Abstract
To investigate the antimicrobial resistance and molecular characterization of gene cassettes from class 1 integrons in Escherichia coli strains isolated from hospitalized patients. Bacterial identification was conducted using the Vitek-2 Compact system, and antimicrobial susceptibility analysis was performed using the Kirby-Bauer method. Class 1 integrons, integrase genes, the variable regions of integrons and promoters from the isolated E. coli were screened by polymerase chain reaction, and subjected to DNA sequencing. In total, 138 E. coli strains were collected from the hospitalized patients, most from urine specimens (41.30%, 57/138). Antimicrobial resistance to ampicillin (89.86%) was most prevalent, with 79.99% of strains being multidrug-resistant (MDR). The class 1 integron integrase intI1 gene was detected in 67.39% of the isolates (93/138). Three gene cassette arrays and 5 antimicrobial resistance gene cassettes were detected in 69 of the class 1 integron-positive strains. The most common gene cassette array was dfrA17-aadA5. Of the 93 intI1-positive strains, 5 different common promoters were detected. The most prevalent common promoter was PcH1, and most isolates contained the dfrA17-aadA5 gene cassette array. In summary, antimicrobial resistance and MDR were prevalent among E. coli isolates in our city Weifang in Shandong Provence China. Gene cassettes of the class 1 integron variable region mostly conferred resistance to traditional antimicrobials. Weak promoters in the variable regions were predominant in this study. Integrons pose a great threat to the treatment of MDR bacterial infections and further investigations are needed.
Keywords: class 1 integrons, Escherichia coli, antimicrobial resistance, antimicrobial-resistance mechanism
Introduction
Escherichia coli, a conditional pathogen, is one of the most common and important pathogens in medical care settings. It is the most prominent cause of enteritis, urinary tract infections, septicemia, and various other clinical infections, including neonatal meningitis. The therapeutic treatment of E. coli infections is threatened by antimicrobial resistance, and the widespread and unreasonable use of antimicrobials has led to antimicrobial-resistant bacteria, a major and ever increasing global health care problem. The resistance rate range for the multiple antimicrobial agents used to treat urinary tract infections is 15.75–80.98% for E. coli.1,2
The World Health Organization's global antimicrobial resistance monitoring report,3 released in 2014, pointed out that E. coli has acquired high-level resistance to third-generation cephalosporins and fluoroquinolones in 114 countries. In China, E. coli resistance rates for ceftazidime, cefotaxime, and ceftriaxone are reportedly 31.3%, 65.6%, and 70%, respectively, and integrons were noted to play important roles in the generation and dissemination of antimicrobial resistance genes in this bacterial species.4,5
Class 1 integrons, mediated by mobile genetic elements, enable the horizontal spread of drug resistance genes among clinical isolates. Class 1 integrons structurally consist of three core elements: an integrase gene (intI1), catalyzing site-specific recombination, a primary recombination site (attI1) and a common promoter (Pc), enabling transcription of the gene cassettes.6 Class 1 integrons play an important role in drug resistance gene transmission among bacteria because they can capture exogenous gene cassettes and express genes within the gene cassette.7–10 The detection rate for class 1 integrons in clinical strains is the highest of all such elements at 50–70%, and this is linked with the generation and spread of antimicrobial-resistant bacteria.11,12
The expression of the gene cassette requires the aid of the integrator promoter Pc.8,11 Variants of the promoters for class 1 integrons have been defined by previous studies according to their spacer sequences and −35 and −10 hexamer sequences.8,13–15 Some class 1 integrons contain not only a Pc promoter but also a P2 promoter that is located ∼90 bp downstream of Pc2. It was shown that the P2 promoter is active when bound to relatively weak Pc promoters, such as PcW or PcH1.8,13–15 Wei et al. showed an inverse correlation between the strength of the Pc promoter and integration efficiency.16
Therefore, integrons may play an important role in the resistance of E. coli, and different promoters affect the expression, integration, and excision of gene cassettes. In the present study, we investigated the molecular characteristics of the variable regions and common promoters of integrons in E. coli.
