Abstract
With a widely attended virtual kickoff event on January 29, 2021, the National Cancer Institute (NCI) and the Department of Energy (DOE) launched a series of 4 interactive, interdisciplinary workshops—and a final concluding “World Café” on March 29, 2021—focused on advancing computational approaches for predictive oncology in the clinical and research domains of radiation oncology. These events reflect 3,870 human hours of virtual engagement with representation from 8 DOE national laboratories and the Frederick National Laboratory for Cancer Research (FNL), 4 research institutes, 5 cancer centers, 17 medical schools and teaching hospitals, 5 companies, 5 federal agencies, 3 research centers, and 27 universities. Here we summarize the workshops by first describing the background for the workshops. Participants identified twelve key questions—and collaborative parallel ideas—as the focus of work going forward to advance the field. These were then used to define short-term and longer-term “Blue Sky” goals. In addition, the group determined key success factors for predictive oncology in the context of radiation oncology, if not the future of all of medicine. These are: cross-discipline collaboration, targeted talent development, development of mechanistic mathematical and computational models and tools, and access to high-quality multiscale data that bridges mechanisms to phenotype. The workshop participants reported feeling energized and highly motivated to pursue next steps together to address the unmet needs in radiation oncology specifically and in cancer research generally and that NCI and DOE project goals align at the convergence of radiation therapy and advanced computing.
INTRODUCTION
In 2016 the NCI and DOE formed the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) collaboration to simultaneously accelerate advances in predictive oncology and computing (1). As a result of this effort, several co-designed pilot projects were developed and are ongoing with an emphasis on discovery and understanding of cancer mechanisms and patient health trajectories (2–4). As a means to explore future areas at the intersection of advanced computing and cancer treatment, the leaders of the collaboration met with the NCI Radiation Research Program and recognized that the synergy, spirit, and energy of the JDACS4C program could be further focused via application to radiation therapy (RT) given its unique position in clinical medicine, bridging physics, patient care, and advanced computing to affect patient outcomes. In the context of the SARS-COV-2 pandemic, the team designed an interactive, bottom-up virtual workshop series focused on developing the ability to make predictions regarding patient outcomes across multiple scales of space and time in radiation oncology that would link the frontiers of clinical care and supercomputing-enabled science.
Radiotherapy is an essential component of effective cancer control and used in the care of about half of all cancer patients (5). The field of radiation therapy is unique in its deep dependence on mathematics, physics, and computing in the practice of medicine and in the use of quantitative modeling in the design of new treatment technologies. The dependence on digital technologies to effect complex, yet safe, personalized radiation delivery has created a data-rich clinical ecosystem that is at the forefront of medicine in the application of computing to personalize cancer intervention. In addition, the field of radiotherapy is actively exploring new technologies and approaches such as circulating biomarkers, advanced medical imaging, combination therapies, radiomics, radiopharmaceutical therapy/theranostics, and digitally enabled patient reported outcomes as shown schematically in Fig. 1. Critically important, radiation therapy is delivered in a controlled, prescribed fashion through multiple fractions over time and, increasingly, the field is measuring the response of patients and exploring image-enabled adaptation strategies to modify intervention for improved individual outcomes. Radiation oncology practice is well-positioned for computationally enabled predictive oncology and its future would benefit from the multidisciplined scientific integration and the joint resources of NCI and DOE. Finally, advances in radiation therapy will impact many other areas of science and medicine such as drug development, computational modeling, surgical practice, survivorship research, late effects research, space and aeronautics research, engineering, national security, radiation safety, radiation biology, mitigation of radiation events, and disaster management (6–13).
FIG. 1.
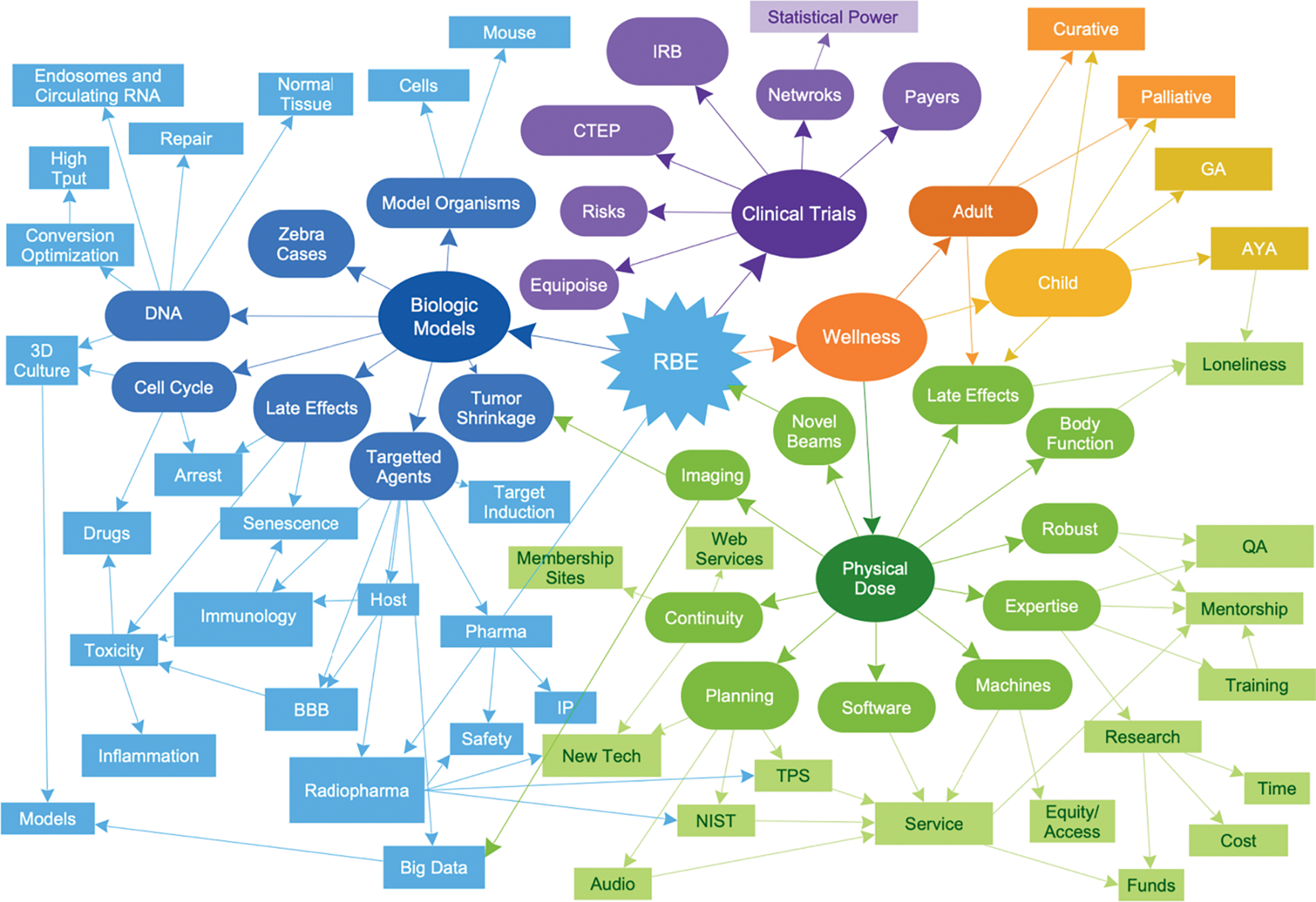
The complexity of scientific and clinical interactions in radiation oncology makes it a good model system to test next generation approaches that can then be applied if successful to all aspects of cancer care and ultimately medicine. These elements apply universally to the overarching medical specialties and not just radiation oncology: imaging, pathology/initial tumor profiling, surgery, medical oncology, hematology/transplantation, immunology, pediatric oncology, infectious disease, and others.
