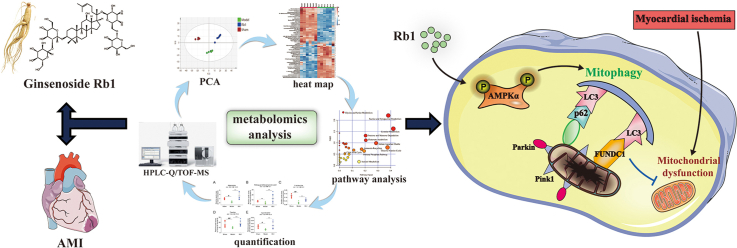
Abstract
Background
Ginsenoside Rb1, a bioactive component isolated from the Panax ginseng, acts as a remedy to prevent myocardial injury. However, it is obscure whether the cardioprotective functions of Rb1 are related to the regulation of endogenous metabolites, and its potential molecular mechanism still needs further clarification, especially from a comprehensive metabolomics profiling perspective.
Methods
The mice model of acute myocardial ischemia (AMI) and oxygen glucose deprivation (OGD)-induced cardiomyocytes injury were applied to explore the protective effect and mechanism of Rb1. Meanwhile, the comprehensive metabolomics profiling was conducted by high-performance liquid chromatography and quadrupole time-of-flight mass spectrometry (HPLC-Q/TOF-MS) and a tandem liquid chromatography and mass spectrometry (LC-MS).
Results
Rb1 treatment profoundly reduced the infarct size and attenuated myocardial injury. The metabolic network map of 65 differential endogenous metabolites was constructed and provided a new inspiration for the treatment of AMI by Rb1, which was mainly associated with mitophagy. In vivo and in vitro experiments, Rb1 was found to improve mitochondrial morphology, mitochondrial function and promote mitophagy. Interestingly, the mitophagy inhibitor partly attenuated the cardioprotective effect of Rb1. Additionally, Rb1 markedly facilitated the phosphorylation of AMP-activated protein kinase α (AMPKα), and AMPK inhibition partially weakened the role of Rb1 in promoting mitophagy.
Conclusions
Ginsenoside Rb1 protects acute myocardial ischemia injury through promoting mitophagy via AMPKα phosphorylation, which might lay the foundation for the further application of Rb1 in cardiovascular diseases.
Keywords: Acute myocardial ischemia, AMPK, Ginsenoside Rb1, Metabolomics, Mitophagy
Graphical abstract
1. Introduction
Acute myocardial ischemia (AMI), a severely cardiovascular disease, causes cardiomyocyte death, cardiac dysfunction and cardiac metabolic disorder due to prolonged nutritional deficiency and hypoxia, and lastly leads to heart failure and death [1]. Contemporarily, angiotensin-converting enzyme inhibitors, β-blockers and calcium antagonists are the main therapeutic strategies. However, clinical treatments provide limited symptomatic relief and the current attempts in drug treatment are far from enough.
Ischemic myocardium is highly dependent on AMPK-mediated mitochondrial aerobic metabolism to restore energy balance for maintenance of both cell viability and contractile function [2]. However, AMI frequently triggers mitochondrial Ca2+ overload, oxidative stress, and opening of the mitochondrial permeability transition pore, which ultimately induce mitochondrial dysfunction and cardiomyocyte death [3]. Meanwhile, Dysfunctional mitochondria have a restrained capacity for ATP synthesis and generate excessive reactive oxygen species (ROS), contributing to the development of ischemic tissue necrosis. Mitophagy, the selective form of macroautophagy, is a mitochondrial quality control machinery, which targets unhealthy mitochondria or aberrant mitochondrial proteins for degradation and removal, plays a vital role in maintaining myocardial homeostasis [4]. Activating mitophagy protects against ischemia/reperfusion injury of liver, brain, kidney and heart through eliminating damaged mitochondria and maintaining mitochondrial homeostasis [[5], [6], [7], [8]]. Additionally, stimulation of AMPK enhanced PINK1/Parkin-dependent beneficial mitophagy of skeletal muscle [9]. Mitochondrial dysfunction is recognized as a precursor of cell death. Therefore, it is necessary to eliminate damaged mitochondria by mitophagy to maintain healthy mitochondrial network.
Panax ginseng is a traditional medicinal herb, and its components, especially ginsenoside Rb1, have received much attention due to their protective effects on the central nervous system, cardiovascular systems and immune systems [10,11]. Previous studies have reported that Rb1 could exert cardioprotection against ischemia/reperfusion (I/R) injury through anti-apoptosis, anti-inflammation and regulation of energy metabolism [12,13]. Moreover, it could attenuate chondriokinesis to restraint the release of MDA and improve circulation by augmented expression of NO [11]. However, the molecular mechanism of Rb1 against AMI remains obscure and further studies are still needed to elucidate potential mechanisms.
Metabolomics, a methodology for comprehensive analysis of small molecular substances in biological systems, has a unique potential to discover biomarkers, which could predict incidence, progression and severity of diseases, and might cast new light on excavating underlying mechanisms [14]. Recently, metabolomics is widely performed to study cardiovascular diseases. The functional metabolomics found that N-acetylneuraminic acid was a potential metabolic marker for CAD progression and silencing its regulatory enzyme, neuraminidase-1, might become an unrecognized therapeutic intervention for CAD [15]. In our previous study, schizandrol A effectively protected AMI mice through upregulation of ecto-5′-nucleotidase, methionine synthase and platelet-derived endothelial cell growth factor based on metabolomics analysis [16].
Whereas, there are few studies on the cardioprotective functions of Rb1 from the perspective of metabolomics. In the present study, metabolomics was applied to clarify the metabolic pathways regulated by Rb1 on the improvement of AMI, and also excavate novel potential cardioprotective mechanism of Rb1 for AMI treatment. Subsequently, its regulatory metabolic network map was constructed, and the potential mechanism was further verified through in vivo and in vitro experiments. Our present study will provide some references for the mechanism elucidation of Rb1 from the perspective of endogenous metabolites.
2. Materials and methods
2.1. Drugs and reagents
Ginsenoside Rb1 was got from Chengdu Pufei De Biotech Co., Ltd (Chengdu, China; purity ≥ 98 %). Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Compound C was obtained from APExBIO Technology (Houston, USA). Cyclosporin A (CSA) was obtained from Aladdin (Shanghai, China). Antibodies against GAPDH and E3 ubiquitin ligase (parkin) were obtained from Bioworld Technology (Shanghai, China). Antibodies against phospho-AMPKα (p-AMPKα), AMPKα and microtubule-associated protein 1 light chain 3 (LC3B) were purchased from Cell Signaling Technology (Boston, MA, USA). Antibodies against PTEN-induced putative kinase 1 (PINK1) was purchased from proteintech (Nanjing, China). Antibodies against ubiquitin-binding protein p62 (p62) and FUN14 domain containing 1 (FUNDC1) were obtained from Abcam (Cambridge, MA, USA).