Materials and Methods
Bacterial strains
Specimens from patients in our hospital were collected from September to November 2018 according to a method reported in the Manual of Clinical Microbiology (11th Edition, 2015) published by the American Society for Microbiology.17 Species were identified by the Vitek-2 Compact system (BioMerieux, France). E. coli DH5α was stored in our laboratory. Proteus mirabilis 47437 (from the Center Clinical Laboratory of Zhejiang Province People's Hospital) and E. coli ATCC25922 were used as antimicrobial resistance control strains. We received and archived the written consent forms from all of the patients included in this study, and we obtained approval by the Hospital Ethics Committee of Weifang People's Hospital for the use of these samples from human patients (NO.:KYLL2018003).
Antimicrobial susceptibility tests
Several antimicrobials were selected as representative drugs. The Kirby-Bauer disk diffusion method was used to test antimicrobial susceptibility according to the Clinical and Laboratory Standards Institute (CLSI).18 The antibiotics chosen were amikacin (AMK), ampicillin (AMP), ampicillin/sulbactam (SAM), ceftazidime (CAZ), ciprofloxacin (CIP), levofloxacin (LEV), cefepime (FEP), ceftriaxone (CRO), cefotetan (CTT), cefotaxime (CTX), cefuroxime (CXM), cefazolin (CZO), cefoxitin (FOX), gentamicin (GEN), tobramycin (TOB), ertapenem (ETP), imipenem (IPM), meropenem (MEM), aztreonam (ATM), cefoperazone/sulbactam (CSL), piperacillin/tazobactam (TZP), and trimethoprim/sulfamethoxazole (SXT). Isolates shown to be resistant to three or more drug classes were defined as multidrug-resistant (MDR).
Bacterial genomic DNA extraction
All strains were streaked onto Luria–Bertani (LB; Oxoid, United Kingdom) agar plates and incubated overnight at 37°C. A single clone from each strain was inoculated into LB liquid medium, and the culture was shaken overnight (200 r/min, 37°C). The next day, 1.5 mL of the bacterial solution was centrifuged (12,000 g, 2 min), the supernatant was discarded, and total genomic DNA was isolated using the EZ-10 Spin Column Bacterial Genomic DNA Miniprep Kit (Bio Basic, Canada).
Polymerase chain reaction assay for screening class 1 integrons
The extracted genomic DNA was used as the polymerase chain reaction (PCR) template to screen for class 1 integrons. Primers intF and P2R (Table 1), which target the class 1 integron integrase (intI1), were used. A blank control (distilled water), a negative control (E. coli DH5α), and a positive control (P. mirabilis 47437) were included in each run.
Table 1.
Polymerase Chain Reaction Primers Used in This Study
| Gene | Description | Sequence (5′–3′) | References |
|---|---|---|---|
| intF | Screening primer for class 1 integrons | CCAAGCTCTCGGGTAACATC | 13 |
| P2R | Screening primer for class 1 integrons | GCCCAGCTTCTGTATGGAAC | 13 |
| 5CS | Screening primer for the variable region of class 1 integrons | GGCATCCAAGCAGCAAG | 13 |
| 3CS | Screening primer for the variable region of class 1 integrons | AAGCAGACTTGACCTGA | 13 |
Identifying the variable regions in class 1 integrons
Primers 3CS and 5CS were used to amplify the variable regions of the class 1 integron-positive isolates. LA Taq DNA polymerase (TaKaRa Biotechnology, Japan) was used for the PCRs. After electrophoresis on a 1% agarose gel, the PCR products were excised and purified using the EZ-10Spin Column Gel Extraction Kit (Bio Basic). All of the PCR products were sequenced by primer walking, starting with the 3CS and 5CS primers (Table 1). Primer design and DNA sequencing were performed by Sangon Biotechnology Co., Ltd. (Shanghai, China). Nucleotide sequences were analyzed and compared using BLAST software (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov).
Statistical analysis
The data were analyzed using SSPS v21.0 (IBM, Armonk, NY). Data from bacterial counts were analyzed using Pearson's chi-square test and were expressed as the chi-square. Comparisons were considered significant at p < 0.05.
Results
Bacterial isolation and antimicrobial resistance
Specimens were collected from patients hospitalized at Weifang People's Hospital in Shandong and, among these, we detected 138 E. coli isolates. Most of the isolates derived from urine (41.30%, 57/138), followed by blood (16.67%, 23/138), pus (15.22%, 21/138), secretions (10.14%, 14/138), sputum (5.80%, 8/138), and other sterile body site specimens (10.87%, 15/138).
Antimicrobial susceptibility testing revealed that the most common resistance was to AMP (89.86%). In addition, the resistance rates to CIP, CRO, CTX, CXM, CZO, LEV, SAM, and SXT were all greater than 50%. MDR strains accounted for 79.99% (109/138) of the isolates. The results are shown in Table 2.
