Abstract
Background
Skin microbiota is important for maintenance of skin homeostasis; however, its disturbance may cause an increase in pathogenic microorganisms. Therefore, we aimed to develop a red ginseng formulation that can selectively promote beneficial bacteria.
Methods
The effects of red ginseng formulation on microorganism growth were analyzed by comparing the growth rates of Staphylococcus aureus, S. epidermidis, and Cutibacterium acnes. Various preservatives mixed with red ginseng formulation were evaluated to determine the ideal composition for selective growth promotion of S. epidermidis. Red ginseng formulation with selected preservative was loaded into a biocompatible polymer mixture and applied to the faces of 20 female subjects in the clinical trial to observe changes in the skin microbiome.
Results
Red ginseng formulation promoted the growth of S. aureus and S. epidermidis compared to fructooligosaccharide. When 1,2-hexanediol was applied with red ginseng formulation, only S. epidermidis showed selective growth. The analysis of the release rates of ginsenoside-Rg1 and -Re revealed that the exact content of Pluronic F-127 was around 11%. The application of hydrogel resulted in a decrease in C. acnes in all subjects. In subjects with low levels of S. epidermidis, the distribution of S. epidermidis was significantly increased with the application of hydrogel formulation and total microbial species of subjects decreased by 50% during the clinical trial.
Conclusion
We confirmed that red ginseng formulation with 1,2-hexanediol can help maintain skin homeostasis through improvement of skin microbiome.
Keywords: Clinical study, Korea red ginseng, Panax ginseng, Skin microbiota
Graphical abstract
1. Introduction
There are various types of microorganisms on our skin that constitute the skin microbiome. The skin microbiome plays an important role in maintaining the health of the skin, similar to intestinal microbes [[1], [2], [3]]. The skin is the largest organ protecting the body by serving as the primary defense line to prevent the invasion of external pathogens. Microorganisms beneficial to the skin are known to be essential to skin immunity [4]. For example, S. epidermidis is known to produce glycerol, which creates an ideal environment for the growth of bacteria in the epithelium of the skin and to create a zone of inhibition against harmful microorganisms such as C. acnes [5]. S. epidermidis has also been reported to prevent skin diseases by inhibiting biofilm formation of S. aureus, which causes various skin infections [6].
Ginseng is a medicinal herb that has long been used as a blood-enriching and tonifying agent in Korea, China, and other parts of East Asia. In 1843, ginseng was first named Panax ginseng Meyer, meaning ‘cure-all’, by the Russian scientist, C.A. Meyer. Depending on the processing method used, ginseng can be broadly categorized into fresh, white, and red ginseng (RG). Fresh ginseng is a natural product that takes 4–6 years to mature and is sourced from the roots of the plant to preserve the unique ginseng compounds. However, because the water content is 70–80%, the product can decay and be damaged easily during distribution. To overcome this difficulty, specialized storage facilities or packaging was designed for long-term storage. White ginseng is produced by sun drying or hot-air drying of 4–6-year-old fresh ginseng either in its original state or after removing the outer layer; its water content is ≤ 14% and is milky-white or pale yellow in color. RG is produced by steam cooking and then drying fresh ginseng; its water content is ≤ 15.5% and is pale red or red-brown in color. In the process of steaming and drying the fresh ginseng to produce RG, changes in the types and concentrations of unique compounds found in ginseng, ginsenosides, occur [7]. Ginseng is known as a medicinal plant that has various biologically active effects on the human body with minimal side effects. Ginseng improves immunity [8,9], blood flow [10], and memory [11]. It also prevents skin damage from ultraviolet rays [12], and has antioxidant effects [13]. In particular, RG can enhance physiological activity to heal skin tissue wounds [14] or prevent skin damage caused by ultraviolet rays [15]. On the other hand, hydrophobic fractionation of RG results in its antibacterial effects of gram-positive bacteria growing in an anaerobic environment, including C. acnes or S. mutans. These effects have been found to be caused by polyacetylene compounds, such as panaxynol and panaxydol [16].
In this study, a RG formulation was designed and characterized to promote bacteria beneficial to the skin. Clinical trials were employed to confirm whether this formulation could cause changes in the skin microbiome. Further, genetic analysis was performed to determine the various microorganisms present on the subject skin and to confirm the overall changes in the skin after application of RG formulation during clinical trials.