2.2. Animals and acute myocardial ischemia (AMI) injury model
ICR male mice were purchased from the Experimental Animal Center of Yangzhou University (Yangzhou, Jiangsu, China, certificate NO. 202019004). The AMI model was produced by the left anterior descending coronary artery ligation (CAL) [16] following the Animal Ethics Committee of China Pharmaceutical University and the Laboratory Animal Management Committee of Jiangsu Province (Approval No.: 220193566). Sham-operated mice carried out the same surgical procedures without ligation. The dose of Rb1 was 6 mg/kg based on the effective dose determined by our previous study [17], and the dose of metoprolol (Met) was 5.14 mg/kg referring to the clinical dose. All drugs were administered intraperitoneally after 20 min of CAL.
2.3. Measurement of myocardial infarct size
The hearts were incubated in 2,3,5-triphenyl tetrazolium chloride (TTC) for 15 min, and pictured timely.
2.4. Enzyme-linked immunosorbent assay (ELISA)
The concentrations of tumor necrosis factor-α (TNF-α), cardiac troponin I (cTn-I) and C-reactive protein (CRP) in serum were measured using commercial kits.
2.5. Histopathological examination
The hearts were stained with hematoxylin-eosin (H&E) or Masson's trichrome, which were applied to investigate the morphological damages and fibrosis levels, respectively.
2.6. Untargeted metabolomics analysis
2.6.1. Serum and urine samples collection and pretreatment
Samples of serum and urine were collected within twenty-four hours after surgery. After a series of processing, the samples were used for later analysis.
2.6.2. HPLC-Q-TOF MS analysis
The untargeted metabolomics analysis of samples was performed using an Agilent Technologies 6530 Accurate-Mass Q-TOF LC/MS (USA).
2.6.3. Data processing and analysis
Data were processed using the R package, then performed multivariate analysis in SIMCA-P 14.1 software (Umetrics, Sweden) and also imported into MetaboAnalyst 4.0 platform (https://www.metaboanalyst.ca/) for univariate analysis.
2.6.4. Metabolite identification and metabolic pathway analysis
For identification of potential metabolites, the structure messages of metabolites were matched in online databases such as HMDB (http://www.hmdb.ca/), Metlin (http://metlin.scripps.edu/) and MassBank (http://www.massbank.jp/). The online platforms, MetaboAnalyst 4.0, STRING (https://string-db.org/) and Metascape (https://www.metascape.org/) were implemented to enrichment & pathway analysis of influential metabolites and related regulated enzymes.
2.6.5. Method validation
The reproducibility and stability of the analytical procedure were assessed using the quality control (QC) sample. The results of unsupervised principal component analysis (PCA) indicated well reproducibility and stability of the analytical procedure as shown in Fig. S1A-D. Meanwhile, the total ion chromatograms (TICs) of the QC samples were overlapped in Fig. S1E-H, which showed that the response intensity and retention time of each chromatographic peak were basically overlapped. Besides, six representative ions were extracted to assess the feasibility of method in Table S1, which showed that the reproducibility of this method was acceptable. These data indicated the good reproducibility of the analytical method and the stability of the equipment.
2.7. Targeted metabolomics analysis
2.7.1. Mice samples and reference & internal standards pretreatment
The urine samples were mixed with Caffeic acid (internal standard), Ketoprofen (internal standard) and methanol, and then centrifuged to remove precipitated protein. The supernatant was conducted for targeted metabolomics analysis.
2.7.2. LC-MS/MS analysis
For identifying and semi-quantifying metabolites, the analysis was performed on a triple quadrupole LC-MS/MS system. Tandem mass spectrometry (MS/MS) analysis and data acquisition were performed with an Agilent Ultivo Triple Quadrupole (QqQ) mass spectrometer (Agilent Technologies Singapore (International) Pte.Ltd) with a Jet Stream ESI source. Data acquisition and processing were performed using Masshunter software.
2.8. Transmission electron microscopy
The ultrathin heart tissues from the margin of infarction area were prepared and and their ultrastructure was detected through transmission electron microscopy (JEM-1001, JEOL Ltd., Tokyo, Japan).
2.9. Immunohistochemistry
The 4-5 μm heart slices were incubated with primary antibodies plus HRP-conjugated secondary antibodies following the protocols in the user manuals.
2.10. Cell preparation and culture
Rat H9c2 cells were obtained from Shanghai Institute of Cell Biology, Chinese Academy of Science (Shanghai, China). Passage 3-9 of cells were used.
2.11. OGD injury model in vitro
Oxygen glucose deprivation (OGD)-injured cell model was established based on a previous study, which mimics the AMI injury in vitro [16]. The OGD injury was induced by incubating cells with none glucose DMEM and a hypoxic environment of 94 % N2, 1 % O2 and 5 % CO2 for 6 h. Cells were treated with Rb1 (10 μM), CCCP (10 μM) or CSA (10 μM) during hypoxia.
2.12. Cell viability and LDH assays
The cell viability was determined by MTT assays as reported previously [16]. The culture supernatants used to detect the release of LDH according to the manufacturer's instructions.
2.13. Determination of mitochondrial membrane potential (ΔψM)
5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide (JC-1, BD) was used to analyze the changes in ΔψM. Cell fluorescence was monitored using a confocal laser scanning microscopy (CLSM, LSM700, Zeiss, Germany).
2.14. Immunofluorescence staining
The cells were incubated with MitoTracker® Deep Red (Molecular Probes) and primary antibody, followed by incubation with Alexa Fluor® 488 conjugated Donkey Anti-Rabbit IgG (H + L) antibody (Thermo Fisher Scientific, USA) and 4′,6-Diamidino-2-phenylindole (DAPI, Beyotime Biotechnology, Shanghai, China).
2.15. Western blot analysis
The expression of proteins associated with mitophagy were measured by western blot analysis according to our previous steps [16]. The bands were visualized using a Bio-Rad Laboratories, and band density was determined by ImageJ software (Bethesda Md, USA).
2.16. Statistical analysis
Statistical analysis was carried out using Student's two-tailed t-test for comparison between two groups, and one-way analysis of variance (ANOVA) followed by Dunnett's test for comparisons between three or more groups. P values of < 0.05 were considered significant. Statistical analysis was performed by the GraphPad Prism 8 software (La Jolla, CA, USA).
