Abstract
Training of the innate immune system with orally ingested bacterial extracts was demonstrated to have beneficial effects on infection clearance and disease outcome. The aim of our study was to identify cellular and molecular processes responsible for these immunological benefits. We used a murine coronavirus (MCoV) A59 mouse model treated with the immune activating bacterial extract Broncho-Vaxom (BV) OM-85. Tissue samples were analysed with qPCR, RNA sequencing, histology, and flow cytometry. After BV OM-85 treatment, interstitial macrophages accumulated in lung tissue leading to a faster response of type I interferon (IFN) signalling after MCoV infection resulting in overall lung tissue protection. Moreover, RNA sequencing showed that lung tissue from mice receiving BV OM-85 resembled an intermediate stage between healthy and viral infected lung tissue at day 4, indicating a faster return to normal tissue homoeostasis. The pharmacologic effect was mimicked by adoptively transferring naive lung macrophages into lungs from recipient mice before virus infection. The beneficial effect of BV OM-85 was abolished when inhibiting initial type I IFN signalling. Overall, our data suggest that BV OM-85 enhances lung macrophages allowing for a faster IFN response towards a viral challenge as part of the oral-induced innate immune system training.
Keywords: Macrophage, Trained immunity, Bacterial extract, Mouse coronavirus
Introduction
Following an exposure to infectious agents or vaccines, trained immunity can mount a faster and greater response to a challenge with the same agent or even heterologous pathogens [1]. Live-attenuated vaccines were already shown to induce a protective trained innate immune system [2]. The use of bacterial extracts was previously demonstrated to have immune activation and immune training properties [3]. Treatment with the approved microbial product Broncho-Vaxom OM-85 (BV OM-85) resulted in protective effects in rhinovirus infection of human bronchial epithelial cells [4] and in animals infected with influenza on initial infection outcome and on prevention of a following bacterial infection [5]. In humans, BV OM-85 is indicated for prophylactic treatment to prevent recurrent respiratory tract infections in both adults and children with a suggested mode of action of triggering immunomodulatory and protective immune responses against different pathogens in vivo, including influenza and respiratory syncytial virus as well as bacterial superinfection following influenza [6, 7]. BV OM-85 is composed of a mixture of lyophilized bacterial compounds connected to lung infections including Haemophilus influenzae, Streptococcus (Diplococcus) pneumoniae, Klebsiella pneumoniae ssp. pneumoniaeund ssp. ozaenae, Staphylococcus aureus, Streptococcus pyogenes and sanguinis (viridans), and Moraxella(Branhamella/Neisseria) catarrhalisthat is applied orally.
We used a murine coronavirus (MCoV) model to determine the effects of oral immune system training. MCoV or mouse hepatitis virus (MHV) was first isolated in 1949 [8]. It is highly contagious in laboratory mice, having been one of the most abundant viruses in mouse colonies [9]. Duration of infection varies depending on the MCoV strain, route of inoculation, and host factors, including age, immunocompetence, passive immunity, genetic strain, and genetic alterations [10]. Initially, MCoV infection via a nasal route leads to infection of the lung with subsequent spreading to other organs, including the liver. Immunocompetent mice that recover from an infection with MCoV are resistant to strain-specific reinfection [11, 12]. The innate immune system acts as a powerful barrier against MCoV infection [13]. Therefore, a proper balance between early activation of the innate immune system to fight off the viral infection and controlling the activation of the immune system to prevent organ damage is required to successfully battle a coronavirus infection [14].
A central role in host defence versus viral infection are interferons (IFNs) and their downstream activation targets as IFNs possess antiviral, anti-proliferative, and immunomodulatory effects [15]. Among the first responses to a viral challenge is the induction of type I IFN including IFNα and IFNβ, mostly secreted from local innate immune cells [16]. Interestingly, evasion tactics of betacoronaviruses include the initial inhibition of especially IFNβ [17]. In contrast, IFNα levels were upregulated after infection with MCoV [18]. Especially the early activation of interferon-alpha/beta receptor alpha chain 1 (IFNAR1), which is the major component of IFN type I signalling, on innate immune cells was described to be crucial in an MCoV mouse model [18]. A late activation of the innate immune system was demonstrated to be detrimental in human SARS-CoV-2 patients, which might be due to delayed activation of the IFNAR1 signalling cascade [19].
The aim of our study was to investigate the mode of action of immune system training via oral bacterial extracts during a viral infection. We used a mouse MCoV model as this was demonstrated to lead to pneumonia in mice when infected via the nasal route. We found that training of the host immune system is beneficial via the increase of lung tissue macrophages.
Materials and Methods
Mice
Animals were bred in the in-house breeding facility of the Centre for Biomedical Research under SPF conditions. Mice were housed in a BSL 2 unit in groups of 3–5 animals randomly grouped by sex in randomly distributed cages with dust free bedding, nesting material, tap water, and pelleted food ad libitum. Room temperature was 21°C (± 2°C) with 55 % (± 10 %) humidity, and 12-h day/12-h night light cycle. During viral infection, humane endpoints were weight loss of 20 % and behavioural changes. The mean weight change of all mice at the end of experiments was 96.23 % (± 6.020 %). 74 mice were divided into two equally sized groups. Treated mice were orally gavaged with 7.2 mg BV OM-85 (dissolved in 200 μL 3% D-mannitol) for 10 days. Control mice were orally gavaged with 200 μL 3% D-mannitol. After treatment regimen, mice were inoculated with 20 μL MCoV (1.5 × 106 TCID50) intranasally under general anaesthesia. Uninfected samples were collected from 7 mice each. On days 2, 4, and 10 post-inoculations, 10 mice of each group were sacrificed by cardiac puncture under general anaesthesia followed by cervical dislocation. Samples were obtained under sterile conditions. General anaesthesia: 0.5 mg/kg medetomidine (Domitor®, Orion Pharma) and 5 mg/kg midazolam (Accord) administered intraperitoneally. Antagonization, if required, was performed with 2.5 mg/kg atipamezole (Antisedan®, Orion Pharma) and 0.5 mg/kg flumazenil (Pharmaselect) subcutaneously. For the adoptive transfer experiments, 6 mice received 410,000 cells/20 μL, harvested from 12 mice, by endotracheal intubation under general anaesthesia using a size 22-G intravenous catheter in combination with a commercial mouse intubation device. After endotracheal intubation, the cell solution was administered directly via the catheter using the previously blunted intravenous catheter needle. A control group of 6 mice received 20 μL Ringer's lactate solution (saline). The following day, the mice were inoculated with MCoV and sacrificed on day 4 described above. For inhibition of IFNAR1, 30 μg Ultra-LEAFTM anti-mouse IFNAR1 (MAR1-5A3, BioLegend) was co-applied with the virus.
