Abstract
Corynebacterium jeikeium is an opportunistic pathogen primarily of immunocompromised (neutropenic) patients. Broad-spectrum resistance to antimicrobial agents is a common feature of C. jeikeium clinical isolates. We studied the profiles of susceptibility of 20 clinical strains of C. jeikeium to a range of antimicrobial agents. The strains were separated into two groups depending on the susceptibility to erythromycin (ERY), with one group (17 strains) representing resistant organisms (MIC > 128 μg/ml) and the second group (3 strains) representing susceptible organisms (MIC ≤ 0.25 μg/ml). The ERY resistance crossed to other members of the macrolide-lincosamide-streptogramin B (MLSb) group. Furthermore, this resistance was inducible with MLSb agents but not non-MLSb agents. Expression of ERY resistance was linked to the presence of an allele of the class X erm genes, erm(X)cj, with >93% identity to other erm genes of this class. Our evidence indicates that erm(X)cj is integrated within the chromosome, which contrasts with previous reports for the plasmid-associated erm(X) genes found in C. diphtheriae and C. xerosis. In 40% of C. jeikeium strains, erm(X)cj is present within the transposon, Tn5432. However, in the remaining strains, the components of Tn5432 (i.e., the erm and transposase genes) have separated within the chromosome. The rearrangement of Tn5432 leads to the possibility that the other drug resistance genes have become included in a new composite transposon bound by the IS1249 elements.
Corynebacterium jeikeium is commonly found colonizing the skin (particularly the inguinal, axillary, and rectal areas) of hospital workers and hospitalized patients (for a review, see reference 2). The highest incidence (up to 82%) of colonization occurs in severely immunocompromised patients. This colonization can lead to infection (sepsis), especially in neutropenic patients and in patients with skin disruption (e.g., by indwelling medical devices). Frequently, infected patients have been on prolonged antibiotic regimens, which often results in disease caused by organisms with broad-spectrum drug resistance, including resistance to macrolides. In these cases, therapeutic options are very limited, essentially relying on vancomycin. C. jeikeium can also cause infections in immunocompetent patients. For example, in a recent study (24), C. jeikeium (and other corynebacteria) was identified as a cause of infections in patients following joint replacement surgery and in patients with open bone fractures.
Although C. jeikeium is a significant opportunistic pathogen primarily of the immunocompromised host, the presence of C. jeikeium in the hospital environment is probably the most clinically important aspect of the natural history of this organism. This is because there is recent evidence that drug resistance genes may have transferred from corynebacteria to a Proprionibacterium sp. clinical isolate (A. Eady, presented at the 100th Gen. Meet. Am. Soc. Microbiol., 2000). Thus, the high incidence of multiply drug-resistant C. jeikeium suggests that this organism may be an important environmental reservoir of drug resistance genes.
The molecular bases for multidrug resistance in C. jeikeium are incompletely understood but may be a consequence of the accumulation of individual and specific genetic events or may involve nonspecific mechanisms, such as increased drug efflux or changes in the permeability of the cell wall. Macrolide resistance in C. jeikeium is of particular interest because it is clearly an acquired phenotype and is common in clinical isolates. Thus, in this report, we present a study of the basis of macrolide resistance in C. jeikeium as a first step in understanding the molecular and genetic basis for multidrug resistance in this clinically important species.
MATERIALS AND METHODS
Antimicrobial agents.
The following agents were obtained as reference powders ampicillin (AMP) (Sigma, St. Louis, Mo.), azithromycin (AZM) (Pfizer Inc., Groton, Conn.), chloramphenicol (CHL) (Sigma), clarithromycin (CLR) (Abbott Laboratories, Abbott Park, Ill.), clindamycin (CLI) (Upjohn Pharmaceuticals, Kalamazoo, Mich.), erythromycin (ERY) (Sigma), kanamycin KAN (Sigma), lincomycin (LIN) (Upjohn), rifampin (RIF) (Sigma), spiramycin (SPI) (Sigma), streptogramin A (dalfopristin [D]), streptogramin B (quinupristin) [Q]), Q-D (Rhone-Poulenc Rorer, Collegeville, Pa.), and tetracycline (TET) (Sigma). Stock solutions were prepared following the suppliers' recommendations. Susceptibility disks (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France) were also used for susceptibility testing against CHL, CLI, ERY, LIN, SPI, TET, and vancomycin.
Bacteria.
Seventeen clinical strains of C. jeikeium were generously provided by R. Leclercq, Hôpital-Henri Mondor, Creteil, France. These strains were isolated from patients attending the Hôpital-Henri Mondor. Antibiograms that were determined at the source laboratory indicated that all 17 strains were multidrug resistant, including ERY resistant. Consequently, we expanded the collection with three ERY-susceptible clinical strains, supplied by D. Bruckner, UCLA Medical Center, Los Angeles, Calif. Species identification was confirmed by the API CORYNE panel (bioMerieux, Inc., St. Louis, Mo.). Routine liquid culture of C. jeikeium was in BHI-YT, i.e., brain heart infusion broth (Difco Laboratories, Detroit, Mich.) supplemented with Tween 80 (0.2%) and yeast extract (0.4%) (Difco). The agar medium used was Mueller-Hinton agar (Difco) supplemented with 5% lysed sheep blood.
We used the following reference strains for susceptibility testing: Escherichia coli ATCC 25923, Staphylococcus aureus ATCC 29213, and a clinical strain of C. diphtheriae (obtained from the Clinical Microbiology Laboratory at Childrens Hospital Los Angeles).
