Abstract
Neutrophil extracellular traps (NETs) have been described as a potential trigger of severe COVID-19. NETs are known as extracellular DNA fibers released by neutrophils in response to infection. If the host is unable to balance efficient clearance of NETs by dornases (DNases), detrimental consequences occur. Elevated levels of NETs in COVID-19 patients are associated with higher risk of morbid thrombotic complications. Here, we studied the level of NET markers and DNase activity in a cohort of COVID-19 patients compared to healthy controls. Our data confirmed an increased level of NET markers in the plasma of COVID-19 patients, with a higher level in male compared to female patients. At the same time, there was an increased DNase activity detectable in COVID-19 patients compared to healthy controls. Importantly, there was a negative correlation of DNase activity with the age of male patients. The antimicrobial peptide LL-37, which is known to stabilize NETs against DNase degradation, is embedded in NETs upon severe acute respiratory syndrome coronavirus-2-infection. The LL-37 plasma level correlates with the NET-marker level in male COVID-19 patients, indicating a potential role of LL-37 in the risk of NET-associated thrombosis in male COVID-19 patients by stabilizing NETs against DNase degradation. In conclusion, our data identify two potential risk factors of elderly male patients which may lead to inefficient NET degradation and a subsequently higher risk of NET-associated thrombosis during COVID-19: reduced DNase activity and an increased LL-37 level.
Keywords: Neutrophil extracellular traps, COVID-19, Elderly male, LL-37
Introduction
The COVID-19 pandemic has had an immense impact on the health care system, worldwide. There is an increased need for new intervention strategies or for possibilities to identify individuals at risk. Infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is associated with an exacerbated host response in patients with severe COVID-19. Neutrophils are known to infiltrate into the infected lung during SARS-CoV-2 infections [1]. Severe COVID-19 is shown to be associated with dysfunctional mature neutrophils that massively release neutrophil extracellular traps (NETs) [2, 3]. NETs consist of a DNA backbone decorated with several antimicrobial components that serve to entrap and kill various pathogens [4]. The host itself is known to produce dornases (DNases) to recycle NET-structures and to avoid tissue-damaging effects of NETs during infectious or noninfectious diseases [5, 6, 7]. However, in the case of an overwhelming release of NETs, they contribute to thrombosis [8, 9], autoimmune diseases such as lupus erythematous [10], acute respiratory distress syndrome (ARDS) [11], and other diseases [12]. In COVID-19 patients, NETs have been shown to contribute to immunothrombosis in ARDS and vascular occlusions [13, 14]. Several authors have demonstrated that plasma levels of NET markers were elevated in patients with COVID-19 relative to healthy controls [15, 16]. Furthermore, amount of NET-marker level have been associated with respiratory support requirement and short-term mortality and declined to level found in healthy individuals 4 months post-infection [17]. An association between circulating markers of NET-formation and clinical outcome was observed, demonstrating a potential role of NET markers in clinical decision-making [17].
Although aggregated NETs are known to promote the resolution of neutrophilic inflammation by degrading cytokines and chemokines and disrupting neutrophil recruitment and activation in gout patients [18, 19], during severe SARS-CoV-2 infections a balanced and immune-protective NET-formation is absent and therefore uncontrollable. Besides a massive induction of NET-formation in the SARS-CoV-2-infected host, a noneffective or deficient host nuclease activity to efficiently degrade NETs might also contribute to COVID-19. The aim of our study was to quantify the level of NET markers and DNase activity in a cohort of COVID-19 patients compared to healthy controls to evaluate if reduced or blocked DNase activity might be a risk factor for COVID-19 with focus on elderly patients and sex differences.
Materials and Methods
Ethical Approval
A human blood sample for neutrophil isolation was collected from a healthy donor with the help of a physician in agreement with the local Ethical Board. The study was approved by the Ethics Committee of Hannover Medical School, Hannover, Germany, and registered under no. 3295–2016. The blood collection followed the local guidelines.
The study with laboratory-confirmed COVID-19 patients was reviewed and approved by the Ethics Committee at the Hamburg State Chamber of Physicians (registration no.: WF-015/21). Retrospective data collection and anonymized analysis by COVID-19 patients and healthy blood donors were conducted in accordance with local government law (HmbKHG. §12) without requiring informed consent. Biopsies of human patients were received based on the ethic votum 308/2020BO2 from the Ethics Committee, University Hospital Tübingen, Germany.
Blood Sample Collection
Samples were collected from the blood of COVID-19 patients (plasma) and control donors (serum). Heparin blood or serum was centrifuged (2,100 g, 15 min at 4°C), and the plasma or serum was collected and stored at −80°C, until the respective assays were performed. From three COVID-19 patients in all assays, serum and plasma samples were analyzed, to compare the influence of the different samples on the read out and to verify that serum and/or plasma led to similar results in our assays. The results are shown in online supplementary Figure 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000521594). In some measurements, not all samples were analyzed due to technical problems, hemolytic serum/plasma, or limitation in blood volume.
Measurement of Free eDNA and Histone Fragments in Plasma and Serum
A Quant-iT TM PicoGreen® assay (P11496, Invitrogen Corp., Carlsbad, CA, USA) was used as described previously [20, 21], to determine the amount of extracellular DNA (eDNA) in the plasma or serum samples. The amount of eDNA in each sample was calculated based on the standard curve.
The number of histone-associated-DNA-fragments (mono- and oligonucleosomes) was quantified with a cell death detection ELISA PLUS kit (Roche Diagnostics GmbH, Mannheim, Germany). The assay was conducted in accordance with the manufacturer's instructions.
Measurement of TT and D-Dimer in COVID-19 Patients
Testosterone (TT) was analyzed in human plasma by electro-chemiluminescence immunoassay. The normal values (reference values of our laboratory) for TT for females are 0.22–2.90 nmol/L and for males are 9.90–27.80 nmol/L. Thus, measured values <0.22 nmol/L for females and <9.90 nmol/L for males are designated as low. D-dimer (cross-linked fibrin cleavage products) are quantitatively determined in plasma using a particle-enhanced immunoturbidimetric assay.
Measurement of LL-37 in Plasma and Serum
LL-37 ELISA Kit (HK321-02, HycultBiotech Inc., Uden, The Netherlands) was used to determine the amount of LL-37 in the plasma or serum samples. The test was performed in accordance with the manufacturer's instructions with 1:20 diluted samples.
Measurement of DNase Activity in Plasma and Serum
DNase I Activity Assay Kit (K429-100, BioVision, Milpitas, CA, USA, Fluorometric) was used to determine the DNase I activity in the plasma or serum samples. The test was performed following the manufacturer's instructions with 25 μL for each sample.
Isolation Human Neutrophils
Human granulocytes were isolated from fresh whole heparin blood of healthy donors using the polymorph prep system (Progen; 1.113 g/mL), as previously described [20]. In short, the fresh blood was layered 1:1 onto polymorph prep solution and centrifuged at room temperature and 472 g for 30 min without a break. Layers of plasma and PBMC were removed, and the layer of neutrophils was transferred, using a fresh plastic Pasteur pipette, into a new falcon tube, filled up with 1 × LPS-free PBS and centrifuged at 472 g for 10 min with a break. Afterward, supernatant was discarded, and the cell pellet was resuspended with 5 mL sterile LPS-free water for erythrocyte lysis for maximum 15 s and immediately filled up with 1 × LPS-free PBS and centrifuged for 10 min at 472 g. Finally, the cell pellet was resuspended in 1 mL RPMI (11835063, ThermoFisher Inc., Waltham, MA, USA) without phenol red, brought to room temperature. The cell count was determined via trypan blue staining and counting in a Neubauer chamber.