(The detailed steps were described in supplementary material)
3. Results
3.1. Rb1 effectively protected AMI-injured mice
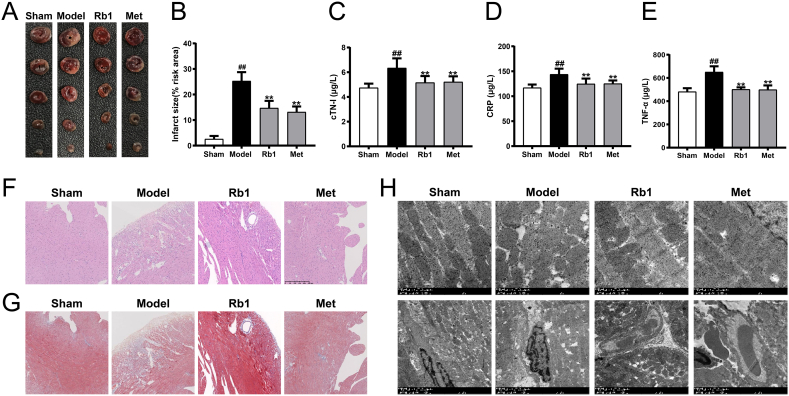
24 h of acute myocardial ischemia resulted in myocardial injury as demonstrated by increased infarct size, increased serum contents of C-reactive protein (CRP), cardiac troponin I (cTN-I) and tumor necrosis factor-α (TNF-α). However, Rb1 treatment developed smaller infarct areas and significantly reduced the contents of CRP, cTN-I and TNF-α (Fig. 1A–E). The histopathological examination of heart tissues in AMI-injured mice exhibited increased left ventricular wall fibrosis, and severely morphological damages including widespread destruction of myocardial structure, increased necrotic and fusion areas, and massive inflammatory cells infiltrated in myocardial tissues. Whereas, Rb1 treatment significantly made morphological features normalized, and reduced myocardial fibrosis and collagen deposition (Fig. 1F and G). Simultaneously, the myocardial ultrastructure in AMI mice exhibited swollen mitochondria with abnormal cristae, distorted sarcomeres in the myofibrils, and disorganized surface of the cardiac microvessels. However, the damaged myocardial ultrastructure tended to be normal or mild after Rb1 and Met treatment (Fig. 1H). These results demonstrated that AMI-injured myocardium was attenuated by Rb1 treatment.
Fig. 1.
Rb1 effectively protected acute myocardial ischemia-injured mice. (A) Representative images of 2,3,5-triphenyl tetrazolium chloride (TTC) staining. (B) The statistical results of TTC staining. The serum contents of (C) cTN-I, (D) CRP and (E) TNF-α. Representative images of (F) H&E and (G) Masson's trichrome staining (200 × ). Scale bar = 250 μm. (H) Myocardial ultrastructure was detected by transmission electron microscopy (5000 × ). Scale bar = 1 μm. Results were presented as mean ± SD. ##P < 0.01 vs. the sham group; ∗∗P < 0.01 vs. the model group, n = 6.
3.2. Multivariate analysis of serum and urine samples
Total ion chromatograms (TICs) of the samples were obtained by Q-TOF system (Fig. S2). The results of unsupervised principal component analysis (PCA) indicated a successfully modeling process and severely metabolic changes in AMI mice (Fig. S3A-D). The high model quality parameters demonstrated that the supervised orthogonal partial least squares discriminant analysis (OPLS-DA) model had good fitness and predictability (Table S2). The results of OPLS-DA indicated that the metabolic profile of the model group was significantly different from that of the Rb1 group (Fig. S3E-H). S-plot of OPLS-DA was used to visually find altered metabolites which significantly contributed to the classification of the model and Rb1 group (Fig. S3I-L). These endogenous metabolites with features of VIP-value over 1 and p-value less than 0.05 will be considered as differential metabolites and performed further identification. 65 metabolites were selected as differential metabolites (Table S3-4). Furthermore, a heat map was used to visualize the relative levels of the endogenous metabolites in AMI mice (Fig. S4).
3.3. Validation and semi-quantification of selected metabolites by LC-MS/MS
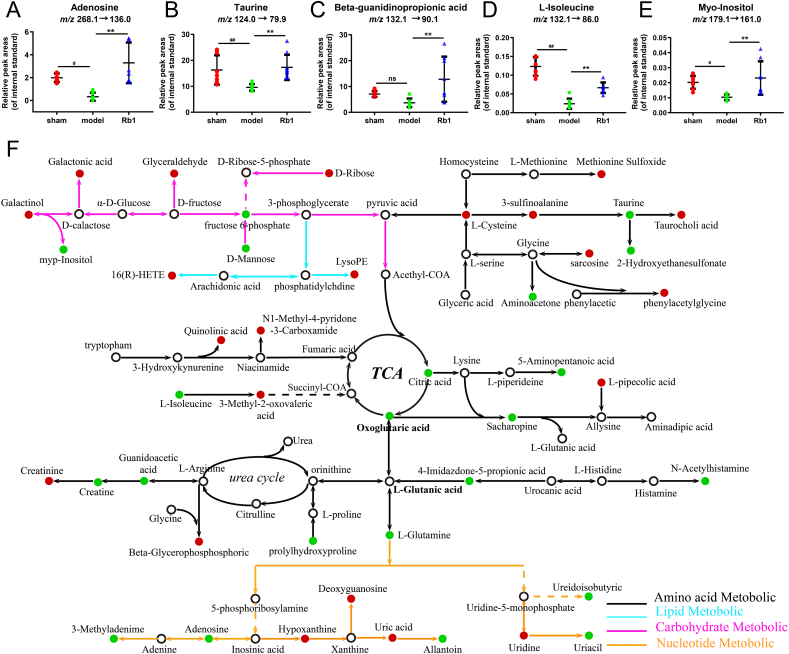
Based on the above results, the urine levels of adenosine, taurine, beta-guanidinopropionic acid (β-GPA), L-isoleucine and myo-Inositol were measured by LC-MS/MS (Fig. S5). The relative peak areas (RPA) of these metabolites were decreased in AMI mice, while the RPA of these metabolites were significantly elevated after Rb1 treatment (near normal level) (Fig. 2A–E). Interestingly, the five significantly changed metabolites were closely linked to mitophagy, suggesting a possible mechanism in cardioprotective effect of Rb1.
Fig. 2.
Validation and semi-quantification of selected metabolites, and the metabolic network map of the differential metabolites. The RPA of (A) adenosine, (B) taurine, (C) β-GPA, (D) L-isoleucine and (E) myo-Inositol, in reference to the retention time of reference standards and the peak area of internal standards. (F) The metabolic network map of the differential metabolites. The red dots represent that metabolites in the model group were up-regulated compared with those in the Rb1 group and the down-regulated metabolites were represented by green dots. Results were presented as mean ± SD. #P < 0.05, ##P < 0.01 vs. the sham group; ∗∗P < 0.01 vs. the model group, n = 8.
3.4. Enrichment analysis of metabolic pathway and regulatory enzymes
According to the fold enrichment of pathways, taurine and hypotaurine metabolism, glycine and serine metabolism, cysteine metabolism, methionine metabolism and purine metabolism were screened out as the most influentially metabolic pathways shown in Fig. S6A-C. The functions of related regulatory enzymes mainly involved cellular amino acid metabolic process, carboxylic acid biosynthetic process, biological oxidations, and nucleotide metabolic process (Fig. S6D). The protein interaction network was constructed via GO enrichment analysis (Fig. S6E). Additionally, KEGG analysis also indicated that the related regulatory enzymes mediated by Rb1 were principally involved in arginine and proline metabolism, glycine, serine and threonine metabolism, and carbon metabolism (Table S5). According to the above results, a metabolic network map of the differential metabolites in both urine and serum was constructed (Fig. 2F). Rb1 could ameliorate AMI injury via affecting the above metabolic pathways.