Virus Propagation
The MCoV MHV-A59 virus was kindly provided from Prof. Burkhard Ludewig. Virus was expanded on the NCTC clone 1469 cell line. In short, a confluent cell layer was infected with 50 μL virus solution for 20 min and cultured in Dulbecco's modified Eagle's medium containing 10 % horse serum for 48 h. Virus was harvested by separating the cell culture supernatant from the cellular debris by centrifugation. Virus was titrated by serial dilution using the Reed-Munch method to a TCID50/ml of 7.52 × 107.
RNA Isolation and qPCR
RNA from the lobus medianus pulmonis dextri was isolated using Maxwell® RSC simplyRNA Tissue Kit (Promega) in combination with a ball mill and reverse transcribed into cDNA using GoScriptTM Reverse Transcription System (Promega). Real-Time-PCR was performed using C1000 Touch Thermal Cycler (Bio-Rad). Primers were designed and assigned to their respective probes using the Roche Universal ProbeLibrary Assay Design Centre (http://www.universalprobelibrary.com/), with the exception of PolyU [20] and Nsp12 [21]. Cq values were normalized by using delta-delta Ct method to β-actin as housekeeper. Virus genes in Figure 1b were further normalized to day 2 values and averaged. In Figure 2 and Figure 3c, values were normalized to naive average value (n = 6; data not shown). Primer sequences are given in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000519699).
Fig. 1.
Immune activation by BV OM-85 reduced virus burden and lung apoptosis. a Schematic overview of treatment and infection scheme. b To measure viral burden, total lung tissue was analysed for the 2 viral mRNAs PolyU and Nsp12 at days 2, 4, and 10 post-infection as indicated in Methods. Values represent individual averages at each time point and condition with individual values and an average ± SD given in the panel (n = 10). c Lung tissue was stained at days 2, 4, and 10 post-infection for the MCoV protein Nsp9 (yellow staining). Scale bar: 400 μm. d Viral distribution was evaluated by counting infected alveoli by a blinded investigator at days 2, 4, and 10. Values represent total infected alveoli per slide with individual values and an average ± SD given in the panel (n = 10). e Cell apoptosis at days 2, 4, and 10 was determined by staining for DNA nicks using TUNEL staining and automatically analysed for TUNEL-positive nuclei using CellProfiler (157,232 cells on average). Identified TUNEL-positive areas are marked with arrows in the upper panels while their overlays are displayed in the lower panels. Scale bar: 50 μm. f Values represent percent cells positive for TUNEL staining per slide with individual values and an average ± SD given in the panel (n = 10). g As a second apoptosis marker, cleaved caspase 3 staining was used at days 2, 4, and 10. Cells positive for cleaved caspase 3 are indicated by white arrows. Scale bar: 50 μm. h Analysis of cleaved caspase 3 positive cells at days 2, 4, and 10 were performed on whole tissue section using CellProfiler (116,248 cells on average). Values represent cleaved caspase 3 positive cells as a percentage of total lung cells with individual values and an average ± standard deviation given in the panel (n = 10). Infected control: MCoV infected and mannitol control; infected treated: MCoV-infected and BV OM-85 treatment. BV, Broncho-Vaxom; MCoV, murine coronavirus; SD, standard deviation.
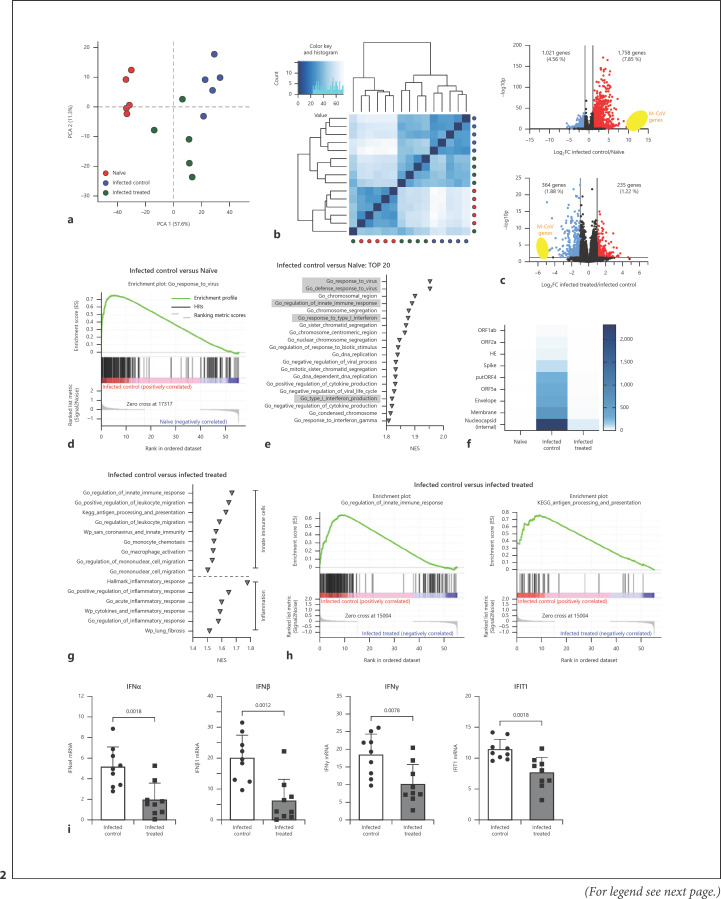
Fig. 2.
