Abstract
DNA gyrase is a target of quinolone antibacterial agents, but the molecular details of the quinolone-gyrase interaction are not clear. Quinolone resistance mutations frequently occur at residues Ser83 and Asp87 of the gyrase A subunit, suggesting that these residues are involved in drug binding. Single and double alanine substitutions were created at these positions (Ala83, Ala87, and Ala83 Ala87), and the mutant proteins were assessed for DNA supercoiling, DNA cleavage, and resistance to a number of quinolone drugs. The Ala83 mutant was fully active in supercoiling, whereas the Ala87 and the double mutant were 2.5- and 4- to 5-fold less active, respectively; this loss in activity may be partly due to an increased affinity of these mutant proteins for DNA. Supercoiling inhibition and cleavage assays revealed that the double mutant has a high level of resistance to certain quinolones while the mutants with single alanine substitutions show low-level resistance. Using a drug-binding assay we demonstrated that the double-mutant enzyme-DNA complex has a lower affinity for ciprofloxacin than the wild-type complex. Based on the pattern of resistance to a series of quinolones, an interaction between the C-8 group of the quinolone and the double-mutant gyrase in the region of residues 83 and 87 is proposed.
DNA gyrase is an essential enzyme that is responsible, in part, for the maintenance of DNA topology within the bacterial cell (1, 20, 25). Gyrase catalyzes the ATP-dependent introduction of negative supercoils into closed circular DNA, as well as ATP-independent relaxation of supercoiled DNA. Differences between gyrase and its eukaryotic equivalent, topoisomerase (topo) II, render gyrase susceptible to the action of quinolones while topo II remains largely resistant (11). This has contributed to the success of quinolones as antibacterial agents with relatively few side effects. However, the emergence of quinolone-resistant pathogens in recent years has increased concern that these drugs may soon be clinically ineffective.
DNA gyrase consists of two proteins, GyrA and GyrB, which form an A2B2 complex in the active enzyme. Gyrase introduces changes in the topology of closed circular DNA by cleaving the helix in both strands, forming a 4-base stagger, passing another segment of DNA through the break, and resealing the broken ends. Quinolones exert their toxicity on the bacterial cell by stabilizing the double-stranded break in DNA created by gyrase so that religation becomes unfavorable. The ternary complex blocks transcription (26) and, more importantly in terms of cell survival, DNA replication (15, 24). It is thought that blocking of DNA polymerase by the quinolone-topoisomerase complex triggers the release of broken DNA ends by an as-yet-undefined mechanism (5).
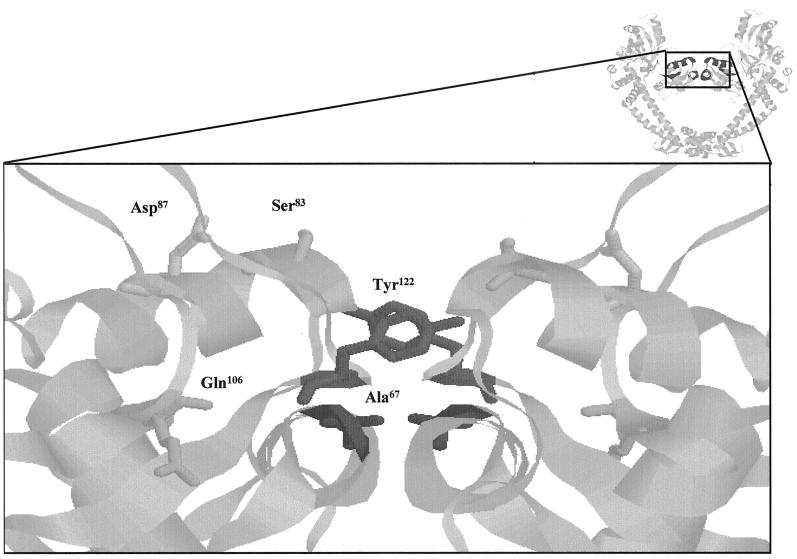
Currently there is no definitive model for the interaction of quinolones with gyrase, although both DNA and Mg2+ are thought to be involved in the complex (14, 19). Mutations in both GyrA and GyrB subunit residues have been found to confer resistance or hypersensitivity to quinolones. The majority of resistant clinical isolates contain substitutions between positions 67 and 106 (inclusive) of GyrA, leading to the categorization of this section as the quinolone resistance-determining region (QRDR) (30). This region is within the N-terminal domain of GyrA, for which a high-resolution structure has been determined (17). QRDR residues are situated close to the catalytic cleavage residue Tyr122, and those that are solvent exposed may be involved in DNA binding (Fig. 1).