2. Materials and methods
2.1. Sample preparation and isolation
Six-year-old fresh ginseng roots were prepared by steaming and drying to produce RG in the RG manufacturing plant of Korea Ginseng Corporation (Buyeo, Chungnam, Korea). Red ginseng water (RGW) extract (dry weight, 0.5 kg) was macerated and extracted twice using 2 L of distilled water (DW) at 70 °C for 8 h to prepare samples of RG extracts. The hydrophobic fraction of RG was extracted using 2 L of 70% ethanol at 25 °C for 8 h and the extract was fractionated separately with a Diaion® HP-20 column (Mitsubishi Chemical Corp., Tokyo, Japan). The ethanol extract of RG was loaded onto the column and washed out with DW. The hydrophobic fraction was then obtained by eluting with 100% ethanol. RGW extract contained ginsenosides Rb1 (6.97 mg/g), Rb2 (2.79 mg/g), Rc (3.06 mg/g), Rd (1.01 mg/g), Re (1.97 mg/g), Rf (1.40 mg/g), Rg1 (1.67 mg/g), Rg2s (1.23 mg/g), Rg3s (2.22 mg/g), and Rh1 (0.77 mg/g) and the hydrophobic fraction of RG contained Rb1 (0.84 mg/g), Rb2 (0.44 mg/g), Rc (0.45 mg/g), Rd (0.27 mg/g), Re (0.15 mg/g), Rf (0.09 mg/g), Rg1 (0.04 mg/g), Rg2s (0.12 mg/g), Rg3s (0.13 mg/g), and Rh1 (0.02 mg/g) as determined by high-performance liquid chromatography (HPLC). The RG formulation was obtained by mixing RGW extract and the hydrophobic fraction of RG according to their yield and then freeze-dried. Potassium sorbate, 1,2-hexanediol (1,2-HD), 1,2-octanediol, 2-phenoxyethanol, sodium benzoate, and sodium propionate were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Pluronic F-127 (molecular weight = 12,600 g/mol) was purchased from Sigma -Aldrich (St. Louis, MO, USA) and sodium hyaluronic acid was obtained from Hyundai Bioland (Osong, Chungbuk, South Korea).
2.2. HPLC analysis of samples
HPLC analysis of RG formulation with 1,2-HD and its ingredients, that is, RGW extract, hydrophobic fractions of RG and 1,2-HD was performed on a Waters 2695 system (Waters, Milford, MA, USA) equipped with a Waters 996 photodiode array detector, autosampler, and degasser. Each sample was separated on a Hypersil GOLD C18 column (250 × 4.6 mm, 5 μm; Thermo Fisher Scientific, OH, USA) using sequential mixtures of acetonitrile and H2O (20 %–90% aqueous acetonitrile). The injection volume was 20 μL, and the flow rate was 1.6 mL/min. The UV chromatogram of RGW extract, hydrophobic fraction of RG, 1,2-HD and RG formulation with 1,2-HD (203 nm) are shown in Fig. S1.
2.3. Microorganisms and culture
The standard strains of C. acnes, which were isolated from acne lesions on human facial skin, (KCTC 3314, Korean Collection for Type Cultures, Jeonbuk, South Korea) were cultured in reinforced clostridial agar medium (Difco Laboratories, Franklin Lakes, NJ, USA). S. aureus, which was isolated from a lesion on a human (NCTC 10788, The National Collection of Type Cultures, Salisbury, UK) and S. epidermidis, which was isolated from human nose (KCTC 3958), were cultured in tryptic soy agar medium (Difco Laboratories). C. acnes was cultured at 37 °C in an anaerobic atmosphere containing 10% (v/v) CO2, 10% (v/v) H2, and 80% (v/v) N2. Other strains were cultured in an aerobic atmosphere. The cultures of microorganisms were serially diluted 1:10, and 1 mL of each diluent was spread-plated. The plates were incubated and the dilution plate that grew 30–300 colonies was used to calculate the colony-forming units (CFU)/mL. The optical density at 600 nm (OD600) of each batch culture was measured and the standard curve of CFU and OD600 was calculated.
2.4. Evaluation of the effect of RG formulation on the growth rate of microorganisms
The RG formulation and fructooligosaccharide (FOS) were dissolved in minimal growth medium (0.25% beef extract (Difco Laboratories) in saline buffer) and diluted sequentially by half. Each microorganism was cultured in nutrient medium and inoculated in the minimum medium with RG formulation, FOS and cultured for 24 h (S. aureus, S. epidermidis) to 72 h (C. acnes). The OD600 of each culture medium was measured using a spectrophotometer (Thermo Fisher Scientific, Vantaa, Finland) and converted into CFU/mL.
2.5. Evaluation of the effects of RG formulation with preservatives on the growth of microorganisms
Each preservative was diluted in the following concentrations in the minimal nutrient medium with 1 mg/mL of RG formulation: potassium sorbate, sodium benzoate, and sodium propionate: 2, 1, 0.5, 0.25 mg/mL, 1,2-HD: 20, 10, 5, 2.5 μL/mL, 2-phenoxyethanol and 1,2-octanediol: 2, 1, 0.5, and 0.25 μL/mL. Three types of microorganisms were inoculated and cultured, and the growth rate was compared with that of the untreated group.
2.6. Preparation of pluronic F-127 hydrogel
For the preparation of the hydrogel containing ginseng, 100 mg of RG formulation and 60 mg of 1,2-HD were mixed with 13.5 mg of the aqueous solution containing sodium hyaluronic acid (0.67 wt%). Then, Pluronic F-127 aqueous solutions were added to form Ginseng Pluronic F-127 hydrogel composites. To find out the optimum composition, 900 mg of Pluronic F-127 aqueous solution with various concentrations (11, 14, 17, and 20 wt%) was used.