RNA sequencing analysis of total lung tissue at day 4 after virus infection with or without treatment. a Plot of the first 2 dimensions of a PCA, based on regularized log2-transformed count data. b Plot of Euclidian distance between groups with dendrogram. Dark blue: samples expected to cluster together. Light blue: samples expected to be more distant from each other. c Volcano plots showing differentially expressed genes of infected control versus naive (upper plot) and infected treated versus infected control (lower plot) lungs. Y-axis displays the negative log10-transformed adjusted p value, and x-axis shows log2-fold changes. Red dots represent 2-fold upregulated and blue dots 2-fold downregulated genes. Threshold for significance was -log10p > 1.3. MCoV genes are highlighted in yellow. d GSEA enrichment plot of GO term “Response to Virus” for infected control versus naive lungs. e Top 20 of all GO terms of GSEA analysis of infected control versus naive lungs. GO terms were ranked according to NES. Highlighted GO terms include viral responses, type 1 IFN signalling, and innate immunity. f Heatmap of MCoV genes from naive lungs, infected control lungs, and infected treated lungs, generated using average of TPMs. g Selected gene sets of GSEA analysis of infected control versus infected treated lungs, as described in Methods. Gene sets were ranked according to NES. h GSEA enrichment plot of GO term “Regulation of Innate Immune Response” and KEGG pathway “Antigen Processing and Presentation” infected control versus infected treated lungs. i IFN α, IFNβ, IFNγ, and IFIT1 mRNA levels in lung tissue from animals with or without BV OM-85 treatment before virus infection. Mean ± SD; n = 9. Naive: uninfected, untreated mice; infected control: MCoV-infected and mannitol control; infected treated: MCoV-infected and BV OM-85 treatment. BV, Broncho-Vaxom; MCoV, murine coronavirus; PCA, principal component analysis; IFNs, interferons; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; GO, gene ontology; GSEA, Gene Set Enrichment Analysis; NES, normalized enrichment score; SD, standard deviation.
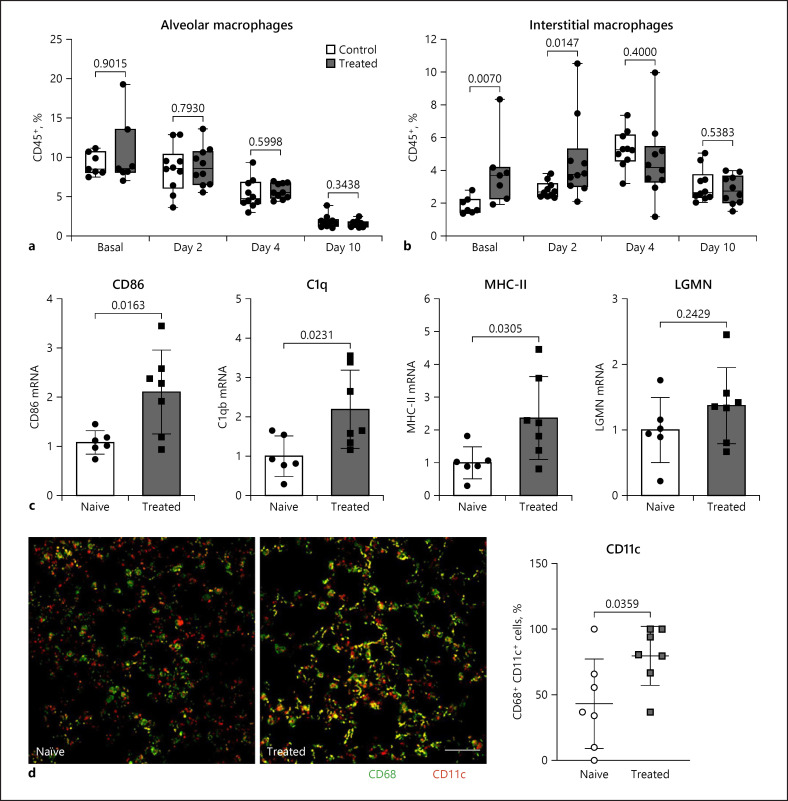
Fig. 3.
Macrophages during treatment and viral infection. a Percentage CD64+ CD170+ alveolar macrophages of CD45+ was determined by flow cytometry at baseline and 2, 4, and 10 days after virus infection (n = 7–10). b Percentage CD64+ CD170− interstitial macrophages of CD45+ was determined by flow cytometry at baseline and 2, 4, and 10 days after virus infection (n = 7–10). c Lobus medianus pulmonis dextri tissue was analysed by qPCR for markers of enhanced antigen presentation after 10 days of treatment with BV OM-85 in comparison to naive lung. Individual values and an average ± SD is given in the panel (n = 6–10). d The activation status of lung macrophages in uninfected, naive mice treated with BV OM-85 compared to naive lungs was determined by co-staining of the macrophage marker CD68 with the activation marker CD11c. Double-stained cells appear as yellow structures and were analysed and quantified using CellProfiler. Values represent CD11c-positive CD68 macrophages and are given in percent of all nucleated cells. Individual levels and an average ± SD are given in the panel. Scale bar: 50 μm; n = 7. Naive: uninfected, untreated mice; treated: uninfected and BV OM-85 treatment. BV, Broncho-Vaxom; MCoV, murine coronavirus; SD, standard deviation.
Histology
Freshly isolated left lung (pulmo sinister) were fixed with 4% formaldehyde, embedded in paraffin, and cut in 5-μm-thick sections. TUNEL stain (Promega) was performed according to manufacturer's protocol. For eosin and haematoxylin staining, deparaffinised, rehydrated slides were stained for 4 min with haematoxylin (Morphisto), rinsed for 4 min with tap water, stained for 30 s with eosin Y (Sigma-Aldrich), and finished by dehydration with ethanol and xylol. For immunofluorescence, lungs were deparaffinised and rehydrated; target retrieval was performed using Dako target retrieval solution. Sections were blocked with 2% bovine serum albumin, 0.5% fish gelatin, and 0.3% Tween-20 for 90 min, followed by blocking with mouse-on-mouse IgG blocking solution (Thermo Fisher), if anti-Nsp9 was used. Primary antibodies were Nsp9 (2C6.H1; Thermo Fisher), cleaved caspase 3 (5A1E; Cell Signaling), CD68 (FA-11; Bio-Rad), and CD11c (N418; BioLegend). Primary antibodies were incubated overnight at 4°C and corresponding secondary antibodies for 2 h at room temperature. All images were scanned on an automated tissue FACS microscopy stage on an Observer Z1 microscope at ×20 magnification (NA 0.5). Automated analysis was performed with CellProfiler [22].