3.5. Rb1 significantly promoted mitophagy and upregulated AMPKα phosphorylation in AMI-injured mice
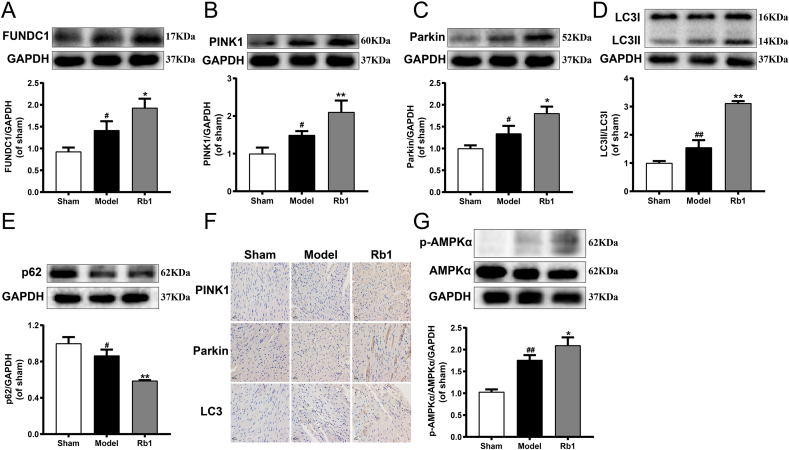
The AMI injury significantly enhanced the expression of FUNDC1, PINK1, Parkin and LC3Ⅱ/LC3Ⅰ, and diminished the expression of p62 compared with the sham group. Nevertheless, Rb1 treatment further induced up-regulation of FUNDC1, PINK1, Parkin and LC3Ⅱ/LC3Ⅰ, and down-regulation of p62 in AMI mice (Fig. 3A–E). Similarly, immunohistochemical analysis indicated that Rb1 significantly promoted the expression of PINK1, Parkin and LC3 (Fig. 3F). Moreover, we also found that the expression of p-AMPKα significantly increased after AMI injury, and Rb1 treatment further upregulated the phosphorylation of AMPKα (Fig. 3G). All of these demonstrated that Rb1 dramatically activated mitophagy in AMI mice, which could be associated with AMPKα phosphorylation.
Fig. 3.
Rb1 significantly promoted mitophagy and upregulated AMPKα phosphorylation in AMI-injured mice. The expression of (A) FUNDC1, (B) PINK1, (C) Parkin, (D) LC3Ⅱ/LC3Ⅰ and (E) p62 were detected using western blot analysis. (F) The expression of PINK1, Parkin and LC3 were detected using immunohistochemistry (400 × ). Scale bar = 50 μm. (G) The expression of p-AMPKα and AMPKα were detected using western blot analysis. All the experiments were performed in triplicate. Results were presented as mean ± SD. #P < 0.05, ##P < 0.01 vs. the sham group; ∗P < 0.05, ∗∗P < 0.01 vs. the model group.
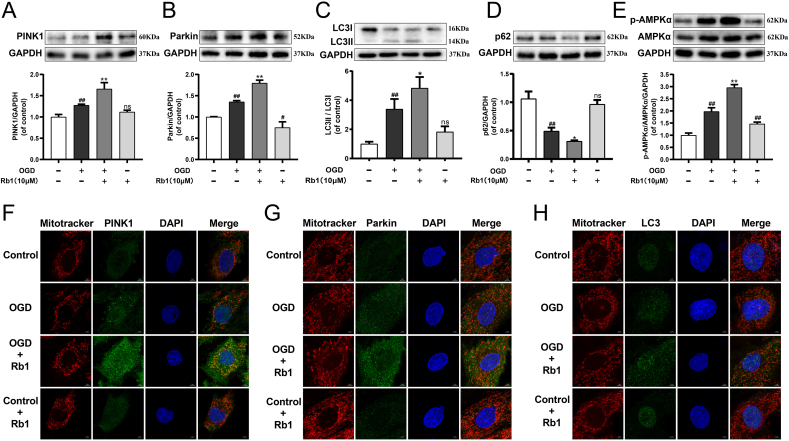
3.6. Rb1 profoundly promoted mitophagy and AMPKα phosphorylation in OGD-injured H9c2 cardiomyocytes
As exhibited in Fig. 4A, the level of p-AMPKα/AMPKα was significantly increased in the OGD group compared with the control group, and Rb1 further up-regulated p-AMPKα/AMPKα in OGD-injured H9c2 cardiomyocytes. Besides, the oxygen glucose deprivation (OGD) injury markedly increased the expression of PINK1, Parkin and LC3II/LC3I, and decreased the expression of p62, while these alterations continued to augment after treatment with Rb1 (Fig. 4B–E). Simultaneously, the OGD challenge noticeably enhanced the colocalization of the mitochondria with PINK1, Parkin and LC3, and Rb1 treatment further augmented the location of PINK1, Parkin and LC3 at mitochondria, while Rb1 had no effect on the H9c2 cardiomyocytes without OGD injury (Fig. 4F–H). The relevant statistical results of immunofluorescence analysis were shown in Fig. S7. Additionally, less fragmentation of mitochondria suggested that Rb1 partly attenuated the OGD-induced change in mitochondrial morphology, and might help maintain mitochondrial function. These results suggested that AMPKα phosphorylation and the levels of mitophagy were enhanced in OGD-injured H9c2 cardiomyocytes, and Rb1 treatment continued to promote mitophagy and AMPKα phosphorylation.
Fig. 4.
Rb1 profoundly promoted mitophagy and AMPKα phosphorylation in OGD-injured H9c2 cardiomyocytes. The expression of (A) PINK1, (B) Parkin, (C) LC3Ⅱ/LC3Ⅰ, (D) p62 and (E) p-AMPKα/AMPKα were detected using western blot analysis. Mitochondrial morphology and the colocalization of the mitochondrial with (F) PINK1, (G) Parkin and (H) LC3 were analyzed using a 63 × oil immersion lens (630 × ). Results were obtained from three independent experiments and were presented as mean ± SD. ##P < 0.01 vs. the control group; ∗P < 0.05, ∗∗P < 0.01 vs. the OGD group.