Table 2.
Antimicrobial Resistance of Escherichia coli Isolates
| Antibiotic | Class 1 integrons positive strains (n = 93) |
Class 1 integrons negative strains (n = 45) |
Total (n = 138) |
χ2 | p | |||
|---|---|---|---|---|---|---|---|---|
| n | Drug resistance rate (%) | number | Drug resistance rate (%) | n | Drug resistance rate (%) | |||
| AMK | 4 | 4.30 | 1 | 2.22 | 5 | 3.62 | 0.016 | 0.899 |
| AMP | 86 | 92.47 | 38 | 84.44 | 124 | 89.86 | 1.354 | 0.245 |
| ATM | 43 | 46.24 | 18 | 40.00 | 61 | 44.20 | 0.489 | 0.489 |
| CAZ | 33 | 35.48 | 10 | 22.22 | 43 | 31.16 | 2.486 | 0.115 |
| CIP | 74 | 79.57 | 28 | 62.22 | 102 | 73.91 | 4.733 | 0.03 |
| CRO | 62 | 66.67 | 25 | 55.56 | 87 | 63.04 | 1.607 | 0.205 |
| CSL | 7 | 7.53 | 7 | 15.56 | 14 | 10.14 | 1.354 | 0.245 |
| CTT | 7 | 7.53 | 4 | 8.89 | 11 | 7.97 | 0 | 1 |
| CTX | 62 | 66.67 | 26 | 57.78 | 88 | 63.77 | 1.037 | 0.407 |
| CXM | 61 | 65.59 | 29 | 64.44 | 90 | 65.22 | 0.795 | 0.373 |
| CZO | 67 | 72.04 | 29 | 64.44 | 96 | 69.57 | 2.808 | 0.094 |
| ETP | 3 | 3.23 | 1 | 2.22 | 4 | 2.90 | 0 | 1 |
| FEP | 24 | 25.81 | 8 | 17.78 | 32 | 23.19 | 1.098 | 0.295 |
| FOX | 18 | 19.35 | 7 | 15.56 | 25 | 18.12 | 0.295 | 0.587 |
| GEN | 40 | 43.01 | 10 | 22.22 | 50 | 36.23 | 5.672 | 0.017 |
| IPM | 3 | 3.23 | 1 | 2.22 | 4 | 2.90 | 0 | 1 |
| LVX | 66 | 70.97 | 25 | 56.56 | 91 | 65.94 | 3.207 | 0.073 |
| MEM | 3 | 3.23 | 1 | 2.22 | 4 | 2.90 | 0 | 1 |
| SXT | 71 | 76.34 | 6 | 13.33 | 76 | 55.07 | 48.81 | <0.001 |
| TOB | 21 | 22.58 | 3 | 6.67 | 24 | 17.39 | 5.346 | 0.021 |
| TZP | 3 | 3.23 | 1 | 2.22 | 4 | 2.90 | 0 | 1 |
| SAM | 62 | 66.67 | 23 | 51.11 | 85 | 61.59 | 3.102 | 0.078 |
| MDR | 82 | 88.17 | 27 | 60.00 | 109 | 79.99 | 14.501 | <0.001 |
AMK, amikacin; AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CSL, cefoperazone/sulbactam; CTT, cefotetan; CTX, cefotaxime; CXM, cefuroxime; CZO, cefazolin; ETP, ertapenem; FEP, cefepime; FOX, cefoxitin; GEN, gentamicin; IPM, imipenem; LEV, levofloxacin; MDR, multidrug resistant; MEM, meropenem; SAM, ampicillin/sulbactam; SXT, trimethoprim/sulfamethoxazole; TOB, tobramycin; TZP, piperacillin/tazobactam.