RNA Sequencing and Data Analysis
Sequencing libraries were prepared at the Core Facility Genomics, Medical University of Vienna using the NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (both New England Biolabs) according to manufacturer's protocols. Libraries were QC-checked on a Bioanalyzer 2100 using a high-sensitivity DNA kit (Agilent) for correct insert size and quantified using Qubit dsDNA HS Assay (Thermo Fisher). Pooled libraries were sequenced on a NextSeq500 instrument (Illumina) in 1 × 75 bp single-end sequencing mode. Approximately 22 million reads were generated per sample. Reads in FASTQ format were aligned to the mouse reference genome version GRCm38 supplemented with the viral genome NC_048217.1 with Gencode mV23 annotations supplemented with virus genome annotations using STAR aligner [23] version 2.6.1a in 2-pass mode. Reads per gene were counted by STAR, and differential gene expression was calculated using DESeq2 [24] version 1.22.2. TPMs were generated by RSEM [25]. Gene set enrichment was calculated using GSEA [26, 27] version 4.1.0, and the Molecular Signatures Databases hallmark gene sets, canonical pathways (BIOCARTA, KEGG, PID, REACTOME, and WikiPathways), and Gene Ontology (GO) gene sets [27, 28, 29].
Flow Cytometry
The lobus cranialis pulmonis dextri and caudalis dextri were cut into small pieces and incubated with 2 mg/mL collagenase IV and 50 U/mL DNase I in HBSS at 37°C on a shaking plate. After 1 h, digested tissue was meshed through a 70-μm cell strainer, washed with PBS, and erythrocytes were lysed with 150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM Na2EDTA for 5 min. Washed cells were stained with antibodies from BioLegend against CD45 (30-F11, 1:50), CD64 (X54-5/7.1, 1:50), and SiglecF (CD170) (S17007L, 1:50) for 15 min and measured with an Attune NxT flow cytometer (Thermo Fisher) and data analysed with Attune NxT Software v3.1.2. For intracellular FACS, washed cells were stained with CD45, CD64, and SiglecF for 15 min and prepared with Intracellular Staining Permeabilization Wash Buffer (BioLegend) according to manufacturer's protocols. Cells were stained with anti-interferon regulatory factor antibody (D5E4, 1:50, Cell Signaling) for 20 min.
Adoptive Cell Transfer
Whole lungs of 12 mice were cut into small pieces and digested for 1 h at 37°C with 2 mg/mL collagenase IV and 50 U/mL DNase I in HBSS at 37°C on a shaking plate. After meshing through a 70-μm cell strainer, cells were pooled, counted, adjusted to 108 cells/mL in EasySep Buffer (Stemcell Technologies), and processed by a positive selection kit with magnetic bead release (17655, Stemcell Technologies) according to manufacturer's protocol, with 2 μg/mL biotinylated CD14 antibody (Clone Sa14-2, BioLegend), 100 μL/mL Positive Selection Cocktail, and 100 μL/mL Release RapidSpheres.
Statistics
Data were analysed using GraphPad 8.0. Normality was tested using Anderson-Darling or, if samples size was smaller than 8, the Kolmogorov-Smirnov test. Data with Gaussian distribution were checked for statistical significance using Student's t test, data without Gaussian distribution were analysed using the Mann-Whitney U test.
Results
To understand if a BV OM-85 trained immune system is beneficial during an MCoV infection, mice were pre-treated with BV OM-85 on 10 consecutive days before viral infection and analysed 2, 4, and 10 days after intranasal MCoV infection (Fig. 1a). Two viral markers, Nsp12 and PolyU, were determined in the lung using qPCR to measure viral load. Our results demonstrated that mice with a trained immune system displayed a massive virus reduction at day 4, which was retained at day 10 (combined score Figure 1b, separate values are given in online suppl. Fig. 1a). Immunofluorescence staining of lung tissue for the MCoV protein Nsp9 showed fewer viral foci 2 days after infection in treated animals. After 4 days, the viral staining was dispersed similarly in both groups, which was then resolved to foci at day 10 with treated animals having fewer viral foci than the control group (Fig. 1c and d, total lung images are given in online suppl. Fig. 1b–e). We did not observe adverse effects in lung pathology in treated mice indicating that the preactivation of the immune system did not magnify pathological processes induced by the virus infection (online suppl. Fig. 1f, g). Virus infections are accompanied by apoptosis of the host cells [30]. Treated animals displayed reduced apoptotic burden in lung tissue at days 4 and 10 compared to the control group (Fig. 1e, f). In addition, treated animals had a significant reduction in cleaved caspase-3 positive cells 10 days after virus infection (Fig. 1g, h).
BV OM-85 treatment led to enhanced viral clearance and reduced apoptotic burden in the lung. To identify changes in the lung tissue of infected BV OM-85-treated versus infected control animals, we performed RNA sequencing on samples of naive lungs, MCoV-infected control lungs, and lung tissue of mice with MCoV infection and training with BV OM-85. Samples of infected lungs for both treated and untreated mice were taken at day 4 as we observed strong reduction of viral load and lung tissue protection in the treated group. Upon principal component analysis, we found that experimental groups formed defined clusters within each other so that the variability between samples of a group was smaller than between groups (Fig. 2a). Clustering visualized as a heatmap of Euclidean distances supported the notion that BV OM-85 pretreatment (infected treated) led to lungs that can be characterized as an intermediate stage between naive and infected control lungs (Fig. 2b). Overall, MCoV virus infection led to a significant upregulation of 1,758 genes and a downregulation of 1,021 genes when comparing infected control lung tissue to naive tissue (Fig. 2c). The strongest differentially expressed features are MCoV transcripts. In general, GO term analysis of differentially expressed genes confirmed the immune response towards viral infection (Fig. 2d). The regulation of innate immune response was a central readout (Fig. 2e, online suppl. Fig. 2a). We could clearly distinguish virus response, innate immunity, and inflammation-related pathways during MCoV infection (online suppl. Fig. 2b). The difference between infected control and infected BV OM-85-treated lung tissue was smaller with a total of 599 differentially expressed genes (Fig. 2c). The strongest reduction of expression in samples with BV OM-85 treatment was observed in virus genes which can also be seen in an enrichment plot (Fig. 2f). When analysing the top 20 changed pathways using GO analysis by comparing infected control mice with BV OM-85-treated infected mice, we found signatures for the aforementioned innate immune response together with changes in type I IFN response (online suppl. Fig. 2c, d). Analysis for pathways specifically linked to innate immune cells and inflammation revealed strong changes in infected control versus infected, BV OM-85-treated animals (Fig. 2g, h). As IFN signalling is crucial during a viral infection, we confirmed the changes in IFN pathway-related genes using qPCR and the 2 type I IFNs, IFNα and IFNβ, the type II IFN IFNγ, and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) (Fig. 2i). Our data therefore support our previous observation of reduced viral stress starting at day 4 in BV OM-85-treated animals.