The hosts for gene transfer were E. coli strain XL1-Blue MRF′ (Stratagene, La Jolla, Calif.) and C. glutamicum strain KO8. The latter organism was derived from ATCC 13032, by introducing an unmarked mutation in the cglIR gene, part of the restriction system of C. glutamicum (17). Briefly, we isolated the cglIR gene by PCR with the primers CGLIR-1 and CGLIR-2 (Table 1) and cloned the 1,480-bp amplification product using the PCR-Script cloning system (Stratagene). Removing the BstEII (Sigma) restriction fragment from the cloned PCR product created a 102-bp deletion in the cglIR gene. We subcloned the mutated cglIR gene into the suicide vector pK19mobsacb (ATCC 87098) (18) and used the construct to transform C. glutamicum strain ATCC 13032 by electroporation (9). The vector, pK19mobsacb, contains the aminoglycoside-phosphotransferase of the transposon Tn5 and the sacB gene of Bacillus subtilis, and these genes are known to confer KAN resistance and sucrose toxicity to C. glutamicum (18). Since pK19mobsacb cannot replicate in C. glutamicum, any derived transformants will represent organisms with the pK19mobsacb:cglIR construct inserted into the genome. We isolated the KAN-resistant transformants and grew them overnight in the absence of selective agent. From these cultures, we selected organisms that were able to grow in the presence of 10% sucrose and were susceptible to kanamycin. This approach identified organisms with double recombination events. We confirmed that the endogenous cglIR gene had been replaced with the mutated gene by amplification of genomic DNA from the isolated variants with primers CGLIR-1 and CGLIR-2. Electroporation of one variant (strain KO8) with plasmid pECM2 (23) (supplied by A. Tauch, University of Bielefeld, Bielefeld, Germany), showed that it was at least 3 orders of magnitude more transformable than the parental strain, ATCC 13032. Transformation of the E. coli and C. glutamicum hosts was by electroporation using previously described protocols (9; Epicurian Coli electroporation-competent cells, 1999 [Stratagene, La Jolla, Calif.]).
TABLE 1.
Primers used for PCR analysis
| Primer | Sequence (5′→3′) | Target | Directiona | Position(s)b |
|---|---|---|---|---|
| CGLIR-1 | CCCCGATGACTTTGAGTTTTTGG | cglIR | → | 1128 |
| CGLIR-2 | GTGTGAGGTTTTACCGCTTTGGAC | cglIR | ← | 2607 |
| Cerm1 | GACACGGCCGTCACGAGCAT | erm(X) | → | 4862 |
| Cerm2 | GGCGGCGAGCGACTTCC | erm(X) | ← | 5251 |
| erm3282 | TGCCCGGCTCCCTTTCA | erm(X) | → | 4813 |
| erm4176 | CTGGTGGATTTCGGTTTTGGTG | erm(X) | ← | 5728 |
| EPR06 | GTCGATGGTGAGGTAGAGAGGAA | erm(X) | ← | 5637 |
| IS1249-1 | CCGCTACACCACCACCAACC | IS1249 | → | 4006, 7145 |
| IS1249-2 | GATCGTCAGGCAGTTCCGTTTTTA | IS1249 | ← | 4460, 7599 |
PFGE.
DNA plugs suitable for pulsed-field gel electrophoresis (PFGE) analysis were prepared from C. jeikeium isolates using a method described previously (11) except that the organisms were pretreated with lysozyme (20 mg/ml) for 1 h before embedding in agarose. Restriction enzyme digestion of the plugs was for 18 to 24 h with 36 U of either DraI, SfiI, or AseI restriction enzyme (New England Biolabs Inc., Beverly, Mass.). For the analysis of intact chromosomal DNA, the plugs were electrophoresed without prior treatment with a restriction enzyme. A CHEF-DR II PFGE system (Bio-Rad, Hercules, Calif.) was used for the PFGE analysis. The run conditions for the undigested, DraI-, and SfiI-digested plugs were 6 V/cm for 20 h with a ramped switch time of 50 to 90s. The run conditions for the AseI-digested, DNA was 6 V/cm for 6 h with a ramped switch time of 0.1 to 10s. The gels were stained with ethidium bromide (0.5 μg/ml) in 0.5× TBE buffer as described elsewhere (16). Isolates were deemed to be distinct if the PFGE patterns differed by more than two bands (10). The DNA was isolated from the PGFE gel using the Qiaex II system (Qiagen, Valencia, Calif.).
Susceptibility testing.
Susceptibility to antimicrobial agents was assessed by either disk diffusion or agar dilution methods according to the guidelines of the National Committee for Clinical Laboratory Standards (12).
To assess for induction of the ERY resistance phenotype, C. jeikeium strains CJ12 and CJ21 were preincubated for 5 h in a range of macrolide-lincosamide-streptogramin B (MLSb) and non-MLSb antimicrobial agents. The drug concentrations used for the preincubation step were 0.1 and 1 μg/ml, with the exception of AMP and KAN, which were used at 1 and 10 μg/ml. Preliminary experiments showed that the highest drug concentration used had a slight, but detectable effect on the growth rates of the C. jeikeium strains. We determined the MIC of ERY for the organisms in these cultures using a broth microdilution-based susceptibility assay as described elsewhere (6).
Nucleic acid extraction and PCR.
Total DNA from C. jeikeium was isolated by the procedure described by Tauch et al. (22). We obtained plasmid DNA from E. coli and C. jeikeium using the Qiagen Plasmid DNA Isolation System (Qiagen). For corynebacteria, we included an initial 2-h incubation at 37°C with buffer P1 containing 20 mg of lysozyme per ml.