2.7. In vitro drug release characteristics
The in vitro drug release characteristics of the mixture were studied using diffusion cells divided into side-by-side chambers with a Spectrum Spectra/Por Regenerated Cellulose Membrane (molecular weight cutoff: 500,000; Fisher Scientific, Rancho Dominquez, CA). To measure the release pattern of RG, 4 mL of the Pluronic F-127 mixture was placed into the donor compartment and 4 mL of DW was added to the receptor cell, a sampling port. To maintain homogeneity of temperature, the experimental setup was placed in an incubator maintained at 37 °C, and both media were stirred with cylindrical magnetic stirring bars. At predetermined time intervals, 2 mL aliquots of the release medium (DW) were withdrawn via a side port of the diffusion cell, and the total receiver solution was replaced with 4 mL of fresh DW at every time point to maintain the sink conditions. To quantify the RG released from the different formulations, representative indicator ingredients of RG, ginsenoside Rg1 and Re, were selected. Analysis of ginsenoside was conducted by HPLC using a Hypersil GOLD C18 column under the same conditions as described above.
2.8. Clinical trials
Clinical trials were conducted after IRB approval (KDRI-IRB-20487) by the Korea Dermatology Research Institute Ethics Committee. The twenty female subjects were tested (age range, 23–54 y; average, 43.8 y). Subjects washed their faces with only water 12 h before their visit, and stayed at constant temperature and humidity (20–24 °C, 40–60% room humidity) for 30 min. A sterilized swab stick was wetted with saline buffer and the bacterial samples were collected from the entire facial area. The Pluronic F-127 hydrogel containing RG formulation with 1,2-HD (test group) and saline buffer with RG formulation with 1,2-HD (control group) was applied twice a day for two weeks each on one side of the face of 20 subjects. The bacterial samples were collected from the subjects in the same manner as in the first sampling, and microbial cluster analysis was conducted. Total skin flora in subject with low S. epidermidis ratio (A) and high S. epidermidis ratio (B) is shown in Tables S2–4.
2.9. DNA extraction, PCR amplification, and illumina sequencing
Total DNA was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer's instructions. PCR amplification (Applied Biosystem ® 2720 Thermal Cycler, Thermo Fisher Scientific, OH, USA) was performed using primers targeting the V3 to V4 regions of the 16S rRNA gene in the extracted DNA. For bacterial amplification, primers of 341F (5′-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3’; underlined sequence indicates the target region primer) and 805R (5′-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGACAG-GACTACHVGGGTATCTAATCC-3′) were used. The amplification was carried out under the following conditions: initial denaturation at 95 °C for 3 min, followed by 25 cycles of denaturation at 95 °C for 30 s, primer annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, with a final elongation at 72 °C for 5 min. Then, secondary amplification for attaching the Illumina Nextera barcode was performed using the i5 forward primer (5′-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXXX-TCGTCGGCAGCGTC-3'; X indicates the barcode region) and i7 reverse primer (5′-CAAGCAGAAGACGGCATACGAGAT-XXXXXXXX-GTCTCGTGGGCTCGG-3′). The conditions of the secondary amplification were the same as before except the amplification cycle was set to 8. The PCR products were confirmed using 1% agarose gel electrophoresis and visualized using a Gel Doc XR + system (Bio-Rad, Hercules, CA, USA). The amplified products were purified using CleanPCR (CleanNA, Waddinxveen, Netherlands). Equal concentrations of purified products were pooled together and short fragments (non-targeted products) were removed using CleanPCR again (CleanNA). The quality and product size were assessed on 2100 Bioanalyzer Instrument (Agilent, Palo Alto, CA, USA) using a DNA 7500 chip. Mixed amplicons were pooled, and the sequencing was carried out at Chunlab Inc. (Seoul, Korea) using the MiSeq Sequencing system (Illumina, USA) according to the manufacturer's instructions.
2.10. MiSeq pipeline
The raw reads were processed by performing quality check and low-quality reads (<Q25) were filtered using Trimmomatic version 0.32 [17]. After quality control pass, paired-end sequence data were merged using VSEARCH [18]. Primers were then trimmed with ChunLab's in-house program at a similarity cutoff of 0.8. Non-specific amplicons that did not encode 16S rRNA were detected using HMMER hmmsearch program [19] with 16S rRNA profiles. Sequences were denoised using DUDE-Seq [20], and non-redundant reads were extracted using UCLUST clustering [21]. The EzBioCloud 16S rRNA database was used for taxonomic assignment using VSEARCH followed by more precise pairwise alignment [22], UCHIME [23], and the non-chimeric 16S rRNA database from EzBioCloud was used to detect chimeric reads with <97% similarity. Reads that were not identified at the species level (with <97% similarity) in the EzBioCloud database were compiled and clustered de novo using UCLUST5 to generate additional operational taxonomic units. Finally, operational taxonomic units with single reads (singletons) were omitted from further analysis. The alpha diversity indices [[24], [25], [26], [27], [28], [29]], rarefaction curves, and rank abundance curves were estimated using in-house codes [30].