RNA sequencing data indicated differences in innate immune response, including antigen processing and presentation, and macrophage activation between treated and untreated mice. Therefore, we determined differences in lung macrophages at baseline and during the course of the virus infection to understand changes in macrophage function. The lung macrophage homeostatic population can be distinguished into 2 main cell populations: alveolar macrophages with embryonic origin and interstitial macrophages deriving from circulating monocytes [31, 32]. Alveolar macrophages were not changed with treatment and were reduced gradually during infection, as expected (Fig. 3a). However, we observed an increase in interstitial macrophages after BV OM-85 treatment and before virus infection which was still retained 2 days after virus infection (Fig. 3b). In addition, BV OM-85 treatment led to a phenotypical change with increased mRNA levels of genes related to antigen presentation already at baseline (Fig. 3c). Similarly, CD68+ lung macrophages of BV OM-85-treated mice showed enhanced levels of CD11c compared to macrophages of naive lungs (Fig. 3d). However, this baseline activation was not associated with a changed baseline inflammatory profile or a different course of inflammation as determined by mRNA levels of IL-6 and TNF-α (online suppl. Fig. 3a, b). We further evaluated the baseline activation state of endothelial cells and their activation profile during MCoV infection. We did not observe differences in VCAM and ICAM mRNA and soluble ICAM protein in circulation at any time point (online suppl. Fig. 3c–e).
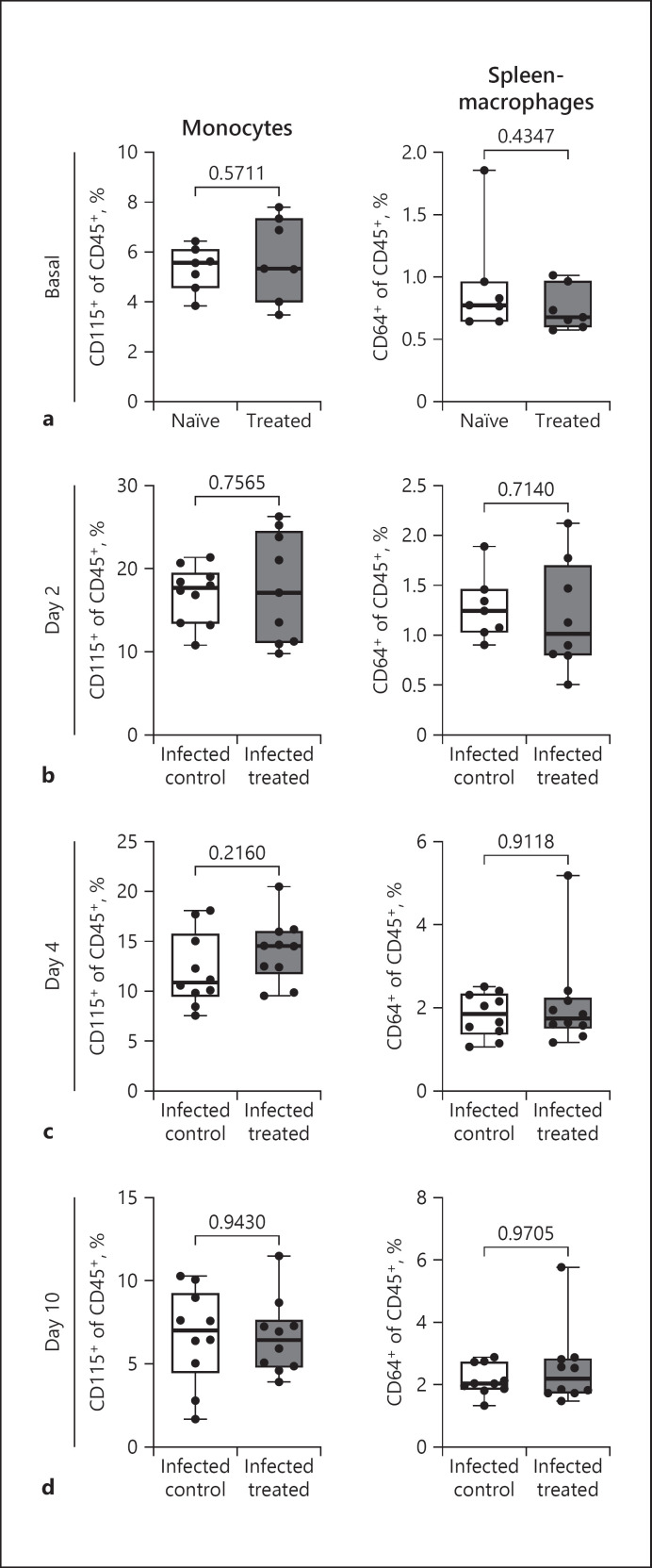
Oral treatment of mice with BV OM-85 leads to a modulation of the immune system of the lung. To determine if these modulations are organ-specific or can be observed in circulation or spleen, we analysed monocytes and spleen macrophages before virus infection and during the course of MCoV infection. We found no difference in monocyte amounts in the circulation or in macrophages in the spleen at baseline (Fig. 4a) or during MCoV infection (Fig. 4b-d), regardless of BV OM-85 treatment therefore indicating a lung-specific accumulation of interstitial macrophages induced by BV OM-85 pretreatment.
Fig. 4.
Changes in monocytes and macrophages with BV OM-85 treatment at baseline and during coronavirus infection. Levels of circulating CD45+ CD115+ monocytes and of CD45+CD64+ spleen macrophages were evaluated at baseline (a) and at day 2, (b) day 4, (c) and day 10 (d) after MCoV infection. Values are given as percent cells per CD45+ cells and an average ± SD is given in the panel (n = 10). Naive: uninfected, untreated mice; treated: uninfected and BV OM-85 treatment; infected control: MCoV-infected and mannitol control; infected treated: MCoV-infected and BV OM-85 treatment. BV, Broncho-Vaxom; MCoV, murine coronavirus; SD, standard deviation.