The primers used in the PCR analysis are summarized in Table 1. All primers were designed using OLIGO software (version 5.0; National Biosciences, Inc., Plymouth, Minn.). The basic 50-μl PCR mixture consisted of 1× PCR buffer (including 1.5 mM MgCl2), 10 pmol of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1.25 U of DNA polymerase, and 0.05 to 1 μg of template DNA. For applications where amplification fidelity was not critical, we used HotStarTaq DNA polymerase (Qiagen) and its supplied buffer, whereas for high fidelity amplification (i.e., for cloning) we used PfuTurbo DNA polymerase (Stratagene) and its supplied buffer. Each PCR was amplified for 35 to 40 cycles (low-fidelity amplifications) or 25 to 30 cycles (high-fidelity amplifications) of 1 min at 94°C, 1 min at 65°C, and 2 min at 72°C using a Perkin-Elmer model 480 thermal cycler. Amplification products were characterized by ethidium bromide fluorescence staining following electrophoresis in 1 to 2% agarose using 0.5× TBE running buffer.
Cloning of the resistance determinant.
Genomic DNA, isolated from C. jeikeium strains CJ12 and CJ21, was digested with BamHI and size selected between 2 and 8 kbp following electrophoresis in a 0.8% agarose gel. The DNA was isolated from the agarose using the Qiaquick Gel Purification system (Qiagen) and then ligated to BamHI-digested, calf intestinal alkaline phosphatase-treated pBluescript II SK(+) (Stratagene). These ligation reactions were used to transform electrocompetent E. coli strain XL1-Blue MRF′ and the transformants were selected on Luria-Bertani (LB) agar containing ampicillin (50 μg/ml). For each transformation reaction, we prepared a pool of the derived colonies. We replated the plasmid pools onto LB agar containing ampicillin (50 μg/ml) and ERY (400 μg/ml). From each pool, we picked eight colonies that grew on the ERY-containing plates and isolated their plasmids. Plasmid isolation was accomplished using the Qiagen Plasmid Mini Kit. For the strain CJ12-derived and CJ21-derived pools, these plasmids were termed pBC12-1 to pBC12-8 and pBC21-1 to pBC21-8, respectively.
In order to define functionally the gene that confers resistance, we cloned PCR products generated from C. jeikeium strain CJ21 DNA using a range of primers (Table 1) and the high-fidelity DNA polymerase, PfuTurbo (Stratagene). These PCR products were cloned using the PCR-Script cloning system (Stratagene) and subcloned into the Corynebacterium-E. coli shuttle vector, pECMT. This vector was derived from pECM2 (23) by inserting a transcription terminator (GTCAAAAGCCTCCGGTCGGAGGCTTTTGAC) at the SmaI site. This insertion prevented transcription from the aminoglycoside phosphotransferase gene through the multiple-cloning site of the plasmid. We used the pECMT constructs to transform E. coli and C. glutamicum hosts. ERY resistance was detected in the transformants by plating the organisms on LB agar containing ERY (400 μg/ml) and either 50 μg of AMP per ml (for pPCR-Script AMP constructs) or 25 μg of KAN per ml (for pECMT constructs).
Nucleotide sequencing.
Nucleotide sequence analysis was performed with the PRISM Ready Reaction Dye Deoxy-Terminator Cycle Sequencing Kit and an ABI, (San Francisco, Calif.) 373A automated sequencer according to the manufacturer's recommended protocols. Open reading frames (ORF) were identified using the computer program MacVector (version 6.5.3; Genetics Computer Group, Madison, Wis.) and sequence homology was analyzed using NCBI BLASTN and BLASTX protocols (www.ncbi.nlm.nih.gov/BLAST/) (1).
Southern blot.
We digested C. jeikeium DNA (1 μg) with the restriction enzymes BamHI, Bsp106I, and NotI (Stratagene). The restricted DNA was analyzed by Southern blotting using standard protocols (16), with UV Duralon nylon membrane (Stratagene) as the transfer membrane. We derived an erm-specific DNA probe from the 916-bp PCR product generated with primers erm3282 and erm4176 (Table 1). In addition, we derived an IS1249-specific DNA probe from the 455-bp PCR product generated with primers IS1249-1 and IS1249-2. The PCR products were fluorescein tagged using the Illuminator Prime-It labeling system (Stratagene). After an overnight hybridization at 42°C in UltraHyb solution (Ambion, Austin, Tex.), we removed excess probe with three high-stringency washes, each for 15 min at 60°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate. We visualized bound probe using the Illuminator Nonradioactive Detection System (Stratagene) and Kodak Biomax Light film.
Computer-generated images.
Digital images of gels and Southern analyses were acquired using a UMAX 1200S scanner connected to a Macintosh PowerBook G3 computer. Image formatting was performed with Abode Photoshop (version 4.0.1), and figures were assembled with Adobe Illustrator (version 9.0).
RESULTS
Strain typing of C. jeikeium isolates.
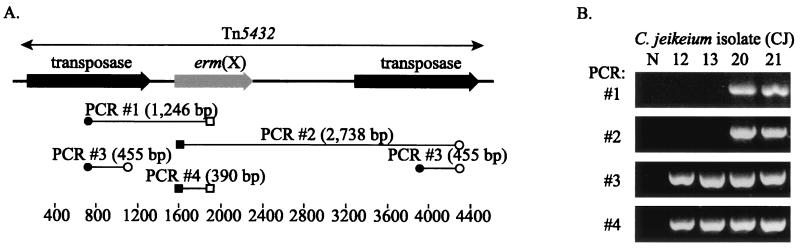
PFGE analysis indicated that the 20 C. jeikeium isolates represented 13 distinct strain types (data not shown), with the three ERY-susceptible strains being distinct from the ERY-resistant strain types. Furthermore, the C. jeikeium strains CJ12, CJ13, CJ20, and CJ21 (see Fig. 2 to 4) were different strains.
FIG. 2.
(A) Arrangement of Tn5432 and the regions spanned by the PCR targets used to investigate this transposon in ERY-resistant C. jeikeium strains. The primers used in the PCR assays were IS1249-1 (●), IS1249-2 (○), Cerm1 (■), and Cerm2 (□). The ruler is in base pairs. (B) Representative results for four C. jeikeium strains (CJ12, CJ13, CJ20, and CJ21) in the four PCR assays outlined in panel A. Lane N, negative control.