2.11. Statistical analysis
Wilcoxon signed ranks test was used for paired data to compare the change in microbiome distribution between before and after the test. p ≤ 0.05 was considered statistically significant. Appropriate statistical analysis was performed using SPSS software.
3. Results and discussion
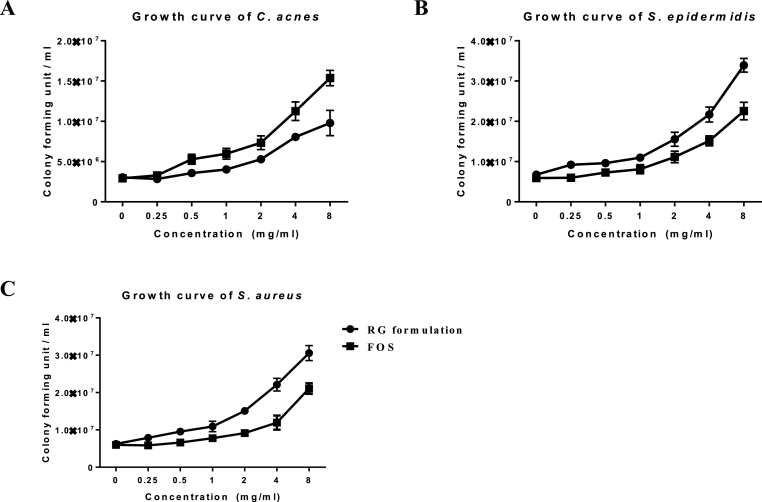
The effects of RG formulation, including RGW and hydrophobic fraction, on the growth of C. acnes, S. epidermidis, and S. aureus, which are the major epithelial bacteria of the skin, were verified against the typical prebiotic component, FOS. RGW extract was treated at concentrations of 0.258 mg/mL. We found that RGW extract promoted the growth of the three species of microorganisms (Data not shown). The RGW extract was divided into 5 fractions with HP-20 resin and the effect of each fraction on the growth of S. epidermidis was tested (Fig. S2). The highly polar HP-20 DW, 30% EtOH fraction improved the growth of S. epidermidis, and the contents and the effects of individual components on the growth of S. epidermidis can be seen in Table S1 and Fig. S3. However, it over-increased the growth of C. acnes, which RGW has the largest population among the skin microbiomes. To solve this problem, the hydrophobic fraction of RG, a growth inhibitor of C. acnes, that was identified in a previous research [16] was also used for treatment. As a result, the RG formulation was found to promote the growth of S. epidermidis without causing C. acnes to proliferate when compared to that by FOS (Fig. 1).
Fig. 1.
The effects of RG formulation and FOS on the growth of C. acnes (A), S. epidermidis (B) and S. aureus (C). Results are presented as means ± SD (n = 3).
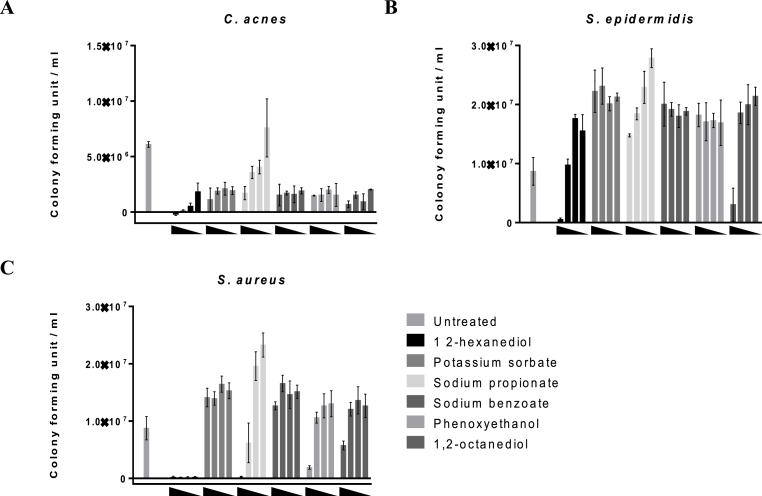
The growth promotion of S. epidermidis and inhibition of C. acnes by RG formulation was desirable, but the growth of S. aureus belonging to the same genus as S. epidermidis, unlike C. acnes, was also promoted by the RG formulation. To solve this problem, we explored the optimal composition for specific antibacterial effect on S. aureus. Six types of preservatives (1,2-HD, potassium sorbate, sodium propionate, sodium benzoate, 2-phenoxyethanol, and 1,2-octanediol) were selected and applied simultaneously to various microorganisms. The concentration of preservatives was selected based on the guidelines from the Ministry of Food and Drug Safety of the Government of Korea. Treatment at less than 10 μL/mL of 1,2-HD with RG formulation inhibited the growth of S. aureus and C. acnes while maintaining the growth promotion effect of S. epidermidis (Fig. 2).
Fig. 2.
