Abstract
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic syndrome/myeloproliferative neoplasm overlap syndrome characterized by monocytic proliferation in the presence of dysplastic bone marrow changes, inflammatory symptoms and propensity for transformation to AML, with a poor prognosis and limited treatment options. Unlike the α and β isoforms, the PI3K-δ signaling protein is predominantly expressed by hematopoietic cells and therefore has garnered interest as a potential target for the treatment of lymphomas and leukemias. We revealed a pattern of increased PIK3CD : PIK3CA ratio in monocytic M5 AML patients and cell lines and this ratio correlated with responsiveness to pharmacological PI3K-δ inhibition in vitro. As CMML is a disease defined by monocytic clonal proliferation, we tested the PI3K-δ inhibitor, umbralisib, as a single agent, and in combination with the JAK1/2 inhibitor, ruxolitinib in CMML. Our ex vivo experiments with primary CMML patient samples showed synergistic inhibition of viability and clonogenicity with this combination. Phospho-specific flow cytometry revealed that dual inhibition had the unique ability to decrease STAT5, ERK, AKT and S6 phosphorylation simultaneously, which offers a mechanistic hypothesis for the enhanced efficacy of the combination treatment. These preclinical data demonstrate promising activity by co-inhibition of PI3K-δ and JAK1/2 and support the use of ruxolitinib + umbralisib combination therapy in CMML under active clinical investigation.
Keywords: Chronic myelomonocytic leukemia, PI3K, JAK/STAT
Category: Malignant Hematopoiesis
Background
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic syndrome/myeloproliferative neoplasm overlap syndrome (MDS/MPN) characterized by monocytic proliferation in the presence of other cytopenias, splenomegaly and propensity for transformation to AML. Prognosis is generally poor, with a mean overall survival between 2–5 years.1 Response to conventional treatment is variable, and the only curative treatment is allogeneic hematopoietic stem cell transplantation (HSCT). Preclinical observations of hypersensitivity to janus kinase/signal transducer and activator of transcription (JAK/STAT)-dependent GM-CSF signaling in CMML have led to successful trials testing JAK-inhibitors in the clinic. 2–4 Compensatory JAK2/STAT5 signaling following PI3K/AKT inhibition has been observed in breast cancer models, spurring interest in inhibiting both signaling pathways simultaneously.5 Co-targeting of the JAK/STAT and phosphatidylinositol-3-kinase/AKT (PI3K/AKT) pathways has been investigated pre-clinically in a range of neoplasms, showing promising results, including in myleofibrosis.6,7 Considerable pharmaceutical effort towards inhibiting the PI3K/AKT pathway have focused on inhibitors which are specific to one or more of the 4 Class I PI3K catalytic subunit isoforms (p110-α, β, δ, γ), and strong in vitro growth inhibitory synergy between a PI3K-δ-specific inhibitor and a JAK1/2 inhibitor has been previously reported in myeloid leukemia patient samples, including a single CMML patient sample.8
Umbralisib (TGR-1202) is a PI3K-δ isoform-selective and casein kinase 1-ε (CK1ε) inhibitor that has shown impressive responses in clinical trials for the treatment of lymphoid malignancies, which are known to be reliant on the PI3K-δ subunit for AKT activation and survival.9 Unlike the ubiquitously expressed PI3K-α and –β isoforms, PI3K-δ is largely confined to hematopoietic cells, and has been identified as the predominant isoform seen in bone marrow aspirates from AML patients, although its role in CMML has yet to be investigated.10,11 An ongoing clinical trial in a related myeloid disease, myelofibrosis, reported that umbralisib augmented the response to ruxolitinib in patients previously resistant to ruxolitinib monotherapy, and resulted in a complete response (CR) in 2 patients (IWG-MRT consensus criteria, response rate of 56.5%).12 Here, we evaluated the efficacy of umbralisib in combination with ruxolitinib in human leukemia cell lines and primary CMML patient samples to provide support for this combination as a new therapeutic strategy in CMML.