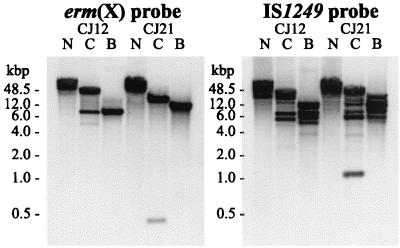
FIG. 4.
Southern analysis of DNA isolated from C. jeikeium strains CJ12 and CJ21 restricted with BamHI (lanes B), Bsp106I (lanes C), and NotI (lanes N) using an erm(X)-specific probe and an IS1249-specific probe.
Antimicrobial susceptibilities of C. jeikeium.
A summary of the susceptibilities of the ERY-resistant and ERY-susceptible C. jeikeium strains to selected antimicrobial agents is shown in Table 2. Thus, the ERY-resistant organisms expressed high-level resistance (>128 μg/ml) to other MLSb agents, whereas the ERY-susceptible organisms were susceptible to the other MLSb agents. The results for susceptibility to ampicillin, kanamycin, and streptogramin A were largely the same for the ERY-resistant and ERY-susceptible strains.
TABLE 2.
Susceptibilities of C. jeikeium strains to selected antimicrobial agents
| Species (n) | ERY phenotype | MIC (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | AZM | CLI | CLR | LIN | Da | Qb | Q-D | AMP | KAN | ||
| C. jeikeium (17) | Resistant | >128 | >128 | >128 | >128 | >128 | 1–8 | >128 | ≤0.5–8 | >128 | ≥128 |
| C. jeikeium (3) | Susceptible | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | 1–8 | ≤1 | ≤1 | >128 | ≥128 |
| C. diphtheriae (1) | Susceptible | ≤0.5 | 1 | ND | ND | 2 | 1 | ≤1 | ≤1 | 0.125 | 2 |
Streptogramin A.
Streptogramin B.
The range of Q-D MICs was wider for the ERY-resistant strains than for the ERY-susceptible strains. The MICs of Q-D at which 50 and 90% of the ERY-resistant strains tested were inhibited were 2 and 4 μg/ml, respectively. In addition, the MICs of Q-D for two ERY-resistant strains were <0.5 μg/ml. Overall, there was no clear association between the susceptibility to Q or D and the susceptibility to Q-D. For example, the Q, D, and Q-D MICs for two ERY-susceptible strains were ≤1, 8, and <0.5 μg/ml, respectively. Similarly, the MICs of Q, D, and Q-D for one of the ERY-resistant were 128, 4, and <0.5 μg/ml. Thus, the apparent differences in the susceptibility to Q-D between the ERY-resistant and ERY-susceptible strains may reflect the differences in the sample size of the two groups rather than being associated with MLSb resistance.
There was considerable strain-to-strain variability in susceptibility to CHL and TET (assessed by disk diffusion), with zone diameters ranging from 6 to 14 mm. There was no correlation between the MIC of ERY and zone diameter for either CHL or TET. All strains were susceptible to vancomycin (zone diameters, >7 mm).
Thus, ERY resistance in C. jeikeium represents an MLSb-resistant phenotype. Furthermore, the large difference in ERY MIC between the resistant and susceptible organisms supports the belief that this phenotype is acquired rather than being just a reflection of strain-to-strain variability in inherent resistance.
A common approach to studying induction of MLSb resistance is by characterizing the zones of inhibition surrounding antimicrobial disks. Unfortunately, there were no zones of inhibition for the ERY-resistant strains when standard susceptibility disks for several MLSb agents (ERY, CLIN, LIN, and SPI) were used. Consequently, we investigated induction of MLSb resistance in C. jeikeium by assessing changes in MIC using a microdilution susceptibility assay. However, this approach is limited by the possibility that induction of resistance can occur within the susceptibility assay, leading to an increase in the apparent ERY MIC for noninduced cells. Despite this, we found that ERY resistance was inducible with MLSb agents, including 14-, 15-, and 16-membered macrolides, lincosamides, and streptogramin B (Q). A summary for two C. jeikeium strains is shown in Table 3. Non-MLSb agents did not induce an increase in the level of ERY resistance. The non-MLSb agents included CHL, KAN, TET, and D, which target the ribosome. Thus, induction of high-level ERY resistance was not simply the consequence of protein synthesis inhibition, nor was it a nonspecific stress response.
TABLE 3.
ERY MICs for C. jeikeium strains preincubated for 5 h in a range of MLSb and non-MLSb agents
| Preincubationa | ERY MIC (μg/ml) for:
|
|
|---|---|---|
| CJ12 | CJ21 | |
| None | 512 | 512 |
| MLSb agents | ||
| AZM | >2,048 | >2,048 |
| ERY | >2,048 | >2,048 |
| CLI | >2,048 | >2,048 |
| LIN | >2,048 | >2,048 |
| SPI | >2,048 | >2,048 |
| Q | >2,048 | >2,048 |
| Non-MLSb agents | ||
| AMP | 512 | 512 |
| KAN | 512 | 512 |
| CHL | 512 | 512 |
| TET | 512 | 512 |
| RIF | 512 | 512 |
| D | 512 | 512 |
Preincubation concentration of AMP and KAN was 10 μg/ml; the concentration of all other agents was 1 μg/ml.
It is possible that the results of these experiments were a consequence of the selection of organisms with constitutive resistance rather than phenotypic induction. In order to address this, we removed samples from wells containing 2,048 μg of ERY per ml and cultured the organisms for 48 h with and without 1 μg of ERY per ml. The ERY MIC for the organisms grown without agent reverted to 512 μg/ml, whereas, the ERY MIC for organisms maintained in ERY was >2,048 μg/ml. These results support the presumption that MLSb resistance is inducible in C. jeikeium.