The effects of RG formulation with preservative on the growth of C. acnes (A), S. epidermidis (B) and S. aureus (C). Untreated means that only RG formulation (1 mg/mL) was treated. Each bar concentration of each preservative was as follows. Potassium sorbate, sodium benzoate, and sodium propionate: 2, 1, 0.5, 0.25 mg/mL, 1,2-HD: 20, 10, 5, 2.5 μL/mL, 2-phenoxyethanol and 1,2-octanediol: 2, 1, 0.5, and 0.25 μL/mL. Results are presented as means ± SD (n = 3).
Upon treatment with RG formulation (1 mg/mL) alone, the effect on microbial growth was limited compared to that in the untreated group. In addition, when treating with 1,2-HD alone, it was difficult to observe significant differences in microbial growth at concentrations below 10 μL/mL, but all microorganisms were inhibited at concentrations of 20 μL/mL (Fig. S4). These results showed that treatment with 1,2-HD or RG formulation alone cannot be expected to have a specific antimicrobial or growth-promoting effect on certain strains. On the other hand, treatment with both RG and 1,2-HD can selectively promote beneficial bacteria.
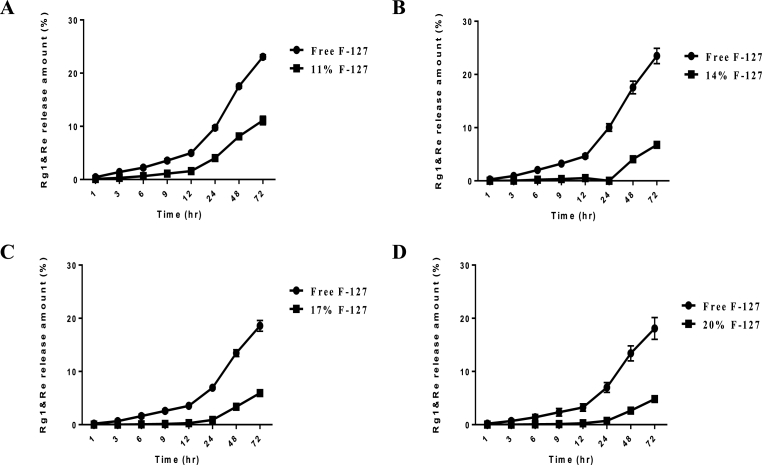
In the case of the skin, especially the face, not only was the formulation exposed to the external environment but could also be washed away by the discharge of sweat and sebum. As a complement to this problem, a hydrogel composed of Pluronic F-127 and hyaluronic acid was selected as the delivery vehicle for RG formulation [31]. Both components are highly biocompatible and controlled release of RG formulation with preservatives is expected rheological behaviors of Pluronic F-127 [[31], [32], [33]]. In this study, the concentration of Pluronic F-127 was adjusted from 11% to 20% to prepare the formulation, and the ginsenoside Rg1 and Re levels were measured to compare the release rate (Fig. 3). Around 11% of Pluronic F-127 was optimal for the hydrogel composition. The hydrogel was not formed properly when less than 11% of Pluronic F-127 was applied (data not shown).
Fig. 3.
Release profile of ginsenosides Rg1 and Re from hydrogels as a function of Pluronic F-127 concentration. Free means RG formulation without hydrogel. (A) 11% Pluronic F-127. (B) 14% Pluronic F-127. (C) 17% Pluronic F-127. (D). 20% Pluronic F-127.
The subjects were divided into two groups according to the quantity of S. epidermidis. The group with low S. epidermidis ratio had a minimum of 0.68%, maximum of 8.8%, and average of 3.22%. On the other hand, groups with high S. epidermidis ratio had a minimum of 15.91% up to 50.84% and an average of 29.22%. There was no significant deviation in the counts of C. acnes and S. aureus and ages between the groups (Table 1).
Table 1.
Age and microbial proportions of subjects in two groups divided by the ratio of S. epidermidis. (A) Group with a low S. epidermidis ratio. (B) Group with a high S. epidermidis ratio.