Methods
Patient Samples.
Experiments were conducted on viably frozen, primary CMML patient samples, which were provided by the Vanderbilt-Ingram Cancer Center Hematologic Malignancy Tumor Bank in accordance with the tenets of the Declaration of Helsinki and approved by the Vanderbilt University Medical Center Institutional Review Board.
Cell lines.
AML cell lines MV-4–11, Kasumi-1, K-562, U-937, Kasumi-3, PL-21, MOLM-16, and THP-1, and leukemia mantle cell lymphoma cell line Z-138 were purchased from the ATCC. OCI-AML3, HEL, F36P, and MOLM-13 cell lines were purchased from Deutsche Sammelung von Mikroorganismen und Zellkulturen (DSMZ). ATCC and DSMZ cell bank cell lines were authenticated by short tandem repeat profiling and cytochrome c oxidase gene analysis. Cultured cells were split every 3 to 4 days and maintained in exponential growth phase. Cell lines were tested for Mycoplasma in 2019 using the Universal Mycoplasma Detection Kit (ATCC). Cells were used for the experiments presented here within 10 to 30 passages from thawing. MV-4–11 cell line was grown in Iscove’s modified Dulbecco’s medium (IMDM), THP-1 cell line was grown in RPMI 1640 supplemented with 10% FBS and 0.05 mM 2-ME, and all other cell lines were cultured in RPMI and supplemented with 10% to 20% fetal bovine. Cells were kept at 37°C in a 5% CO2 incubator. All media was supplemented with 100U/mL penicillin and 100ug/mL streptomycin.
Protein extracts and Western blotting analysis.
Cell lines were grown in their respective media before total protein lysates were extracted in Laemmli sample buffer (Bio-Rad), sonicated, and boiled at 95°C for 10 minutes. The samples were loaded in a 10% sodium dodecyl sulfate polyacrylamide gel (3.6 μg protein/well for patient samples and 1.67 × 105 cells/well for cell lines). Cryopreserved CMML patient samples were resuspended in RPMI 1640 supplemented with 10% FBS and immediately exposed to RIPA lysis buffer then lysates were extracted with RIPA extraction buffer and boiled at 95°C for 10 minutes. Western blot analysis was performed according to standard protocol with antibodies to PI3K-α (C73F8 clone, Cell Signaling Technology), PI3K-β (C33D4 clone, Cell Signaling Technology), Pi3k-δ (D1Q7R clone, Cell Signaling Technology), PI3K-γ (D55D5 clone, Cell Signaling Technology) and Actin (Sigma-Aldrich). Relative density of bands was measured using Li-COR Image Studio Lite version 5.2.
Gene Expression Data sets.
Cell line AML subtype gene expression data were analyzed from microarray data (log2 expression) from the Hemap data set (http://hemap.uta.fi/).13 The data were generated using RNA extracted from the mononuclear cell fraction of AML patients. For healthy monocytes, gene expression data were used only from experimental controls without manipulation with drugs or cytokines. Cell line microarray data were gathered from the Cancer Cell Line Encyclopedia (CCLE), whose creators bear no responsibility for our analysis or interpretation.14 For analysis of CMML gene expression, log2-transformed expression values of Affymetrix Human Genome U133 Plus 2.0 Array probe sets were downloaded from GEO (GSE42731, GSE61804). Heatmaps were generated using the Heatmapper resource (http://heatmapper.ca/expression/) using median log2 gene expression values from the Hemap dataset.15 Color shading represents the relative abundance of gene expression across each sample type, represented as the Z-score.
In vitro growth inhibition assays.