The formal mission statement of the workshop series was to “explore emerging and futuristic opportunities among DOE, NCI and partner institutions to advance radiation therapy via 1. personalized, adaptive treatment through understanding and development of mechanism-based, computationally-enabled modeling and 2. advanced computing to achieve dynamic, multiscale, data-informed, clinically actionable predictions and decision making.” The expected outcomes of the workshops were, first, to create the scope and goals for potential new NCI–DOE collaborative projects and science and, second, to identify opportunities to engage and collaborate with cross-domain researchers and clinicians (https://events.cancer.gov/cbiit/radonc2021). Key outcomes include a set of 12 critical questions and corresponding testable ideas. These critical questions formed the basis for vigorous multi-day workshops on actionable next steps to advance computational oncology in the science and practice of radiation oncology.
METHODS OF THE WORKSHOPS
The workshop series was designed to operate over 3 months with an initial event to stimulate engagement and dialog across diverse communities followed by 4 virtual, highly interactive 4-h workshops tackling different areas of synergy across disciplines, specifically: 1. The Biological Machinery for Advancing Radiation Oncology, 2. The Frontiers of Computational Modeling and Simulations in Multiscale Radiation Oncology, 3. Learning from Care Delivery: The How and Why of Multi-omics, Biomarkers, and Prediction for Radiation Oncology, and 4. Multimodal Patient Trajectories: Individual Predictive Modeling. The final 6-h “World Café” workshop then fused the directions and key questions raised into a finite set of questions. In total, 3,870 human hours of virtual engagement were achieved over the 3-month period with representation from 9 national laboratories, 4 institutes, 5 cancer centers, 17 medical schools and teaching hospitals, 5 companies, 5 federal agencies, 3 research centers, and 27 universities. This report captures the narrative and resulting actionable recommendations.
Two parallel tracks took place over the series. The primary focus was the development of twelve critical questions that need to be addressed to enable predictive radiation oncology derived from focused, team-led interaction with presentations, scoring, and voting processes empowered by an advanced virtual platform and driven by national subject matter experts who led focused small group discussions and bottom-up processes. The secondary focus was the generation of a set of collaboration topics generated by initial spontaneous contribution by individuals in a chatroom format followed by dynamic, interactive maturation by the attendees without formal intervention. The collaboration topics are described in more detail in Supplementary Materials (https://doi.org/10.1667/RADE-22-00012.1.S1).
RESULTS
The workshop series’ primary result was a set of priority questions to enable predictive radiation oncology, formed from interactive top-down and bottom-up processes. The secondary result formed from spontaneous collaboration efforts during the workshops in a chatroom space called the “Collaboration Corner” that was truly bottom-up. We discuss the primary set of questions here and the secondary results from the collaboration corner in the Supplementary Materials (https://doi.org/10.1667/RADE-22-00012.1.S1).
The Primary Generated Questions
A total of 42 discussion questions were developed across the 4 half-day sessions which were summarized and refined into the top 10 to 12 questions in the final World Café Workshop. Participants had been asked to consider both near term and visionary, long-term (called “Blue Sky”) gaps and next steps related to these questions and to define the reason the questions were important and their likely impacts. The following top 12 final questions emerged based on real-time electronic voting by all participants:
Can we leverage advances in machine learning to build predictive, mechanistic models of radiation oncology outcomes?
What is the optimal use of computation and digitization to develop more continuous and dynamic monitoring of patient response to inform the design of future interventions and enable mechanistic modeling of disease and response?
How can mathematical and computational approaches help define valid biomarkers across physical (imaging, circulating, systemic) and temporal (hours, days, months) scales? (Tumor vs, normal, population versus individual, early biomarkers for late outcomes, etc.)
How do we combine multiscale images (e.g., radiology and pathology) and physical models (e.g., biomechanical, physiological) to increase collaboration across disciplines and to remove data and investigator-associated bias in the resulting models?
How do we effectively prioritize the process and type of measurement (radiomics, genomics, etc.) for an individual patient and integrate those data into a patient model or avatar? (Supercomputer-defined biomarkers.)
What is the formalism for integrating computational decision-making to guide radiation and systemic therapy in clinical practice? (e.g., How to select systemic therapy as well as implementation, education, and rollout.)
What do we need to track and accommodate to address uncertainties in the modeling and the data?
How can we more accurately measure the “health” or “freedom from cancer burden” in a fashion that reflects the complexity and nuance of an individual patient? (Many different “normals” exist.)
What is the opportunity and process for establishing machine-human hybrid approaches in cancer care? (Interpreting data from models and how to then feed human interpretation back into the models.)
How can advanced computing help us not only to analyze data but to also facilitate the featurization/semantic interoperability of the data?
How can computational approaches help reduce the diversity of biomarkers to more defined patient specific factors that lead to accurate, validated, and computationally tractable model formation of cancer and response to treatment?
How can we develop computational models to simulate how radiation kills a cell, affects a group of cells, transforms a tumor, and impacts a patient—and how does underlying patient-specific heterogeneity affect this process at multiple scales?
Short-Term Foci and Blue Sky Themes
The World Café final workshop addressed a continuum of opportunities within radiation oncology that may impact the next two decades of patient care. The opportunities developed in the discussion were notable both for existing NCI and DOE science and for new science that can only be developed via multidisciplinary collaboration. The short-term areas upon which to dedicate energy and resources were broken down into four main foci.
First, bringing people from very different scientific “worlds” together to collaborate closely is critical. Senior leaders in the fields of aerospace and meteorology at multiple points along the timeline of the workshops noted that our project has clear parallels to their projects and that the lessons already learned in those very different fields would have immense impact on moving predictive radiation oncology forward. A critical point is that all leaders and participants in the workshops said they felt a shared common mission and from this a clear sense of community and energy emerged from the workshop series.
Second, both data availability and data aggregation are critical for advancement and for equity. Participants and leadership felt strongly that the process needed to start immediately, and that “imperfect” data now was far better than “better” data that might be obtained later given the critical need to help our patients avoid toxicities and achieve better outcomes. Throughout the workshops, data conversations were part of nearly all discussions and represent a common denominator of the proposed new endeavor between NCI and DOE focused on precision/predictive oncology. In various contexts throughout the workshops, many participants suggested the need for increased investment in methods to collect, anonymize, store, and validate data. From the start of the workshop series, participants identified and discussed the critical need for the data to represent the global population fairly, not simply large data sets from a few geographic regions or centers. Discussions and planning for data infrastructures are ongoing in many hospitals, cancer centers, universities, and national laboratories, these are often in isolation and not coordinated across institutes. With limited talent in this space and limited data in each individual institute, workshop participants discussed the need and potential for coordinated and concerted efforts to develop data standards, data warehouses, and data infrastructures.
The third, short-term focus from the workshops is that critical state variables need to be identified from the “physics” or biochemistry of the patient to rapidly allow modeling to link to the physical reality of the clinic (14). For example, using very simple models repeatedly updated with longitudinal data, weather forecasting was able to mimic models of immense complexity once the critical variables were identified and placed in relatively simple models. The same is likely true in cancer care with the caveat that the models will need to be adaptable and ideally placed into digital twins to empower patients and their doctors to make more informed decisions along their clinical journeys.
Finally, the fourth short-term focus was to create new energy—and a new research space—in the field. At the conclusion of the World Café meeting, it was clear that those present, representing a broad set of skill sets and backgrounds, supported the intention of the workshop series: to make prediction of patient outcomes an overall goal and driving focus of our fields moving forward. Participants also enthusiastically agreed that radiation oncology is a good ecosystem within which to find a path forward due to its inherent focus on four-dimensional treatment data. Participants wanted additional workshops and opportunities to work together and expressed a shared commitment to create research momentum and discovery from the seeds planted by the workshops.
Shifting to long-term or “Blue Sky” themes, the workshop series developed several that were less defined than the short-term foci but also more transformative.