| A | ||||
|---|---|---|---|---|
| Group with a low S. epidermidis ratio | ||||
| Ages | C. acnes (%) | S. epidermidis (%) | S. aureus (%) | |
| 1 | 23 | 89.67 | 1.51 | 7.28 |
| 2 | 33 | 76.96 | 1.75 | 0.09 |
| 3 | 39 | 73.23 | 6.91 | 0.01 |
| 4 | 46 | 91.91 | 1.13 | 0.00 |
| 5 | 47 | 58.07 | 8.80 | 1.64 |
| 6 | 47 | 84.00 | 0.68 | 0.07 |
| 7 | 48 | 79.14 | 1.71 | 0.00 |
| 8 | 49 | 79.28 | 1.98 | 0.06 |
| 9 | 52 | 93.47 | 1.98 | 0.00 |
| 10 | 54 | 64.58 | 5.71 | 0.05 |
| Average | 43.8 | 79.03 | 3.22 | 0.92 |
| B | ||||
|---|---|---|---|---|
| Group with a high S. epidermidis ratio | ||||
| Ages | C. acnes (%) | S. epidermidis (%) | S. aureus (%) | |
| 11 | 28 | 73.43 | 21.49 | 0.59 |
| 12 | 32 | 25.62 | 30.96 | 0.02 |
| 13 | 44 | 74.86 | 20.27 | 0.01 |
| 14 | 45 | 69.22 | 21.25 | 1.04 |
| 15 | 46 | 32.47 | 47.58 | 0.00 |
| 16 | 47 | 47.06 | 50.84 | 0.03 |
| 17 | 47 | 64.53 | 26.20 | 0.00 |
| 18 | 47 | 59.31 | 38.32 | 0.01 |
| 19 | 48 | 76.51 | 15.91 | 0.25 |
| 20 | 52 | 80.76 | 16.27 | 0.00 |
| Average | 43.6 | 60.38 | 28.91 | 0.20 |
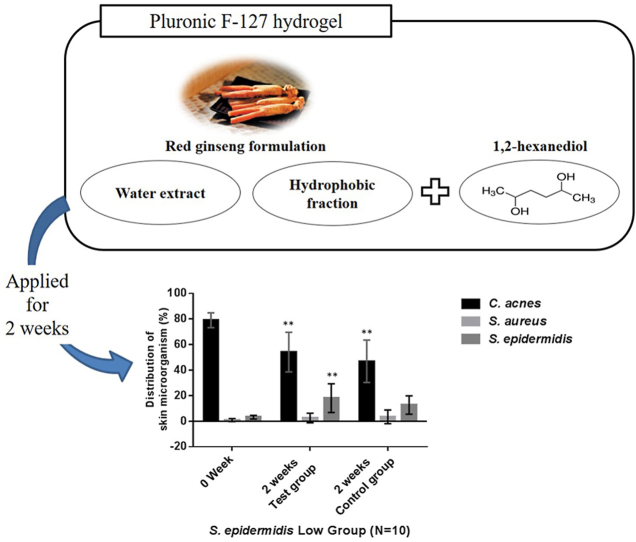
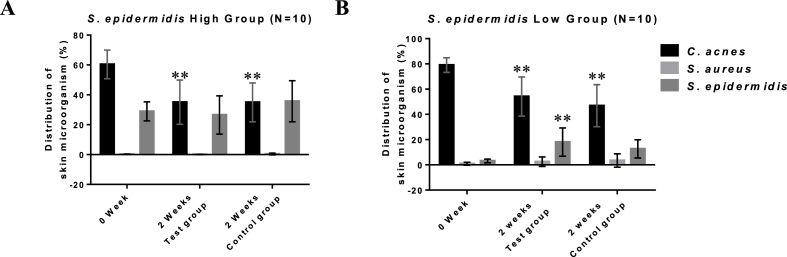
The ratio of C. acnes in most subjects decreased, regardless of hydrogel application (Fig. 4). On the other hand, S. epidermidis increased significantly only in groups with hydrogel application. These results demonstrate that growth-promoting effect is related to exposure to the active ingredient for a long period, rather than to the antibacterial effect, indicating that the application of the appropriate delivery system is important. Moreover, S. epidermidis and C. acnes compete with each other for growth because of their common use of carbon energy sources such as glycerol [35,36]. However, the proportion of C. acnes decreased in control groups that did not show a significant increase in S. epidermidis, which may have been caused by the components of the polyacetylene family inhibiting the growth of C. acnes [16].
Fig. 4.
Changes in three major skin microbes during the clinical trial in the group with a low S. epidermidis ratio (A) and a high S. epidermidis ratio (B). Results are presented as means ± SD (n = 10). ∗∗ Significantly different from week 0 (p < 0.05).
In addition, the growth-promoting effect of the RG formulation with 1,2-HD was not significant in the group with high S. epidermidis, whereas growth enhancement was confirmed in the group with low S. epidermidis. These results indicate that the RG formulation with 1,2-HD increased enhanced cell growth up to a certain limit in case of low S. epidermidis. The effect on S. aureus was difficult to measure because its retention ratio in the control subjects versus the test subjects was small.
Prior to clinical treatment, 490 species of bacteria were detected on the skin on average in the subjects, but after treatment, the number of species detected decreased significantly in all groups (Table 2A). Further research is required to observe if these results are beneficial or not. However, when skin and scalp microbiomes were studied with age changes, species richness was significantly lower in younger groups (21–37) than in older groups (60–76). Based on a prior study, it is highly likely that the decrease in the diversity of species plays a positive role in skin health [34].
Table 2.