The high ERY MIC determined for the noninduced organisms suggests that induction causes only a minor change in phenotype. However, this is misleading since the level of growth in the susceptibility assay for the organisms induced with ERY at 512 μg/ml was only marginally less than the growth in medium alone. Obviously, there was no growth of the noninduced organisms with ERY at 512 μg/ml. Furthermore, overnight incubation in high ERY concentrations (tested up to 100 μg/ml) was able to induce an increase in ERY resistance in C. jeikeium strains CJ12 and CJ21 (data not shown). Thus, the high ERY MIC determined for noninduced organisms may reflect the expected phenotype induction within the susceptibility assay. However, the lack of a zone of inhibition for MLSb agents, as seen in a disk diffusion assay, suggests that ERY-resistant C. jeikeium may express a high level of MLSb resistance constitutively.
Identification of the gene that confers MLSb resistance.
As a first step in elucidating the molecular basis of macrolide resistance, we analyzed C. jeikeium DNA by erm(X)-specific PCR (primers Cerm1 and Cerm2). Abundant amplification products of slightly less than 400 bp were generated from DNA isolated from the 17 MLSb-resistant strains, whereas no amplification products were generated with the DNA isolated from the three susceptible strains (data not shown). The PCR products were consistent with the predicted size of 390 bp. Furthermore, the DNA sequences of the amplification products showed >95% identity to the erm(X) gene isolated from a C. xerosis strain, erm(X)cx or ermCX (GenBank accession no. U21300). Thus, MLSb resistance in C. jeikeium is associated with the presence of an allele, erm(X)cj, of the class X erm genes, according to the nomenclature proposed by Roberts et al. (15).
In order to isolate the complete gene that confers MLSb resistance, we prepared plasmid libraries of BamHI-digested DNA isolated from C. jeikeium strains CJ12 and CJ21. Within each library, we isolated eight clones that conferred increased resistance to E. coli strain XL1-Blue MRF′. The ERY MIC for the transgenic E. coli was >2,048 μg/ml, compared to 32 μg/ml for the parental organism. We analyzed the plasmids that conferred ERY resistance by BamHI restriction mapping and found that all plasmids containing DNA isolated from C. jeikeium strain CJ12 had an insert of ∼6 kbp, whereas all those plasmids containing DNA from strain CJ21 contained an insert of ∼8 kbp (data not shown). We amplified the insert DNA using an erm(X)-specific PCR (primers Cerm1 and Cerm2) and confirmed the presence of the putative erm gene in all 16 plasmids (data not shown). Consequently, we limited subsequent analysis to the two plasmids, pBCJ12-5 and pBCJ21-3, derived from strains CJ12 and CJ21, respectively.
We sequenced the cloned fragments of plasmids pBC12-1 and pBC21-3 in the region of the putative erm gene, and located ORF homologous to the erm(X)cx gene. The first 215 amino acids of the predicted polypeptides for strains CJ12 and CJ21 are 93.5 and 98.6% identical to Erm(X)cx (Fig. 1), the Erm protein from C. xerosis (translated from GenBank accession no. U21300). The major difference between the two Erm(X)cj polypeptides and the Erm(X)cx polypeptide is a frame shift within codon 216. This results in the Erm(X)cj polypeptides being 31 amino acids longer than Erm(X)cx.
FIG. 1.
(A) The predicted Erm(X)cj polypeptide sequences of C. jeikeium strains CJ12 and CJ21 aligned with Erm(X)cx translated from the erm(X)cx gene (22) (GenBank accession no. U21300). Amino acid identities are boxed, with shading indicating amino acid similarities. (B) Alignment of the leader regions (DNA) of the erm(X)cj genes of strains CJ12 and CJ21. Probable promoter elements (−35 and −10 regions), a conserved region coding for a leader peptide (lp), and the first four codons of the erm(X)cj gene are indicated by a lines above the sequences. (C) Alignment of the theoretical leader peptides from strains CJ12 and CJ21. The amino acid and DNA sequences for strains CJ12 and CJ21 are available in the GenBank database (accession no. AF338705 and AF338706).
To confirm that the apparent frame shift in the predicted peptide chain of Erm(X)cj was not the result of sequencing errors, we cloned two fragments of the erm gene from C. jeikeium strain CJ21. These fragments were generated by high-fidelity PCR using the 5′ primer erm3282 and either erm4176 or EPRO6 as the 3′ primer (Table 1). Primer erm4176 anneals 3′ to our predicted termination codon, whereas, primer EPRO6 anneals 5′ to our termination codon and 3′ to the predicted termination codon of the erm(X)cx. We cloned these fragments into the vector pPCR-Script AMP under transcriptional control of the lacZ promoter and used the constructs to transform E. coli strain XL1-Blue MRF′. Transformants containing the erm3282-erm4176 fragment were resistant to ERY concentrations of 400 μg/ml, whereas transformants containing the erm3282-EPRO6 fragment were unable to grow in the presence of ERY concentrations of 400 μg/ml. This result supports the location of the predicted termination codon of the erm(X)cj gene.
We confirmed these results in corynebacteria by transforming C. glutamicum strain KO8 with two constructs cloned into the plasmid pECMT. One construct contained the PCR product generated from C. jeikeium strain CJ21 DNA using primers IS1249-1 and erm4176, and the other construct contained the PCR product generated using primers IS1249-1 and EPRO6. Using IS1249-1 as the upstream primer ensured that the erm(X) promoter was included in the PCR products. The C. glutamicum transformant containing the IS1249-1–erm4176 product was able to grow in the presence of ERY at 400 μg/ml, whereas the transformant containing the IS1249-1–EPRO6 product could not.