The first “Blue Sky” theme was to increase the capacity to be nimble and prepared for rapid change as technology allows discoveries in biology to be more akin to step functions than gradual progress. As corollaries to this overall need to be nimble and willing to move to a new ground truth rapidly, participants felt it was critical not to waste time developing theories or science based on newly erroneous or “bad” data. The fact that the data are “bad” in this context represents the fact that with rapid change in knowledge and techniques, the data believed to be “ground truth” today could very rapidly be found to be non-informative. Therefore, a willingness to recognize both this new set of facts and the need to stop using old facts is vital for the field to progress and provide greater impact to patients.
The second “Blue Sky” theme was that of dynamics change as opposed to static diagnosis and categorization. Presently tumors are analyzed at a point in time for location (images), lab values (biomarkers), and genetic and other “omic” datasets (pathology and other lab values). These all represent points in time and fail to fully describe either untreated tumor evolution or treated tumor response patterns in general. In very rare cases a before state and an after state are collected, but the very complex dynamics of the ”during” state make it nearly impossible to collect and analyze data. Therefore, few data exist for the “during” state even if it were possible or proper to change midtherapeutic course.
Predictive radiation oncology requires interdisciplinary expertise and multi-scale scientific data and models; and will require clinical granular data and models to be merged with dynamical theory to predict what we should see and what we should or can do to achieve the best outcomes for patients. This represents a large, long-term research, clinical and cultural shift. It requires that we adapt the way we think, design experiments, test hypotheses, and use computer power to ask new scientific questions (in new ways). Since the completion of the workshop series this new perspective has been presented as a focus of a new NCI radiation research program opportunity (15, 16).
The third “Blue Sky” theme was that the proposed computational and data science approach to create predictive radiation oncology is a long-term endeavor. In this context, computational development, computer architecture, and coding standards and tools would be very closely associated and would be co-developed to work optimally with biological science. To achieve the overarching workshop goals and this new synergistic focus, there is a pressing need to invest in a sustainable, scalable ecosystem that will merge/integrate the compute system with the data. Starting with imperfect models and data and iterating together to improve the data and models on an ongoing basis is a critical component for this long-term goal. If successful, data would advance from being “over the fence” to being attached “to a pipeline,” to ultimately becoming inherent in the compute infrastructure. This would allow a patient care path to optimal health before, during and after a cancer therapy event, as shown in Fig. 2. Along with the ever-present data focus the workshop participants identified the need to deploy DOE resources to explore the very physics of life alongside the biologists. Consensus among participants was that this would reveal primary truths as the process iterates over time and would help define scales and compute design. Participants even conjectured that the very nature of life and biology would lend itself to being studied in part via quantum computers rather than current types of supercomputers.
FIG. 2.
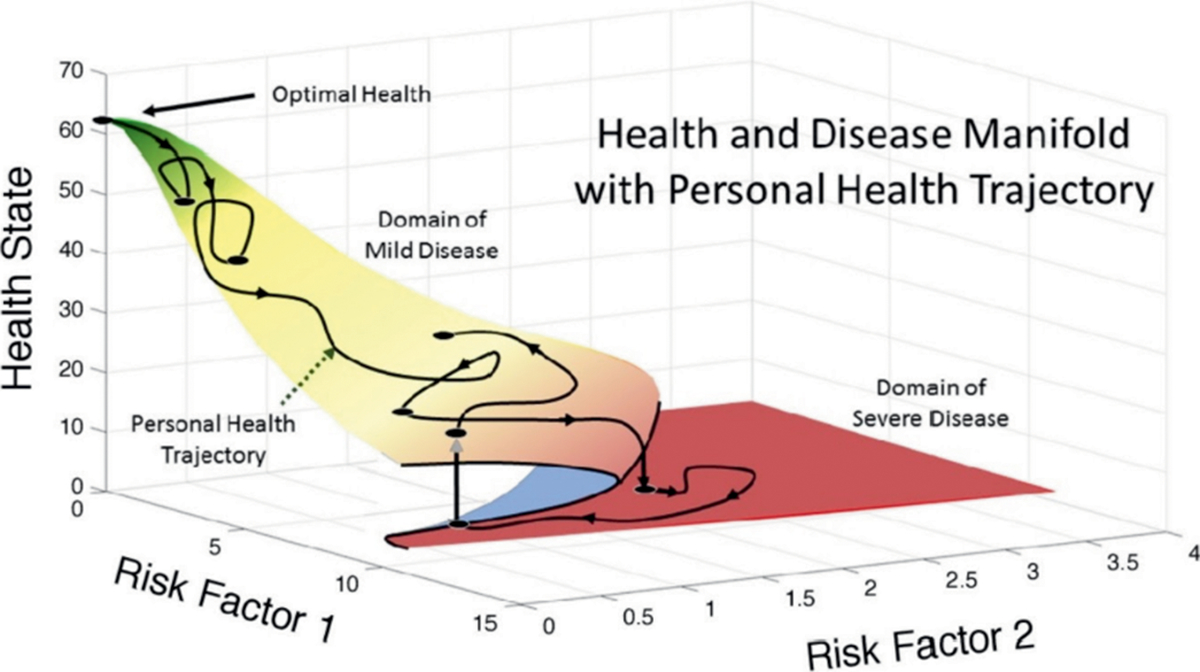
Multimodal patient trajectories demonstrate the way a person’s health can vary over time and with each “risk factor” potentially contributing one dimension in an ultimately n-dimensional problem. Here n = 2 to simply the figure.
DISCUSSION
Critical Nature of Collaboration and Mentorship – the Keys to Success Overall
That a workshop of this complexity is distilled to 2 groups of 12 questions and ideas is a remarkable achievement (17–19). The Supplementary Table S1 (https://doi.org/10.1667/RADE-22-00012.1.S1) includes the questions and the Supplementary Table S2 (https://doi.org/10.1667/RADE-22-00012.1.S1) lists various tools and skills. To bring this together for clinical and research application requires obvious collaboration across skill sets. While the taxonomy of some fields is very much a foreign language to others, the underlying concepts can be understood with a collaboratively designed team approach such as the NCI–DOE Collaboration. The rapidly changing understanding of artificial intelligence/machine or deep learning (AI/ML) includes the type and quality of information/data needed, how to make models more interpretable, and willingness to accept the short lifespan of any technology given the rapid rate of development, evolution, and improvement to technology for understanding biology. In addition, the fundamental understanding of molecules, cells and organisms not only changes rapidly, but often disproves a previous “paradigm” (e.g., “junk DNA” that turn out to be anything but junk). Mentoring, patience and communication must be multidirectional across fields and multi-generational to balance exuberance for new technology with wisdom gained from experience with other rapid changes and disruptive paradigm shifts that create an entirely new scientific landscape (ahh, yes, I remember when we used to. . .”).
To have confidence in this proposed NCI–DOE Predictive Radiation Oncology initiative, we must first demonstrate that it works in positive controls. This must happen from the outset, starting with validation of approaches. This is critical to answer the “who cares” question. We might start with some model murine systems with multi-parameter studies and some clinical scenarios for which there are long-term data, e.g., breast cancer and prostate cancer to understand risk factors and diseases for which we have even modest biology and cure rates of about 50% so there are successes and failures (e.g., oropharyngeal cancer and subsets on non-Hodgkin’s lymphoma where imaging guides therapy.) Multi-author “big science” papers seen in fundamental physics can link mentors, mentees, and divergent fields into shared success.
The workshop participants summated the 88 collaboration corner questions and the subsequent documents generated and developed a list of expertise believed to be necessary to move the goals of the workshop ahead. This is shown in Fig. 3 as a double helix to symbolize the belief of many workshop participants that the pairing of these fields, perhaps in permutations yet to be conceived, will enable and drive advances in radiation oncology.
FIG. 3.