Total microbial species in skin flora of subjects (A). Distribution of major skin flora in subjects with low S. epidermidis counts (B) and high S. epidermidis counts (C).
| A | |||
|---|---|---|---|
| 0 week | 2 weeks Test group | 2 weeks Control group | |
| Group with a low S. epidermidis | 481 | 210 | 178 |
| Group with a high S. epidermidis | 499 | 254 | 201 |
| Average | 490 | 232 | 189.5 |
| B | |||
|---|---|---|---|
| Group with a low S. epidermidis ratio | |||
| Taxon Name | 0 week | 2 weeks Test group | 2 weeks Control group |
| Cutibacterium acnes | 79.0% | 54.2% | 46.9% |
| Staphylococcus epidermidis | 3.1% | 18.1% | 12.7% |
| Klebsiella aerogenes | 0.1% | 17.3% | 20.9% |
| FM873692_s | 8.7% | 3.9% | 4.5% |
| Acinetobacter pittii | 0.0% | 0.5% | 6.7% |
| AY225604_s | 1.9% | 0.4% | 0.3% |
| Leclercia adecarboxylata | 0.0% | 0.0% | 1.7% |
| Staphylococcus aureus | 0.9% | 2.5% | 3.4% |
| Streptococcus salivarius | 1.4% | 0.0% | 0.0% |
| Total | 95.3% | 96.8% | 97.0% |
| C | |||
|---|---|---|---|
| Group with a high S. epidermidis ratio | |||
| Taxon Name | 0 week | 2 weeks Test group | 2 weeks Control group |
| Cutibacterium acnes | 60.4% | 35.1% | 35.0% |
| Staphylococcus epidermidis | 28.9% | 26.5% | 35.7% |
| Klebsiella aerogenes | 0.0% | 16.7% | 11.1% |
| Serratia marcescens | 0.0% | 6.1% | 7.2% |
| Cutibacterium granulosum | 4.1% | 5.4% | 2.9% |
| AY225604_s | 0.6% | 0.2% | 0.4% |
| Pantoea eucrina | 0.0% | 5.0% | 0.6% |
| Streptococcus salivarius | 0.3% | 0.5% | 0.3% |
| Staphylococcus hominis | 0.3% | 0.1% | 0.2% |
| Total | 94.6% | 95.6% | 93.3% |
In addition, it was observed that unlike intestinal bacteria, the bacteria on the skin were not very diverse as the top ten species accounted for more than 90% of the total skin microbiota (Table 2B) [4]. Further, we identified a variety of skin microorganisms in addition to C. acnes, S. epidermidis, and S. aureus, which constitute the skin microbiome. In particular, Klebsiella aerogenes, which is known to cause nosocomial infections [37], was found to be significantly increased by the RG formulation with 1,2-HD treatment (Table 2B–C). Further investigation is required to determine whether K. aerogenes is beneficial or not, because S. epidermidis is also known as an infectious agent in the hospital but plays a beneficial role on the skin [38].
4. Conclusion
We confirmed that RG formulation with 1,2-HD that can selectively promote beneficial bacteria, S. epidermidis. And we also demonstrated that 11% of Pluronic F-127 hydrogel could be an appropriate delivery system for RG formulation via extending the exposure time to the active ingredient. After treatment with RG formulation together with 1,2-HD, the number of species of skin microbiota significantly decreased and some microorganisms, including K. aerogenes, statistically increased. Further research needs to be carried out to clarify the effects of these results on the human skin.
Declaration of competing interest
None declared.
Acknowledgement
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2019R1A6A1A03031807).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2021.12.002.
Contributor Information
Soon Hong Yuk, Email: shyuk@korea.ac.kr.
Ki Yong Lee, Email: kylee11@korea.ac.kr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Scharschmidt T.C., Fischbach M.A. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome? Drug Discov Today Dis Mech. 2013;10(3–4):e83–e89. doi: 10.1016/j.ddmec.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y., Segre J.A. Dialogue between skin microbiota and immunity. Science. 2014;346(6212):954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 3.Grice E.A. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res. 2015;25(10):1514–1520. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 5.Schemann L.G., Knox G., Sher D., Rothman S. The role of bacteria in the formation of free fatty acids on the human skin surface. J Invest Dermatol. 1960;34:171–174. [PubMed] [Google Scholar]
- 6.Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., Agata T., Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465(7296):346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 7.Hyun S.H., Kim S.W., Seo H.W., Youn S.H., Kyung J.S., Lee Y.Y., In G., Park C.K., Han C.K. Physiological and pharmacological features of the non-saponin components in Korean Red Ginseng. J Ginseng Res. 2020;44(4):527–537. doi: 10.1016/j.jgr.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohn E.H., Kim J.H., Choi H.S., Park H.J., Kim B.O., Rhee D.K., Pyo S.N. Immunomodulatory effects of non-saponin red ginseng components on innate immune cells. J Ginseng Res. 2008;32(1):67–72. [Google Scholar]
- 9.Youn S.H., Lee S.M., Han C.K., In G., Park C.K., Hyun S.H. Immune activity of polysaccharide fractions isolated from Korean red ginseng. Molecules. 2020;25(16):3569. doi: 10.3390/molecules25163569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W.M., Kamruzzaman S., Song Y.B., Cho J.Y., Park H.J., Rhee M.H. Inhibitory activities of red ginseng acidic polysaccharide in platelet aggregation. J Ginseng Res. 2008;32(1):73–78. [Google Scholar]