Cryopreserved CMML bone marrow (BM) mononuclear cells (MNC) were resuspended in STEMSPAN serum-free expansion media supplemented with 10ng/mL of GM-CSF (PEPROTech). To determine the efficacy of umbralisib independently and in combination with ruxolitinib, cell lines or patient samples were plated into a 384-well plate and treated for 72 hours at 37°C and 5% CO2 with umbralisib (provided by TG Therapeutics) or ruxolitinib (Selleckchem) at concentrations ranging from 3nM to 10μM. Cell viability was measured using CellTiter-Glo reagent (Promega Corp.), and the relative luminescence units (RLU) were measured with a micro-plate reader (BioTek). Viability was defined as the percent of RLU of each well compared to the RLU of cells treated with a DMSO vehicle. Data are shown as mean ± SD from 3 replicates for cell line experiments and 2 replicates in the case of patient samples.
Drug combination analysis and calculation of synergy.
The effects of the combinations were calculated using the Zero Interaction Potency (ZIP) model, which compares observed and expected combination effects.16 A detailed description and interpretation guide are included in the supplementary materials.
Colony formation assays.
CMML BMMCs were resuspended as above and added to 3 mL of MethoCult H4230 methylcellulose (STEMCELL Technologies) to a final concentration of 1–3×105 cells/mL containing GM-CSF (10 ng/mL) and either DMSO, 30nM ruxolitinib, 3.3μM umbralisib, or the combination. Next, 1.1 mL of the MethoCult solution was plated into 35 ×10-mm Petri dishes, in duplicate. Plates were incubated for 14 days at 37°C and 5% CO2, and colony (defined as a cluster of >50 cells) counts were then determined under a microscope at 10X magnification.
Flow Cytometry.
CMML BMMCs were resuspended as above. Cells were treated with DMSO (<0.1% by volume), 100 nM ruxolitinib, 10 μM umbralisib or their combination for 24 or 48 hours in an incubator at 37°C and 5% CO2. CD33+CD45+ subpopulation measurements were taken at the 24 hour time point, while CD34+CD38− viability is reported from the 48 hour time point measured in separate experiments. This was done to preserve adequate cell counts in the CD33+CD45+ population which decreased in number more rapidly than the CD34+CD38− population. Cells were stained with a combination of antibodies against human CD33-PE/Cy7 (Clone P67.6, BioLegend), CD45-APC (Clone 2D1, BioLegend), CD34-PE-Dazzle 594 (Clone 581, BioLegend), CD38-APC/Cy7 (Clone 90, BioLegend), AnnexinV/ DAPI Apoptosis Detection Kit (BD Pharmingen) and subjected to flow-cytometric analysis using a 4-laser LSR Fortessa (Becton Dickinson).
Phospho-specific flow cytometry.
Phospho-specific flow cytometry with fluorescence cell barcoding were performed as has previously been reported.17 Briefly, cells (either patient samples or cell lines) were plated at 1×106 cells/mL and incubated with either DMSO, umbralisib (Selleck Chemicals, S8194) (20 μM), ruxolitinib (Selleck Chemicals, S1378) (60 nM), or the combination for 30 min. Cells in the stimulated condition were subsequently stimulated with 20 ng/mL GM-CSF (Peprotech, 300–03-20ug) for 15 min and all were then exposed to 50x Ax700 (Invitrogen, A20010) viability stain. Cells were fixed in 1.6% PFA (Fisher Scientific, 50–980-487) in dark at RT for 10 min, washed with PBS, and permeabilized with MeOH (Fisher Scientific, A4121) and stored in −80 °C. Cells were washed twice with PBS before barcoding with serial dilutions of Pacific Orange (1:2.5) (Invitrogen, P30253) and Pacific Blue (1:2.4) (Invitrogen, P10163) dyes and combining in a single tube.18,19 Cells were stained with pSTAT5-PE/Cy7 (Clone 47, BD:560117), pERK-Ax488 (Clone 6B8B69, Biolegend:369508), pAKT-Ax488 (Clone M89–61, Company BD:560404 and Clone D9E, CST:5315S) and pS6-Ax647 (Clone D57.2.2E, Company CST:4851S) in the dark at RT for 30 min. Cells were washed with 1mL PBS/BSA and resuspended in 0.2 mL PBS/BSA and run a 4-laser LSR Fortessa (Becton Dickinson).