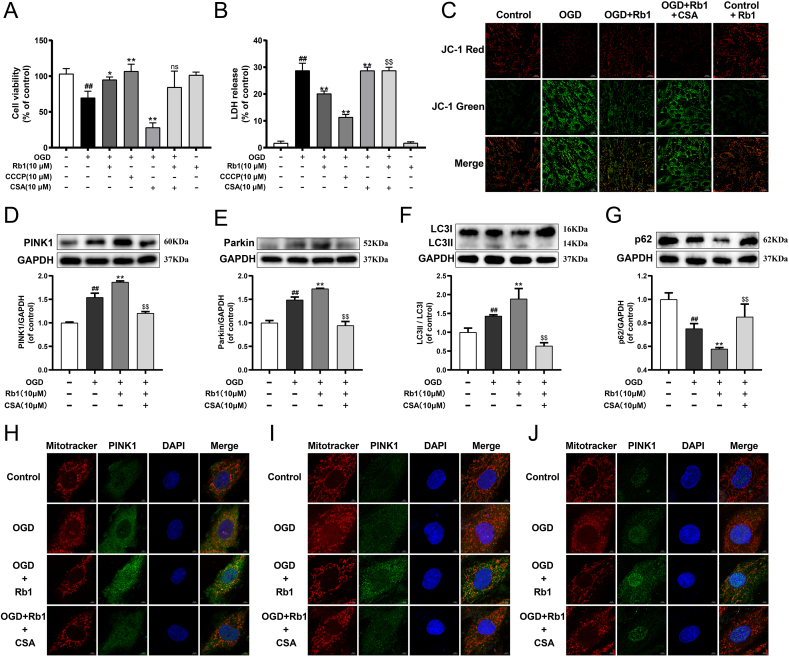
3.7. Rb1 attenuated OGD-induced H9c2 cardiomyocytes injury through mitophagy
Whether mitophagy exerts an essential role in the protection of Rb1 against myocardial ischemia injury was further verified. As illustrated in Fig. 5A and B, OGD injury led to a significant decrease in cell viability and increase in LDH release, and Rb1 markedly mitigated the change of these indicators. The mitophagy inhibitor cyclosporin A (CSA) reduced cell viability and enhanced LDH release in OGD-injured cardiomyocytes, while the mitophagy agonist carbonyl cyanide 3-chlorophenylhydrazone (CCCP) showed the dramatically opposite effects, which suggested that activating mitophagy effectively protected OGD-injured cardiomyocytes. Moreover, Rb1 treatment observably enhanced the mitochondrial membrane potential (ΔΨm) dissipation induced by OGD injury. While treatment with a combination of CSA and Rb1 destroyed the improvement effect of Rb1 on ΔΨm, suggesting that Rb1 modulated mitochondrial function via mitophagy (Fig. 5C). Besides, Rb1 treatment further induced the activation of mitophagy in OGD-injured H9c2 cardiomyocytes. But following treatment with a combination of CSA and Rb1, the expressions of parkin, PINK1 and LC3II/LC3I and the colocalization of the mitochondria with PINK1, Parkin and LC3 were markedly inhibited, and the expression of p62 was significantly increased (Fig. 5D–J). The relevant statistical results of immunofluorescence analysis were shown in Fig. S8. In addition, the CSA also noticeably reversed the improvement of Rb1 for damaged mitochondrial morphology. These results demonstrated that Rb1 protected OGD-injured H9c2 cardiomyocytes by promoting mitophagy.
Fig. 5.
Rb1 attenuated OGD-induced H9c2 cardiomyocytes injury through mitophagy. (A) The cell viability of H9c2 cardiomyocytes. (B) The release of LDH in culture medium of H9c2 cardiomyocytes. (C) Mitochondrial membrane potential (ΔΨm) was assessed by probe JC-1. Red fluorescence represents intact ΔΨm. Green fluorescence represents dissipation of ΔΨm. The expression of (D) PINK1, (E) Parkin, (F) LC3II/LC3I and (G) p62 were detected using western blot analysis. Mitochondrial morphology and the colocalization of the mitochondrial with (H) PINK1, (I) Parkin and (J) LC3 were analyzed using a 63 × oil immersion lens (630 × ). Results were obtained from three independent experiments and were presented as mean ± SD. ##P < 0.01 vs. the control group; ∗P < 0.05, ∗∗P < 0.01 vs. the OGD group; $$P < 0.01 vs. the OGD + Rb1 group.
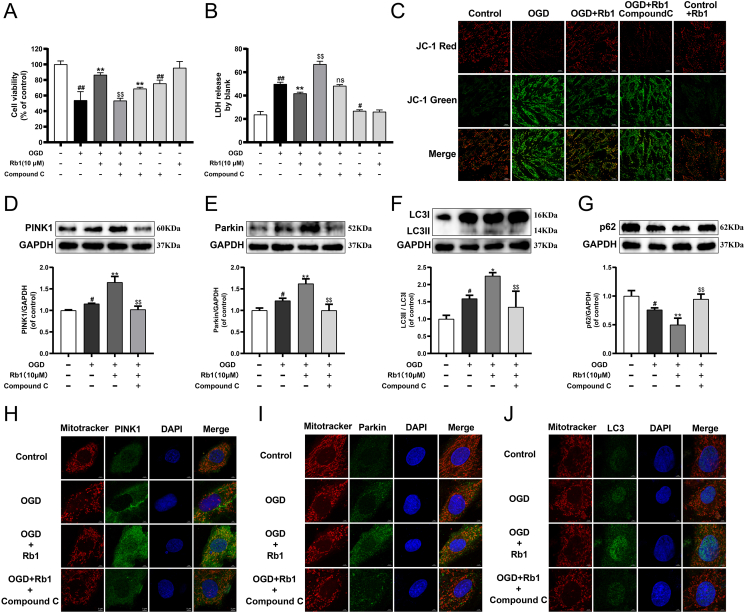
3.8. Rb1 effectively improved myocardial ischemia through AMPKα mediated mitophagy
Treatment with a combination of Compound C and Rb1 significantly diminished the expression of p-AMPKα compared with the Rb1-treated alone group (Fig. S9). Compound C observably reduced cell viability in OGD-injured cardiomyocytes, and partially attenuated the cardioprotective effect of Rb1 (Fig. 6A and B). Meanwhile, ΔΨm analysis revealed that Compound C dramatically attenuated the improvement of Rb1 for OGD-damaged mitochondrial function (Fig. 6C). Moreover, compared with the OGD + Rb1 group, the expression of PINK1, Parkin and LC3II/LC3I were markedly down-regulated, and the colocalization of the mitochondrial with PINK1, Parkin and LC3 were declined in the OGD + Rb1+Compound C group, while the p62 expression was higher (Fig. 6D–J). The relevant statistical results of immunofluorescence analysis were shown in Fig. S10. Besides, Compound C noticeably inhibited the protection of Rb1 for damaged mitochondrial morphology. These results indicated that AMPK mediated the mitophagy of Rb1 for the protection of ischemia injury.
Fig. 6.
Rb1 effectively improved myocardial ischemia through AMPKα mediated mitophagy. (A) The cell viability of H9c2 cardiomyocytes. (B) The release of LDH in culture medium of H9c2 cardiomyocytes. (C) ΔΨm was assessed by probe JC-1. The expression of (D) PINK1, (E) Parkin, (F) LC3II/LC3I and (G) p62 were detected using western blot analysis. Mitochondrial morphology and the colocalization of the mitochondrial with (H) PINK1, (I) Parkin and (J) LC3 were analyzed using a 63 × oil immersion lens (630 × ). Results were obtained from three independent experiments and were presented as mean ± SD. #P < 0.05, ##P < 0.01 vs. the control group; ∗P < 0.05, ∗∗P < 0.01 vs. the OGD group; $$P < 0.01 vs. the OGD + Rb1 group.