NET Induction Assay
A 96-well plate with glass bottom (P96G-1.5-5-F, MatTek Corporation, Ashland, MA, USA) was used for the NET induction. Each well was coated in accordance with the manufacturer's instructions with poly-L-lysine (0.01% solution P4707, Sigma-Aldrich GmbH, Munich, Germany) and handled afterward as previously described [20]. In each well, 1 × 105 neutrophils/50 μL were seeded. The total volume with stimulus was 100 μL. As a negative control, RPMI 1640 medium (11835063, ThermoFisher Inc.) was added. As a positive control, 50 μL phorbol 12-myristate 13-acetate (25 nM final concentration, 524400 Sigma-Aldrich) was added. The neutrophils were stimulated with 25 μL of serum from COVID-19 patients. The plate was incubated at 37°C, 5% CO2. After a 2-h incubation period, the plate was centrifuged (370 g, 5 min). Samples were fixed with paraformaldehyde (4% final concentration, Science Services GmbH, E15710-25, Munich, Germany) and the plate was wrapped with parafilm and stored at 4°C until the staining was conducted.
Eight mm glass cover slides were added in a 48-well suspension cell plate (Greiner Bio-one GmbH, Kremsmünster, Austria). The glass slides were coated with poly-L-lysin as stated above. In each well 2 × 105 neutrophils/100 μL were seeded. For stimulation, 100 μL of a 1:20,000 dilution from SARS-CoV-2 stock (TCID50/mL: 106.83) derived from SARS-CoV-2/Germany/Hamburg/01/2020; ENAstudy PRJEB41216 and sample ERS5312751 [22] were added to the cells and incubated at 37°C, 5% CO2 for 2 h. After incubation, the plate was centrifuged as stated above and fixed with paraformaldehyde (4% final concentration).
NET Staining and Analysis
NETs were stained after the permeabilization and the blocking of the samples [21]. These were incubated with the first antibodies: a mouse monoclonal antibody (IgG1) against LL-37/CAP-18 (HM2070, 100 μg/mL diluted 1:25) and a rabbit antihuman myeloperoxidase (MPO) (Dako A0398, 3.3 mg, 1:300), for 1 h at room temperature. For the isotype controls murine IgG1 (from murine myelom, M5284–0.2 mg/mL, Sigma Aldrich) and rabbit IgG (from rabbit serum, Sigma Aldrich, I5006, 1.16 mg) were used. As secondary antibodies goat anti-mouse Alexa 488 Plus (1:500, Invitrogen) and a goat anti-rabbit Alexa 633 (Thermo Scientific, 2 mg, 1:500) were used and incubated for 1 h at room temperature in the dark. Afterward, the samples were washed three times with 1 × PBS and embedded in 20 μL ProlongGold (with DAPI, Invitrogen). Samples were stored at 4°C in the dark until analysis.
The respective isotype control (50 μL phorbol 12-myristate 13-acetate stimulated) was used for the microscope settings. The pictures were taken by the Leica TCS SP5 AOBS confocal inverted-base fluorescence microscope with an HCX PL APO ×40 0.75–1.25 oil immersion objective.
NETs from direct SARS-CoV-2 infection experiments were stained as follows: subsequent to permeabilization and blocking of samples, first antibodies, mouse monoclonal anti-DNA-Histone-1 complexes (MAB 3864, Millipore, 1:300) and anti-neutrophil elastase rabbit pAB (#481001, Millipore, 1:300) were added for a 1-h incubation period. As isotype controls murine IgG2a murine myeloma (Sigma, M5409-1MG, 1:100) and IgG from rabbit serum (Sigma, I5006, 1:60) were used. After washing, secondary antibodies, Alexa Fluor 488 Plus (Goat anti-mouse IgG, A32723 Invitrogen, 1:1,000) and Alexa Fluor 633 (goat anti-rabbit IgG, A21070, Thermo Scientific, 1:1,000) were added to the sample and incubated in the dark at room temperature for 45 min. Next, samples were washed and embedded on ProlongGold with DAPI on microscope slides and stored at 4°C in the dark until further analysis. Images were taken as stated above.
DNA-Histone-1 Complexes/LL-37/MPO Examination in Paraffin Sections
For DNA-Histone-1 complexes/LL-37/MPO detection in lung tissue, paraffin sections of lungs from COVID-19 patients and control patients (healthy lung) were analyzed. The immunofluorescence staining of paraffin section was performed as previously described [23] with the following changes: first mouse IgG2a anti-DNA/histone-1 complexes (Millipore MAB3864, 0.55 mg; 1:100) or mouse IgG1 anti-LL-37 CAP-18 (HM2070, 100 μg/mL diluted 1:25), and rabbit antihuman MPO antibodies (Dako A0398, 3.3 mg, 1:300) in blocking buffer were incubated overnight at 4°C. As secondary antibodies, a goat anti-mouse antibody (Alex488PLUS, Thermo Fisher Scientific) and goat-anti rabbit antibody (Alexa568, Thermo Fisher Scientific) were used diluted 1:500 in blocking buffer. Isotype controls were included as described above. Finally, all samples were processed with TrueVIEW autofluorescence quenching kit (Vector Laboratories Inc., Burlingame, CA, USA) in accordance with the manufacturer's instructions.
Samples were recorded using a Leica TCS SP5 AOBS confocal inverted-base fluorescence microscope with an HCX PL APO ×40 0.75–1.25 oil immersion objective or HCX PL APO 63 × 1.40 oil objective. The settings were adjusted using isotype control antibodies in separate preparations.
Using ImageJ 1.51q software several images from each patient were analyzed for the integrated intensity (IntDen) in separate channels (blue = DNA and green = LL-37). For each single image, the green to blue ratio (%) was calculated, and the mean from all images per patient was used for statistical analysis.
Statistical Analysis
Data were analyzed using Excel 2019 (Microsoft), GraphPad Prism 8.3 (GraphPad Software), and IBM® SPSS® Statistics 27. Data were analyzed as indicated in each figure legend and presented with mean ± SD and the differences between groups were analyzed as described in the figure legends (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). In the online supplementary Table 1, continuous variable (age) is expressed as median with 1st to 3rd quantile, and the categorical variable (sex) is given as absolute and relative (%) numbers.
Results and Discussion
The reasons why some patients exhibit severe symptoms in COVID-19 are still not entirely understood. Especially age and sex are one of the main factors for people at risk [24, 25]. Interestingly, elevated levels of NETs in patients hospitalized with COVID-19 are associated with higher risk of morbid thrombotic complications. Thus, NETs are highly discussed to contribute to the severity of COVID-19. Since physiologically the host produces DNases to keep a balance between detrimental and beneficial effects of NETs, we studied the presence of NET markers and DNase activity in COVID-19 patients compared to healthy controls and in correlation to age and sex.
