioideae species, which is their antioxidant capacities that were higher for Wolffia borealis (IC50 11.17 μg mL−1) and Wolffiella caudata (IC50 15.27 μg mL−1) (Table 2), with more C-glycosylation compounds in these species likely conferring a higher antioxidant activity. Furthermore, the higher proportion of luteolin-8-C-glucoside-6-C-rhamnoside (6) (7.4 μg mg−1 DM) and apigenin-8-C-glucoside-6-C-glucoside (11) (10.4 μg mg−1 DM) in L. punctata and luteolin-8-C-glucoside-6-C-rhamnoside (6) (5.65 μg mg−1 DM) in W. caudata contributed for their higher antioxidant levels (Tables 1 and 2). Thus, besides the species from the Wolffioideae family, L. punctata could also be an interesting candidate species for the human diet.
Some of the identified compounds have been previously described for duckweeds, as in S. polyrhiza where the major secondary metabolites are orientin (9) and vitexin (12).18 In the present study, orientin and luteolin-8-O-glucoside accounted for 13.9 μg mg−1, while vitexin was found in relatively low concentration (2.3 μg mg−1 DM). Also, compounds 13, 14, 15, and 16 were already identified for Spirodela polyrhiza.18,37 Besides S. polyrhiza, L. punctata and W. globosa have been reported as rich sources of luteolin, and apigenin derivatives.50,65 The compounds 5-O-(E)-caffeoylquinic acid (1), 3-O-(E)-coumaroylquinic acid (2), luteolin-7-O-glucoside-C-glucoside (3), 4-O-(E)-coumaroylquinic acid (4), luteolin-6-C-glucoside-8-C-rhamnoside (5), and luteolin-8-C-glucoside-6-C-rhamnoside (6) are described here for the first time in duckweeds.
The diversity of flavonoids in the five duckweed species revealed as major constituents luteolin and its derivatives (36.4 μg mg−1 DM), except for L. punctata (Table 1). In L. punctata the apigenin content constituted 56.4% of the quantified compounds (Table 1), which positively impacted the antioxidant activity when compared to the other species evaluated from the Lemnoideae subfamily (Table 2).
The flavonoid class distribution does not seem significant for the phylogenetic relationship between Lemnaceae and other plant families. However, inside the duckweed family, is found a correlation among species, genera, and chemical compounds.11 During the evolution of the Wolffioideae species (W. caudata and W. borealis), there seems to have been the loss of the root system and body reduction accompanied by the reduction of secondary metabolites diversity.11 The ethanolic extract of W. caudata led to the identification of only two compounds characterized as luteolin-7-O-glucoside-C-glucoside (3) and luteolin 8-C-glucoside-6-C-rhamnose (6) (Table 1), common to L. punctata, L. gibba, and W. borealis. Nevertheless, W. borealis – the more derived species within the duckweeds – displayed the same compounds found in W. caudata plus three luteolin derivatives (5, 7, and 9) (Table 1). PCA separates the duckweed genera regarding its phenolic profile, with a higher diversity in Lemnoideae (S. polyrhiza, L. punctata, and L. gibba) than in Wolffioideae (W. caudata and W. borealis) (Fig. 3). PC1 segregated Spirodela polyrhiza due to the chlorogenic derivatives (1, 2, and 4) and L. punctata regarding luteolin-6-C-glucoside-8-C-rhamnose (5) and apigenin-8-C-glucoside-6-C-glucoside (11) exclusivity. PC2 grouped Wolffioideae with L. gibba based on the occurrence of luteolin-7-O-glucoside-C-glucoside (3), luteolin-8-C-glucoside-6-C-rhamnose (6), and luteolin-8-C-glucoside (9) (Fig. 3). The proximity of L. gibba can also be explained by the fact that this species is closer to the Wolffioideae subfamily than L. punctata and S. polyrhiza through the evolution trend (Spirodela > Landoltia > Lemna > Wolffiella > Wolffia) described by Les et al. (2002).3
Among our findings, Landoltia punctata is an interesting addition to the human diet due to the presence of a high concentration of apigenin, which has been widely reported as a potent anticancer adjuvant.66 Vitexin, whose C-glucosylated version we found in L. punctata, has recently been proposed as an agent to treat non-small lung cancer.67
Conclusions
The phenolic profile of the five species of duckweeds separate duckweed genera and seems to corroborate what is known about the evolution of the group. S. polyrhiza was the only species displaying significant chlorogenic acid derivatives, which could enhance the use of the plant extract for human health applications. The Wolffioideae species studied here may be attractive candidates as luteolin derivative bioresources, whereas L. punctata displays higher content for apigenin derivatives. Duckweeds have higher proportions of C-glycosyl-flavonoids that could benefit the human diet due to their favorable antioxidant and anticancer capacity. Besides that, L. punctata could be used as supplements of antioxidants products. We conclude that duckweeds could be an economical source for valuable additives to the human diet, and their potential should be further explored.