FIG. 1.
QRDR of GyrA. The image at the top right shows a ribbon representation (RasMol) of the 59-kDa N-terminal fragment of GyrA (17). The QRDR is in dark gray. The main image is an expanded version of this region showing the active-site tyrosine (Tyr122) and residues of the QRDR. Ser83 and Asp87 are solvent exposed in this structure and have been mutated to Ala in this study.
It seems likely that residues of the GyrB subunit are also involved in stabilizing the interaction with quinolones; spontaneous quinolone resistance mutations at positions 426 and 447 have been identified and characterized (31). The Asp426-to-Asn mutant is resistant to all quinolones tested, but the Lys447-to-Glu mutant exhibits resistance to acidic quinolones, such as nalidixic acid, and hypersensitivity to amphoterics, such as ciprofloxacin. It is possible that the GyrA QRDR residues and a region of GyrB including these mutations interact with one another to form one drug-binding pocket per GyrA-GyrB dimer. This would be consistent with drug-binding experiments that suggest a stoichiometry of 2 drug molecules per complex (6). The assembly of such a drug-binding pocket would require large conformational changes in the enzyme to bring the relevant region of GyrB close to the QRDR. Evidence for such changes in conformation comes from the crystal structures of a 92-kDa domain of yeast topo II (2, 9). This region has sequence similarity with the C-terminal domain of GyrB and the N-terminal domain of GyrA and contains residues equivalent to those at positions 426 and 447 and in the QRDR. The two structures show the protein in two conformational states involving movement of the GyrB residues relative to the QRDR.
Spontaneous quinolone resistance mutations (including those in clinical isolates) are most frequently found at Ser83 (e.g., to Leu or Trp) and Asp87 (e.g., to Asn or Val) in GyrA (7, 18, 30, 32). Site-directed mutations have also been made at these positions and shown to confer quinolone resistance (13, 28, 29). We have targeted these residues, as it would appear that they play a role in drug binding. Single and double alanine substitutions were introduced at these positions in order to monitor the effects of a small, nonpolar residue on gyrase activity and quinolone sensitivity. Alanine mutations infrequently occur at positions 83 and 87, although it was recently reported that 1 of 18 resistant clinical isolates of Escherichia coli had a Ser83-to-Ala substitution (23). (In addition, it is worth noting that the amino acid equivalent to Ser83 in mycobacterial species [gram positive] is Ala [4].) The aim of this study was to assess the role of these residues in the quinolone-gyrase interaction via in vitro experiments with reconstituted gyrase.
MATERIALS AND METHODS
Preparation of gyrase mutants.
E. coli GyrA mutations Ala87 and Ala83Ala87 were created using the QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. The plasmid template for PCR, pPH3, contains a full-length copy of wild-type (WT) gyrA (12). Primers with the appropriate base substitutions were synthesized by Perkin-Elmer Applied Biosystems: 87AF (5′-GGT GAC TCG GCG GTC TAT GCG ACG ATT GTC CGC ATG GCG-3′) and 83A87AF (5′-GGT GAC GCG GCG GTC TAT GCG ACG ATT GTC CGC ATG GCG-3′) (representing nucleotides 241 to 279 of the E. coli gyrA sequence, together with complementary strands). Sequencing of purified plasmid was performed following the QuikChange procedure, and also after the overexpression stage, by the Protein and Nucleic Acid Chemistry Laboratory (University of Leicester). Plasmid DNA containing base substitutions was transformed into Epicurian coli Solopack Gold competent cells according to the manufacturer's instructions (Stratagene). Overexpression of WT and mutant GyrA was also carried out in this strain, followed by ammonium sulfate precipitation and fast protein liquid chromatography (FPLC) purification, as described previously (12). GyrA containing the Ala83 mutation was expressed from plasmid pPH311.1 as described previously (13); the sequence of the QRDR was confirmed and the plasmid was used for protein overexpression, as above. Full-length WT GyrB protein was overexpressed from plasmid pAG111 and purified in two steps by heparin-Sepharose column chromatography and FPLC, as described previously (12). Protein concentrations were estimated by the method of Bradford (3).
Supercoiling assays.