Molecular characterization of class 1 integrons and comparison of antimicrobial resistance between integron-positive and integron-negative isolates
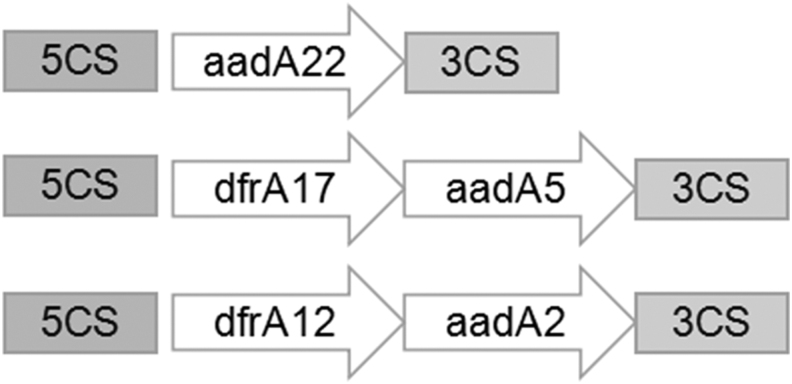
Next, we performed molecular characterization of class 1 integrons and compared antimicrobial resistance between integron-positive and integron-negative isolates. Class 1 integrons were detected in 93 (67.39%, 93/138) of the isolates. PCR amplification of the variable regions in the class 1 integrons identified 69 intI1-positive isolates, but amplification failed in 24 of the isolates. Among the 69 intI1-positive isolates, only partial variable region sequences could be amplified and sequenced. Three gene cassette arrays were detected in these isolates, the most prevalent of which was dfrA17-aadA5 (detected in 55 strains). The dfrA12-aadA2 and aadA22 cassettes were detected in nine and five isolates, respectively.
Five gene cassettes were detected, including dfrA12/17 and aadA2/5/22, with the most prevalent antimicrobial-resistant gene cassettes being dfrA17 (55/93), conferring resistance to trimethoprim, aadA5 (55/93), conferring resistance to streptomycin and spectinomycin, dfrA12 (9/93), conferring resistance to trimethoprim, and aadA2 (9/93), conferring resistance to streptomycin and spectinomycin. The structural characteristics and translation direction of the variable-region gene cassettes are shown in Fig. 1.
FIG. 1.
Genetic environment of the variable regions of the class 1 integrons.
Comparison of the drug sensitivity of the intI1-positive strains and intI1-negative strains revealed that the antimicrobial resistance rates for CIP, GEN, SXT, and TOB in the integron-positive isolates were higher than those in the integron-negative isolates, and this difference was significant (p < 0.05). MDR among the intI1-positive and intI1-negative strains showed prevalence rates of 88.7% (82/93) and 60% (27/45), respectively, and this difference was significant (Table 2).
Molecular characterization of promoters in the class 1 integrons
Next, we performed molecular characterization of the promoters in class 1 integrons. Among the 93 intI1-positive strains, 5 different types of common promoters were detected. The most prevalent common promoter was PcH1. Fifty-five of these isolates contained dfrA17-aadA5, and in 9 of these, amplification of the variable region failed. The second most prevalent common promoter was PcWTGN-10. For nine of the isolates containing dfrA12-aadA2, and the other four isolates, amplification of the variable region failed. The active P2 promoter was detected in seven strains and all were combined with PcH1 (PcH1-P2). Five of the isolates harbored aadA22, and for two of them, the variable region was unable to be amplified. PcS and PcW were detected in six and three class 1 integrons, respectively (Table 3).
Table 3.
Promoters of the Class 1 Integron Variable Region
Discussion
Antimicrobial therapy is the method of choice to treat bacterial infections, but the continuous emergence and spread of MDR strains in recent years has compromised the effectiveness of antimicrobial drugs.19 E. coli is one of the most commonly isolated bacteria clinically, but its increasing resistance to antimicrobials in recent years is making treatment more difficult.20 In this study, we collected clinical E. coli isolates in our hospital over a 3-month period. Antimicrobial sensitivity tests on these isolates showed that the antimicrobial resistance rates for 9 of the 22 tested antimicrobials exceeded 50%, with most belonging to the fluoroquinolones, cephalosporins, and β-lactam family of antibiotics.
Similar findings have been reported by Mfoutou Mapanguy et al. and Chen et al.20,21 In our investigation, the rate of detection of MDR strains was nearly 80%, which was lower than that found by Lee et al.3 and similar to that found by Masoud et al.22 This may be related to geographical differences or differences in specimen collection.
To date, integrons associated with mobile genetic elements have been classified into five types based on the sequence of the integrase (classes 1 to 5). Class 1 integrons act as evolutionary platforms for bacteria by capturing gene cassettes and expressing cassette-harbored genes that enable bacteria to adapt to environmental changes.4 What is more, integrons can transmit between bacteria through associations with transposons and plasmids, as reviewed by Mazel,10 which allows for the horizontal transfer of multidrug resistance genes.10
In this study, 67.39% (93/138) of E. coli isolates were found to harbor class 1 integrons, which was similar to the results of Wei et al.4 This indicated that the dissemination of integrons in E. coli was extremely high in China. Sequence analysis showed that the most prevalent gene cassette of the integron variable region was dfrA17-aadA5 in this study.