We have identified probable promoter elements and a coding region for a conserved theoretical leader peptide within the leader regions of erm(X)cj for strains CJ12 and CJ21 (Fig. 1B and C). We could not identify any clear secondary structures within the leader region that were consistent with the attenuation of transcription or attenuation of translation models of regulation that have been described for other erm genes (4, 8). However, this does not rule out the role of posttranscriptional attenuation in the regulation of erm(X)cj expression.
Is erm(X)cj associated with a mobile element?
The sequence analysis of the DNA upstream from the erm(X)cj gene was very different in C. jeikeium strains CJ12 and CJ21 (Fig. 1B). In strain CJ21, we identified the IS1249 insertion element, consistent with erm(X)cj being within the transposon, Tn5432. However, ∼700 bp upstream from the erm(X)cj gene in strain CJ12 was an ORF 72% identical to the theoretical gene, Rv1112 (a probable GTP-binding protein), within the chromosome of Mycobacterium tuberculosis (GenBank accession no. AL021897). Thus, erm(X)cj was not adjacent to an upstream IS1249 element in strain CJ12. To assess if the insertion element was local (i.e., within 4 or 5 kbp) to erm(X) in strain CJ12, we digested plasmid pBC12-5 with restriction enzymes BamHI and Bsp106I and isolated the 5.5-kbp fragment. Since the erm(X)cj gene has an internal Bsp106I site, the BamHI-Bsp106I fragment should contain most of the erm(X)cj gene and ∼4.7 kbp of upstream DNA. We analyzed this fragment by PCR using the erm(X)-specific primers, Cerm1 and Cerm2, and the IS1249-specific primers, IS1249-1 and IS1249-2 (Table 1). An amplification product was detected with the erm-specific PCR of the expected size (390 bp); however, no product was detected with the IS1249-specific PCR (data not shown). As controls, we included amplification reactions containing either total DNA isolated from strain CJ12 or the plasmid pBCJ21-5. The latter contains the erm(X)cj and IS1249 elements isolated from strain CJ21. These control reactions generated amplification products of the expected size with both the erm(X)-specific (390-bp) and IS1249-specific (455-bp) primer pairs. To support these results and rule out a partial IS1249 element within the cloned DNA, we sequenced 700 bp of the 5.5-kbp BamHI and Bsp106I DNA fragment at the end distal to erm(X)cj. In this region (GenBank accession no. AF343961), we found no evidence for a partial IS1249 element in this cloned fragment. Thus, the erm(X)cj gene of strain CJ12 appears to be >4.7 kbp from an upstream IS1249 element.
To explore the association between erm(X)cj, IS1249 and Tn5432 further, we used a series of PCR assays that spanned the individual elements of this transposon (Fig. 2A). We chose 10 MLSb-resistant strains at random for PCR mapping of Tn5432. Representative results for four strains are presented in Fig. 2B. In four strains (including strains CJ20 and CJ21), erm(X)cj was associated with upstream and downstream IS1249 elements consistent with Tn5432. The sizes of the PCR products were consistent with the sizes predicted by the archetypal Tn5432 sequence (GenBank accession no. U21300). In the remaining strains (including strains CJ12 and CJ13) the amplification reactions that span from the upstream and from the downstream IS1249 elements to erm(X) failed to generate products (PCR 1 and 2). Despite this, the IS1249-specific (PCR 3) and erm(X)-specific (PCR 4) PCR assays amplified products from all 10 strains. These results are important for several reasons. Firstly, the generation of PCR products with PCR assays 3 and 4 rules out the possibility that the failures of PCR assays 1 and 2 were a result of primer mismatches. Secondly, the presence of IS1249 elements in the strains in which erm(X)cj is not associated with Tn5432 suggests that rearrangement of this transposon has occurred. However, erm(X)cj may be still associated with IS1249 elements in a much larger composite transposon than Tn5432.
To assess whether erm(X)cj is associated with extrachromosomal DNA, we attempted to transform C. glutamicum strain KO8 with plasmids isolated from several ERY-resistant C. jeikeium strains (including CJ12 and CJ21). No ERY-resistant C. glutamicum transformants were generated with any plasmid preparations. However, large plasmids (>50 kbp) tend to be underrepresented in standard plasmid preparations; therefore, we also attempted to transform C. glutamicum with high-molecular-weight (average size, >100 kbp) total cellular DNA, which also failed.
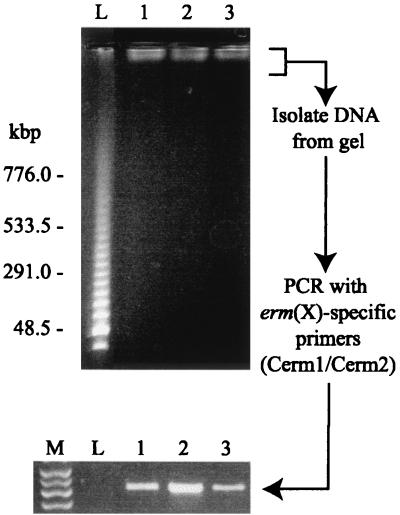
Although the transformation results suggested that erm(X)cj is not associated with a plasmid, we sought to confirm this by an alternative approach, i.e., PFGE analysis of intact chromosomal DNA. Figure 3 presents a pulsed-field gel of the DNA from three C. jeikeium strains (CJ12, CJ13, and CJ21) and shows the chromosomal DNA (at the top of the gel, adjacent to the wells). There was no evidence for high-molecular-weight (>50-kbp) extrachromosomal DNA. Southern analysis, with an erm(X)-specific probe, of PFGE-separated C. jeikeium DNA proved to be difficult to reproduce; however, the only hybridization bands seen corresponded with the chromosomal DNA (data not shown). In order to provide further support for this observation, we gel purified the chromosomal DNA from a pulsed-field gel and used the material in an erm(X)-specific PCR assay (Fig. 3). Amplification products were generated with the C. jeikeium DNA (Fig. 3, lanes 1 to 3) but not the DNA isolated from the lambda concatemer lane (Fig. 3, lane L). This confirms that erm(X)cj is integrated within the C. jeikeium chromosome.