WT or mutant GyrA was mixed in a ratio of 1:2 with WT GyrB and stored at a concentration of 600 nM GyrA monomer. Supercoiling and drug inhibition assay mixtures, contained (final concentrations) 10 μg of relaxed pBR322 DNA/ml, 9 μg of tRNA/ml, 35 mM Tris · HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 1 mM ATP, 0.1 mg of bovine serum albumin/ml, and 6.5% (wt/vol) glycerol. Reaction mixtures were incubated at 37°C for 5 min before addition of gyrase (A2B2), followed by incubation for as long as 1 h at 37°C. Assays were terminated following the addition of 0.5 volume of STEB (20% [wt/vol] sucrose, 0.05 M Tris · HCl [pH 7.5], 0.05 M EDTA, 50 μg of bromophenol blue/ml [final concentrations]) and 2 volumes of chloroform-isoamyl alcohol (24:1), and reactions were analyzed on 1.1% agarose gels. Except where stated, quinolones were dissolved in a 1:5 molar ratio of drug to KOH; enoxacin required equimolar NaOH. Quinolones used were kind gifts from Parke-Davis (PD0117962, PD0164488, PD0129603, PD0163449, and PD0117731), Bayer (ciprofloxacin), Merck (norfloxacin), Dainippon (enoxacin), and Hoechst (ofloxacin).
Cleavage assays.
Quinolone- and Ca2+-stabilized cleavage reactions were carried out under supercoiling conditions, except that ATP was omitted and reaction mixtures were incubated at 25°C for 30 min. In Ca2+-stabilized cleavage reactions, 4 mM CaCl2 replaced 4 mM MgCl2. The final concentration of enzyme was 50 nM (monomer A) in all cleavage experiments. Linear product was revealed by the addition of sodium dodecyl sulfate (SDS) to 0.2% (wt/vol) and proteinase K to 0.1 mg/ml, followed by a 30-min incubation at 37°C. Reaction products were prepared for electrophoresis by the addition of STEB and chloroform-isoamyl alcohol as before. Cleavage products were analyzed on 1.1% agarose gels containing 5 μg of chloroquine/ml. In cleavage specificity reactions, 0.5 U of EcoRI was added following gyrase cleavage and restriction digestion was allowed to proceed for 15 min at 37°C. SDS and proteinase K were added as before and products were analyzed on a 1.35% agarose gel.
Drug-binding assays.
Reaction mixtures were prepared as described above for cleavage assays, in a final volume of 80 μl. Radiolabeled ciprofloxacin ([3H]piperazine ciprofloxacin; Amersham) was added to reaction mixtures, together with gyrase (130 nM GyrA monomer), following a preincubation of 5 min at 25°C. Reactions were incubated for ∼1 h at 25°C. Residual buffer was drained from NICK spin columns (Amersham Pharmacia Biotech), which were then equilibrated with 1 ml of ice-cold buffer (as for cleavage assays) at 4°C for 5 min. Columns were centrifuged at 750 × g for 4 min at 4°C before 75 μl of each reaction product was loaded. A further 75 μl of reaction buffer (without bovine serum albumin) was loaded before recovery of cleaved complexes by centrifugation of columns as before. The total amount of [3H]ciprofloxacin per reaction eluate was quantified by scintillation counting. Control experiments containing DNA but no enzyme showed that a significant amount of drug bound to the plasmid DNA. Results for DNA alone were therefore subtracted from those for cleavage reactions to give final data points.
RESULTS
DNA supercoiling.
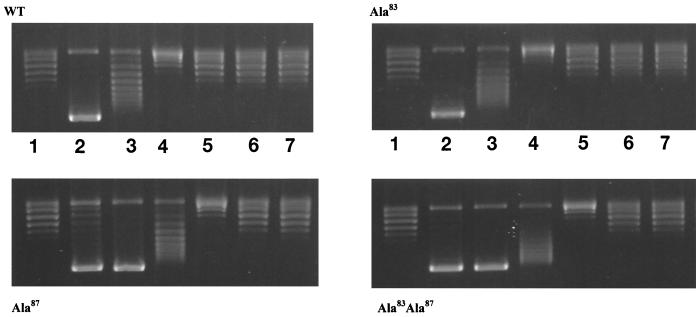
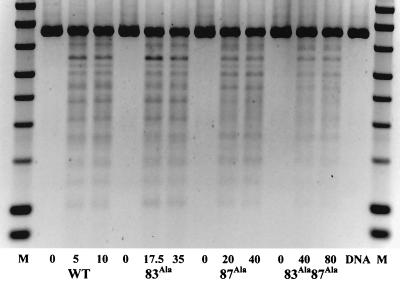
WT and mutant GyrA proteins were purified to near-homogeneity and assayed for DNA supercoiling in the presence of a twofold excess of GyrB. The GyrA Ala83 mutant exhibited activity comparable to that of the WT, whereas the Ala87 mutant and the double mutant (Ala83 Ala87) showed 40 and 20 to 25% of WT activity, respectively (data not shown). The enzymes were also assayed for supercoiling in the presence of a range of NaCl concentrations (Fig. 2). We found that the salt dependence of the Ala83 mutant was comparable to that of the WT enzyme but that both the Ala87 mutant and the double mutant had supercoiling activities that were more salt stable. This suggests that the Ala87 mutation leads to reduced supercoiling activity through stabilization of the DNA-protein complex.