NET Marker in Plasma and Biopsies of COVID-19 Patients
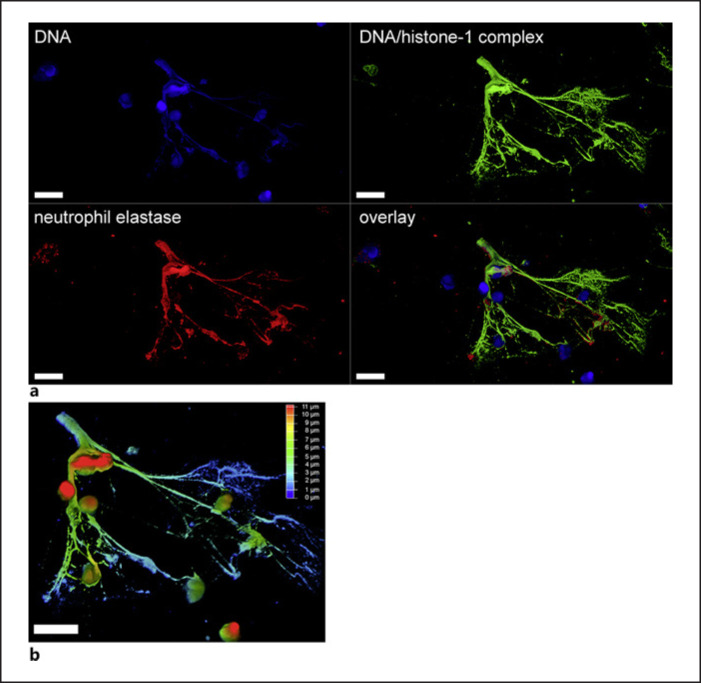
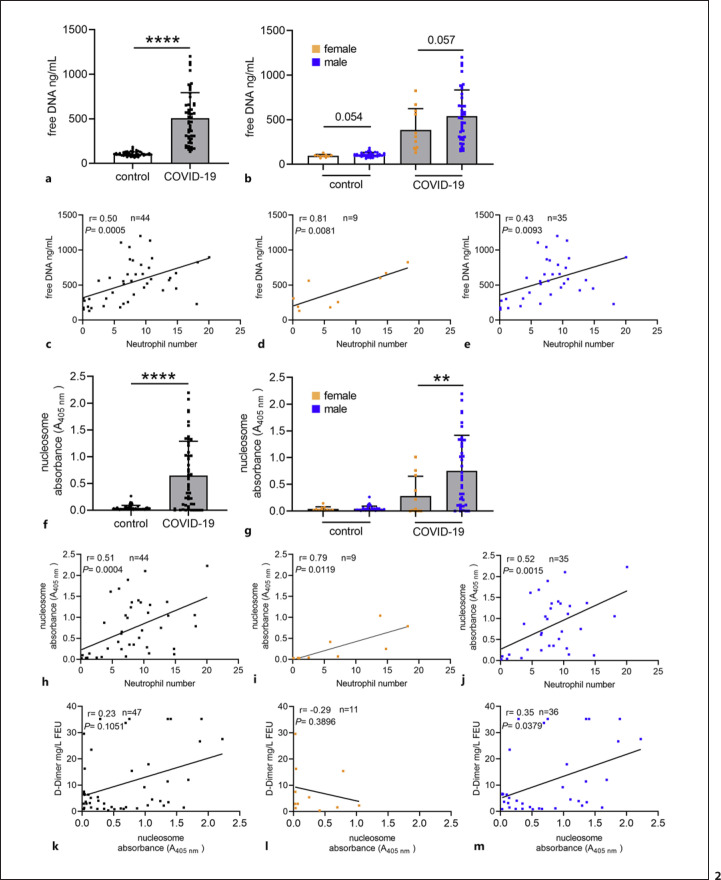
As shown in Figure 1, human blood-derived neutrophils release NETs in response to SARS-CoV-2, consisting of DNA-histone-1 complexes and neutrophil elastase (shown in Fig. 1a) with a thickness of up to 14 μm (shown in Fig. 1b). These data confirm previously published data [26, 27]. Our data presented in Figure 2 confirm increased NET markers in COVID-19 patients compared to healthy controls. As NETs consist of DNA, the amount of free eDNA was evaluated by a Pico Green assay in the plasma/serum. A tendency for more eDNA was detectable in male compared to female patients (shown in Fig. 2b). Hereby, it needs to be considered that eDNA in serum might originate from sources other than neutrophils [28, 29]. However, Figure 2c-e shows that eDNA correlates with the number of neutrophils, strongly supporting that it might mainly derive from neutrophils. In accordance with these data, also the nucleosome level as an additional NET marker besides eDNA [30] was significantly increased in COVID-19 patients, with highly significant alterations seen for male patients (p < 0.01) (shown in Fig. 2g). Again, nucleosome values correlate with neutrophil numbers (shown in Fig. 2h-j). A limitation regarding the comparison of NET markers between healthy controls and COVID-19 patients is that the analysis was performed with serum from controls and plasma from patients. In online supplementary Figure 1, we, therefore, show control data from measurements of NET markers in plasma and serum of three COVID-19 patients, indicating that measurement of NET markers in plasma and serum yield similar results. Only the graph with nucleosome data (in online suppl. Fig. 1) shows a clear tendency for higher nucleosome levels in serum. Since measurements of nucleosome levels in plasma may underestimate the level in serum, the difference in nucleosome levels between controls and COVID-19 patients may, in fact, be even greater. This goes in parallel with a recent study that investigated two independent cohorts of COVID-19 patients from Belgium and Germany and detected that circulating nucleosomes may serve as potential marker to monitor COVID-19 progression [31].
Fig. 1.
Human blood-derived neutrophils release NETs in response to SARS-CoV-2. NETs were released after 2-h infection of human neutrophils with SARS-CoV-2 and detected by confocal immunofluorescence microscopy. The settings were adjusted to a respective isotype control. aA representative image is presented, showing an eDNA-fiber positive for DNA/histone-1 complex (green) and neutrophil elastase (red) (scale bar = 10 μm). bFor this representative image, a 3D reconstruction of z-stacks was constructed with LAS X 3D Version 3.1.0 software (Leica) (17.37 μm consisting of 70 sections). Of this 3D volume, a depth coding analysis was conducted and is presented with a depth coding scale bar.
Fig. 2.
Increased free eDNA and nucleosome level as NET marker were detected in COVID-19 patients compared to healthy controls. The amount of free eDNA and nucleosome in blood was determined by Pico Green and cell death detection analysis. aSignificantly higher amount of free eDNA was determined in COVID-19 patients. bThe data in both groups were divided into male and female blood donors. A remarkably higher amount of free eDNA was detected between male and female in both groups. c–eIn the COVID-19 blood donors, a positive correlation between free eDNA and neutrophil numbers was observed, confirming that eDNA may derive from neutrophils. The neutrophil number is presented in 109/L. fA significantly higher amount of free eDNA and nucleosome was determined in COVID-19 patients. gA significantly higher amount of nucleosome was detected between males and females in the COVID-19 group. h–jIn the COVID-19 blood donors, a positive correlation between nucleosome and neutrophil numbers was observed, confirming that nucleosome may derive from neutrophils. k–mIn the COVID-19 blood donors, a slightly positive correlation between D-dimers and nucleosome was observed in male patients. Each dot represents one blood donor, and data are presented ±SD (black dots = all blood donors in the group, orange dots = female blood donors, blue dots = male blood donors). In (a, f) (ncontrol = 49, nCOVID-19 = 50); in (b, g) (ncontrol male = 38; ncontrol female = 11; nCOVID-19 male = 39; nCOVID-19 female = 11). Data in (a, b, f, g) were analyzed with an unpaired, one-tailed Student's t test or the Mann-Whitney test if not normally distributed (**p < 0.01, ****p < 0.0001), the values in (c–e) and (h–m) were calculated with a Pearson's r analysis of correlation coefficients (two-tailed).
In parallel to increased NET-formation, severe infection with SARS-CoV-2 exhibits more obvious dysregulated coagulation function compared with mild cases [32]. This is indicated here by a slight correlation of D-Dimer with an increased nucleosome level in plasma of male COVID-19 patients as shown in Figure 2k-m. These data confirm previous published data, showing that NETs and fibrin form a composite network within thrombin that triggers the risk of thrombosis in COVID-19 patients [33].