Next-Generation Sequencing.
For next-generation sequencing (NGS), CMML patient bone marrow aspirates were obtained and DNA was isolated using a DNA midi-prep (Qiagen) for NGS in a panel of commonly mutated regions of myeloid neoplasia-associated genes across the genome. The analytic targets included in the OnkoSight™ NGS Myeloid Malignancies gene panel (Illumina) include exonic regions across each of the following genes: SRSF, U2AF1, TET2, IDH2, DNMT3A, RUNX1, TP53, BCOR, BCORL1, ETV6, NPM1, GATA2, WT1, ASXL1, EZH2, JAK2, FLT3, FBXW7, CBL, KRAS, NRAS, SETBP1, ABL1, CSF3R, PTEN, PTPN11, SRSF2, TP53, ZRSR2, PHF6, MYD88, IDH1, HRAS, CALR, BRAF, and CDKN2A. The panel of validated genes consisted of therapeutic markers, as well as genes with diagnostic and prognostic utility in myeloid and other hematologic tumors.
Statistical Analysis.
Statistical comparison of gene expression levels between myeloid leukemia subtypes was done using two-tailed Wilcoxon test followed by Benjamini-Hochberg adjustment of P-values, and fold change was computed to compare gene expression levels for PIK3CA, PIK3CB, PIK3CD, PIK3CG using the NIH GEO2R web-based tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/). For the in vitro growth inhibition assays and the flow cytometry analysis of viability, differences in viability relative to control between the treated groups were analyzed with a paired t-test using GraphPad software analysis (Prism 6.0h). Dose response curves were generated with GraphPad Prism version 6.0h and the 50% growth inhibition concentration (GI50) values were determined using linear regression of double-log transformed data. For comparison of colony forming ability among treatment arms, a multiple regression analysis was performed comparing the relative viability of each single agent and the combination across the 7 samples tested using Stata (Ver 14).
Results
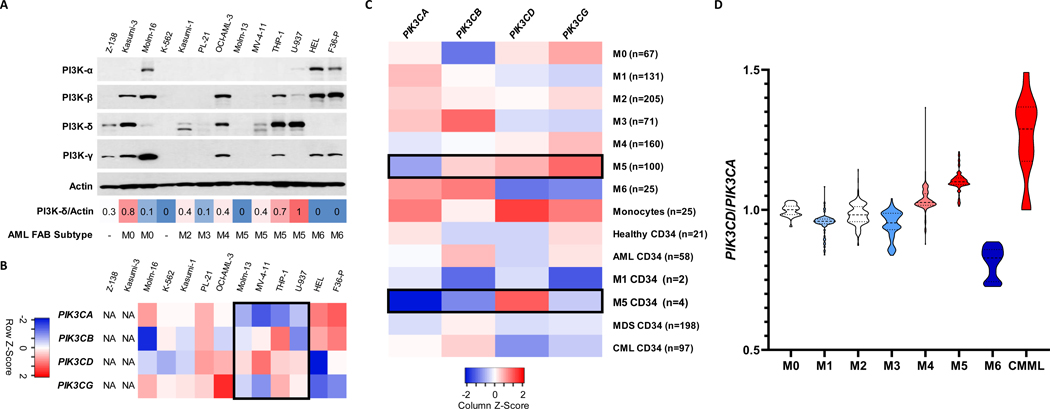
We first sought to assess the expression pattern of the PI3K isoforms in leukemic cell lines (Figure 1A). There was a range of expression levels across the 12 myeloid leukemia and 1 mantle cell lymphoma cell lines tested for all 4 PI3K subunit isoforms. The PI3K-δ protein was most abundant in myelomonocytic and monocytic AML cell lines, histologically defined with the French-American-British (FAB) classification as M4 or M5. There was less expression in both erythroid leukemias (M6) and primitive leukemia cell lines (M0-M2 and the CML blast crisis line, K562). There were notable exceptions to this pattern of protein expression, however, the gene expression data generated a more consistent picture. Comparing publicly available microarray datasets from these cell lines revealed, again, a common pattern of decreased PIK3CA and increased PIK3CD in cell lines derived from M5 AML patients (Figure 1B).14 An unsurprising result given that PI3K-δ has been previously reported to be predominately regulated at the level of transcription.20
Figure 1. PI3K catalytic subunit expression patterns in myeloid leukemias.