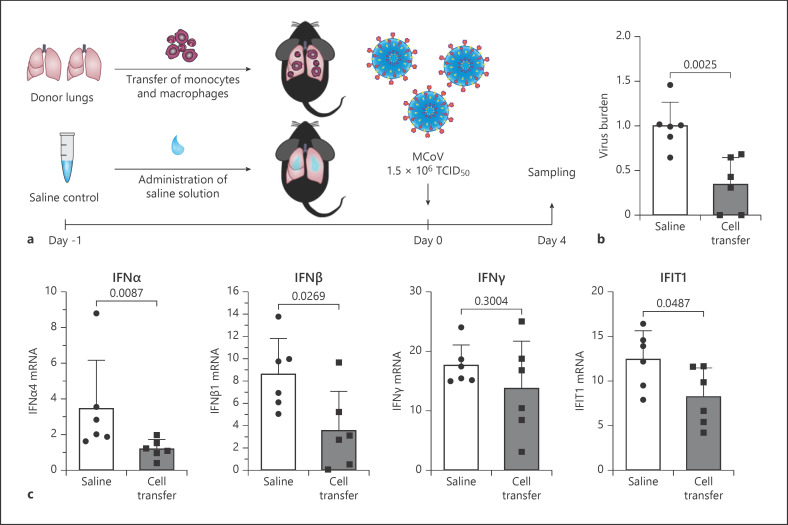
To test whether increased accumulation of lung macrophages is already sufficient to reduce MCoV infection, we isolated monocytes and macrophages by CD14+ magnetic bead selection from lung tissue from naive lungs and adoptively transferred them into recipient mice (2 donor animals for each recipient animal, Fig. 5a). Mice were then infected with MCoV and samples were analysed 4 days later. Similar to BV OM-85 treatment, mice receiving a macrophage cell transfer showed 53.91% reduced lung viral burden compared to saline treated mice (Fig. 5b). In addition, the reduced viral burden was also accompanied by a reduction in IFN pathway-related genes (Fig. 5c), indicating a faster IFN signalling and thereby faster viral resolution.
Fig. 5.
Adoptive transfer of macrophages protects lung tissue from virus infection. a Schematic representation of the adoptive cell transfer and infection experiment. b Viral burden in total lung tissue of mice with or without adoptive cell transfer was analysed for the 2 viral mRNAs PolyU and Nsp12 and a combined analysis is presented for day 4 as indicated in Methods. Values represent averages with individual values and an average ± SD given in the panel (n = 6). c IFN α, IFNβ, IFNγ, and IFIT1 mRNA levels were determined by qPCR in lung tissue from saline or adoptive cell transfer treated animals as indicated in Methods. Individual values and average ± SD is given. (n = 6). IFNs, interferons; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; SD, standard deviation.
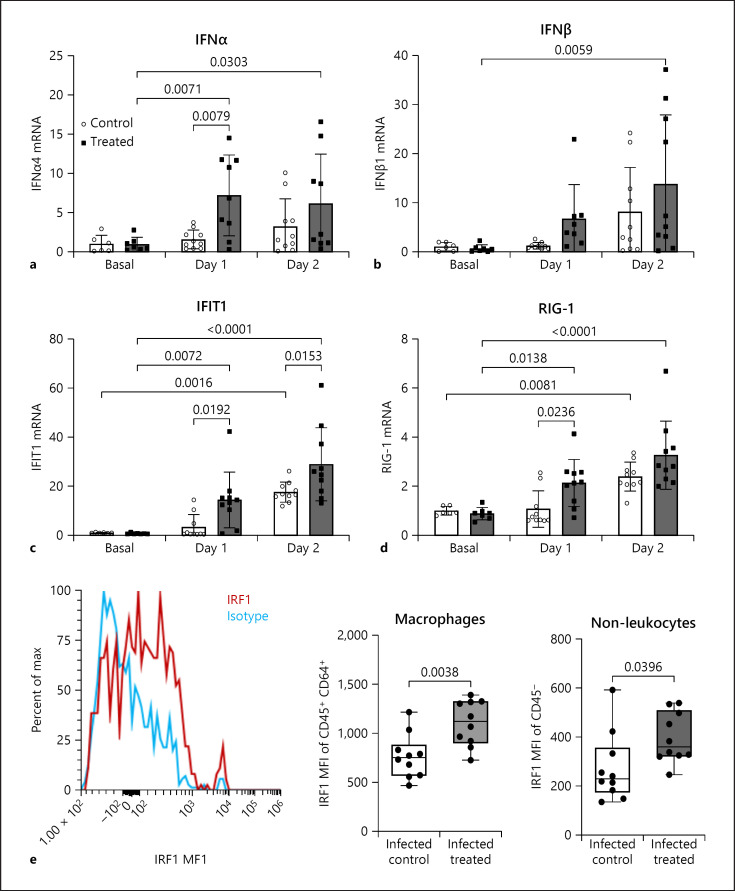
Our data indicate that pretreatment with BV OM-85 enhanced the initial response towards the virus infection and led to faster clearance of MCoV probably via the induction of type I IFN. Therefore, we determined baseline values and values of IFN pathway activation at days 1 and 2 after infection. BV OM-85 treatment did not increase baseline IFNα levels but we found a strong and significant induction of IFNα at day 1 in BV OM-85-treated animals (Fig. 6a). IFNβ was significantly induced in treated animals at day 2 (Fig. 6b). Similar to IFNα, we found a strong induction of IFIT1 with BV OM-85 pretreatment that was still in effect at day 2 after virus infection as compared to the control group (Fig. 6c). Finally, we checked if the IFN response element retinoic acid-inducible gene 1 (RIG-1), an intracellular receptor to identify RNA viruses, was also altered with BV OM-85 treatment. Again we observed no baseline differences but a significant increase at day 1 in comparison to the control group (Fig. 6d). Our results indicate a strong upregulation of type I IFN responses at day 1 after BV OM-85 treatment. On a cellular level, we found that especially CD45+CD64+ macrophages and CD45- non-leukocytes showed significantly more interferon-regulatory factor 1 protein at day 1 in BV OM-85-treated animals than control animals as identified by flow cytometry (Fig. 6e).
Fig. 6.
Early interferon signalling after virus infection. Levels of IFNα (a), IFNβ (b), IFIT1 (c), and RIG-1 (d) were determined by qPCR in lung tissue from animals receiving MCoV infection with or without treatment. Values represent averages. Values represent individual averages at each time point and condition an average ± SD given in the panel. e To evaluate IRF1 protein levels in lung tissue, we used intracellular flow cytometry on isolated lung cells as indicated in Methods. Values are given as MFI per CD45+CD64+ or CD45- cells and an average ±SD is given in the panel. Basal: uninfected mice; Day 1/Day 2: days post-MCoV infection; control: mannitol control; treated: BV OM-85-treated. BV, Broncho-Vaxom; MCoV, murine coronavirus; IFNs, interferons; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; IRF1, interferon-regulatory factor 1; MFI, mean fluorescence intensity; RIG-1, retinoic acid-inducible gene 1; SD, standard deviation.