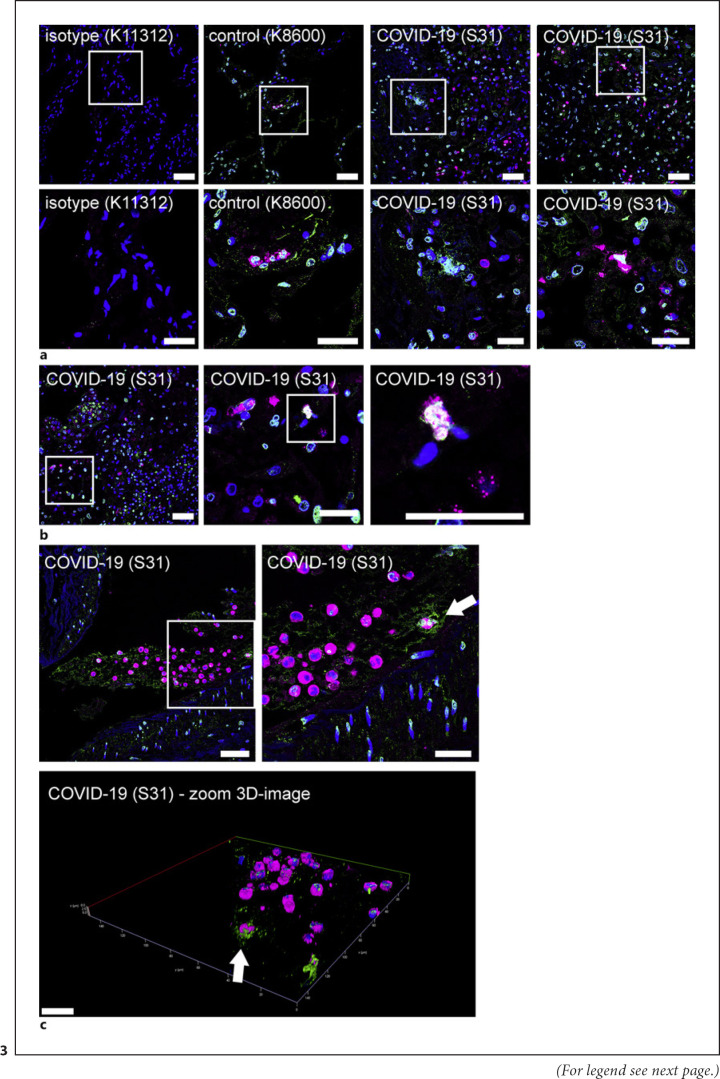
Finally, we also confirm that lung tissue biopsies stained positive for NET markers (shown in Fig. 3). Lung sections from a non-SARS-CoV-2-infected patient (K8600) and SARS-CoV-2-infected patient (S31) were analyzed by confocal immunofluorescence microscopy for NET structures. Representative images are presented, and inside the lung sections, MPO-positive cells were identified. Furthermore, areas with signals for MPO and DNA/histone-1 complexes in proximity were observed inside the alveolar septa of the lung from the COVID-19 patient. High-resolution imaging shows that NET-releasing neutrophils are positive for MPO inside the nucleus confirmed by a co-localization of DAPI signal (blue) and MPO (red), resulting in white staining (shown in Fig. 3b). This is a typical feature shown by NET-releasing neutrophils that undergo NETosis. It describes a specific cell death associated with translocation of MPO and elastase to the nucleus, the subsequent degradation of histones, rupture of nuclear membrane, and release of eDNA fibers [34, 35], a phenotype typical for the process of NET-formation.
Fig. 3.
Detection of NET markers in lung tissue biopsies of a SARS-CoV-2 infected patient. Lung sections from a non-SARS-CoV-2 infected patient (K8600) and SARS-CoV-2 infected patient (S31) were analyzed by confocal immunofluorescence microscopy for NET structures. The settings were adjusted to a respective isotype control. Representative images are presented (blue = DNA, green = DNA/histone-1 complex, magenta = MPO). aInside the lung sections MPO-positive cells were identified. Furthermore, areas with signals for MPO and DNA/histone-1 complexes in proximity were observed inside the alveolar septa of the COVID-19 patient (scale bar upper panel = 50 μm, lower panel = 25 μm). bThe zoom image presents a MPO and DNA/histone-1 complexes releasing cells in the alveolar septa of patient S31 (scale bar left image = 50 μm, middle, and right image = 25 μm). cInside the alveoli of the COVID-19 patient a DNA/histone-1 complex positive area was observed with numerous embedded MPO-positive cells. The arrow marks a MPO positive cell within a DNA/histone-1 complex area. Representative 3D image of z-stacks (5.87 μm consisting of 31 sections) was constructed with LAS X 3D Version 3.1.0 software (Leica) of this area (arrow marks orientation). Scale bar 2D image left side = 50 μm, right side = 25 μm, scale bars in 3D image = 20 μm.
DNase Activity in COVID-19 Patients Compared to Healthy Controls
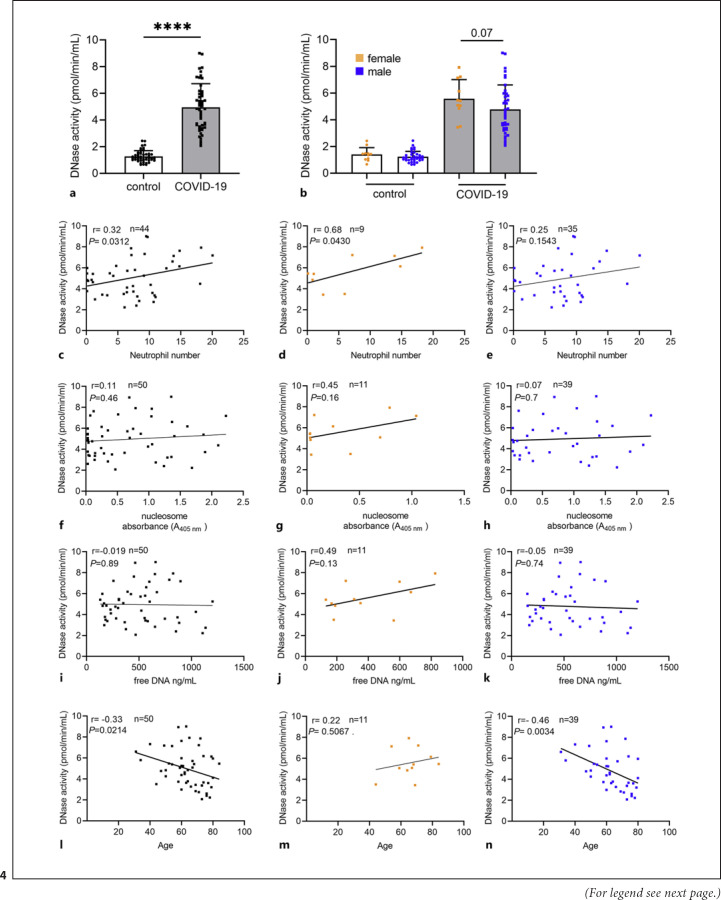
As NET-formation is often associated with an increased release of DNases by the host to maintain a balance between NET-formation and NET-elimination [36], the DNase activity was measured in the same samples as NET markers. As shown in Figure 4, a significant difference between samples from COVID-19 patients compared to healthy controls was detectable. However, only a remarkable (p = 0.07) difference between male and female COVID-19 patients was observed. The DNase activity data also partially correlate with the neutrophil number (Fig. 4c-e). A correlation between DNase activity and the two NETs markers cell-free eDNA, and nucleosome was not identified (Fig. 4f-k).
Fig. 4.
Increased DNase activity as response to NET release in COVID-19 patients compared to healthy controls. DNase activity was measured in an enzymatic kinetic assay over 90 min. aA significantly higher DNase activity was detected in COVID-19 patients (ncontrol = 49, nCOVID-19 = 50). bNo difference in DNase activity in male compared to female individuals was observed. However, a remarkably lower DNase activity was detected in the male COVID-19 patients (ncontrol male = 38; ncontrol female = 11; nCOVID-19 male = 39; nCOVID-19 female = 11). c–eIn the COVID-19 blood donors, a positive correlation between DNase activity and neutrophil numbers was observed, which was stronger in the female group (d). f–kIn the COVID-19 blood donors, no correlation between DNase activity and free eDNA or nucleosome was observed. l–nA negative correlation between DNase activity and age of patients, especially in the case of male patients (n) was identified, indicating that elderly male patients have an impaired DNase activity and subsequently an impaired ability to get rid of NETs during infection. This explains the higher NET marker values in male patients seen in Fig. 2. Each dot represents one blood donor, and data are presented ±SD (black dots = all blood donors in the group, orange dots = female blood donors, blue dots = male blood donors). Data in (a, b) were analyzed with an unpaired, one-tailed Mann-Whitney test (****p < 0.0001), the values in (c–h) were calculated with a Pearson's r analysis of correlation coefficients (two-tailed).