Funding
This work was supported by the Instituto Nacional de Ciência e Tecnologia do Bioetanol – INCT does Bioethanol (FAPESP 2008/57908-6 and CNPq 574002/2008-1). D. P. (CAPES 88882.377113/2019-1). C. P. (FUSP/2910). A. G. (FAPESP 2019/13936-0). The support by a travel grant to E. L. by the U.S. Fulbright-Brazil Scholar Mobility Program (2014) to travel to the laboratory of M. B. to jump-start this project in 2014–2015 is gratefully acknowledged. M. J. P. F. and M. B. are fellow researchers of CNPq. The authors thank the Fulbright Foundation for the fellowship to E. L.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank Dr Eny Iochevet Segal Floh for the use of laboratory facilities. This work was supported by the Instituto Nacional de Ciência e Tecnologia do Bioetanol – INCT does Bioethanol (FAPESP 2014/50884-5) and CNPq 465319/2014-9). DP (CAPES 88882.377113/2019-1). AG (FAPESP 2019/13936-0). The support by a travel grant to EL by the U.S. Fulbright-Brazil Scholar Mobility Program (2014) to travel to the laboratory of MB to jump-start this project in 2014-2015 is gratefully acknowledged. MJPF and MB are fellow researchers of CNPq. The authors thank the Fulbright Foundation for the fellowship to EL.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06741e
Notes and references
- Landolt E. J. Ariz.-Nev. Acad. Sci. 1992;26:10–14. [Google Scholar]
- Appenroth K. J. Borisjuk N. Lam E. Chin. J. Appl. Environ. Biol. 2013;19:1–10. [Google Scholar]
- Les D. H. D. Crawford D. J. D. Landolt E. Gabel J. D. J. Kimball R. T. R. Syst. Bot. 2002;27:221–240. [Google Scholar]
- Ziegler P. Adelmann K. Zimmer S. Schmidt C. Appenroth K.-J. J. Plant Biol. 2014;17:1–9. doi: 10.1111/plb.12184. [DOI] [PubMed] [Google Scholar]
- Cheng J. J. Stomp A. M. Clean: Soil, Air, Water. 2009;37:17–26. [Google Scholar]
- Ekperusi A. O. Nwachukwu E. O. Sikoki F. D. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-65389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomp A.-M. M. Biotechnol. Annu. Rev. 2005;11:69–99. doi: 10.1016/S1387-2656(05)11002-3. [DOI] [PubMed] [Google Scholar]
- Liu Y. Chen X. Wang X. Fang Y. Huang M. Guo L. Zhang Y. Zhao H. Sci. Rep. 2018;8:1–9. doi: 10.1038/s41598-018-27944-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenroth K. J. Sree K. S. Fakhoorian T. Lam E. Plant Mol. Biol. 2015;89:647–654. doi: 10.1007/s11103-015-0396-9. [DOI] [PubMed] [Google Scholar]
- Oron G. Wildschut L. R. Porath D. Water Sci. Technol. 1985;17:803–817. [Google Scholar]
- McClure J. W. Alston R. E. Am. J. Bot. 1966;53:849–860. [PubMed] [Google Scholar]
- Avci U. Peña M. J. O'Neill M. A. Planta. 2018;247:953–971. doi: 10.1007/s00425-017-2837-y. [DOI] [PubMed] [Google Scholar]