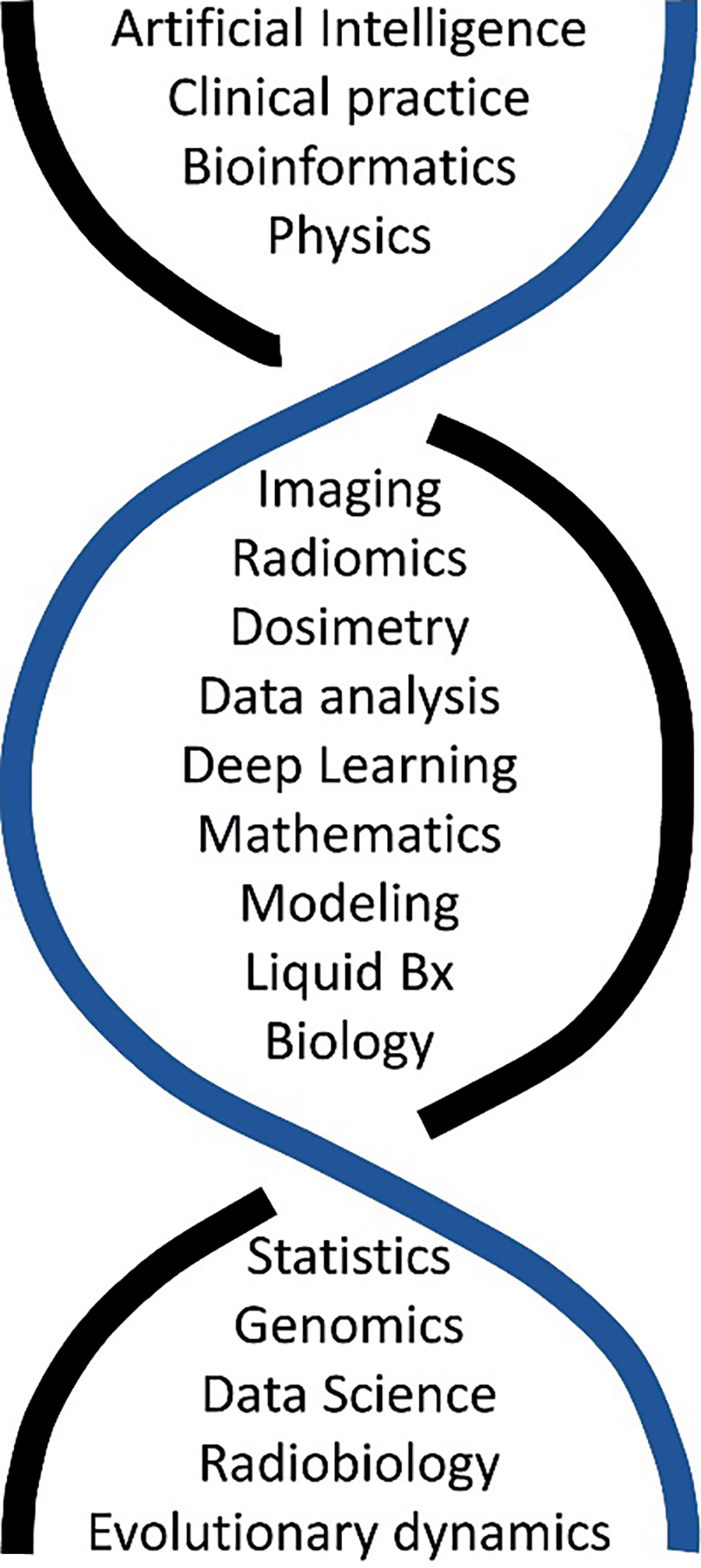
The merging of expertise that is needed to perform and develop predictive oncology in radiation oncology based on the workshop attendees’ inputs from 88 collaboration corner input documents.
Data Curation and National/International Change to Support the Future
Critical to the development of a predictive oncology infrastructure are interdisciplinary expertise and multiscale data that are validated and accessible. The workshop participants universally focused on this point, and is something that touches on every other scientific, logistic, and moral issue associated with the workshop’s focus. NCI and DOE have invested in this space. NCI has created the Cancer Research Data Commons (CRDC, https://datascience.cancer.gov/data-commons) to address data curation issues in the cancer space and the DOE National Center for Computational Science (NCCS) has created a secure data zone in its high performance computing infrastructure called CITADEL (https://bit.ly/3rdJOxK) that can hold non-deidentified patient data securely.
The needs for curation and support go beyond hardware and national infrastructure, however. Significant human talent is needed to develop and maintain the data optimally. The field of data science is growing and will be a critical component of cancer research moving forward. NCI focused resources include the NCI Center for Biomedical Informatics and Information Technology (CBIIT; https://datascience.cancer.gov/) and the Informatics Technology for Cancer Research (ITCR, https://itcr.cancer.gov) program. Many other NIH initiatives are emerging in this area, such as Bridge to Artificial Intelligence (Bridge2AI; https://commonfund.nih.gov/bridge2ai).
To support widespread scientific cross-disciplinary collaboration highlighted at the workshops, standards are needed for safe, federated data exchange. These standards can be established by the data hubs and centers being created across the U.S. and internationally. Research and investment in development of standards is currently underway (across many different fields). Optimized data analysis and storage are also vital to support the computational modeling and data evaluation. Some data, such as free text, will need to be imported. Other factors, such as spelling mistakes and colloquialisms will also need to be addressed.
The Path to Understanding is Multimodal-Multiscale
The concept of multimodality (different forms of measurement) and multiscale (spatial and temporal) data permeated all dimensions of the workshop. The participants highlighted the need for broad application of systems thinking that bridges physical, chemical, biological, physiological, and structural aspects. It will be essential to integrate the diverse information collected to help identify the right intervention to improve patient care and treatment outcomes (20). Currently, drugs are often designed to affect targets along biochemical pathways and combinations of drugs are tested based on their theoretical capacity to synergize. This theoretical capacity to create synergy with other drugs is based on data from the cellular scale that is, in turn, based on the underlying assumption that the systems are non-adaptable and static. Tumors in these patients are measured by imaging with tens and hundreds of billions of cells (centimeters) and biological responses that cross time scales from microseconds to years. Causalities from parent to self to society account for the default state-of-the-art clinical data in hand today which is intrinsically multiscale, as shown in Fig. 4 (21). While it is impossible today to go from a cell to a society in a model, this may be possible or even commonplace in the future. In addition, the pursuit of multi-modal, multiscale approaches requires bridging ontologies to allow different disciplines to collaborate.
FIG. 4.