Interestingly, a negative correlation with the age of male patients was observed (shown in Fig. 4l-n). These data indicate that elderly male patients have an impaired DNase activity and, subsequently, an impaired ability to get rid of NETs during infection. This explains the higher NET-marker values in male patients seen in Figure 2. It is quite important to consider that NETs are still excessively formed in severe COVID-19 patients despite the presence of increased DNase activity seen in this cohort. These data indicate that the host DNase is not able to efficiently degrade and eliminate NETs completely during severe SARS-CoV-2 infection as also seen for other infections e.g., Streptococcus suis meningitis [37]. Thus, it might be speculated that additional NET-stabilizing factors play a role in this process.
Stabilization of NETs by the Cathelicidin LL-37 during SARS-CoV-2 Infection
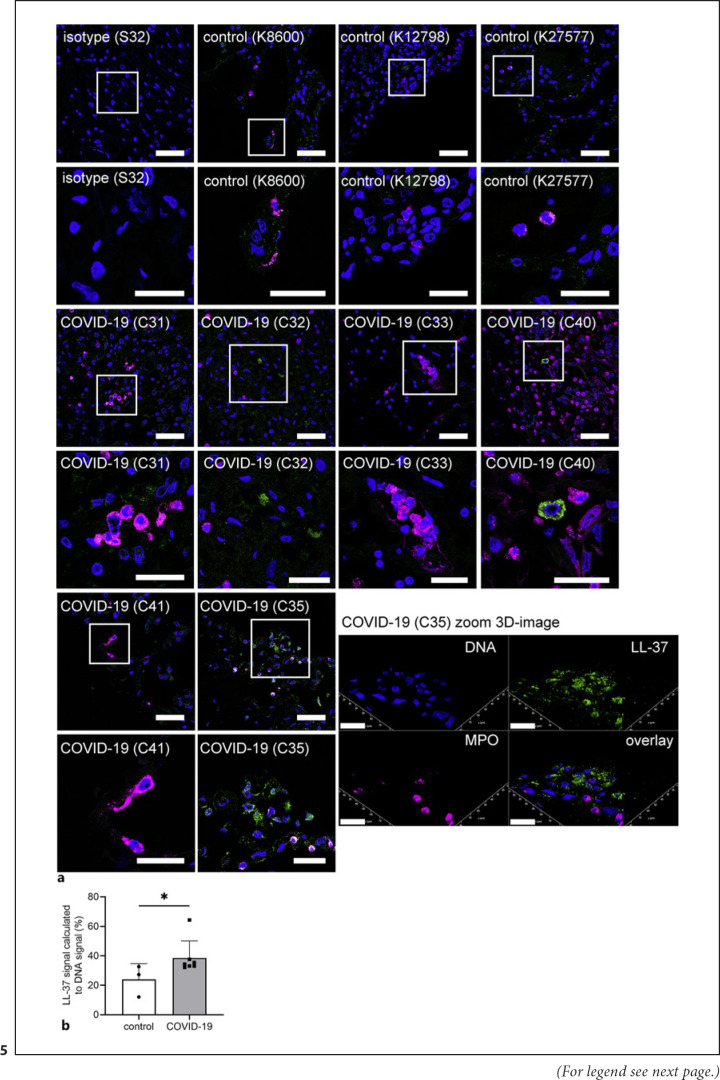
It is known that NETs are stabilized against DNase degradation by cationic peptides as, for example, the human cathelicidin LL-37 [38], which can be produced by neutrophils among several other cells [39]. Therefore, lung sections from three non-SARS-CoV-2-infected patients (control) and six SARS-CoV-2-infected patients (COVID-19) were analyzed by confocal immunofluorescence microscopy for LL-37 in combination with the neutrophil marker MPO (shown in Fig. 5a). Inside the lung sections, MPO-positive cells and LL-37-positive cells were identified more frequently in the COVID-19 patients. A quantitative measurement of integrated density in the histological slices revealed a significantly higher LL-37 signal in COVID-19 patients compared to healthy controls (Fig. 5b). Furthermore, LL-37 was partially observed extracellularly, in different quantities. In conclusion, LL-37 in NETs is confirmed in biopsies from COVID-19 patients indicating that LL-37-stabilized NETs might explain the presence of NETs despite DNase activity in the COVID-19 patients (shown in Fig. 3, 4).
Fig. 5.
Detection of MPO and LL-37 in lung tissue biopsies of SARS-CoV-2-infected patients. aLung sections from three non-SARS-CoV-2-infected patients (control) and six SARS-CoV-2-infected patients (COVID-19) were analyzed by confocal immunofluorescence microscopy for MPO and LL-37. The settings were adjusted to a respective isotype control. Representative images are presented always in the upper panel as overview and the lower panel as zoom of the area marked with the white square (blue = DNA, green = LL-37, magenta = MPO). The zoom image of the COVID-19 patient C35 is presented as a representative 3D image of z-stacks (5.16 μm consisting of 42 sections) with extracellular LL-37 and MPO-positive cells. This reconstruction was performed with LAS X 3D Version 3.1.0 software (Leica). Scale bar overview image = 50 μm, zoom image = 25 μm, 3D zoom image = 20 μm. bThe integrated density in the blue (DNA) and green (LL-37) channel was measured in several pictures of each patient. The ratio was calculated and is presented as percentage. Each dot presents the mean of each individual patient. Data are analyzed with an unpaired, one-tailed Mann-Whitney test, control n = 3, COVID-19 n = 7 (*p < 0.05).
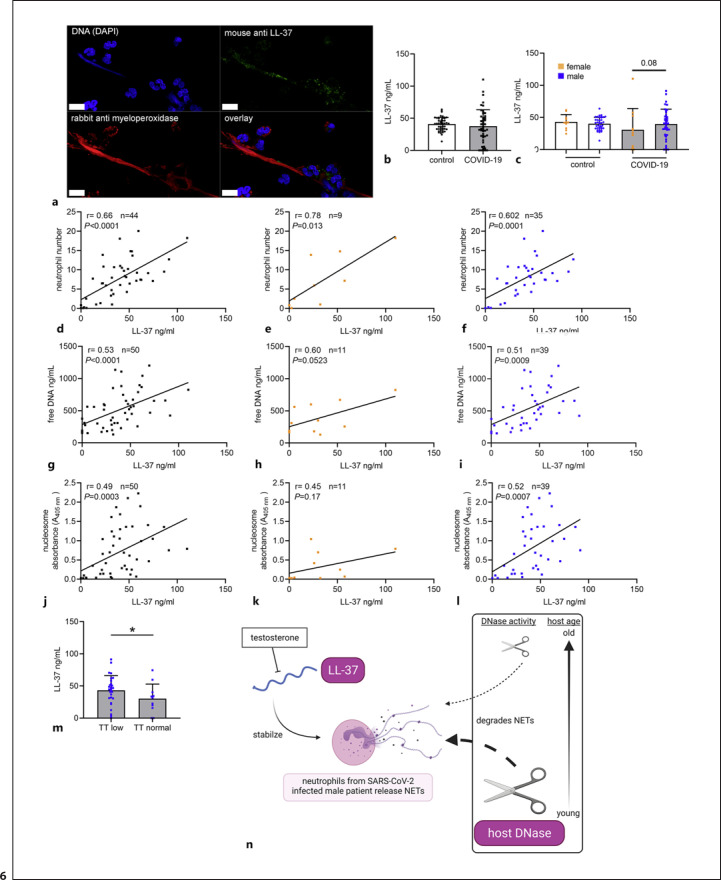
As additional confirmation that LL-37 plays a role in the stabilization of NETs in COVID-19 patients, human blood-derived neutrophils were incubated with serum of a COVID-19 patient and analyzed for NETs by confocal immunofluorescence microscopy for LL-37 and MPO. A previous study identified NET release after incubating neutrophils in serum from COVID-19 patients [40]. MPO can be found in NETs [34]. A representative image is presented in Figure 6a, showing an eDNA-fiber positive for LL-37 (green) and MPO (red). These data show that NETs are positive for LL-37 after treatment of human blood-derived neutrophils with serum from a COVID-19 patient.