(A) Western blot analysis of 11 myeloid and 1 lymphoid neoplasm cell lines to determine protein levels of PI3K-δ. Actin was used as a loading control. PI3K-δ/Actin ratio describes relative density of western blot band as measured by Image Studio Lite ver 5.2. French-American-British (FAB) subtyping differentiates AML by cell maturity and origin based on histological evaluation. (B) Heatmap of the microarray median gene expression of PI3K catalytic subunit genes in available cell lines in the CCLE. NA = not available (C) Heatmap of the median gene expression for each AML FAB class, healthy samples, healthy monocytes, and CD34+ cells from AML, MDS, CML and healthy donor samples in the Hemap dataset. (D) Violin plot of PIK3CD to PIK3CA median log2 gene expression ratio by AML FAB subtype from Hemap data set. CMML data gathered from Gelsi-Boyer et al.21
This pattern also held true for gene expression data from primary human AML blasts, with M5 AMLs expressing more PIK3CD and less PIK3CA than other AML subtypes, including in isolated M5 CD34+ hematopoietic progenitor cells (Figure 1C). We next analyzed a pair of datasets containing both CMML and AML patient samples and found an increase in PIK3CD expression in CMML compared to AML samples, though PIK3CA was equivalent in this case (Figure S1, Table S1). The differences of both genes across disease types are best visualized by the PIK3CD:PIK3CA ratio, with M5 AML and CMML showing the greatest ratio of delta to alpha (Figure 1D), but a statistical comparison of individual gene mean log fold differences across leukemia subtype was also performed and were consistent with this representation (Figure S1, Table S1). The ratio of PIK3CD to PIK3CA gene expression has previously been shown to be correlated with response to PI3K-δ subunit inhibition in the clinic.22 No other AML subtypes, nor other myeloid malignancies such as MDS or chronic myeloid leukemia followed this pattern of PI3K isoform gene expression (Table S2).
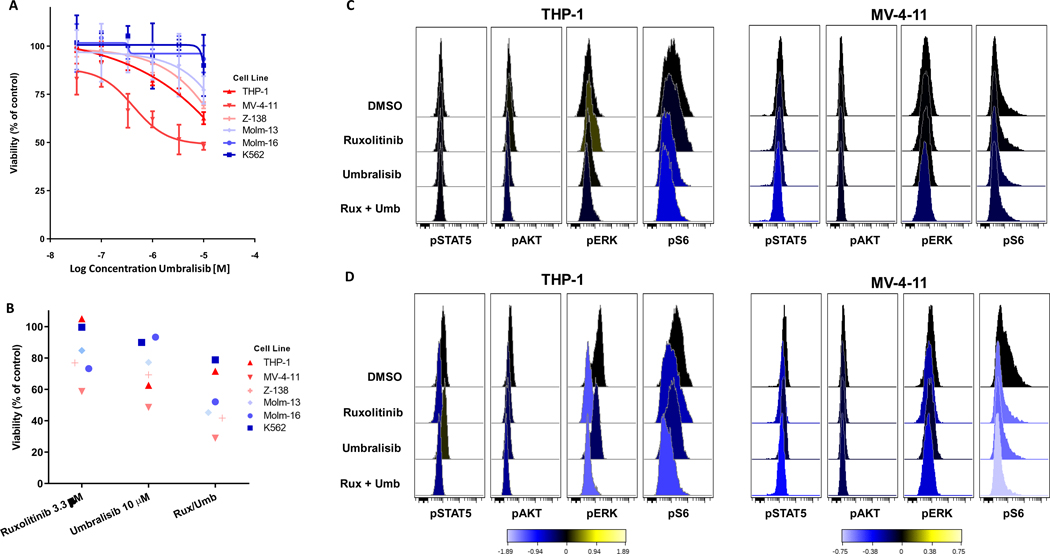
We next assessed the efficacy of umbralisib against select cell lines with varying PI3K-δ protein levels. Myeloid leukemia cell lines were largely insensitive to umbralisib as a solo agent, similar to the observed activity of the related compound, idelalisib, in AML samples (Figure 2A).11 However, a trend emerged, wherein cell lines with increased PI3K-δ expression (THP-1, MV-4–11, Z-138) had the greatest dose response to umbralisib. The myelomonocytic (B-myeloid) leukemia cell line, MV-4–11, was the most sensitive with 50% growth inhibition (GI50) reached at 5.7 μM and is also the only cell line tested where the PI3K-δ protein was the only isoform detected by western blot. The combination