FIG. 3.
PFGE analysis of high-molecular-weight DNA isolated from three ERY-resistant strains of C. jeikeium: lane 1, CJ12; lane 2, CJ13; and lane 3, CJ21. The DNA molecular weight marker (lane L) is a lambda ladder. DNA was isolated from the gel within 1 cm of each well (including lane L) and analyzed by PCR using the erm(X)-specific primers, Cerm1 and Cerm2. Lane M contains a DNA marker, showing (from top to bottom) bands of 500, 400, 300, and 200 bp.
Since erm(X)cj is within the C. jeikeium chromosome, we wanted to determine how many copies of this gene were present per genome. To assess this we analyzed restricted high-molecular-weight DNA (isolated from strains CJ12 and CJ21) by Southern blot and hybridization with an erm(X)-specific probe and an IS1249-specific probe (Fig. 4). The three restriction enzymes used all cut C. jeikeium DNA relatively infrequently; the mean fragment sizes for BamHI, Bsp106I (an isoschizomer of ClaI), and NotI were approximately 4 to 8 kbp, 10 to 20 kbp, and >20 kbp, respectively. Furthermore, there is a BamHI site within Tn5432 (between the erm gene and the downstream IS1249 element), and there is a Bsp106I site within the erm(X) gene.
The Southern analysis with the erm-specific probe (Fig. 4) shows only a single band in each of the BamHI and NotI digests. However, it is difficult to rule out the presence of multiple bands in the NotI digest because of the poor resolution of agarose gel electrophoresis of such large DNA fragments (>48.5 kbp). The second band in the Bsp106I digests is the result of the site for this enzyme within the erm(X)cj gene. Thus, the Southern analysis indicates that in both C. jeikeium strains, erm(X)cj is present only as a single copy.
The Southern analysis with the IS1249-specific probe (Fig. 4) indicates that the IS1249 element is present in more than the two copies predicted by the structure of Tn5432. This suggests that the IS1249 element may be associated with composite mobile elements other than Tn5432.
DISCUSSION
In this report, we describe the genetic elements responsible for the acquisition of macrolide resistance in C. jeikeium, as part of the phenomenon of multidrug resistance observed in this species. Macrolide resistance was found to cross to the other members of the MLSb group, and the acquisition of macrolide resistance was separate from resistance to other non-MLSb agents. These observations suggest that broad-spectrum resistance in C. jeikeium is the result of the accumulation of several independent genetic changes. MLSb resistance in C. jeikeium is conferred by alleles of the erm gene class X.
Recently, Roberts et al. (15) proposed a logical nomenclature for erm genes, and grouped the corynebacterial genes in erm class X. Thus, MLSb resistance in C. jeikeium is conferred by alleles of the erm gene class X. However, despite the high degree of homology between the erm(X) alleles isolated from different corynebacteria (i.e., C. diphtheriae, C. jeikeium, and C. xerosis), the alleles were found in different genetic contexts. The C. diphtheriae erm(X) gene (originally termed ermCd) was located within a 14.5-kbp plasmid pNG2 (7), whereas the C. xerosis erm(X) gene (originally termed ermCX) was located within transposon Tn5432, which itself was carried by the 50-kbp R plasmid, pTP10 (22). In contrast, our evidence indicates that erm(X)cj is integrated within the C. jeikeium chromosome. This is consistent with a previous report by Pitcher et al. (13), which suggests that macrolide resistance in C. jeikeium is not plasmid associated. Evidence for chromosomal association of macrolide resistance determinants has also been reported for C. striatum (14).
Despite the differences in the carriage of the erm(X) genes of C. jeikeium and C. xerosis, in 40% of MLSb-resistant C. jeikeium strains erm(X) is associated with the transposon Tn5432. In the remaining 60% of MLSb-resistant C. jeikeium strains erm(X) is not associated with Tn5432, although these strains do possess all the component parts of the transposon. Since Tn5432 is highly conserved between C. jeikeium and C. xerosis, we believe that all MLSb-resistant C. jeikeium strains originally acquired erm(X)cj within Tn5432 but subsequently the transposon became rearranged within the chromosome. This phenomenon may indicate the presence of an integron (5). This leads to the possibility that the IS1249 elements have formed new composite transposons containing other drug resistance genes. This point is particularly troubling since the transposase of IS1249 is known to be able to insert Tn5432 (and presumably other IS1249-containing transposons) into the genomes of unrelated bacteria (22).
The source organisms from which the corynebacterial erm genes were characterized were isolated from geographically distinct locations. The original pNG2-containing C. diphtheriae and the C. striatum were isolated from patients from the northwestern United States (3, 14), whereas the pTP10-containing C. xerosis was isolated in Japan (22). The C. jeikeium organisms used in this study were isolated from patients in France. This distribution of erm(X) suggests that the corynebacteria acquire the erm gene from a common source, possibly an organism that can be found colonizing skin or mucosa. The fact that the mode of carriage of the erm genes is different suggests that the reservoir is not readily transferred from organism to organism, i.e., the erm gene either is on a plasmid with a narrow host range or is chromosome associated. It is interesting that the ermCd-containing plasmid (pNG2) is similar to plasmids isolated from a range of skin-colonizing corynebacteria (20) and appears to have a broad host range, including E. coli (19, 21). Thus, pNG2 is unlikely to be the environmental reservoir element for all the corynebacterial erm genes. The transposon, Tn5432, is a more likely candidate as it is common to two Corynebacterium species, is integrated within the chromosome of one species, and is known to be mobile (22).