Fig. 6.
Cationic antimicrobial peptide LL-37, known to bind NETs and stabilize them against DNase degradation, was higher in male COVID-19 patients with low TT. aHuman blood-derived neutrophils were incubated with serum of a COVID-19 patient and analyzed for NETs by confocal immunofluorescence microscopy. The settings were adjusted to a respective isotype control (presented in online suppl. Fig. 2). A representative image is presented, showing an eDNA-fiber positive for LL-37 (green) and MPO (red) (scale bar = 10 μm). bNo differences in LL-37 level were detectable in COVID-19 patients compared to controls (ncontrol = 49, nCOVID-19 = 50). cNo differences in the LL-37 level were detectable between male and female individuals (ncontrol male = 38; ncontrol female = 11; nCOVID-19 male = 39; nCOVID-19 female = 11). d–fIn the COVID-19 blood donors, a positive correlation between LL-37 and neutrophil numbers was observed. g–iIn the COVID-19 blood donors, a positive correlation between LL-37 and free DNA was observed. j–lIn the COVID-19 blood donors, a positive correlation between LL-37 and nucleosome was observed. mAs known from literature that TT blocks LL-37 expression, male COVID-19 patients with low TT (<9.90 nmol/L) showed higher LL-37 amounts in blood (nCOVID-19 male low TT = 28; nCOVID-19 male normal TT = 11). nA summary of identified risk parameters is presented. Each dot represents one blood donor, and data are presented ±SD (black dots = all blood donors in the group, orange dots = female blood donors, blue dots = male blood donors). This figure was created with BioRender.com. Data in (a, b, m) were analyzed with an unpaired, one-tailed Student's t test or Mann-Whitney test if not normally distributed (*p < 0.05). The values in (d–l) were calculated with a Pearson's r analysis of correlation coefficients (two tailed). Data in (h–j) were analyzed with an unpaired, one-tailed Student's t test (**p < 0.01).
To confirm the results from the analysis of histological slices and the ex vivo neutrophil assay, we performed further quantification of LL-37 in the blood of COVID-19 patients compared to healthy controls. As shown in Fig. 6b-c, no differences in LL-37 level were detectable in COVID-19 patients compared to controls. However, a slight tendency (p = 0.08) of higher LL-37 levels was detectable in male COVID-19 patients compared to female patients.
Interestingly, the LL-37 level correlates with neutrophil numbers and both NET markers free DNA as well as nucleosomes in COVID-19 patients, especially in male COVID-19 patients (Fig. 6d-l). Furthermore, positive correlations with the D-Dimer level and DNase activity (online suppl. Fig. 2) were observed again in male COVID-19 patients, indicating a potential additional role of LL-37 in the risk of NET-associated thrombosis of male COVID-19 patients by stabilizing NETs against DNase degradation. As it is known from literature that TT blocks LL-37 expression, male patients with low TT show higher LL-37 expression [41]. Interestingly, low serum levels of TT are speculated to predispose men, especially elderly men, to poor prognosis or death during COVID-19 [42]. In line with these aforementioned publications, a significantly higher LL-37 level was detected in the male COVID-19 cohort with a low TT level (Fig. 6m), leading to the hypothesis that a high LL-37 level besides low DNase activity may be an additional risk factors in COVID-19 patients when stabilizing massive NET-formation in the infected host (Fig. 6n).
However, as an oral application of LL-37 producing Lactococcus lactis was tested in COVID-19 patients as a new treatment strategy and published as a preliminary study [43], and antimicrobial peptides like LL-37 are widely discussed in the COVID-19 pathogenesis [44, 45], further studies are needed to clarify the influence of LL-37 on SARS-CoV-2 infections in different patient groups including age and sex. Furthermore, dose-dependent effects of LL-37 are conceivable.
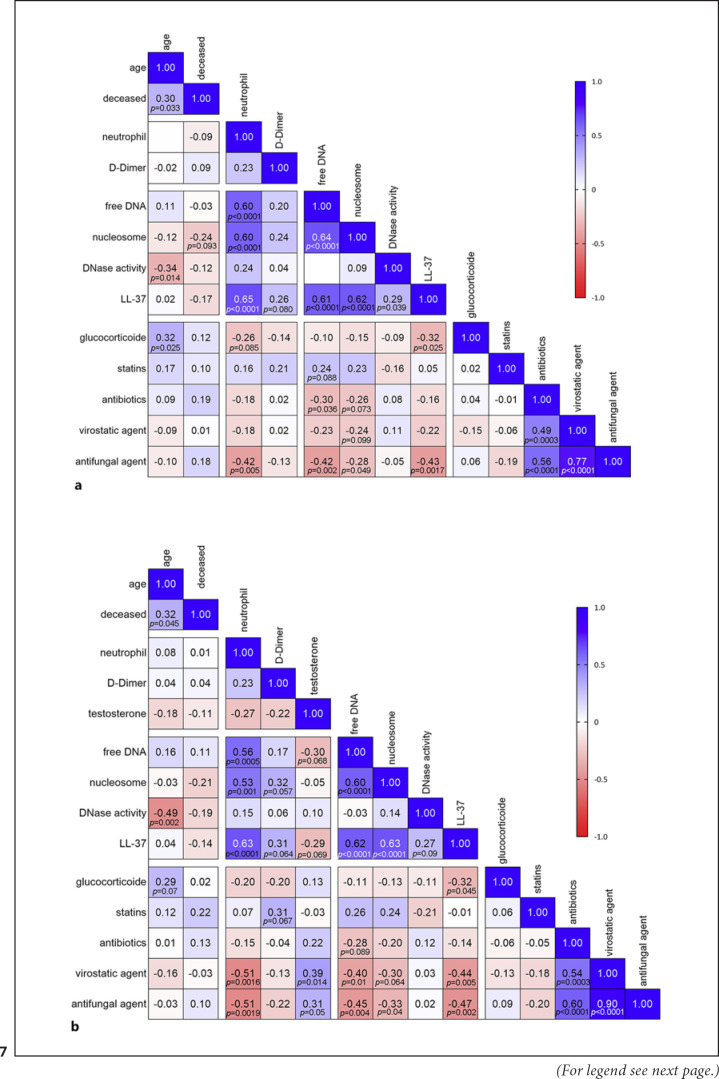
Finally, to give an overview of the correlations between all recorded clinical data including the medication as additional parameter (online suppl. Table 2) and all measured laboratory values, correlation matrixes summarize the overall results again (Fig. 7 and online suppl. Fig. 3, 4): several significant correlations identified in male COVID-19 patients were less clearly identified in female COVID-19 patients and not observed in the control patients. Further studies are needed with higher female patient numbers to identify possible risk factors therein. Regarding the medication, treatment of COVID-19 patients with antibiotics, virostatic, or antifungal agents showed a distinct inverse correlation with NET markers, e.g., free DNA or nucleosomes, which directly reflect the therapeutic effects of these drugs by minimizing cytotoxic effects of the pathogens. Interestingly, a significant inverse correlation of LL-37 and glucocorticoid treatment was observed in male COVID-19 patients (Fig. 7), indicating that glucocorticoid treatment decreases LL-37 expression. Similar results were also shown for salivary concentrations of LL-37 in patients with oral lichen planus when systemically treated with corticosteroids [46]. However, further studies are needed to characterize the effects of specific glucocorticoids on LL-37 and its stabilization of NETs during SARS-CoV-2-infections.