- Pichersky E. Gang D. R. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- Samanta A. Das G. Das K. S. Das S. K. Int. J. Pharm. Sci. 2011;6:12–35. [Google Scholar]
- Schmidt T. J. Khalid S. A. Romanha A. J. Alves T. M. A. Biavatti M. W. Brun R. Da Costa F. B. de Castro S. L. Ferreira V. F. de Lacerda M. V. G. Lago J. H. G. Leon L. L. Lopes N. P. das Neves Amorim R. C. Niehues M. Ogungbe I. V. Curr. Med. Chem. 2012;19:2128–2175. doi: 10.2174/092986712800229023. [DOI] [PubMed] [Google Scholar]
- Plazonić A. Bucar F. Maleŝ Z.^. Mornar A. Nigović B. Kujundẑić N. Molecules. 2009;14:2466–2490. doi: 10.3390/molecules14072466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashaei R. Int. J. Food Sci., Nutr. Diet. 2016;5:300–304. [Google Scholar]
- Qiao X. He W. N. Xiang C. Han J. Wu L. J. Guo D. A. Ye M. Phytochem. Anal. 2011;22:475–483. doi: 10.1002/pca.1303. [DOI] [PubMed] [Google Scholar]
- Akhtar T. A. Lees H. A. Lampi M. A. Enstone daryl. Brain R. A. Greenberg B. M. Plant, Cell Environ. 2010;33:1205–1219. doi: 10.1111/j.1365-3040.2010.02140.x. [DOI] [PubMed] [Google Scholar]
- Orhan D. D. Özçelik B. Özgen S. Ergun F. Microbiol. Res. 2010;165:496–504. doi: 10.1016/j.micres.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Azizah M. Pripdeevech P. Thongkongkaew T. Mahidol C. Ruchirawat S. Kittakoop P. Antibiotics. 2020;9:1–34. doi: 10.3390/antibiotics9090606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Annu. Rev. Nutr. 1996;16:33–50. doi: 10.1146/annurev.nu.16.070196.000341. [DOI] [PubMed] [Google Scholar]
- Atmani D. Chaher N. Atmani D. Berboucha M. Debbache N. Boudaoud H. Curr. Nutr. Food Sci. 2009;5:225–237. [Google Scholar]
- Siddiqui I. A. Afaq F. Adhami V. M. Ahmad N. Mukhtar H. Antioxid. Redox Signaling. 2004;6:571–582. doi: 10.1089/152308604773934323. [DOI] [PubMed] [Google Scholar]
- Wiseman S. A. Balentine D. A. Frei B. Crit. Rev. Food Sci. Nutr. 1997;37:705–718. doi: 10.1080/10408399709527798. [DOI] [PubMed] [Google Scholar]
- Minussi R. C. Rossi M. Bologna L. Cordi L. Rotilio D. Pastore G. M. Durán N. Food Chem. 2003;82:409–416. [Google Scholar]
- López-Vélez M. Martínez-Martínez F. Del Valle-Ribes C. Crit. Rev. Food Sci. Nutr. 2003;43:233–244. doi: 10.1080/10408690390826509. [DOI] [PubMed] [Google Scholar]
- Sales-medina D. F. Ferreira L. R. P. Romena L. M. D. Gonçalves K. R. Guido R. F. C. Courtemanche G. Buckeridge M. S. Durigon É. L. Moraes C. B. Freitas-Junior L. H. bioRxiv. 2020 doi: 10.1101/2020.07.09.196337. [DOI] [Google Scholar]
- Appenroth K. J. Sree K. S. Böhm V. Hammann S. Vetter W. Leiterer M. Jahreis G. Böhm V. Hammann S. Vetter W. Leiterer M. Jahreis G. Food Chem. 2017;217:266–273. doi: 10.1016/j.foodchem.2016.08.116. [DOI] [PubMed] [Google Scholar]