FIG. 2.
DNA supercoiling in the presence of NaCl. Increasing amounts of NaCl were added to standard supercoiling reaction mixtures containing WT or mutant gyrases. Lanes 1, DNA substrate only; 2, no added NaCl; 3, 100 mM NaCl; 4, 150 mM NaCl; 5, 200 mM NaCl; 6, 250 mM NaCl; 7, 300 mM NaCl. WT and mutant GyrA subunits were added at a concentration of 11 nM. Differences in the supercoiling activities of mutants were accounted for by varying incubation times: for WT, 3 min; for Ala83, 2.5 min; for Ala87, 7 min; and for Ala83 Ala87, 10 min.
Ca2+-induced cleavage.
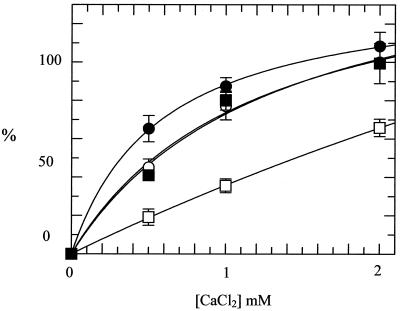
In the presence of Ca2+ the covalent bond between DNA gyrase and DNA can be stabilized, and cleaved DNA is revealed if reaction products are treated with SDS and proteinase K (21). In this way it is possible to determine the effects of the alanine mutations on double-stranded cleavage of DNA, a reaction that is part of the supercoiling cycle. All three mutant enzymes were able to cleave DNA in the presence of Ca2+ (Fig. 3); the Ala83 mutant was slightly more active in this reaction, whereas the Ala87 mutant was less active. The double mutant had cleavage activity comparable to that of the WT. These data show that all three mutants are active in DNA cleavage and that Ca2+-induced cleavage is not simply correlated with supercoiling activity.
FIG. 3.
Ca2+-induced DNA cleavage by DNA gyrase. CaCl2 was added to reaction mixtures containing WT or mutant gyrases (final concentrations, 25 nM) and relaxed pBR322 DNA. The amount (shown as a percentage) of cleaved product (linear pBR322) was quantified. Results are averages from three separate experiments and are normalized such that the intensity of the linear band in the WT cleavage reaction is 100% at 2 mM CaCl2. Symbols: ○, WT; ●, Ala83; □, Ala87; ■, Ala83 Ala87.
Inhibition of supercoiling by quinolones.
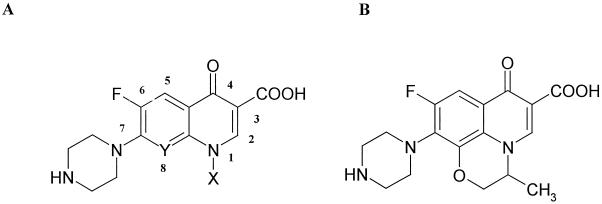
The 50% inhibitory concentrations (IC50) of a range of quinolones were determined for the WT enzyme and the three mutants (Table 1). Results for the mutant enzymes are expressed as the IC50 for the mutant divided by the IC50 for the WT. Most of the quinolones tested had IC50 for the Ala83 mutant that were ∼3-fold greater than those for the WT, i.e., this mutation generally confers a threefold increase in resistance. The only exceptions were norfloxacin and enoxacin, to which the mutant was slightly more resistant (4.6- and 5.8-fold, respectively). In the case of the Ala87 mutant, the IC50 were generally three- to five-fold greater than those for the WT. Again, the exceptions were norfloxacin and enoxacin, to which the mutant was 6.6- and 15.1-fold resistant, respectively. In the case of the double mutant (Ala83 Ala87) there was wide variation in relative IC50. The IC50 of PD0129603 and PD0163449, which have Cl and Br, respectively, at C-8, were ninefold greater than those for the WT, whereas the IC50 of PD0117962 and PD0164488, which have F and OEthyl, respectively, at C-8, were 29- and 25-fold greater than those for the WT. Ciprofloxacin (with H at C-8) has an even greater relative IC50 (38.3-fold). From these data there appears to be a correlation between the substituent at C-8 and the relative IC50. Indeed, enoxacin (where C-8 is replaced by a ring N atom) has the highest relative IC50 of all (>2,000). By contrast, changes at N-1 have only a modest effect on the relative IC50; for example, compare PD0117962 with PD0117731, and ciprofloxacin with norfloxacin.