4. Discussion
There have been an increasing number of studies demonstrating the effectiveness of Rb1 in the treatment of cardiovascular diseases. Rb1 prevented oxidative stress-induced cardiomyocytes apoptosis through activating glutathione reductase, alleviated myocardial ischemia/reperfusion (MI/R) injury via down-regulated production of ROS from mitochondrial complex I, and improved diabetic cardiomyopathy through regulation of protein O-GlcNAcylation-mediated calcium signaling [[18], [19], [20]]. Additionally, as a potential cardioprotective candidate for clinical trials of myocardial infarction, Rb1 attenuated MI/R injury via antioxidantion, antiapotosis, anti-inflammation, promoting angiogenesis and improving circulation [21]. However, the potential mechanism of Rb1 in the treatment of MI is not yet clear and still needs further clarification, especially from a comprehensive metabolomics profiling perspective. Currently, metabolomics, a method for comprehensive analysis of small molecular substances in biological systems, is gradually conducted to elucidate disease mechanisms and potential new drug targets [14].
In the present study, we firstly confirmed that Rb1 could effectively protect acute myocardial ischemia (AMI)-injured mice and oxygen glucose deprivation (OGD)-injured cardiomyocytes. Then we revealed the therapeutic mechanism of Rb1 through exploring the differential metabolites in AMI-injured mice. The levels of hypoxanthine, uric acid, homocysteinesulfinic acid (HSCA), methionine sulfoxide and creatinine in the Rb1 group were lower than those in the model group. Whereas, the glutamine level in the Rb1 group was higher than that in the model group. Previous atudies have shown that hypoxanthine and uric acid could be used as potential metabolites associated with the further onset of CAD in T2DM patients [22]. And the HSCA level was enhanced among patients at risk for AMI or stroke [23,24]. Furthermore, the high levels of methionine sulfoxide and methionine induced changes in redox status and adenine nucleotide hydrolysis in platelets and serum of rats [25]. Meanwhile, in the general population, moderately elevated plasma creatinine was associated with the higher risk of myocardial infarction, ischemic heart disease, and death early [26]. Additionally, glutamine was applied to maintain cardiac function and promote glycogen metabolism via enhancing mitochondrial function [27]. All of these metabolites indicated a close association with the development of myocardial disease and suggested the therapeutic effects of Rb1 for AMI.
Subsequently, semi-quantification of five key metabolites were applied to excavate novel mechanism of Rb1 in the treatment of AMI. Rb1 treatment significantly augmented the urine levels of adenosine, taurine, beta-guanidinopropionic acid (β-GPA), L-isoleucine and myo-Inositol compared with model group. Adenosine exerts cardioprotection through regulation of energy intake and life activities of cardiomyocytes to reduce ATP requirement under hypoxic or ischemic stress conditions [28]. Moreover, taurine, a ubiquitous aminoethane sulfonic acid with high concentration in heart, indirectly regulated oxidative stress and improved energy metabolism, and its deficiency limited the degradation of damaged mitochondria via mitophagy [29,30]. Meanwhile, β-GPA, an analog of creatine, was reported to diminish phosphocreatine and ATP content in vivo [31]. Previous studies have verified that β-GPA-treated C2C12 muscle cells enhanced AMPK signal, the levels of autophagy-related proteins, and the density of autolysosomes, lysosomes, and autophagosomes [32]. L-isoleucine, an essential branched-chain amino acid, is finally metabolized into acetyl-CoA and succinyl-CoA, which are consumed in mitochondria through tri-carboxylic acid cycle [33]. Besides, myo-Inositol and its derivatives play many relevant biological functions, including modulation of glucose metabolism, calcium release in cell signaling gene transcription and proliferation. Treatment with myo-Inositol or other Inositol isomers has been proven to induce appreciable clinical results in different diseases such as cancer, respiratory distress syndrome and neurological disorders [34]. The changes in the content of five urine-based differential metabolites provided us with the new inspiration, whether Rb1 may protect ischemic myocardium by clearing damaged mitochondria via mitophagy and maintaining mitochondrial homeostasis.
Additionally, the related pathway of differential metabolites and regulatory enzymes were analyzed, and the main metabolic pathways of Rb1 included taurine and hypotaurine metabolism, glycine and serine metabolism, cysteine metabolism, methionine metabolism. The taurine and hypotaurine metabolism were the most concerned pathway which included differential metabolites such as L-cysteine, 3-sulfinoalanine and taurine. Taurine, generated through 3-sulfinoalanine and L-cysteine, is associated with mitophagy [30]. Glycine, the precursor of taurine, protected hypoxia-ischemic injured brain through regulating AMPK-mediated mitophagy [35] and protected H9c2 cardiomyocytes from hypoxia/reoxygenation injury by improving mitochondrial quality [36]. Moreover, methionine metabolism was involved in the pathogenesis of AMI and atherosclerosis [37]. Methionine restriction, dietary manipulation of methionine, could extend cellular lifespan by promoting the autophagic recycling of mitochondria [38]. All of these again suggested that the cardioprotection of Rb1 on ischemic myocardium might be associated with mitophagy.
Mitophagy, controlling mitochondrial quality, exerts an essential role in protecting myocardial ischemia injury [39]. Being mostly present in the cytosol under normal conditions, Parkin quickly translocates into mitochondria upon loss of mitochondrial membrane potential and promotes protein ubiquitination. The p62 protein, a junction protein, binds ubiquitinated proteins to LC3 [40], then LC3I and LC3II interact with p62 to tether damaged mitochondria to the autophagosome, which leads to mitophagy [41]. Previous studies have demonstrated that the deficiency of Parkin results in accumulating dysfunctional mitochondria and exacerbating cardiac injury following myocardial infarction [42]. Mice deficient for PINK1 are more prone to heart failure induced by pressure overload and ischemia/reperfusion injury [43,44]. FUNDC1, a mitophagy receptor, regulates mitochondrial homeostasis by enhancing the interaction between dephosphorylated FUNDC1 and LC3, and protects against I/R-induced myocardial injury [8]. In our present study, we found that Rb1 treatment increased the expression of PINK1, Parkin, LC3II/LC3I and FUNDC1, decreased the expression of p62, and enhanced the location of PINK1, Parkin and LC3 at mitochondria, which pointed to the involvement of PINK1/Parkin and FUNDC1 pathway in Rb1-induced elevation of mitophagy. Interestingly, the protection of Rb1 for OGD-injured cardiomyocytes was attenuated by mitophagy inhibitor. AMPK, a low ATP sensor that restores ATP homeostasis, regulates various aspects of mitochondrial biology and homeostasis through regulation of autophagy and mitophagy [45]. Some studies have demonstrated that the activation of AMPK signaling protects H9c2 cardiomyocytes from chronic hypoxia injury via enhancing mitophagy, and regulates autophagy to maintain the life of eukaryotic cells in nutrient scarcity situations [46,47]. In this study, we also found that Rb1 treatment further augmented the phosphorylation level of AMPKα in AMI-injured mice. In addition, Compound C diminished the level of mitophagy activated by Rb1 in OGD-injured cardiomyocytes, partially attenuated the cardioprotective effect of Rb1, and observably reversed the improvement of Rb1 for damaged mitochondrial morphology and mitochondrial function. All above results revealed that Rb1 protects acute ischemic myocardium through stimulation of AMPKα-mediated mitophagy.