The dfrA12-aadA2 and aadA22 cassettes were detected in nine and five isolates, respectively. In total, five gene cassettes were detected, including dfrA12/17 and aadA2/5/22, which was similar to previous studies.4,20,23 Therefore, domestic and foreign studies show high similarity in the gene cassettes carried in the variable region of E. coli integrons. Besides, the reason that several strains were failed to amplify variable region may due to the absence of 3CS, which requires further investigation.
Our statistical analysis showed that the antimicrobial resistance rates of the integron-positive strains for CIP, GEN, SXT, and TOB were higher than those of the negative strains, and this difference was statistically significant (p < 0.05). However, in our study, aadA and dfr were the most common antimicrobial resistance gene cassettes found in clinical E. coli isolates, which conferred resistance to traditional antimicrobials, such as trimethoprim, streptomycin, and spectinomycin. These drugs are now less widely used in clinic, so the prevalence of these gene cassettes may reflect historic selection pressure when these drugs were previously widely used.4,23,24
The sul1 gene carried within the integron structure confers resistance to SXT. Resistance to CIP and AMP may be related to the plasmid or transposon, in which the integron is located. Thus, the drug resistance rate of integron-positive strains has been shown to be significantly higher than that of integron-negative strains,25,26 and this will be the subject of further studies.
In this study, the most prevalent promoter was PcH1, a relatively weak promoter. Previous studies have shown that the integrators, where such promoters reside, have strong gene integration capabilities, while expression of the gene cassette is relatively weak.13–15 Therefore, although most of the resistance genes carried by the E. coli integron variable region were to traditional drugs in this study, the strong integration ability of the integrons implies that they will capture gene cassettes in response to environmental changes, especially under the selection pressure of antimicrobial agents.
Conclusions
The data presented here indicates that E. coli resistance and MDR strains are prevalent in our city Weifang in Shandong Provence China. The gene cassettes of the class 1 integron variable region mostly conferred resistance to traditional antimicrobials. Weak promoters in the variable regions were predominant in this study. Integrons have a strong ability to integrate gene cassettes, indicating the importance of integrons for enhancing bacterial resistance. In summary, integrons pose a great threat to the treatment of MDR bacterial infections and further investigations are needed.
Acknowledgments
We thank Sandra Cheesman, PhD, from Liwen Bianji (Edanz), for editing the English text of a draft of this article. We received and archived the written consent forms from all of the patients included in this study, and we obtained approval by the Hospital Ethics Committee of Weifang People's Hospital for the use of these samples from human patients.
Authors' Contributions
H.W. designed the study. W.L., M.L., and J.M. conducted the study, collected the data, and prepared the article. X.S. provided advice and edited the article. All authors approved the final version of the article.
Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by research grant funding from the Health Commission of Shandong Province (No. 2018WS078) and the Health Commission of Weifang (No. wfwsjs_2018_110).