TABLE 1.
Inhibition of supercoiling by WT and mutant gyrase with a range of quinolones
| Compounda
|
Yb | Xc | IC50 (μM) for WT | Fold increase in IC50 for mutantd
|
|||
|---|---|---|---|---|---|---|---|
| No. | Name | Ala83 | Ala87 | Ala83 Ala87 | |||
| 1 | Ciprofloxacin | CH | C3H5e | 1.1 ± 0.23 | 3.0 ± 0.7 | 4.6 ± 0.4 | 38.3 ± 0.9 |
| 2 | PD0117962 | CF | C3H5 | 0.7 ± 0.06 | 2.7 ± 0.3 | 4.0 ± 0.5 | 29 ± 2.8 |
| 3 | PD0164488 | COEth | C3H5 | 1.7 ± 0.24 | 3.7 ± 0.6 | 4.1 ± 0.4 | 25 ± 0.8 |
| 4 | PD0129603 | CCl | C3H5 | 0.8 ± 0.07 | 2.9 ± 0.3 | 3.1 ± 0.6 | 9 ± 1.2 |
| 5 | PD0163449 | CBr | C3H5 | 0.9 ± 0.12 | 3.1 ± 0.2 | 3.5 ± 0.1 | 9 ± 1.3 |
| 6 | Norfloxacin | CH | C2H5 | 4.8 ± 1.06 | 4.6 ± 1.0 | 6.6 ± 1.0 | 78 ± 1.8 |
| 7 | Enoxacin | N | C2H5 | 4.4 ± 0.62 | 5.8 ± 1.1 | 15.1 ± 2.9 | >2,000 |
| 8 | PD0117731 | CF | C2H5 | 2.0 ± 0.38 | 2.7 ± 0.1 | 3.6 ± 0.3 | 31 ± 2.1 |
| 9 | Ofloxacin | 3.5 ± 0.60 | 2.7 ± 0.1 | 3.1 ± 0.3 | 17 ± 1.0 | ||
Compounds 1 through 8 have structure A, while compound 9 has structure B.
C-8.
N-1.
Relative to that for WT; calculated as IC50 for mutant divided by IC50 for WT.
Cyclopropyl form.
Quinolone-induced cleavage.
An alternative way to assess in vitro susceptibilities of gyrase to quinolones is to examine the quinolone-induced cleavage of DNA (10, 22). Using three of the quinolones (ciprofloxacin, norfloxacin, and PD0163449) we determined the amount of drug required to induce 50% maximal DNA cleavage for each of the mutants; results are expressed relative to those for the WT (Table 2). The data for the single mutants (Ala83 and Ala87) are very similar (within a factor of ∼2) to the data achieved for inhibition of supercoiling (Table 1). Although the double mutant shows an increase over the single mutants in the relative amount of drug required to give 50% cleavage, this increase is not as great as that for the inhibition of supercoiling.
TABLE 2.
Quinolone-induced DNA cleavage by gyrase
| Compound | Relative amt of drug required to induce 50% maximal DNA cleavage for mutanta
|
||
|---|---|---|---|
| Ala83 | Ala87 | Ala83 Ala87 | |
| Ciprofloxacin | 3.6 ± 0.2 | 3.9 ± 1.6 | 8.9 ± 1.8 |
| PD0163449 | 1.4 ± 0 | 3.1 ± 0.4 | 4.3 ± 1.4 |
| Norfloxacin | 4.8 ± 1.0 | 4.4 ± 0.8 | 20.5 ± 1.6 |
Calculated as the amount of drug required to induce 50% maximal cleavage for the mutant divided by the amount required to induce 50% maximal cleavage for the WT.
Another aspect of the quinolone-stabilized cleavage reaction is the frequency with which cleavage occurs at certain sites on the DNA. This might be affected by the alanine mutations; therefore the cleavage specificity of the mutants was investigated. A DNA cleavage reaction that reveals specific double-stranded break sites relative to the unique EcoRI site on plasmid pBR322 was performed. Concentrations of enzyme that gave an approximately equivalent amount of ciprofloxacin-induced cleavage for each mutant were used. Figure 4 shows cleavage specificity reactions for WT, Ala83, Ala87, and Ala83 Ala87 enzymes. Some subtle differences in the amounts of cleavage at certain sites are observed, but generally the results suggest that the sites for ciprofloxacin-stabilized cleavage are not greatly altered by these mutations; it appears that the total amount of cleavage is reduced for the Ala87 and Ala83 Ala87 enzymes.