Fig. 7.
Correlation matrix of clinical and laboratory parameters from COVID-19 patients. aData show all COVID-19 patients. The correlation was conducted with n = 50 in all data sets, except neutrophil data with only n = 44 and D-Dimer n = 47. bData show male COVID-19 patients. The correlation was conducted with n = 39 in all data sets, except neutrophil data with only n = 35 and D-Dimer n = 36. All data were analyzed with a nonparametric two-tailed Spearman correlation matrix. The matrix presents p values lower than 0.1.
Conclusion
In summary, our data identified two potential risk factors of male elderly patients: Insufficient DNase activity and/or increased LL-37 level which may lead to inefficient NET degradation and subsequent higher risk of NET-associated detrimental effects. Based on the increasing evidence that NETs contribute to severe COVID-19 cases, DNase treatment of COVID-19 patients to degrade NETs is widely discussed as a potential therapeutic strategy. Since increasing age (especially >60 years) may be the most important risk factor for severe outcomes [25], it might be considered that a therapeutic treatment of DNase 1) might be useful especially in this group of patients. Currently, there are ongoing clinical trials with DNase in COVID-19 patients that aim to characterize the activity of aerosolized intratracheal DNase 1 administration on the severity and progression of ARDS in COVID-19 patients [47, 48].
However, more data are needed to verify our hypothesis. On the one hand, it is considered that therapeutic DNase treatment might be helpful to prevent the detrimental effects of massive NET-formation during COVID-19. On the other hand, DNase treatment can impact the growth of certain pathogenic bacteria, e.g., the lung pathogen Haemophilus influenzae, which can efficiently use degraded NETs as a growth factor [21, 49]. Finally, a better understanding of the role of NETs and their stabilization or degradation in the pathogenesis of COVID-19 seems to be a key element for identifying new treatment strategies for severe and mild cases. With this gained knowledge in the future, new patient individual treatment strategies could be implemented.
Statement of Ethics
A human blood sample for neutrophil isolation was collected from a healthy donor with the help of a physician in agreement with the local Ethical Board. The study was approved by the Ethics Committee of Hannover Medical School, Hannover, Germany, and registered under no. 3295–2016. The blood collection followed the local guidelines. The study with laboratory-confirmed COVID-19 patients was reviewed and approved by the Ethics Committee at the Hamburg State Chamber of Physicians (registration no.: WF-015/21). Retrospective data collection and anonymized analysis by COVID-19 patients, and healthy blood donors were conducted in accordance with local government law (HmbKHG. §12) without the requirement for informed consent. Biopsies of human patients were received based on the ethics votum 308/2020BO2 from the Ethics Committee, University Hospital Tübingen, Germany.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This study was in part supported by the Ministry of Science and Culture of Lower Saxony in Germany (14 − 76,103-184 CORONA-15/20).
Author Contributions
N.B., A.C.P., T.H., M.S., B.S., K.K., and M.K.-B. performed blood analysis; M.S., B.S., K.K., D.J., A.N., J.H., S.P., S.S.-B., and S.K. recruited patient cohorts including clinical data and collected samples; N.B., T.H., and M.K.-B. performed the NET in vitro experiments and NET histological analysis; N.B, A.C.P., T.H., M.S., K.K., G.G., and M.K.-B. analyzed data and interpreted the results; N.B and A.C.P. analyzed the data statistically; N.B. and M.K.-B. drafted the first version of the manuscript; N.B., G.G., and M.K.-B. developed the concept of the study; all authors contributed to the final version of the manuscript.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the published article. Raw data were generated in all Departments and Institutes of the submitting authors. Derived data supporting the findings of this study are available from the corresponding authors G.G. and M.K.-B. on request.
Supplementary Material
Supplementary data
Acknowledgement
This publication was supported by the Deutsche Forschungsgemeinschaft and University of Veterinary Medicine Hannover, Foundation within the funding programme Open Access Publishing.
References
- 1.Reusch N, De Domenico E, Bonaguro L, Schulte-Schrepping J, Baßler K, Schultze JL, et al. Neutrophils in COVID-19. Front Immunol. 2021 Mar;12:652470. doi: 10.3389/fimmu.2021.652470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020 Sep;182((6)):1419–40.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aschenbrenner AC, Mouktaroudi M, Krämer B, Oestreich M, Antonakos N, Nuesch-Germano M, et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021 Dec;13((1)):7. doi: 10.1186/s13073-020-00823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004 Mar;303((5663)):1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 5.Hakkim A, Furnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci U S A. 2010 May;107((21)):9813–8. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiménez-Alcázar M, Rangaswamy C, Panda R, Bitterling J, Simsek YJ, Long AT, et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017 Dec;358((6367)):1202–6. doi: 10.1126/science.aam8897. [DOI] [PubMed] [Google Scholar]
- 7.Meng W, Paunel-Görgülü A, Flohé S, Witte I, Schädel-Höpfner M, Windolf J, et al. Deoxyribonuclease is a potential counter regulator of aberrant neutrophil extracellular traps formation after major trauma. Mediators Inflamm. 2012;2012:149560. doi: 10.1155/2012/149560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, Kelly MM, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007 Apr;13((4)):463–9. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs TA, Brill A, Duerschmied D, Schatzberg D, Monestier M, Myers DD, et al. Extracellular DNA traps promote thrombosis. Proc Natl Acad Sci U S A. 2010 Sep;107((36)):15880–5. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieterse E, van der Vlag J. Breaking immunological tolerance in systemic lupus erythematosus. Front Immunol. 2014 Apr;5:164. doi: 10.3389/fimmu.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Su X, Pan P, Zhang L, Hu Y, Tan H, et al. Neutrophil extracellular traps are indirectly triggered by lipopolysaccharide and contribute to acute lung injury. Sci Rep. 2016 Dec;6((1)):37252. doi: 10.1038/srep37252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallés J, Lago A, Santos MT, Latorre AM, Tembl JI, Salom JB, et al. Neutrophil extracellular traps are increased in patients with acute ischemic stroke: prognostic significance. Thromb Haemost. 2017;117((10)):1919–29. doi: 10.1160/TH17-02-0130. [DOI] [PubMed] [Google Scholar]
- 13.Meegan JE, Yang X, Coleman DC, Jannaway M, Yuan SY. Neutrophil-mediated vascular barrier injury: role of neutrophil extracellular traps. Microcirculation. 2017 Apr;24((3)):e12352. doi: 10.1111/micc.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Wu Z, Long Q, Huang J, Hong T, Liu W, et al. Insights into immunothrombosis: the interplay among neutrophil extracellular trap, von Willebrand factor, and ADAMTS13. Front Immunol. 2020 Dec;11:610696. doi: 10.3389/fimmu.2020.610696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leppkes M, Knopf J, Naschberger E, Lindemann A, Singh J, Herrmann I, et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020 Aug;58:102925. doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020 Sep;136((10)):1169–79. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng H, Havervall S, Rosell A, Aguilera K, Parv K, von Meijenfeldt FA, et al. Circulating markers of neutrophil extracellular traps are of prognostic value in patients with COVID-19. Arterioscler Thromb Vasc Biol. 2021 Feb;41((2)):988–94. doi: 10.1161/ATVBAHA.120.315267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer C, Janko C, Munoz LE, Zhao Y, Kienhöfer D, Frey B, et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med. 2014 May;20((5)):511–7. doi: 10.1038/nm.3547. [DOI] [PubMed] [Google Scholar]