- 11.Zhang G., Liu A., Zhou Y., San X., Jin T., Jin Y. Panax ginseng ginsenoside-Rg2 protects memory impairment via anti-apoptosis in a rat model with vascular dementia. J Ethnopharmacol. 2008;115(3):441–448. doi: 10.1016/j.jep.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Lee H.J., Kim J.S., Song M.S., Seo H.S., Moon C., Kim J.C., Kim S.H. Photoprotective effect of red ginseng against ultraviolet radiation-induced chronic skin damage in the hairlessmouse. Phytother Res. 2009;23(3):399–403. doi: 10.1002/ptr.2640. [DOI] [PubMed] [Google Scholar]
- 13.Kang K.S., Yokozawa T., Yamabe N., Kim H.Y., Park J.H. ESR study on the structure and hydroxyl radical-scavenging activity relationships of ginsenosides isolated from Panax ginseng C. A. Meyer. Biol Pharm Bull. 2007;30(5):917–921. doi: 10.1248/bpb.30.917. [DOI] [PubMed] [Google Scholar]
- 14.Kimura Y., Sumiyoshi M., Kawahira K., Sakanaka M. Effects of ginseng saponins isolated from Red Ginseng roots on burn wound healing in mice. Br J Pharmacol. 2006;148(6):860–870. doi: 10.1038/sj.bjp.0706794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M.J., Won C.H., Lee S.R., Kim J.S., Oh I.G., Hwang E., Kim N., Kang B.C., Chung J.H. Oral administration of KTNG0345 prepared from red ginseng extracts reduces UVB-induced skin wrinkle formation in hairless mice. J Ginseng Res. 2008;32(1):48–56. [Google Scholar]
- 16.Hou J.H., Shin H.J., Jang K.H., Park C.K., Koo B.S., Shin H., Yuk S.H., Lee K.Y. Anti-acne properties of hydrophobic fraction of red ginseng (Panax ginseng C.A. Meyer) and its active components. Phytother Res. 2019;33(3):584–590. doi: 10.1002/ptr.6243. [DOI] [PubMed] [Google Scholar]
- 17.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy S.R. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B., Moon T., Yoon S., Weissman T. DUDE-Seq: fast, flexible, and robust denoising for targeted amplicon sequencing. PLoS One. 2017;12(7) doi: 10.1371/journal.pone.0181463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers E.W., Miller W. Optimal alignments in linear space. Computer applications in the biosciences. CABIOS. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao A., Lee S.M. Estimating the number of classes via sample coverage. J Am Stat Assoc. 1992;87(417):210–217. [Google Scholar]
- 24.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987:783–791. [PubMed] [Google Scholar]
- 25.Burnham K.P., Overton W.S. Robust estimation of population size when capture probabilities vary among animals. Ecology. 1979;60:927–936. [Google Scholar]
- 26.Magurran A.E. John Wiley & Sons; 2013. Measuring biological diversity. [Google Scholar]
- 27.Chao A., Shen T.J. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ Ecol Stat. 2003;10(4):429–443. [Google Scholar]
- 28.Faith D.P. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 29.Heck K.L., Belle G., Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56(6):1459–1461. [Google Scholar]
- 30.Whittaker R.H. Dominance and diversity in land plant communities. Science. 1965;147(3655):250–260. doi: 10.1126/science.147.3655.250. [DOI] [PubMed] [Google Scholar]
- 31.Oh K.S., Kim J.Y., Yoon B.D., Lee M., Kim H., Kim M., Seo J.H., Yuk S.H. Sol-gel transition of nanoparticles/polymer mixtures for sustained delivery of exenatide to treat type 2 diabetes mellitus. Eur J Pharm Biopharm. 2014 Nov;88(3):664–669. doi: 10.1016/j.ejpb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Diniz I.M.A., Chen C., Xu X., Ansari S., Zadeh H.H., Marques M.M., Shi S., Moshaverinia A. Pluronic F-127 hydrogel as a promising scaffold for encapsulation of dental-derived mesenchymal stem cells. J Mater Sci Mater Med. 2015;26(3):153. doi: 10.1007/s10856-015-5493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibagaki N., Suda W., Clavaud C., Bastien P., Takayasu L., Iioka E., Kurokawa R., Yamashita N., Hattori Y., Shindo C., Breton L., Hattori M. Aging related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci Rep. 2017;7(1):10567. doi: 10.1038/s41598-017-10834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbruzzese F., Basoli F., Costantini M., Giannitelli S.M., Gori M., Mozetic P., Rainer A., Trombetta M. Hyaluronan: an overview. J Biol Regul Homeost Agents. 2017;31:9–22. [PubMed] [Google Scholar]
- 35.Shu M., Wang Y., Yu J., Kuo S., Coda A., Jiang Y., Gallo R.L., Huang C.M. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0055380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivakanesan R., Dawes E.A. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. J Gen Microbiol. 1980;118(1):143–157. doi: 10.1099/00221287-118-1-143. [DOI] [PubMed] [Google Scholar]
- 37.Davin-Regli A., Lavigne J.P., Pagès J.M. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32(4):e00002–e00019. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otto M. Staphylococcus epidermidis – the “accidental” pathogen. Nat Rev Microbiol. 2009;7(8):555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.