of 3.3 μM ruxolitinib and 10 μM umbralisib, our highest doses tested, showed moderate ability to reduce growth across the cell lines, with MV-4–11 having the greatest growth inhibition in all three conditions. Umbralisib offered modest additive growth inhibition to ruxolitinib in cases where it had almost no activity as a solo agent, such as with Molm-16 and K-562 (Figure 1B). Interestingly, THP-1 was completely insensitive to ruxolitinib as a solo agent, in stark contrast to MV-4–11, the other PI3K-δ-predominant myeloid leukemia cell line tested.
Figure 2. Ruxolitinib/umbralisib combination treatment of leukemia cell lines.
(A-B) The viability of treated cell lines compared with DMSO control using the CellTiter-Glo assay was plotted for (A) the indicated concentration gradient of solo agent umbralisib and (B) at selected concentrations of both ruxolitinib, umbralisib and the combination. Each symbol represents the mean of three experimental replicates. The color associated with each cell line relates to the relative expression of PI3K-δ as determined by western blot analysis. (C-D) Histograms from a representative experimental replicate (n = 2) of phospho-specific flow cytometry analysis of cell signaling responses of THP-1 and MV-4–11 cell lines to the combination treatment in the absence (C) or presence (D) of GM-CSF stimulation. Values represent arcsinh of the median fold change from DMSO treated cells.
To investigate this further, we examined cell signaling changes in both THP-1 and MV-4–11 in their typical cell culture conditions as described above, without the addition of cytokines (Figure 2C). As expected, umbralisib decreased pAKT and pS6 signaling in THP-1 cells. Interestingly, there was a paradoxical increase in pERK signaling when THP-1 cells were exposed to ruxolitinib alone, a pattern that was reversed when ruxolitinib was combined with umbralisib. MV-4–11 signaling changes were comparable to THP-1 with the solo agents, however, the combination demonstrated a unique ability to decrease all four cell signaling molecules including pSTAT5. Due to our interest in CMML and its known hypersensitivity to GM-CSF stimulation, we also investigated in vitro cell signaling responses in the presence of this cytokine (Figure 2D). In both cell lines, ruxolitinib decreased pSTAT5 to basal levels and pERK to a lesser extent. Umbralisib reduced pAKT and pS6 but neither pERK or pSTAT5 were significantly affected. The combination again was unique in its ability to reduce all four signaling molecules to near basal levels. Experimental replicates with their corresponding dot plots and each cell line’s response to GM-CSF stimulation can be found in the supplementary materials (Figure S2).