This study also had some limitations. Although the above results have demonstrated that Rb1 might ameliorate AMI injury through enhancement of mitophagy via modulating the AMPKα signaling, further validation is required using AMPK-deficient mice. Additionally, the application of mitophagy inhibitors in AMI mice needs to be further used to verify the effect of mitophagy in Rb1 protection against AMI injury.
5. Conclusions
In summary, the present study firstly constructed a regulatory metabolic network map of Rb1 in the improvement of AMI. Importantly, a novel potential protection mechanism of Rb1, activating mitophagy, was excavated and verified. Rb1 exerts cardioprotective functions partly through activation of mitophagy via AMPKα pathway, which might provide some references for the further application of Rb1 in cardiovascular diseases.
Author contributions
BYY, JPK and FL designed study and revised manuscript; JGH, LZ1, FF, TL and LZ2 carried out experiments and wrote the manuscript; FL, JPK and QL advised experimental design and data interpretation; JGH, LZ1, and FF analyzed the data and interpreted the results. All authors read and approved the manuscript.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgments
This research work was supported by the National Science Foundation of China (No. 81973506, No. 81603328, No. 81774150, No. 81573719), Natural Science Foundation of Jiangsu Province (BK20160761). Project funded by China Postdoctoral Science Foundation (2016M600456, 2017T100425), and supported by Double First-Class University project (CPU2018GF06, CPU2018GF07).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2021.06.011.
Contributor Information
Junping Kou, Email: junpingkou@cpu.edu.cn.
Fang Li, Email: lifangcpu@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shimokawa H., Yasuda S. Myocardial ischemia: current concepts and future perspectives. J Cardiol. 2008;52:67–78. doi: 10.1016/j.jjcc.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nature Reviews. Molecular Cell Biology. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez-Resendiz S., Prunier F., Girao H., Dorn G., Hausenloy D.J. Targeting mitochondrial fusion and fission proteins for cardioprotection. J Cell Mol Med. 2020;24:6571–6585. doi: 10.1111/jcmm.15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chistiakov D.A., Shkurat T.P., Melnichenko A.A., Grechko A.V., Orekhov A.N. The role of mitochondrial dysfunction in cardiovascular disease: a brief review. Ann Med. 2018;50:121–127. doi: 10.1080/07853890.2017.1417631. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y., Tang Y., Lu J., Zhang W., Zhu Y., Zhang S., Ma G., Jiang P., Zhang W. PINK1-mediated mitophagy protects against hepatic ischemia/reperfusion injury by restraining NLRP3 inflammasome activation. Free Radic Biol Med. 2020;160:871–886. doi: 10.1016/j.freeradbiomed.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Lan R., Wu J.T., Wu T., Ma Y.Z., Wang B.Q., Zheng H.Z., Li Y.N., Wang Y., Gu C.Q., Zhang Y. Mitophagy is activated in brain damage induced by cerebral ischemia and reperfusion via the PINK1/Parkin/p62 signalling pathway. Brain Res Bull. 2018;142:63–77. doi: 10.1016/j.brainresbull.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 7.Tang C., Han H., Yan M., Zhu S., Liu J., Liu Z., He L., Tan J., Liu Y., Liu H., et al. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy. 2018;14:880–897. doi: 10.1080/15548627.2017.1405880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W., Siraj S., Zhang R., Chen Q. Mitophagy receptor FUNDC1 regulates mitochondrial homeostasis and protects the heart from I/R injury. Autophagy. 2017;13:1080–1081. doi: 10.1080/15548627.2017.1300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seabright A.P., Lai Y.-C. Regulatory roles of PINK1-Parkin and AMPK in ubiquitin-dependent skeletal muscle mitophagy. Frontiers in Physiology. 2020;11 doi: 10.3389/fphys.2020.608474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng M., Xin Y., Li Y., Xu F., Xi X., Guo H., Cui X., Cao H., Zhang X., Han C. Ginsenosides: a potential neuroprotective agent. Biomed Res Int. 2018 doi: 10.1155/2018/8174345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Q., Bao X.Y., Zhu P.C., Tong Q., Zheng G.Q., Wang Y. Ginsenoside Rb1 for myocardial ischemia/reperfusion injury: preclinical evidence and possible mechanisms. Oxid. Med. Cell. Longev. 2017 doi: 10.1155/2017/6313625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., Qian W., Zhao C. Analyzing the anti-ischemia-reperfusion injury effects of ginsenoside Rb1 mediated through the inhibition of p38α MAPK. Can. J. Physiol. Pharmacol. 2016;94:97–103. doi: 10.1139/cjpp-2014-0164. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y.C., Pan C.S., Yan L., Li L., Hu B.H., Chang X., Liu Y.Y., Fan J.Y., Sun K., Li Q., et al. Ginsenoside Rb1 protects against ischemia/reperfusion-induced myocardial injury via energy metabolism regulation mediated by RhoA signaling pathway. Sci. Rep. 2017;7:44579. doi: 10.1038/srep44579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newgard C.B. Metabolomics and Metabolic Diseases: where do we stand? Cell Metab. 2017;25:43–56. doi: 10.1016/j.cmet.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Wei T.T., Li Y., Li J., Fan Y., Huang F.Q., Cai Y.Y., Ma G., Liu J.F., Chen Q.Q., et al. Functional metabolomics characterizes a key role for N-acetylneuraminic acid in coronary artery diseases. Circulation. 2018;137:1374–1390. doi: 10.1161/CIRCULATIONAHA.117.031139. [DOI] [PubMed] [Google Scholar]
- 16.Lai Q., Yuan G.Y., Wang H., Liu Z.L., Kou J.P., Yu B.Y., Li F. Exploring the protective effects of schizandrol A in acute myocardial ischemia mice by comprehensive metabolomics profiling integrated with molecular mechanism studies. Acta Pharmacol. Sin. 2020;41:1058–1072. doi: 10.1038/s41401-020-0377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F., Fan X., Zhang Y., Pang L., Ma X., Song M., Kou J., Yu B. Cardioprotection by combination of three compounds from ShengMai preparations in mice with myocardial ischemia/reperfusion injury through AMPK activation-mediated mitochondrial fission. Sci. Rep. 2016;6:37114. doi: 10.1038/srep37114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan H.J., Tan Z.B., Wu Y.T., Feng X.R., Bi Y.M., Xie L.P., Zhang W.T., Ming Z., Liu B., Zhou Y.C. The role of ginsenoside Rb1, a potential natural glutathione reductase agonist, in preventing oxidative stress-induced apoptosis of H9C2 cells. J Ginseng Res. 2020;44:258–266. doi: 10.1016/j.jgr.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L., Yin X., Chen Y.H., Chen Y., Jiang W., Zheng H., Huang F.Q., Liu B., Zhou W., Qi L.W., et al. Proteomic analysis reveals ginsenoside Rb1 attenuates myocardial ischemia/reperfusion injury through inhibiting ROS production from mitochondrial complex I. Theranostics. 