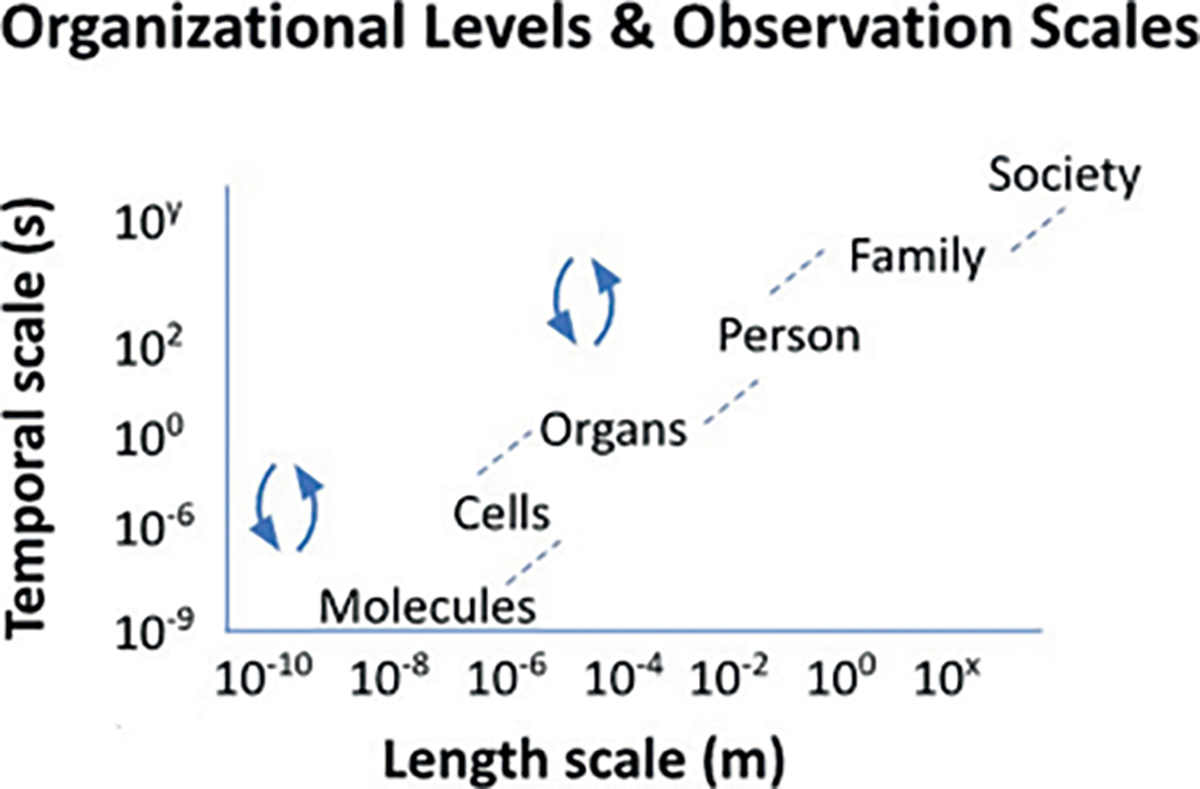
The multiscale aspect of cancer research and therapy is shown. The partnership between NCI and DOE will hopefully accelerate discovery in this space as the partnership’s science can better address the complexity by working together across scales.
Linking Physical Models to Computational Tools – The Key Variables
Physical models of human disease represent laboratory experiments generally employing cultured cells or animal models. These models can be genetically and biochemically manipulated to permit controlled experiments that expand our understanding of disease mechanisms and the potential impact of new therapeutics. Such models are difficult, time consuming and relatively expensive to create and maintain. In contrast, in silico models, if sufficiently accurate in their representation of biological processes, permit a much more complete exploration of a problem domain at a much lower cost and in a fraction of time. The issue, of course, is the creation of sufficiently accurate mathematical/computational models.
Mathematical oncology (22–24) is a rapidly advancing field dedicated to the creation of multi-scale computational models of cancer-related biological processes. Detailed models of the kinetics of cancer metabolism at the biochemical level (25) or models of cancer growth and response to therapy parameterized to predict the resulting MR images (26) are illustrative examples. These models can make accurate and reproducibility predictions of cancer progression and response. Specifically for radiation oncology, integrated mathematical models have identified optimal radiation schedules for individual cancers (27), radiation doses that optimally synergize with different immunotherapeutics (28), and innovative approaches to temporally feather radiation to escalate radiation dose to the target and decrease cumulative doses to organs at risk (29, 30). Different conceptual approaches are emerging on how to integrate mechanistic mathematical and computational models into radiation oncology decision making (31–33).
Data driven modeling using machine learning and AI techniques is also expanding rapidly as a new computational modality of cancer research. Such techniques require large, high-quality training and testing data sets from which to learn how to diagnose cancer or predict therapeutic response. Deep learning algorithms are proving particularly useful in this regard but require detailed analysis of the bases upon which they draw their conclusions (34). This “explainability” problem in AI is in fact an opportunity to generate new mechanistic hypotheses and to link machine learning models not only to the underlying biology but also to computational models of that biology (35, 36). Participants felt it was critical to define the key variables needed to allow models to become both simpler and remain fully predictive and relevant, making discovery of these key variables and their measurement a focus for the field. This was achieved in weather forecasting and allowed otherwise complex non-linear systems to be successfully modelled.
Radiation Biology Models and Mechanisms
Radiation biologists often uniquely combine specialized expertise from multiple fields to address fundamental mechanistic questions and application to the study of relationships between an absorbed radiation dose and subsequent biological responses. In the context of radiation oncology, research is directed at developing this understanding to enable improvements in the therapeutic ratio between tumor control relative to normal tissue damage. A typical strategy for an experimentalist is to test a hypothesis using biological model systems designed to interpret results with less ambiguity.
As was highlighted in the workshops, biological responses to radiation treatment are multiscale in both time and space. Advances in medical physics and instrumentation have brought greater precision in radiation dose delivery for cancer treatment. Characterization of the factors that summate as an individual’s therapeutic response to radiation treatment currently remains relatively crude by comparison. The notion of free radicals in biological signaling was born out of the studies that linked ionizing radiation’s mechanism of action to reactive oxygen species. Radiation has been extensively studied for its lesioning action at the level of DNA and subsequent capacity of cells to repair DNA damage. While the radiation biology field has developed a relatively firm understanding of radiation at the atomic to molecular scale, the radiation oncology field continues to struggle to link these events to an individual patient’s response to radiotherapy.
To address this the development of new capacities in both computational and biological (wet lab) modeling holds promise for coalescing data that decodes how signals become the patterns that govern radiation treatment responses from molecular, cellular, tissue, organ and systems levels. In this context, experimentalists need to focus on methods to prioritize which variables and modulator levers to design in the model systems that will best inform, and importantly, predict how an individual cancer patient may optimally benefit from a particular radiotherapy regime.
Sensing, PROs and Super-Granular Data Collection
Harvesting of high-granularity data from ambiently collected, on body peripheral biochemical monitoring devices and wearable sensing systems—whether in the clinical or ambulatory setting—can enable another source of temporally rich objective information. Further coupling of a patient’s status to mechanisms of reporting (e.g., from mobile apps, telemedicine, other), albeit more subjective, to large and semi-/continual data streams from sensors (e.g., wearables, at-home monitoring, other) adds the context necessary to help validate what was going on with a patient outside the clinic. To date, there have been both small and large studies to discover clinically actionable, and broadly applicable, signals from the noise from these types of devices and tools (37–39). Specifically, these include atrial fibrillation detection and many more measures of patient performance status (objective measures of ECOG), frailty, diabetes management, neurological disorders (40–42).
A myriad of devices have evolved over the last decade, from simple mechanical sensors that measure inertial movement to physiological sensors which employ optical, electrical, acoustic, or thermal sensing components that measure more specific biological signals. The physiological sensors measure vital signs (e.g., heart rate, blood oxygen saturation, temperature, blood pressure), bodily functions such as respiratory and gut activity, and bioelectrical activity. Furthermore, active or passive biochemical sampling (from eccrine sweat or interstitial fluid) is a readily evolving area for wearable devices. Currently there are approved devices for continual glucose monitoring (e.g., Dexcom, Abbott, Biolinq) and other analytes at much earlier stages (lactate, cortisol, electrolytes, cytokines, drugs and more) (43).
This is a rapidly expanding area which could offer much information to the pre-/during/post-treatment status of the patient and enable more robust symptom management or the earlier mitigation of side effects and adverse events from novel therapeutic combinations. Ultimately the medical value of worn devices that provide high-fidelity measures of biological signals will be to offer a coupling of data generated with robust clinical assays (single points in time) to a deluge of behavioral data generated between clinical visits. These high-fidelity, real-time assessments will provide advanced tools for analysis and understanding of individual disease dynamics, and ultimately prediction of more specific patient trajectories and prognosis (by way of models that utilize the totality of medical evidence available). Throughout the workshops, participants highlighted the evolving field of wearables as a promising area for further investigation in radiation oncology and care.