- Bhanthumnavin K. Mcgarry M. G. Nature. 1971;232:495. doi: 10.1038/232495a0. [DOI] [PubMed] [Google Scholar]
- de Beukelaar M. F. A. Zeinstra G. G. Mes J. J. Fischer A. R. H. Food Qual. Prefer. 2019;71:76–86. [Google Scholar]
- Rusoff L. L. Blakeney E. W. Culley D. D. J. Agric. Food Chem. 1980;28:848–850. doi: 10.1021/jf60230a040. [DOI] [PubMed] [Google Scholar]
- Song W. Y. Choi J. H. J. Life Sci. 2017;27:180–186. [Google Scholar]
- Tao X. Fang Y. Huang M. J. Xiao Y. Liu Y. Ma X. R. Zhao H. BMC Genomics. 2017;18:166. doi: 10.1186/s12864-017-3559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenroth K. J. Sowjanya Sree K. Bog M. Ecker J. Seeliger C. Böhm V. Lorkowski S. Sommer K. Vetter W. Tolzin-Banasch K. Kirmse R. Leiterer M. Dawczynski C. Liebisch G. Jahreis G. Front. Chem. 2018;6:1–13. doi: 10.3389/fchem.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Lee I. Seo J. Jung M. Kim Y. Yim N. Bae K. Phyther. Res. 2010;24:1543–1548. doi: 10.1002/ptr.3186. [DOI] [PubMed] [Google Scholar]
- Ren D. Han B. Xin Z. Liu W. Ma S. Liang Y. Yi L. Sep. Purif. Technol. 2016;165:160–165. [Google Scholar]
- Seo C. S. Lee M. Y. Shin I. S. Lee J. A. Ha H. Shin H. K. Immunopharmacol. Immunotoxicol. 2012;34:794–802. doi: 10.3109/08923973.2012.656273. [DOI] [PubMed] [Google Scholar]
- Molyneux P. Songklanakarin J. Sci. Technol. 2003;26:211–219. [Google Scholar]
- Samanta A. Das G. Das S. K. Int. J. Pharm. Sci. 2011;6(1):12–35. [Google Scholar]
- Ghasemzadeh A. Ghasemzadeh N. J. Med. Plants Res. 2011;5:6697–6703. [Google Scholar]
- Ncube E. N. Mhlongo M. I. Piater L. A. Steenkamp P. A. Dubery I. A. Madala N. E. Chem. Cent. J. 2014;8:1–10. doi: 10.1186/s13065-014-0066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng P. Sun J. Zhang M. Li X. Harnly J. M. Chen P. J. Mass Spectrom. 2016;51:914–930. doi: 10.1002/jms.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyckens F. Claeys M. J. Mass Spectrom. 2004;39:1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- Ferreres F. Silva B. M. Andrade P. B. Seabra R. M. Ferreira M. A. Phytochem. Anal. 2003;14:352–359. doi: 10.1002/pca.727. [DOI] [PubMed] [Google Scholar]
- Singh A. Kumar S. Bajpai V. Reddy T. J. Rameshkumar K. B. Kumar B. Rapid Commun. Mass Spectrom. 2015;29:1095–1106. doi: 10.1002/rcm.7202. [DOI] [PubMed] [Google Scholar]
- Hostetler G. L. Ralston R. A. Schwartz S. J. Adv. Nutr. Int. Rev. J. 2017;8:423–435. doi: 10.3945/an.116.012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H. H. Wang N. N. Mi J. Yang T. Fang D. M. Wu L. W. Zhao H. Li G. Y. Fitoterapia. 2018;124:211–216. doi: 10.1016/j.fitote.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Wallace J. W. Alston R. E. E. Plant Cell Physiol. 1966;7:699–700. [Google Scholar]