FIG. 4.
Cleavage specificity assay. The cleavage of EcoRI-linearized pBR322 by WT and mutant enzymes is shown. Ciprofloxacin was used to stabilize cleavage, and the amount of drug used is indicated (in micromolar concentrations). The enzyme concentration was 50 nM in terms of the GyrA monomer. M, marker; DNA, linear pBR322 DNA.
Quinolone binding.
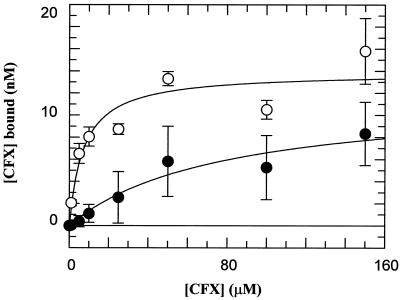
In order to determine whether the Ala substitutions at positions 83 and 87 result in changes in drug binding, we assessed the binding of ciprofloxacin to the WT and double-mutant complexes. Using rapid gel filtration and [3H]ciprofloxacin, we found that the apparent affinity (Kdapp) for the complex involving the double-mutant protein was ∼10-fold less than that for the WT protein (Fig. 5). This difference correlates with the relative amount of ciprofloxacin required to give 50% cleavage (∼9-fold [Table 2]).
FIG. 5.
Quinolone binding to the gyrase-DNA complex. Spin column chromatography data from a number of experiments show the binding of ciprofloxacin (CFX) to WT and mutant (Ala83 Ala87) gyrase-DNA complexes. Reaction mixtures containing [3H]-CFX were passed through Sephadex spin columns, and the amount of drug bound was estimated by scintillation counting. The enzyme concentration was 130 nM in terms of the GyrA monomer. Data were fitted to simple 1:1 ligand binding curves and yielded the following Kdapp values: 7.3 ± 3.1 μM for the WT (○) and 78.0 ± 35.6 μM for the mutant (●).
DISCUSSION
We have investigated gyrase-quinolone interaction by generating amino acid substitutions at two residues considered important in quinolone resistance, Ser83 and Asp87 of GyrA. The results show that the Ser83-to-Ala mutation in the gyrase A protein has little or no effect on supercoiling. In contrast, the Asp87-to-Ala mutation leads to some loss in activity (∼2.5-fold) and the double mutant shows 4- to 5-fold-reduced activity. The reduced activity of the Ala87 and Ala83 Ala87 mutants may be due, in part, to an increased affinity of these enzymes for DNA, as measured by salt tolerance (Fig. 2). The Ca2+-induced cleavage activity of the double mutant is similar to that of the WT, suggesting that the formation of a double-stranded break in DNA is unaffected by the mutations (Fig. 3). The cleavage activity of the Ala87 mutant is reduced, while the Ala83 mutant is slightly more active in cleavage. These data suggest that the effects on DNA supercoiling are not a direct consequence of the effects on DNA cleavage. A cleavage specificity assay (Fig. 4) suggests that there is no significant change in cleavage specificity for enzymes containing these mutations.
Enzymes containing either a single mutation or the double mutation are sufficiently active to allow determination of the quinolone susceptibility in supercoiling and cleavage reactions (Tables 1 and 2). We found that the single Ala83 or Ala87 mutation generally led to a three- to fivefold increase in IC50 for all fluoroquinolones tested (with the exception of norfloxacin and enoxacin). Of the two single mutants, the Ala87 mutant tended to show slightly higher levels of resistance than the Ala83 mutant. The double mutation (Ala83 Ala87) led to significantly increased IC50 that depended, in part, on the identity of the C-8 group in the quinolone drug (Table 1). Supercoiling inhibition results for ciprofloxacin, norfloxacin, and PD0163449 were mirrored by quinolone-stabilized cleavage assays (Table 2).