Challenges are still present with these devices and tools. For example, the tools do not typically measure robust biomarkers (e.g., antibodies, cells, etc.), but more often perturbations to temporally rich data streams of less specific data (e.g., vitals, movement, systemic biochemical concentrations/fluctuations, and more). An additional complication is deciphering the perturbations that are clinically meaningful versus ongoing, ‘normal’ biological processes and this must be addressed in studies utilizing these tools. To this end, most studies to date have required large patient populations to validate, were supported from modern data analytic methods such as deep learning, and have focused on clinical applications with substantial, continual real-world data collection (e.g., glucose monitoring with long histories of day-to-day data across large populations) or that could be easily crowdsourced to generate large, super-granular data sets (e.g., atrial fibrillation detection within the Health eHeart study). The workshop participants highlighted both performance status and sepsis as current evidence to support the use and utility of wearable devices in radiation oncology. Moreover, participants noted with enthusiasm the potential of utilizing out-of-clinic sensing tools for model validation, as secondary real-world measures of efficacy, and to support secondary trial endpoints. All of these are promising areas to explore.
Imaging Science, Biomarkers, and Clinical Radiation Oncology
Within the evolving practice of medicine in an increasingly digital and technological world, imaging data has become central for clinical decision making. In radiation oncology today, imaging information is used ubiquitously across the care pathway, including initial assessment and staging, decision making to treat a patient with radiotherapy, radiation treatment planning and delivery, and treatment response assessment. Advances in imaging have introduced novel approaches to characterizing tissues with greater complexity (44, 45). However, our current use of imaging data, which largely relies on qualitative or semiquantitative, manual analysis only scratches the surface of the full potential of quantitative imaging for personalized treatment of cancer patients. In parallel, advances in molecular biomarkers, measured in tumor tissues or biological fluids, are increasing our ability to predict radiation responses and measure them during treatment (44–46).
Some of the major challenges in clinical translation of these novel imaging approaches and biomarkers include, first, better understanding of the underlying biological mechanisms that are being revealed through these imaging measures to allow for actionability and, second, the need for tools and approaches to enable consistent, quantitative imaging measurements within broad clinical settings. Highlighted by the goals of this workshop, these challenges can be overcome through transdisciplinary collaboration to pursue shared efforts and research to connect the quantitative imaging developments with consistent, quantitative clinical outcomes and with the genomic, molecular tissue characterization. This would allow for multiscalar interpretation while capturing and employing the temporal dynamics of consistent, quantitative imaging measurements.
A unique opportunity available for radiation oncology comes from the investment in understanding radiation injury to normal tissues and post-exposure mitigators of damage supported by the National Institute of Allergy and Infectious Diseases (NIAID) and the HHS Biomedical Advanced Research and Development Authority of the Office of the Assistant Secretary for Preparedness and Response. There are biomarkers being investigated to extensive whole-body dose and for organ-specific injury. Given that radiation is anatomically focused it may be possible to develop biomarkers that assess the tumor and the surrounding normal tissue—and to tailor treatment based on the evolution of a patient’s biomarkers during a course of treatment.
Advanced Computing Linkage to Medicine, the Future and the Past
Advanced computing has been a cornerstone for research advancement, scientific innovation, competitive advantage, and economic prosperity (46). Furthermore, the Department of Energy’s Leadership Computing Facilities have a long history of enabling researchers to accelerate scientific discovery and deliver practical breakthroughs for some of the most computationally challenging problems across many disciplines, including medicine. Using advanced computational methods and supercomputers, biomedical scientists have expanded the scale and scope of their research, solving complex problems in new ways and in less time. Examples include early disease diagnosis, drug discovery, and the development of personalized medical treatments based on next-generation sequencing data.
In the past decade, the convergence of massive computational power, big data, and novel algorithms has ushered a new era in which artificial Intelligence is revolutionizing both biomedical discovery and healthcare delivery. While machines are not designed nor trained to replace physicians and nurses, they do have enormous potential to assist health professionals and clinicians with time-critical decisions for the smart delivery of healthcare. Going forward, additional studies will inform human-computer interactions as we use AI to augment and improve human performance.
At the core of these activities is the need for synergistic advances in both hardware and software. As we are approaching the end of Moore’s Law (47), we will continue to experience growth in heterogeneous architectures, application-specific accelerators, and alternative forms of computing such as neuromorphic and quantum (48). Although both are further away from delivering practical applications in medicine, they both hold a lot of promise. Still, extreme heterogeneity poses new challenges with memory systems, storage systems, interconnects, and software. The community will need to embrace a paradigm shift building upon the concept of hardware/software / application co-design.
Advanced computing capacity has a synergistic relationship with advanced computational modeling techniques. Current efforts focus on efficient implementation of models based on sets of differential equations or convolutional neural networks. These models are implemented using parallel programming techniques and contemporary programming languages and rely on extensive, well tested libraries of software. As the computational platforms move to more esoteric and powerful forms of computing (e.g., quantum computing), these software techniques will be inadequate or, if supported, likely too limiting. Thus, three interlinked development paths must advance together: hardware, software, and mathematical modeling techniques.
Patient Involvement: Being Part of the Process
The presence of three patient advocates and a broad mix of backgrounds in the participant matrix enabled rich discussion on topics regarding the patient perspective and in a related vein equity overall. Participants emphasized the need to optimize the methods and tools to a patient’s unique values and not an assumed set of values. For example, financial toxicity and the capacity to use advanced computing could be used to optimize treatment costs from hotel availability, parking costs, food costs, and avoidance of missed workdays due to predicted toxicity. This was a common theme in several discussions on the primary questions and in the collaboration corner context (49).
Participants highlighted the need for patients to get information from the health ecosystem in an understandable, transparent fashion and that the use of advanced computing, artificial intelligence, and machine learning to minimize bias and optimize understandability is “a very needed” near-term goal. Several patient advocates and staff present emphasized that having data in understandable chunks would increase clinical trial participation, build trust, and help match patient expectations to outcomes to avoid dissatisfaction and confusion. The capacity of advanced computing to match a patient’s background and life context to data presentation methods was identified as an unexplored area and a real gap to be addressed (50).
Personalization, Digital Twin
Over the course of the workshops, participants noted opportunities to use patient-specific data, including biomarkers and multi-omics, to predict individual disease or treatment trajectories. Other ideas in both the primary generated questions and in the collaboration corner that centered around the ability to test all potential options or combinations of treatments to determine an optimized course of treatment for an individual patient that includes built-in feedback loops. All these ideas point to the concept of a cancer patient digital twin (CPDT) or patient-tailored model incorporating multi-omic, clinical, environmental, and social data to predict individual patient trajectories that can inform shared decision making between patient and doctors (51, 52). The vision for the CPDT is to evaluate potential screening, preventive, and/or therapeutic plans at the individual level thus helping to prioritize a treatment plan to meet personalized objectives. CPDTs of the future must continuously integrate new data and knowledge across spatiotemporal scales to iteratively improve the accuracy of predictions in an active/continuous learning loop. Thus, a potential framework for the CPDT is a continuous life cycle that includes multiscale/multimodal patient data intake; creation and exploration of the CPDT leveraging statistical, data-driven, and mechanistic modeling at scale using high performance computing; CPDT prediction integration at point of care; treatment options visualization; and continuous data and knowledge accumulation and feedback. Ultimately, digital twin cohorts will be created and continuously updated as the continuous life cycle becomes more robust over time. The CPDT provides a paradigm shift in oncology because it will enable data-driven simulation models to be integrated into routine clinical care—a key step toward the goals of precision oncology: to uniquely and continuously tailor treatment to each individual patient over time.