- Wang N. Fang Y. Yang T. Li G. Zhao H. Chin. J. Appl. Environ. Biol. 2014;20:1016–1019. [Google Scholar]
- Wallace J. W. Mabry T. J. Alston R. E. Phytochemistry. 1969;8:93–99. [Google Scholar]
- Li S. S. Wu J. Chen L. G. Du H. Xu Y. J. Wang L. J. Zhang H. J. Zheng X. C. Wang L. S. PLoS One. 2014;9(10):1–11. doi: 10.1371/journal.pone.0108860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne J. B. Phytochemistry. 1965;4:107–120. [Google Scholar]
- Courts F. L. Williamson G. Crit. Rev. Food Sci. Nutr. 2015;55:1352–1367. doi: 10.1080/10408398.2012.694497. [DOI] [PubMed] [Google Scholar]
- Heim K. E. Tagliaferro A. R. Bobilya D. J. J. Nutr. Biochem. 2002;13:572–584. doi: 10.1016/s0955-2863(02)00208-5. [DOI] [PubMed] [Google Scholar]
- Lozovaya V. V. Lygin A. V. Zernova O. V. Li S. Widholm J. M. Hartman G. L. Plant Dis. 2006;90:77–82. doi: 10.1094/PD-90-0077. [DOI] [PubMed] [Google Scholar]
- Elfstrand M. Sitbon F. Lapierre C. Bottin A. Von Arnold S. Planta. 2002;214:708–716. doi: 10.1007/s00425-001-0681-5. [DOI] [PubMed] [Google Scholar]
- Sarkanen K. V. Ludwig C. H. J. Polym. Sci., Part B: Polym. Lett. 1972;vol. 10:228–230. [Google Scholar]
- Ithal N. Recknor J. Nettleton D. Maier T. Baum T. J. Mitchum M. G. Mol. Plant-Microbe Interact. 2007;20:510–525. doi: 10.1094/MPMI-20-5-0510. [DOI] [PubMed] [Google Scholar]
- Zhao X. Moates G. K. K. Wellner N. Collins S. R. a. R. A. A. Coleman M. J. J. Waldron K. W. W. Carbohydr. Polym. 2014;111:410–418. doi: 10.1016/j.carbpol.2014.04.079. [DOI] [PubMed] [Google Scholar]
- Yu C. Sun C. Yu L. Zhu M. Xu H. Zhao J. Ma Y. Zhou G. PLoS One. 2014;9:1–15. doi: 10.1371/journal.pone.0115023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. Zhang N. Phillips G. C. Xu J. Bioresour. Technol. 2012;124:485–488. doi: 10.1016/j.biortech.2012.08.050. [DOI] [PubMed] [Google Scholar]
- Pagliuso D. Grandis A. Igarashi E. S. Lam E. Buckeridge M. S. Front. Chem. 2018;6:1–10. doi: 10.3389/fchem.2018.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. Crit. Rev. Food Sci. Nutr. 2017;57:1874–1905. doi: 10.1080/10408398.2015.1032400. [DOI] [PubMed] [Google Scholar]
- Wang N. Xu G. Fang Y. Yang T. Zhao H. Li G. Molecules. 2014;19:6623–6634. doi: 10.3390/molecules19056623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X. Qi M. Li P. Zhan Y. Shao H. Cell Biosci. 2017:1–16. doi: 10.1186/s13578-017-0179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Jiang Q. Liu H. Luo S. Biol. Res. 2019:1–7. doi: 10.1186/s40659-019-0214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.