Clearly, more studies are needed to characterize the clinical importance of multidrug-resistant corynebacteria, such as C. jeikeium, as reservoirs of resistance genes within the hospital environment.
ACKNOWLEDGMENTS
We thank Clark B. Inderlied for the use of laboratory resources and for his comments in preparation of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1977;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyle M B, Lipsky B A. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin Microbiol Rev. 1990;3:227–246. doi: 10.1128/cmr.3.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyle M B, Minshew B H, Bland J A, Hsu P C. Erythromycin and clindamycin resistance in Corynebacterium diphtheriae from skin lesions. Antimicrob Agents Chemother. 1979;16:525–527. doi: 10.1128/aac.16.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gryczan T J, Grandi G, Hahn J, Grandi R, Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980;8:6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall R M, Collis C M, Kim M J, Partridge S R, Recchia G D, Stokes H W. Mobile gene cassettes and integrons in evolution. Ann N Y Acad Sci. 1999;870:68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x. [DOI] [PubMed] [Google Scholar]
- 6.Hindler J. Antimicrobial susceptibility testing. In: Isenberg H D, editor. Clinical microbiology procedures handbook. Vol. 1. Washington, D.C.: American Society for Microbiology; 1992. pp. 5.2.1–5.2.29. [Google Scholar]
- 7.Hodgson A L, Krywult J, Radford A J. Nucleotide sequence of the erythromycin resistance gene from the Corynebacterium plasmid pNG2. Nucleic Acids Res. 1990;18:1891. doi: 10.1093/nar/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak J H, Choi E C, Weisblum B. Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J Bacteriol. 1991;173:4725–4735. doi: 10.1128/jb.173.15.4725-4735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebl W, Bayerl A, Schein B, Stillner U, Schleifer K H. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett. 1989;53:299–303. doi: 10.1016/0378-1097(89)90234-6. [DOI] [PubMed] [Google Scholar]
- 10.Maslow J M, Slutsky A M, Arbeit R D. Application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 11.Nash K A, Inderlied C B. Genetic basis of macrolide resistance in Mycobacterium avium isolated from patients with disseminated disease. Antimicrob Agents Chemother. 1995;39:2625–2630. doi: 10.1128/aac.39.12.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Suggested grouping for U.S. FDA-approved antimicrobial agents that should be considered for routine testing and reporting on fastidious organisms by clinical microbiology laboratories. In: Jorgensen J H, Craig W A, Doern G V, Ferraro M J, Finegold S M, Fung-Tome J, Hansen S L, Hindler J, Preston D A, Reller L B, Swenson J M, Tenover F C, Wilker M A, Wilson W R, editors. Performance standards for antimicrobial susceptibility testing. 15, no. 14. Wayne, Pa: National Commitee for Clinical Laboratory Standards; 1995. p. 1. [Google Scholar]
- 13.Pitcher D, Johnson A, Allerberger F, Woodford N, George R. An investigation of nosocomial infection with Corynebacterium jeikeium in surgical patients using a ribosomal RNA gene probe. Eur J Clin Microbiol Infect Dis. 1990;9:643–648. doi: 10.1007/BF01964264. [DOI] [PubMed] [Google Scholar]
- 14.Roberts M C, Leonard R B, Briselden A, Schoenknecht F D, Coyle M B. Characterization of antibiotic-resistant Corynebacterium striatum strains. J Antimicrob Chemother. 1992;30:463–474. doi: 10.1093/jac/30.4.463. [DOI] [PubMed] [Google Scholar]
- 15.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, n.y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Schafer A, Schwarzer A, Kalinowski J, Puhler A. Cloning and characterization of a DNA region encoding a stress-sensitive restriction system from Corynebacterium glutamicum ATCC 13032 and analysis of its role in intergeneric conjugation with Escherichia coli. J Bacteriol. 1994;176:7309–7319. doi: 10.1128/jb.176.23.7309-7319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 19.Serwold-Davis T M, Groman N, Rabin M. Transformation of Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium glutamicum, and Escherichia coli with the C. diphtheriae plasmid pNG2. Proc Natl Acad Sci USA. 1987;84:4964–4968. doi: 10.1073/pnas.84.14.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serwold-Davis T M, Groman N B. Mapping and cloning of Corynebacterium diphtheriae plasmid pNG2 and characterization of its relatedness to plasmids from skin coryneforms. Antimicrob Agents Chemother. 1986;30:69–72. doi: 10.1128/aac.30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serwold-Davis T M, Groman N B, Kao C C. Localization of an origin of replication in Corynebacterium diphtheriae broad host range plasmid pNG2 that also functions in Escherichia coli. FEMS Microbiol Lett. 1990;54:119–123. doi: 10.1016/0378-1097(90)90268-u. [DOI] [PubMed] [Google Scholar]
- 22.Tauch A, Kassing F, Kalinowski J, Puhler A. The Corynebacterium xerosis composite transposon Tn5432 consists of two identical insertion sequences, designated IS1249, flanking the erythromycin resistance gene ermCX. Plasmid. 1995;34:119–131. doi: 10.1006/plas.1995.9995. [DOI] [PubMed] [Google Scholar]
- 23.Tauch A, Kirchner O, Wehmeier L, Kalinowski J, Puhler A. Corynebacterium glutamicum DNA is subjected to methylation-restriction in Escherichia coli. FEMS Microbiol Lett. 1994;123:343–347. doi: 10.1111/j.1574-6968.1994.tb07246.x. [DOI] [PubMed] [Google Scholar]
- 24.von Graevenitz A, Frommelt L, Punter-Streit V, Funke G. Diversity of coryneforms found in infections following prosthetic joint insertion and open fractures. Infection. 1998;26:36–38. doi: 10.1007/BF02768750. [DOI] [PubMed] [Google Scholar]