Equity and Equality – Responsibility and Opportunity
Related to direct patient interaction enhancement tools and communication process improvement, the formal review and expansion of the representation of training data sets on patient focused AI/ML tools was noted. In a pre-workshop call for just students, the fact that AI data sets (people) are usually generated from only a handful of locations was of concern. This was echoed by several days of discussions during the workshops and was a final topic question in the final “World Café” workshop and in the “Collaboration Corner.” This is a focus of numerous contemporaneous publications and meetings but in the context of this workshop expands into the concepts of radiation therapy and possible real differences in outcomes across different populations on racial and geographic bases (53).
Patient input using natural language processing (NLP) was also discussed. NLP could be used to analyze transcripts from tumor boards to evaluate the biases of local caregivers relative to others across the planet to see if this aspect of bias can be addressed to both improve outcomes and identify the bias to educate caregivers of its existence and to ultimately eliminate it. The theme for patients was that the technologies and advanced computing methods include their unique care path and backgrounds, social and genetic, to make care more equitable and data-based conclusions/predictions according to their unique features, not to those of a small group of different people from another location that may not apply to them (54).
The nature of our society is changing and the new consumers of this effort, the current generations post the rise of social media and the internet of things, are ever more comfortable with computational aids and data movement in ways that would have made such an effort challenging to achieve, tolerate or fathom in previous generations. If this “Blue Sky” vision is achieved, computers will evolve from machines that calculate to enablers of cancer care discovery. This would be a primary long-term goal of data science in the context of NCI and DOE collaborative science. Notably, experts in the room felt it was achievable.
CONCLUSIONS
The workshop series created a new scientific space with energy and immense collaborative creativity with critical integration between mission critical computer science, medical science, and social science. The participants felt that “we,” meaning all areas of science represented by the participants, need to build upon this work with access to validated, multiscale data and continued research collaborations between DOE, NCI, our scientific communities, and our patients that is both equitable and nimble. Key to the success of this new scientific space is the team science or collaborative focus and the shared mission. Additional key success factors include the essential human factors: talent, resources, wisdom, energy, and the clear, shared desire to develop as a priority the ability to precisely predict patients’ outcomes and from that to optimize treatment paths. The radiation oncology space is well-suited for this effort, with a clear capacity to extend the science across the patient care continuum in time—and to drive the resulting paradigm shifts forward to improve care equitably with computational science sharing full membership in the mission of cancer care for the planet.
Supplementary Material
ACKNOWLEDGMENTS
The authors with to thank Petrina Hollingsworth of Frederick National Lab for Cancer Research for her tireless help with logistics and meeting organization emails. We also wish to thank the leadership of both NCI and DOE for support and feedback. The opinions presented in this paper reflect those of the authors and do not necessarily reflect those of their employers.
Footnotes
Editor’s note: The online version of this article (DOI: https://doi.org/10.1667/RADE-22-00012.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Bhattacharya T, Brettin T, Doroshow JH, Evrard YA, Greenspan EJ, Gryshuk AL, et al. AI Meets Exascale Computing: Advancing Cancer Research With Large-Scale High Performance Computing. Front Oncol. 2019; 9:984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NCI-DOE Collaboration 2020 Virtual Ideas Lab: Toward Building a Cancer Patient “Digital Twin”: NIH; 2020. [Available from: https://events.cancer.gov/cbiit/dtwin2020.
- 3.Berzins M Joint Design of Advanced Computing for Cancer (JDACS4C) - A Brief Overview and Perspective: Depart of Energy; 2018. [PDF of a presentation given in 2018]. Available from: https://science.osti.gov/-/media/ascr/ascac/pdf/meetings/201809/20180915_ASCAC_MB.pdf?la=en&hash=8618A60B3C749D4825E61A487173ED67CA995B28.
- 4.Peterson A, Cooke M. CANDLE Illuminates New Pathways in Fight Against Cancer: U.S. Department of Energy; 2019. [Available from: https://www.energy.gov/articles/candleilluminates-new-pathways-fight-against-cancer.
- 5.Atun R, Jaffray DA, Barton MB, Bray F, Baumann M, Vikram B, et al. Expanding global access to radiotherapy. Lancet Oncol. 2015; 16(10):1153–86. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed MM, Coleman CN, Mendonca M, Bentzen S, Vikram B, Seltzer SM, et al. Workshop Report for Cancer Research: Defining the Shades of Gy: Utilizing the Biological Consequences of Radiotherapy in the Development of New Treatment Approaches-Meeting Viewpoint. Cancer Res. 2018; 78(9):2166–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedict SH, Hoffman K, Martel MK, Abernethy AP, Asher AL, Capala J, et al. Overview of the American Society for Radiation Oncology-National Institutes of Health-American Association of Physicists in Medicine Workshop 2015: Exploring Opportunities for Radiation Oncology in the Era of Big Data. Int J Radiat Oncol Biol Phys. 2016; 95(3):873–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman CN, Prasanna PGS, Bernhard EJ, Buchsbaum JC, Ahmed MM, Capala J, et al. Accurate, Precision Radiation Medicine: A Meta-Strategy for Impacting Cancer Care, Global Health, and Nuclear Policy and Mitigating Radiation Injury From Necessary Medical Use, Space Exploration, and Potential Terrorism. Int J Radiat Oncol Biol Phys. 2018; 101(2):250–3. [DOI] [PubMed] [Google Scholar]
- 9.Schofield PN, Kulka U, Tapio S, Grosche B. Big data in radiation biology and epidemiology; an overview of the historical and contemporary landscape of data and biomaterial archives. Int J Radiat Biol. 2019; 95(7):861–78. [DOI] [PubMed] [Google Scholar]
- 10.Vikram B, Coleman CN, Deye JA. Current status and future potential of advanced technologies in radiation oncology. Part 1. Challenges and resources. Oncology (Williston Park). 2009; 23(3):279–83. [PubMed] [Google Scholar]
- 11.Vikram B, Coleman CN, Deye JA. Current status and future potential of advanced technologies in radiation oncology. Part 2. State of the science by anatomic site. Oncology (Williston Park). 2009; 23(4):380–5. [PubMed] [Google Scholar]
- 12.Citrin DE, Prasanna PGS, Walker AJ, Freeman ML, Eke I, Barcellos-Hoff MH, et al. Radiation-Induced Fibrosis: Mechanisms and Opportunities to Mitigate. Report of an NCI Workshop, September 19, 2016. Radiat Res. 2017; 188(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon DJJ, Tay LM, Ho D, Chua MLK, Chow EK, Yeo ELL. Improving the therapeutic ratio of radiotherapy against radioresistant cancers: Leveraging on novel artificial intelligence-based approaches for drug combination discovery. Cancer Lett. 2021; 511:56–67. [DOI] [PubMed] [Google Scholar]
- 14.Revett K, editor Examination of the Parameter Space of a Computational Model of Acute Ischaemic Stroke Using Rough Sets 2007; Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- 15.Buchsbaum JC, Espey MG. Radiation Oncology-Biology Integration Network “ROBIN” U54 Program RFA Concept: U.S. National Cancer Institute; 2021. [Available from: https://deainfo.nci.nih.gov/advisory/bsa/0321/Espey-Buchsbaum.pdf.
- 16.Espey MG, Buchsbaum JC, Vikram B. Notice of Intent to Publish a Funding Opportunity Announcement for Radiation Oncology-Biology Integration Network (ROBIN) Centers (U54 Clinical Trial Required): U.S. National Cancer Institute; 2021. [Available from: https://grants.nih.gov/grants/guide/notice-files/NOT-CA-21-045.html.