References
- 1. Cao, X., Zhang Z., Shen H., et al. 2014. Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. APMIS 122:1088-1095. [DOI] [PubMed] [Google Scholar]
- 2. Wang, Y., Zhao S., Han L., et al. 2014. Drug resistance and virulence of uropathogenic Escherichia coli from Shanghai, China. J. Antibiot (Tokyo) 67:799-805. [DOI] [PubMed] [Google Scholar]
- 3. Lee, Y.Q., Ahmad Kamar A., R.D. Velayuthan, C.W. Chong, and C.S.J. Teh. 2021. Clonal relatedness in the acquisition of intestinal carriage and transmission of multidrug resistant (MDR) Klebsiella pneumoniae and Escherichia coli and its risk factors among preterm infants admitted to the neonatal intensive care unit (NICU). Pediatr. Neonatol. 62:129–137. [DOI] [PubMed] [Google Scholar]
- 4. Wei, Q., Jiang X., Li M., et al. 2013. Diversity of gene cassette promoter variants of class 1 integrons in uropathogenic Escherichia coli. Curr. Microbiol. 67:543–549. [DOI] [PubMed] [Google Scholar]
- 5. Blancarte-Lagunas, M.I., Castro-Escarpulli G., Navarro-Ocaña A., et al. 2020. Commensal and virulent Escherichia coli strains of vaginal origin are reservoirs of resistance cassettes in class 1 integrons. J. Infect. Dev. Ctries, 14:48-58 . [DOI] [PubMed] [Google Scholar]
- 6. Gillings, M.R. 2014. Integrons: past, present, and future. Microbiol. Mol. Biol. Rev. 78:257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall, R.M., and Stokes H.W.. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115–132. [DOI] [PubMed] [Google Scholar]
- 8. Collis, C.M., and Hall R.M.. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hall, R.M., and Collis C.M.. 1998. Antibiotic resistance in Gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1:109–119. [DOI] [PubMed] [Google Scholar]
- 10. Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608–620. [DOI] [PubMed] [Google Scholar]
- 11. Fluit, A.C., and Schmitz F.J.. 1999. Class 1 integrons, gene cassettes, mobility and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761–770. [DOI] [PubMed] [Google Scholar]
- 12. Collis, C.M., and Hall R.M.. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lévesque, C., Brassard S., Lapointe J., et al. 1994. Diversity and relative strength of tandem promoters for antibiotic-resistance genes of several integrons. Gene 142:49–54. [DOI] [PubMed] [Google Scholar]
- 14. Papagiannitsis, C.C., L.S. Tzouvelekis, and Miriagou V.. 2009. Relative strength of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob. Agents Chemother. 53:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jové, T., Da Re S., Denis F., et al. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 6:e1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei, Q., Jiang X., Li M., et al. 2011. Transcription of integron-harboured gene cassette impacts integration efficiency in class 1 integron. Mol. Microbiol. 80:1326–1336. [DOI] [PubMed] [Google Scholar]
- 17. American Society for Microbiology. 2015. Manual of Clinical Microbiology. 11th ed. Washington, DC: ASM Press. [Google Scholar]
- 18. Clinical and Laboratory Standards Institute. 2019. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Ninth Informational Supplement. CLSI Documents M100-S29. CLSI, Chicago. [Google Scholar]
- 19. Obodoechi, L.O., Carvalho I., N.S. Chenouf, et al. 2021. Antimicrobial resistance in Escherichia coli isolates from frugivorous (Eidolon helvum) and insectivorous (Nycteris hispida) bats in Southeast Nigeria, with detection of CTX-M-15 producing isolates. Comp. Immunol. Microbiol. Infect. Dis. 75:101613. [DOI] [PubMed] [Google Scholar]
- 20. Chen, M., Wu Y., Yu S., et al. 2019. Drug resistance and integron genes in Escherichia coli isolated from urinary tract infection. J. Nanosci. Nanotechnol. 19:5989–5993. [DOI] [PubMed] [Google Scholar]
- 21. Mfoutou Mapanguy, C.C., Adedoja A., L.G.V. Kecka, et al. 2021. High prevalence of antibiotic-resistant Escherichia coli in Congolese students. Int. J. Infect. Dis. 103:119–123. [DOI] [PubMed] [Google Scholar]
- 22. Masoud, S.M., R.M. Abd El-Baky, S.A. Aly, and Ibrahem R.A.. 2021. Co-existence of certain ESBLs, MBLs and plasmid mediated quinolone resistance genes among MDR E. coli isolated from different clinical specimens in Egypt. Antibiotics (Basel) 10:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Escudero, J.A., Loot C., Nivina A., and Mazel D.. 2015. The integron: adaptation on demand. Microbiol. Spectr. 3:MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 24. Mi, L., Jie M., Wei J., et al. 2020. Antimicrobial resistance and molecular characterization of gene cassettes from Class 1 integrons in Pseudomonas aeruginosa strains. Microbial. Drug Resist. 26:670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalantari, M., Sharifiyazdi H., Asasi K., and Abdi-Hachesoo B.. 2021. High incidence of multidrug resistance and class 1 and 2 integrons in Escherichia coli isolated from broiler chickens in South of Iran. Vet. Res. Forum 12:101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdel-Rhman, S.H., R.M. Elbargisy, and D.E. Rizk. 2021. Characterization of integrons and quinolone resistance in clinical Escherichia coli isolates in Mansoura City, Egypt. Int. J. Microbiol. 2021:6468942. [Epub ahead of print]; DOI: 10.1155/2021/6468942 [DOI] [PMC free article] [PubMed] [Google Scholar]