- 19.Euler M, Hoffmann MH. The double-edged role of neutrophil extracellular traps in inflammation. Biochem Soc Trans. 2019 Dec;47((6)):1921–30. doi: 10.1042/BST20190629. [DOI] [PubMed] [Google Scholar]
- 20.de Buhr N, von Köckritz-Blickwede M. Detection, visualization, and quantification of neutrophil extracellular traps (NETs) and NET markers. Methods Mol Biol. 2020;2087:425–42. doi: 10.1007/978-1-0716-0154-9_25. [DOI] [PubMed] [Google Scholar]
- 21.de Buhr N, Bonilla MC, Pfeiffer J, Akhdar S, Schwennen C, Kahl BC, et al. Degraded neutrophil extracellular traps promote the growth of Actinobacillus pleuropneumoniae. Cell Death Dis. 2019 Sep;10((9)):657. doi: 10.1038/s41419-019-1895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker K, Beythien G, de Buhr N, Stanelle-Bertram S, Tuku B, Kouassi NM, et al. Vasculitis and neutrophil extracellular traps in lungs of golden syrian hamsters with SARS-CoV-2. Front Immunol. 2021 Apr;12:640842. doi: 10.3389/fimmu.2021.640842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennig-Pauka I, Imker R, Mayer L, Brügmann M, Werckenthin C, Weber H, et al. From stable to lab: investigating key factors for sudden deaths caused by streptococcus suis. Pathogens. 2019 Nov;8((4)):249. doi: 10.3390/pathogens8040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallapaty S. What's the risk of dying from a fast-spreading COVID-19 variant? Nature. 2021 Feb;590((7845)):191–2. doi: 10.1038/d41586-021-00299-2. [DOI] [PubMed] [Google Scholar]
- 25.Wingert A, Pillay J, Gates M, Guitard S, Rahman S, Beck A, et al. Risk factors for severity of COVID-19: a rapid review to inform vaccine prioritisation in Canada. BMJ Open. 2021 May;11((5)):e044684. doi: 10.1136/bmjopen-2020-044684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, de Lima M, Nascimento DC, et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020 Dec;217((12)):e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcanjo A, Logullo J, Menezes CCB, de Souza Carvalho Giangiarulo TC, Dos Reis MC, de Castro GMM, et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19) Sci Rep. 2020;10((1)):19630. doi: 10.1038/s41598-020-76781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc. 2018 Aug;93((3)):1649–83. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 29.Butt AN, Swaminathan R. Overview of circulating nucleic acids in plasma/serum. Ann N Y Acad Sci. 2008 Aug;1137((1)):236–42. doi: 10.1196/annals.1448.002. [DOI] [PubMed] [Google Scholar]
- 30.Bang OY, Chung JW, Cho YH, Oh MJ, Seo WK, Kim GM, et al. Circulating DNAs, a marker of neutrophil extracellular traposis and cancer-related stroke. Stroke. 2019 Oct;50((10)):2944–7. doi: 10.1161/STROKEAHA.119.026373. [DOI] [PubMed] [Google Scholar]
- 31.Cavalier E, Guiot J, Lechner K, Dutsch A, Eccleston M, Herzog M, et al. Circulating nucleosomes as potential markers to monitor COVID-19 disease progression. Front Mol Biosci. 2021 Mar;8:600881. doi: 10.3389/fmolb.2021.600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackman N, Antoniak S, Wolberg AS, Kasthuri R, Key NS. Coagulation abnormalities and thrombosis in patients infected with SARS-CoV-2 and other pandemic viruses. Arterioscler Thromb Vasc Biol. 2020;40((9)):2033–44. doi: 10.1161/ATVBAHA.120.314514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrissey SM, Geller AE, Hu X, Tieri D, Ding C, Klaes CK, et al. A specific low-density neutrophil population correlates with hypercoagulation and disease severity in hospitalized COVID-19 patients. JCI Insight. 2021 May;6((9)):e148435. doi: 10.1172/jci.insight.148435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191((3)):677–91. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007 Jan;176((2)):231–41. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonawane S, Khanolkar V, Namavari A, Chaudhary S, Gandhi S, Tibrewal S, et al. Ocular surface extracellular DNA and nuclease activity imbalance: a new paradigm for inflammation in dry eye disease. Investig Opthalmol Vis Sci. 2012 Dec;53((13)):8253. doi: 10.1167/iovs.12-10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meurer M, Öhlmann S, Bonilla MC, Valentin-Weigand P, Beineke A, Hennig-Pauka I, et al. Role of bacterial and host DNases on host-pathogen interaction during streptococcus suis meningitis. Int J Mol Sci. 2020 Jul;21((15)):5289. doi: 10.3390/ijms21155289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann A, Völlger L, Berends ETM, Molhoek EM, Stapels DAC, Midon M, et al. Novel role of the antimicrobial peptide LL-37 in the protection of neutrophil extracellular traps against degradation by bacterial nucleases. J Innate Immun. 2014;6((6)):860–8. doi: 10.1159/000363699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995 Oct;374((1)):1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- 40.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, et al. Neutrophil extracellular traps in COVID-19. JCI insight. 2020 Apr;5((11)):e138999. doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olmos-Ortiz A, García-Quiroz J, López-Marure R, González-Curiel I, Rivas-Santiago B, Olivares A, et al. Evidence of sexual dimorphism in placental vitamin D metabolism: testosterone inhibits calcitriol-dependent cathelicidin expression. J Steroid Biochem Mol Biol. 2016 Oct;163:173–82. doi: 10.1016/j.jsbmb.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Giagulli VA, Guastamacchia E, Magrone T, Jirillo E, Lisco G, De Pergola G, et al. Worse progression of COVID‐19 in men: is testosterone a key factor? Andrology. 2021 Jan;9((1)):53–64. doi: 10.1111/andr.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Zhao Y, Jiang X, Zhao Y, Li Y, Li C, et al. Preliminary evaluation of the safety and efficacy of oral human antimicrobial peptide LL-37 in the treatment of patients of COVID-19, a small-scale, single-arm, exploratory safety study. medRxiv. 2020;2020:20064584. [Google Scholar]
- 44.Ghosh SK, Weinberg A. Ramping up antimicrobial peptides against severe acute respiratory syndrome coronavirus-2. Front Mol Biosci. 2021 Jun;8:620806. doi: 10.3389/fmolb.2021.620806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pahar B, Madonna S, Das A, Albanesi C, Girolomoni G. Immunomodulatory role of the antimicrobial LL-37 peptide in autoimmune diseases and viral infections. Vaccines. 2020 Sep;8((3)):1–19. doi: 10.3390/vaccines8030517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidopoulou S, Theodoridis H, Nazer K, Kessopoulou E, Menexes G, Kalfas S. Salivary concentration of the antimicrobial peptide LL-37 in patients with oral lichen planus. J Oral Microbiol. 2014 Jan;6((1)):26156. doi: 10.3402/jom.v6.26156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okur HK, Yalcin K, Tastan C, Demir S, Yurtsever B, Karakus GS, et al. Preliminary report of in vitro and in vivo effectiveness of dornase alfa on SARS-CoV-2 infection. New Microbes New Infect. 2020 Sep;37:100756. doi: 10.1016/j.nmni.2020.100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desilles JP, Gregoire C, Le Cossec C, Lambert J, Mophawe O, Losser MR, et al. Efficacy and safety of aerosolized intra-tracheal dornase alfa administration in patients with SARS-CoV-2-induced acute respiratory distress syndrome (ARDS): a structured summary of a study protocol for a randomised controlled trial. Trials. 2020 Dec;21((1)):548. doi: 10.1186/s13063-020-04488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Buhr N, von Köckritz-Blickwede M. The balance of neutrophil extracellular trap formation and nuclease degradation: an unknown role of bacterial coinfections in COVID-19 patients? MBio. 2021 Feb;12((1)):e03304–20. doi: 10.1128/mBio.03304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the published article. Raw data were generated in all Departments and Institutes of the submitting authors. Derived data supporting the findings of this study are available from the corresponding authors G.G. and M.K.-B. on request.