2021;11:1703–1720. doi: 10.7150/thno.43895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin L., Wang J., Zhao R., Zhang X., Mei Y. Ginsenoside-Rb1 improved diabetic cardiomyopathy through regulating calcium signaling by alleviating protein O-GlcNAcylation. J Agric Food Chem. 2019;67:14074–14085. doi: 10.1021/acs.jafc.9b05706. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q., Bao X.Y., Zhu P.C., Tong Q., Zheng G.Q., Wang Y. Ginsenoside Rb1 for myocardial ischemia/reperfusion injury: preclinical evidence and possible mechanisms. Oxid Med Cell Longev. 2017:6313625. doi: 10.1155/2017/6313625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omori K., Katakami N., Yamamoto Y., Ninomiya H., Takahara M., Matsuoka T.A., Bamba T., Fukusaki E., Shimomura I. Identification of metabolites associated with onset of CAD in diabetic patients using CE-MS analysis: a pilot study. J Atheroscler Thromb. 2019;26:233–245. doi: 10.5551/jat.42945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A., Choi Y., Back J.H., Lee S., Jee S.H., Park Y.H. High-resolution metabolomics study revealing l-homocysteine sulfinic acid, cysteic acid, and carnitine as novel biomarkers for high acute myocardial infarction risk. Metabolism. 2020;104:154051. doi: 10.1016/j.metabol.2019.154051. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y., Khan A., Hong S., Jee S.H., Park Y.H. A metabolomic study on high-risk stroke patients determines low levels of serum lysine metabolites: a retrospective cohort study. Mol Biosyst. 2017;13:1109–1120. doi: 10.1039/c6mb00732e. [DOI] [PubMed] [Google Scholar]
- 25.Soares M.S.P., da Silveira de Mattos B., Ávila A.A., Spohr L., Pedra N.S., Teixeira F.C., Bona N.P., Oliveira P.S., Stefanello F.M., Spanevello R.M. High levels of methionine and methionine sulfoxide: impact on adenine nucleotide hydrolysis and redox status in platelets and serum of young rats. J Cell Biochem. 2018;120:2289–2303. doi: 10.1002/jcb.27554. [DOI] [PubMed] [Google Scholar]
- 26.Sibilitz K.L., Benn M., Nordestgaard B.G. Creatinine, eGFR and association with myocardial infarction, ischemic heart disease and early death in the general population. Atherosclerosis. 2014;237:67–75. doi: 10.1016/j.atherosclerosis.2014.08.040. [DOI] [PubMed] [Google Scholar]
- 27.Salabei J.K., Lorkiewicz P.K., Holden C.R., Li Q., Hong K.U., Bolli R., Bhatnagar A., Hill B.G. Glutamine regulates cardiac progenitor cell metabolism and proliferation. Stem Cells. 2015;33:2613–2627. doi: 10.1002/stem.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borea P.A., Gessi S., Merighi S., Vincenzi F., Varani K. Pharmacology of adenosine receptors: the state of the art. Physiol Rev. 2018;98:1591–1625. doi: 10.1152/physrev.00049.2017. [DOI] [PubMed] [Google Scholar]
- 29.Schaffer S.W., Shimada-Takaura K., Jong C.J., Ito T., Takahashi K. Impaired energy metabolism of the taurine-deficient heart. Amino Acids. 2016;48:549–558. doi: 10.1007/s00726-015-2110-2. [DOI] [PubMed] [Google Scholar]
- 30.Jong C.J., Ito T., Schaffer S.W. The ubiquitin-proteasome system and autophagy are defective in the taurine-deficient heart. Amino Acids. 2015;47:2609–2622. doi: 10.1007/s00726-015-2053-7. [DOI] [PubMed] [Google Scholar]
- 31.Oudman I., Clark J.F., Brewster L.M. The effect of the creatine analogue beta-guanidinopropionic acid on energy metabolism: a systematic review. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crocker C.L., Baumgarner B.L., Kinsey S.T. β-guanidinopropionic acid and metformin differentially impact autophagy, mitochondria and cellular morphology in developing C2C12 muscle cells. J Muscle Res Cell Motil. 2020;41:221–237. doi: 10.1007/s10974-019-09568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha A.K., Xu X.J., Lawson E., Deoliveira R., Brandon A.E., Kraegen E.W., Ruderman N.B. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and insulin resistance in rat skeletal muscle. Diabetes. 2010;59:2426–2434. doi: 10.2337/db09-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bizzarri M., Fuso A., Dinicola S., Cucina A., Bevilacqua A. Pharmacodynamics and pharmacokinetics of Inositol(s) in health and disease. Expert Opin Drug Metab Toxicol. 2016;12:1181–1196. doi: 10.1080/17425255.2016.1206887. [DOI] [PubMed] [Google Scholar]
- 35.Cai C.C., Zhu J.H., Ye L.X., Dai Y.Y., Fang M.C., Hu Y.Y., Pan S.L., Chen S., Li P.J., Fu X.Q., et al. Glycine protects against hypoxic-ischemic brain injury by regulating mitochondria-mediated autophagy via the AMPK pathway. Oxid Med Cell Longev. 2019:4248529. doi: 10.1155/2019/4248529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Su W., Zhang Q., Xu J., Liu H., Luo J., Zhan L., Xia Z., Lei S. Glycine protects H9C2 cardiomyocytes from high glucose- and hypoxia/reoxygenation-induced injury via inhibiting PKCβ2 activation and improving mitochondrial quality. J Diabetes Res. 2018 doi: 10.1155/2018/9502895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhar I., Lysne V., Seifert R., Svingen G.F.T., Ueland P.M., Nygard O.K. Plasma methionine and risk of acute myocardial infarction: effect modification by established risk factors. Atherosclerosis. 2018;272:175–181. doi: 10.1016/j.atherosclerosis.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 38.Plummer J.D., Johnson J.E. Extension of cellular lifespan by methionine restriction involves alterations in central carbon metabolism and is mitophagy-dependent. Front Cell Dev Biol. 2019;7:301. doi: 10.3389/fcell.2019.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulek A.R., Anzell A., Wider J.M., Sanderson T.H., Przyklenk K. Mitochondrial quality control: role in cardiac models of lethal ischemia-reperfusion injury. Cells. 2020;9:214. doi: 10.3390/cells9010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubli D.A., Zhang X., Lee Y., Hanna R.A., Quinsay M.N., Nguyen C.K., Jimenez R., Petrosyan S., Murphy A.N., Gustafsson A.B. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y., Lee H.Y., Hanna R.A., Gustafsson Å.B. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:1924–1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Billia F., Hauck L., Konecny F., Rao V., Shen J., Mak T.W. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H., Liu B., Li T., Zhu Y., Luo G., Jiang Y., Tang F., Jian Z., Xiao Y. AMPK activation serves a critical role in mitochondria quality control via modulating mitophagy in the heart under chronic hypoxia. Int J Mol Med. 2018;41:69–76. doi: 10.3892/ijmm.2017.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamargo-Gómez I., Mariño G. AMPK: regulation of metabolic dynamics in the context of autophagy. Int J Mol Sci. 2018;19:3812. doi: 10.3390/ijms19123812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.