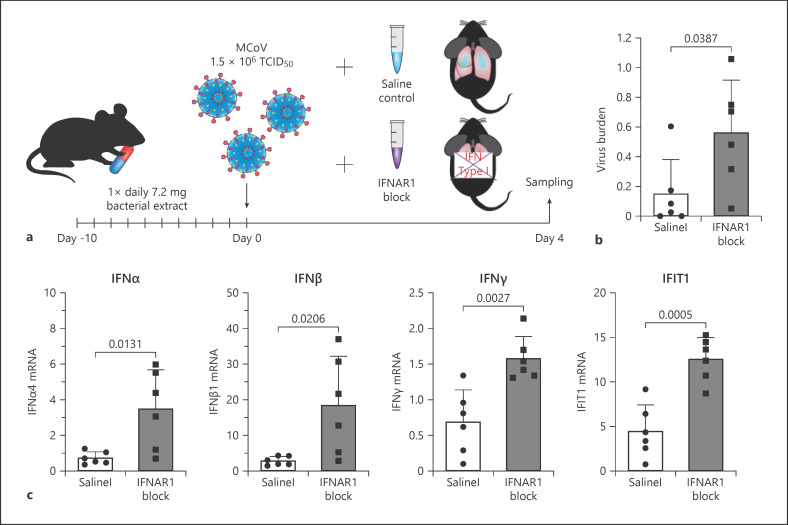
Finally, to test if the faster induction of IFN and its downstream targets is the main mode of action in BV-treated lung tissue viral defence, we used a blocking antibody to IFNAR1 (anti-IFNAR1) to inhibit early IFN signalling. The antibody was applied together with the virus infection in a single dose in BV OM-85 pre-treated animals (Fig. 7a) and lung tissue was analysed at day 4 after viral infection. Virus load was significantly increased in animals receiving anti-IFNAR1 treatment (Fig. 7b). Similar to our previous results, animals receiving anti-IFNAR1 after BV OM-85 also displayed a later activation of IFN-related pathways over animals receiving BV OM-85 treatment prior to virus infection only (Fig. 7c). Of note, viral burden and IFN pathway activation was comparable in the BV OM-85 anti-IFNAR1-treated animals and control infected animals (respective data from Fig. 1, 2, 7 visually combined in online suppl. Fig. 4).
Fig. 7.
Early interferon inhibition during MCoV infection after BV OM-85 treatment. a Schematic representation of the IFNAR1 inhibition experimental setup. b Viral burden in total lung tissue of mice with BV OM-85 pretreatment and saline or IFNAR1 blockade were analysed for MCoV viral burden at day 4 as indicated in Methods. Values represent individual parameters with the average ± SD given in the panel (n = 6). c IFNα, IFNβ, IFNγ, and IFIT1 mRNA levels were determined by qPCR in lung tissue from saline or IFNAR1 blockade treated animals as indicated in Methods. Individual values and average ± SD is given (n = 6). BV, Broncho-Vaxom; MCoV, murine coronavirus; IFNs, interferons; IFIT1, interferon-induced protein with tetratricopeptide repeats 1; SD, standard deviation; IFNAR1, interferon-alpha/beta receptor alpha chain 1; SD, standard deviation.
Discussion/Conclusion
The innate immune system is a first line of defence during infection. We were able to demonstrate that training of the immune system using the bacterial extract BV OM-85 is beneficial during a virus infection by increasing macrophage counts within the lung. This macrophage increase allowed a faster activation of the IFN system. As a result, pretreatment was associated with reduced lung cell apoptosis. We therefore suggest that an enhanced innate immune system is able to speed up the virus clearance and reduce lung damage.
Macrophages during viral infection act like a double-edged sword providing early activation of phagocytosis and IFN responses but when consistently activated can induce tissue damage via NLRP3-dependent activation of IL-1β [33]. BV treatment was shown to prime innate immune cells, keeping them in a preactivated state [34]. In addition, macrophage infection seems to be a crucial strategy of immune system evasion of coronaviruses in both mice and humans [35]. Our data demonstrate that BV OM-85 is capable of enhancing lung macrophage function. Previous reports suggested additional alterations of innate immune cells within the spleen [5]; however, we suggest that the direct effects in the lung might be crucial for the enhanced viral clearance in contrast to the modulation of the spleen, which might be more important for activation of the adaptive immune system. Upon immune cell activation, we found increased expression of antigen presentation-associated proteins and activated macrophages within the lung after preactivation but before virus infection. However, transplantation of naive macrophages suggested that the overall number is more essential than an additional preactivation. We therefore conclude that the increased presence of macrophages before a viral infection is beneficial to combat the virus resulting in advanced viral clearance. Furthermore, even though BV leads to preactivated macrophages, we did not detect adverse effects or a pro-inflammatory environment, suggesting the requirement for a second trigger for full macrophage activation.
To prove the importance of increased macrophage numbers in the lung as beneficial to combat a viral infection, we isolated CD14+ cells from virus-naive mice and transferred them into naive recipients. Indeed, transfer of macrophages led to enhanced viral clearance and thus a significant reduction of IFNα, IFNβ, and IFIT1 in lung tissue at day 4 after virus infection. We therefore suggest that the presence of these macrophages, similar as the ones pharmacologically enhanced with BV OM-85, already boosts the initial immune system barrier allowing a faster recovery.
To verify the crucial role of increased and faster type I IFN response caused by the increased number of macrophages in the lung tissue, we applied a blocking antibody to IFNAR1, the receptor for both IFNα and IFNβ. After BV treatment, the antibody was applied together with the infection to inhibit the initial IFN type I response and to monitor immune system recovery. Indeed, after inhibiting IFNAR1, we found an increased viral load compared to BV-treated control animals. Of note, the viral load in anti-IFNAR1 treated animals at day 4 was comparable to the viral load in untreated mice. Similarly to untreated mice at day 4, anti-IFNAR1 treated animals displayed an upregulation of IFN pathway genes compared to BV-treated animals.
We therefore conclude that BV leads to an increase in the macrophage population in the lung resulting in an enhanced and faster activation of IFN and its downstream targets allowing for a faster viral clearance together with reduced tissue damage. We used a mouse MCoV model that has similarities with human COVID-19 [36, 37]. It is therefore tempting to speculate that similar mechanism could be beneficial during the SARS-CoV-2 pandemic. Overall, oral training of the immune system using BV OM-85 is beneficial during a viral infection due to enhancing lung macrophages.