- 17.Vapiwala N, Thomas CR Jr., Grover S, Yap ML, Mitin T, Shulman LN, et al. Enhancing Career Paths for Tomorrow’s Radiation Oncologists. Int J Radiat Oncol Biol Phys. 2019; 105(1):52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford H Big Data and Small: Collaborations between ethnographers and data scientists. Big Data & Society. 2014; 1(2):2053951714544337. [Google Scholar]
- 19.Rivera DR, Lee JSH, Hsu E, Khoury MJ, Meng F, Olivero O, et al. Harnessing the Power of Collaboration and Training Within Clinical Data Science to Generate Real-World Evidence in the Era of Precision Oncology. Clin Pharmacol Ther. 2019; 106(1):60–6. [DOI] [PubMed] [Google Scholar]
- 20.Zanin M, Chorbev I, Stres B, Stalidzans E, Vera J, Tieri P, et al. Community effort endorsing multiscale modelling, multiscale data science and multiscale computing for systems medicine. Brief Bioinform. 2019; 20(3):1057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silverman EK, Schmidt H, Anastasiadou E, Altucci L, Angelini M, Badimon L, et al. Molecular networks in Network Medicine: Development and applications. Wiley Interdiscip Rev Syst Biol Med. 2020; 12(6):e1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rockne RC, Hawkins-Daarud A, Swanson KR, Sluka JP, Glazier JA, Macklin P, et al. The 2019 mathematical oncology roadmap. Phys Biol. 2019; 16(4):041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson AR, Quaranta V. Integrative mathematical oncology. Nat Rev Cancer. 2008; 8(3):227–34. [DOI] [PubMed] [Google Scholar]
- 24.Altrock PM, Liu LL, Michor F. The mathematics of cancer: integrating quantitative models. Nat Rev Cancer. 2015; 15(12):730–45. [DOI] [PubMed] [Google Scholar]
- 25.Roy M, Finley SD. Computational Model Predicts the Effects of Targeting Cellular Metabolism in Pancreatic Cancer. Front Physiol. 2017; 8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hormuth DA 2nd, Jarrett AM, Feng X, Yankeelov TE. Calibrating a Predictive Model of Tumor Growth and Angiogenesis with Quantitative MRI. Ann Biomed Eng. 2019; 47(7):1539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leder K, Pitter K, LaPlant Q, Hambardzumyan D, Ross BD, Chan TA, et al. Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell. 2014; 156(3):603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poleszczuk J, Enderling H. The Optimal Radiation Dose to Induce Robust Systemic Anti-Tumor Immunity. Int J Mol Sci. 2018; 19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez Alfonso JC, Parsai S, Joshi N, Godley A, Shah C, Koyfman SA, et al. Temporally feathered intensity-modulated radiation therapy: A planning technique to reduce normal tissue toxicity. Med Phys. 2018; 45(7):3466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsai S, Qiu RLJ, Qi P, Sedor G, Fuller CD, Murray E, et al. In vivo assessment of the safety of standard fractionation Temporally Feathered Radiation Therapy (TFRT) for head and neck squamous cell carcinoma: An R-IDEAL Stage 1/2a first-in-humans/feasibility demonstration of new technology implementation. Radiother Oncol. 2021; 163:39–45. [DOI] [PubMed] [Google Scholar]
- 31.Enderling H, Alfonso JCL, Moros E, Caudell JJ, Harrison LB. Integrating Mathematical Modeling into the Roadmap for Personalized Adaptive Radiation Therapy. Trends Cancer. 2019; 5(8):467–74. [DOI] [PubMed] [Google Scholar]
- 32.Caudell JJ, Torres-Roca JF, Gillies RJ, Enderling H, Kim S, Rishi A, et al. The future of personalised radiotherapy for head and neck cancer. Lancet Oncol. 2017; 18(5):e266–e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahid MU, Mohsin N, Mohamed ASR, Caudell JJ, Harrison LB, Fuller CD, et al. Forecasting Individual Patient Response to Radiation Therapy in Head and Neck Cancer With a Dynamic Carrying Capacity Model. Int J Radiat Oncol Biol Phys. 2021; 111(3):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabol P, Sincak P, Hartono P, Kocan P, Benetinova Z, Blicharova A, et al. Explainable classifier for improving the accountability in decision-making for colorectal cancer diagnosis from histopathological images. J Biomed Inform. 2020; 109:103523. [DOI] [PubMed] [Google Scholar]
- 35.Ba D, Babadi B, Purdon PL, Brown EN. Convergence and Stability of Iteratively Re-weighted Least Squares Algorithms. IEEE Transactions on Signal Processing. 2014; 62(1):183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzcar J, Wang Y, Heiland R, Macklin P. A Review of Cell-Based Computational Modeling in Cancer Biology. JCO Clin Cancer Inform. 2019; 3:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo S, Sung D, Kim S, Koo J. A review of wearable biosensors for sweat analysis. Biomed Eng Lett. 2021; 11(2):117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andreozzi E, Centracchio J, Punzo V, Esposito D, Polley C, Gargiulo GD, et al. Respiration Monitoring via Forcecardiography Sensors. Sensors (Basel). 2021; 21(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao W, Ota H, Kiriya D, Takei K, Javey A. Flexible Electronics toward Wearable Sensing. Acc Chem Res. 2019; 52(3):523–33. [DOI] [PubMed] [Google Scholar]
- 40.Jacobsen M, Dembek TA, Ziakos AP, Gholamipoor R, Kobbe G, Kollmann M, et al. Reliable Detection of Atrial Fibrillation with a Medical Wearable during Inpatient Conditions. Sensors (Basel). 2020; 20(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman SA, Adjeroh DA. Deep Learning using Convolutional LSTM estimates Biological Age from Physical Activity. Sci Rep. 2019; 9(1):11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cappon G, Cossu L, Boscari F, Bruttomesso D, Sparacino G, Facchinetti A. An Integrated Mobile Platform for Automated Data Collection and Real-Time Patient Monitoring in Diabetes Clinical Trials. J Diabetes Sci Technol. 2021:19322968211024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn J, Kidzinski L, Runge R, Witt D, Hicks JL, Schussler-Fiorenza Rose SM, et al. Wearable sensors enable personalized predictions of clinical laboratory measurements. Nat Med. 2021; 27(6):1105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busato A, Fumene Feruglio P, Parnigotto PP, Marzola P, Sbarbati A. In vivo imaging techniques: a new era for histochemical analysis. Eur J Histochem. 2016; 60(4):2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He J, Zhang F, Xie G, Yao S, Feng Y, Bastos DCA, et al. Comparison of multiple tractography methods for reconstruction of the retinogeniculate visual pathway using diffusion MRI. Hum Brain Mapp. 2021; 42(12):3887–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Office of Science, Department of Energy, US Government. High Performance Computing 2021. [Available from: https://www.energy.gov/science/initiatives/high-performance-computing.
- 47.Leiserson CE, Thompson NC, Emer JS, Kuszmaul BC, Lampson BW, Sanchez D, et al. There’s plenty of room at the Top: What will drive computer performance after Moore’s law? Science. 2020; 368(6495). [DOI] [PubMed] [Google Scholar]
- 48.Mohamed KS. Neuromorphic Computing and Beyond. 1 ed: Springer International Publishing; 2020. p. 1–13. [Google Scholar]
- 49.Vollmer S, Mateen BA, Bohner G, Kiraly FJ, Ghani R, Jonsson P, et al. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ. 2020; 368:l6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross P, Spates K. Considering the Safety and Quality of Artificial Intelligence in Health Care. Jt Comm J Qual Patient Saf. 2020; 46(10):596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masison J, Beezley J, Mei Y, Ribeiro H, Knapp AC, Sordo Vieira L, et al. A modular computational framework for medical digital twins. Proc Natl Acad Sci U S A. 2021; 118(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laubenbacher R, Sluka JP, Glazier JA. Using digital twins in viral infection. Science. 2021; 371(6534):1105–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castelvecchi D Prestigious AI meeting takes steps to improve ethics of research. Nature. 2021; 589(7840):12–3. [DOI] [PubMed] [Google Scholar]
- 54.Johnson SLJ. AI, Machine Learning, and Ethics in Health Care. J Leg Med. 2019; 39(4):427–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