In most cases, with the C-8 halides PD0163449 (compound 5) and PD0129603 (compound 4) being the exceptions, the resistance of the double mutant is greater than the sum or product of the resistances of two single mutants. However, quinolone-stabilized cleavage values for double-mutant resistance are much closer to the single-mutant cleavage results. This difference may stem from the differential effects of the mutations on the individual reactions of gyrase. The supercoiling activity of the double mutant (four- to five-fold reduced compared with that of the WT) is more severely affected than cleavage, and this seems to be reflected in the extent to which quinolones inhibit the different reactions. However, it should be pointed out that supercoiling and Ca2+ cleavage reactions will not necessarily be correlated, as Ca2+-induced cleavage is likely to be an aberrant reaction of the enzyme. In addition, we measured supercoiling activity using time courses and cleavage activity at a fixed time with varying Ca2+ concentrations. Nonetheless, the pattern of drug resistance remains the same, such that the magnitude of the reduction in drug activity caused by mutation is greatest for norfloxacin, intermediate for ciprofloxacin, and least for PD0163449 in both types of experiment.
We showed that double-mutant resistance to ciprofloxacin correlates with decreased affinity of the drug for the enzyme through spin column chromatography (Fig. 5). However, this does not appear to be a simple case of binding inhibition through, for example, loss of a negative charge, which might have the same effect on all drugs. Resistance patterns among the supercoiling inhibition data show that there is no correlation with WT IC50, suggesting that the mode of drug binding within the double-mutant ternary complex has been altered. For example, compare results for ciprofloxacin (compound 1) and PD0163449 (compound 5): IC50 for the WT are similar, yet the double mutant is 4 times more resistant to ciprofloxacin than to PD0163449. In the case of the Ala83 mutation, earlier work (27) showed a ∼3-fold drop in norfloxacin binding, which correlates with the 4.6-fold increase in the IC50 (Table 1) and the 4.8-fold increase in the amount of drug required to give 50% maximal cleavage (Table 2).
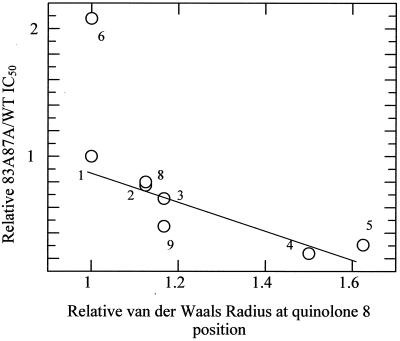
The C-8 halides (CBr, CCl, or CF at position 8) form a relatively strong ternary complex with WT enzyme and have similar IC50 for the WT. However, the double mutant is able to distinguish between a chlorine or bromine atom (∼10-fold more resistant than the WT) and a fluorine at C-8 (∼40-fold more resistant than the WT). This led to speculation that the atomic radius of the atom at position 8 may be related to double-mutant resistance patterns. A plot of van der Waals radius (of the C-8-associated atom) against relative mutant resistance shows that a roughly linear relationship does indeed exist (Fig. 6). Norfloxacin (compound 6 in Table 1) and enoxacin (not shown) do not fit this relationship; the N-1 ethyl group accounts for increased double-mutant resistance to norfloxacin, and it is possible that this side group also lies close to the quinolone-protein interface. The very high level of double-mutant resistance to enoxacin indicates that this drug, with its ring-integrated nitrogen atom at position 8, has quite distinct properties. It is interesting that other work involving C-8 methoxy fluoroquinolones has established the importance of the C-8 substituent in drug potency (8, 16, 33).
FIG. 6.
Plot demonstrating the possible relationship between the quinolone resistance of the double mutant and the van der Waals radius (relative to ciprofloxacin) of the atom associated with position 8 of the quinolone. Numbers correlate with compound numbers given in Table 1. Enoxacin is not included in this figure.
A direct comparison of drugs that are identical except at position N-1 (i.e., cyclopropyl or ethyl groups at N-1) shows that there is a difference in double-mutant resistance between norfloxacin and ciprofloxacin, but this is not the case with C-8 halides. Specifically, compare drug 1 with drug 6 and drug 2 with drug 8 in terms of relative double-mutant resistance (Table 1). We cannot, therefore, rule out the possibility that the N-1 has an effect on ternary complex stability, although the extent of the N-1 effect would seem to depend on the nature of the C-8 atom.
The data in this paper support the idea that Ser83 and Asp87 are important contacts in the gyrase-quinolone-DNA complex. Mutation of Ser83 (to Ala) is largely without effect on enzyme activity, whereas mutation of Asp87 leads to loss of supercoiling activity, which can be attributed to stabilization of enzyme-DNA interactions. Mutation of either residue may lead to the loss of enzyme-drug interactions (e.g., hydrogen bonds). However, we suggest that mutation of both residues leads to an alteration in the mode of drug interaction whereby the C-8 group on the drug makes a more significant contribution to the interaction with the protein (and/or DNA). One possibility is that conversion of Ser83 and Asp87 to Ala allows an alternative mode of binding of the drug with the enzyme, for example, enabling contacts between the drug and the peptide backbone. Thus we propose that removal of both Ser83 and Asp87 causes a rearrangement of the bound drug such that new contacts are made. However, as the structure of the ternary complex is presently unknown, it is not yet possible to identify the nature of these altered contacts.