Statement of Ethics
Experiments were approved by the Animal Care and Use Committee of the Medical University of Vienna and the Austrian Federal Ministry of Education, Science and Research (BMBWF 2020-0.466.172).
Conflict of Interest Statement
P.J.H. is a consultant for OM Pharma.
Funding Sources
This work was supported by OM Pharma with a restricted research grant to P.J.H.
Author Contributions
M.S. performed and designed experiments, analysed and interpreted data, and wrote the manuscript. P.H. performed and designed experiments, analysed and interpreted data, and contributed to the manuscript. C.K., M.B., T.L., and ReP performed experiments. B.H. propagated virus. S.D. analysed RNA sequencing. B.P., C.H., W.S.S., and J.W. interpreted experiments and contributed to the manuscript. R.O.P. performed and designed experiments, interpreted data, contributed to the manuscript, and developed and established the in vivo virus infection model. P.J.H. supervised all research, performed and designed experiments, analysed and interpreted results, and wrote the manuscript.
Data Availability Statement
Data are available from the corresponding author upon reasonable request. The RNAseq data are available under GEO record: GSE167749.
Supplementary Material
Supplementary data
Acknowledgements
The MCoV virus MHV-A59 was a kind gift from Prof. Burkhard Ludewig (Institute of Immunobiology, Kantonsspital St. Gallen, 9007 St. Gallen, Switzerland). RNA sequencing was performed at the Core Facilities of the Medical University of Vienna, a member of VLSI.
References
- 1.Divangahi M, Aaby P, Khader SA, Barreiro LB, Bekkering S, Chavakis T, et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol. 2021;22:2–6. doi: 10.1038/s41590-020-00845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chumakov K, Avidan MS, Benn CS, Bertozzi SM, Blatt L, Chang AY, et al. Old vaccines for new infections: exploiting innate immunity to control COVID-19 and prevent future pandemics. Proc Natl Acad Sci USA. 2021;118((21)):e2101718118. doi: 10.1073/pnas.2101718118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricci R, Palmero C, Bazurro G, Riccio AM, Garelli V, Di Marco E, et al. The administration of a polyvalent mechanical bacterial lysate in elderly patients with COPD results in serological signs of an efficient immune response associated with a reduced number of acute episodes. Pulm Pharmacol Ther. 2014;27:109–13. doi: 10.1016/j.pupt.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Roth M, Pasquali C, Stolz D, Tamm M. Broncho vaxom (OM-85) modulates rhinovirus docking proteins on human airway epithelial cells via Erk1/2 mitogen activated protein kinase and cAMP. PloS One. 2017;12:e0188010. doi: 10.1371/journal.pone.0188010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasquali C, Salami O, Taneja M, Gollwitzer ES, Trompette A, Pattaroni C, et al. Enhanced mucosal antibody production and protection against respiratory infections following an orally administered bacterial extract. Front Med. 2014;1:41. doi: 10.3389/fmed.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardinale F, Lombardi E, Rossi O, Bagnasco D, Bellocchi A, Menzella F. Epithelial dysfunction, respiratory infections and asthma: the importance of immunomodulation. A focus on OM-85. Expert Rev Respir Med. 2020;14:1019–26. doi: 10.1080/17476348.2020.1793673. [DOI] [PubMed] [Google Scholar]
- 7.Esposito S, Jones MH, Feleszko W, Martell JAO, Falup-Pecurariu O, Geppe N, et al. Prevention of new respiratory episodes in children with recurrent respiratory infections: an expert consensus statement. Microorganisms. 2020;8((11)):1810. doi: 10.3390/microorganisms8111810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheever FS, Daniels JB, Pappenheimer AM, Bailey OT. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. J Exp Med. 1949;90:181–210. doi: 10.1084/jem.90.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homberger FR, Zhang L, Barthold SW. Prevalence of enterotropic and polytropic mouse hepatitis virus in enzootically infected mouse colonies. Lab Anim Sci. 1998;48:50–4. [PubMed] [Google Scholar]
- 10.Korner RW, Majjouti M, Alcazar MAA, Mahabir E. Of mice and men: the coronavirus mhv and mouse models as a translational approach to understand SARS-CoV-2. Viruses. 2020:12. doi: 10.3390/v12080880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barthold SW, Smith AL. Duration of challenge immunity to coronavirus JHM in mice. Arch Virol. 1989;107:171–7. doi: 10.1007/BF01317914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barthold SW, Smith AL. Virus strain specificity of challenge immunity to coronavirus. Arch Virol. 1989;104:187–96. doi: 10.1007/BF01315542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Channappanavar R, Kanneganti TD. Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41:1083–99. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vardhana SA, Wolchok JD. The many faces of the anti-COVID immune response. J Exp Med. 2020;217((6)):e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 16.Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Perlman S. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol. 2007;81:568–74. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cervantes-Barragan L, Kalinke U, Zust R, Konig M, Reizis B, Lopez-Macias C, et al. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J Immunol. 2009;182:1099–106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 19.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–9. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci USA. 2020;117:8094–103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89:1523–36. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205:3065–77. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–51. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneberger D, Aharonson-Raz K, Singh B. Monocyte and macrophage heterogeneity and Toll-like receptors in the lung. Cell Tissue Res. 2011;343:97–106. doi: 10.1007/s00441-010-1032-2. [DOI] [PubMed] [Google Scholar]
- 33.Guo S, Yang C, Diao B, Huang X, Jin M, Chen L, et al. The NLRP3 inflammasome and IL-1β accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog. 2015;11:e1005155. doi: 10.1371/journal.ppat.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang AT, Pasquali C, Ludigs K, Guarda G. OM-85 is an immunomodulator of interferon-β production and inflammasome activity. Sci Rep. 2017;7:43844. doi: 10.1038/srep43844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boumaza A, Gay L, Mezouar S, Bestion E, Diallo AB, Michel M, et al. Monocytes and macrophages, targets of SARS-CoV-2: the clue for Covid-19 immunoparalysis. J Infect Dis. 2021;224((3)):395. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason RJ. Pathogenesis of COVID-19 from a cell biology perspective. Eur Respir J. 2020;55:2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFadyen JD, Stevens H, Peter K. The emerging threat of (micro) thrombosis in COVID-19 and its therapeutic implications. Circ Res. 2020;127:571–87. doi: 10.1161/CIRCRESAHA.120.317447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
Data are available from the corresponding author upon reasonable request. The RNAseq data are available under GEO record: GSE167749.