ACKNOWLEDGMENTS
We thank Karl Drlica and Jonathan Heddle for comments on the manuscript and Mike Sutcliffe for suggestions regarding drug-protein interactions.
We thank Parke-Davis (Ann Arbor, Mich.) for financial support.
REFERENCES
- 1.Bates A D, Maxwell A. DNA topology. Oxford, United Kingdom: IRL Press; 1993. [Google Scholar]
- 2.Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure at 2.7 Å resolution of a 92K yeast DNA topoisomerase II fragment. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Caron P R. Appendix: compendium of DNA topoisomerase sequences. In: Bjornsti M-A, Osheroff N, editors. DNA topoisomerase protocols. DNA topology and enzymes. Vol. 94. Totowa, N.J: Humana Press; 1999. pp. 279–316. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-R, Malik M, Snyder M, Drlica M. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 6.Critchlow S E, Maxwell A. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry. 1996;35:7387–7393. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 7.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterisation of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth survival and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fass D, Bogden C E, Berger J M. Quaternary changes in topoisomerase II may direct orthogonal movements of two DNA strands. Nat Struct Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 10.Gellert M, Mizuuchi K, O'Dea M H, Itoh T, Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootz T D, Barrett J F, Sutcliffe J A. Inhibitory effects of quinolone antibacterial agents on eucaryotic topoisomerases and related test systems. Antimicrob Agents Chemother. 1990;34:8–12. doi: 10.1128/aac.34.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallett P, Grimshaw A J, Wigley D B, Maxwell A. Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene. 1990;93:139–142. doi: 10.1016/0378-1119(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 13.Hallett P, Maxwell A. Novel quinolone resistance mutations of the Escherichia coli DNA gyrase A protein: enzymatic analysis of the mutant proteins. Antimicrob Agents Chemother. 1991;35:335–340. doi: 10.1128/aac.35.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heddle J G, Barnard F M, Wentzell L M, Maxwell A. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids. 2000;19:1249–1264. doi: 10.1080/15257770008033048. [DOI] [PubMed] [Google Scholar]
- 15.Hiasa H, Yousef D O, Marians K J. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J Biol Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 16.Lu T, Zhao X, Drlica K. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob Agents Chemother. 1999;43:2969–2974. doi: 10.1128/aac.43.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morais Cabral J H, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Structure of the DNA breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 18.Oram M, Fisher M. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:387–389. doi: 10.1128/aac.35.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palumbo M, Gatto B, Zagotto Z, Palu' G. On the mechanism of action of quinolone drugs. Trends Microbiol. 1993;1:232–235. doi: 10.1016/0966-842x(93)90138-h. [DOI] [PubMed] [Google Scholar]
- 20.Reece R J, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 21.Reece R J, Maxwell A. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J Biol Chem. 1989;264:19648–19653. [PubMed] [Google Scholar]
- 22.Sugino A, Peebles C L, Kruezer K N, Cozzarelli N R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavio M D, Vila J, Ruiz J, Martin-Sanchez A M, de Anta M T J. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J Antimicrob Chemother. 1999;44:735–742. doi: 10.1093/jac/44.6.735. [DOI] [PubMed] [Google Scholar]
- 24.Wentzell L M, Maxwell A. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J Mol Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 25.Wigley D B. Structure and mechanism of DNA gyrase. Nucleic Acids Molecular Biol. 1995;9:165–176. [Google Scholar]
- 26.Willmott C J R, Critchlow S E, Eperon I C, Maxwell A. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J Mol Biol. 1994;242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 27.Willmott C J R, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces the binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonezawa M, Takahata M, Banzawa N, Matsubara N, Watanabe Y, Narita H. Analysis of the NH2-terminal 83rd amino acid of Escherichia coli GyrA in quinolone-resistance. Microbiol Immunol. 1995;39:243–247. doi: 10.1111/j.1348-0421.1995.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 29.Yonezawa M, Takahata M, Banzawa N, Matsubara N, Watanabe Y, Narita H. Analysis of the NH2-terminal 87th amino acid of Escherichia coli GyrA in quinolone-resistance. Microbiol Immunol. 1995;39:517–520. doi: 10.1111/j.1348-0421.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida H, Kojima T, Yamagishi J, Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]