Given the pattern of PI3K catalytic subunit expression observed in cells with a monocytic histological phenotype, we moved to study the activity of umbralisib in CMML patient samples, both as a solo agent and in combination with another emerging therapy, ruxolitinib. All studies were conducted on viably frozen BMMNCs from 14 CMML patients, with a diverse range of BM blast percentage and molecular aberrations as detected by a next generation sequencing panel (Table 1, Figure S3). Congruent with previous reports, ruxolitinib effectively repressed growth of CMML samples in vitro with a majority of samples demonstrating GI50 values between 2 and 550 nM (Table 1).2 Umbralisib demonstrated comparable efficacy in CMML patient samples as was shown in the MV-4–11 cell line, with a majority of GI50 values between 0.3 and 14 μM (Table 1). Umbralisib has previously been shown to induce no cytotoxicity in healthy hematopoietic progenitor cells at concentrations greater than 10 μM, which is also a clinically achievable plasma concentration in humans treated with umbralisib.23,24
Table 1. CMML patient characteristics and response to individual agents.
Clinical information corresponding to CMML patient samples. Karyotype and blast % values are from the time of diagnostic bone marrow biopsy. Samples were tested with targeted next generation sequencing of 37 myeloid genes. Viably frozen CMML BMMNCs were treated ex vivo with threefold dilutions of ruxolitinib, umbralisib and GI50 at 72 hours was calculated.
| Pt ID | Symbol | GENDER | Aqe at Dx | WHO Category | Blast% BMBx | Karyotype at Diagnosis | Molecular Abberations | GI50 Ruxolitinib(μM) | GI50 Umbralisib(μM) |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| CMML 001 | ▼ | M | 67 | CMML-1 | 2.5 | 46XY | ASXL1, EZH2, TET2 | 0.095 | 11.87 |
| CMML002 | ♦ | F | 67 | CMML-2 | 10.0 | 46XX del(9) (q!3,q32) | RUNX1, SRSF2 | 0.021 | 2.257 |
| CMML 003 | ● | M | 62 | CMML-2 | 10.0 | 45XY | ASXL1, IDH2, SRSF2 | 67,067 | >100 |
| CMML 004 | ○ | F | 63 | CMML-1 | 5.0 | 46XX del(7) (q22,q32) | JAK2 | 2.39 | 0.354 |
| CMML005 | • | M | 72 | CMML-1 | 9,5 | 46XY | ASXL1, RUNX1 | 0.549 | 18.2 |
| CMML006 | X | F | 66 | CMML-1 | 7.0 | 46XX | ASX LI, IMR AS, ETV6, SRSF2, SETBP1 | 0.549 | >100 |
| CMML 007 | ■ | M | 65 | CMML-2 | 17.5 | 46XY | ASXL1, EZH2, RUNX1, IDH2, PHF6 | 0.126 | 5.969 |
| CMML 008 | ⎕ | M | 74 | CMML-1 | 5.0 | 46XY | None detected | 0.509 | 14.10861999 |
| CMML 009 | + | M | 59 | CMML-1 | 5.0 | 46XY | ASXL1, RUNX1, JAK2, IDH2, SRSF2 | 0.102 | 2.289 |
| CMML010 | ▽ | M | 73 | CMML-2 | 12.0 | 46XY t(6;13) (p23,ql4) | ASXL1, NR AS, SETBP1, GATA2 | 0.291 | 56.176 |
| CMML011 | ▲ | F | 76 | CMML-1 | 0.8 | 46XX | DNMT3A SF3B1, TET2 | 0.002 | 8.136 |
| CMML012 | ✶ | M | 66 | CMML-1 | <5% | 45XY | ASXL1, TET2, EZH2 | 58.092 | >100 |
| CMML013 | • | F | 65 | CMML-0 | 2.0 | 46XX | KRAS. TET2. SRSF2 | NA | NA |
| CMML014 | △ | F | 57 | CMML-1 | 10.0 | 46XX inv(2) (p11.2q13) | ASXL1, FLT3-ITD, SETBP1 | NA | NA |
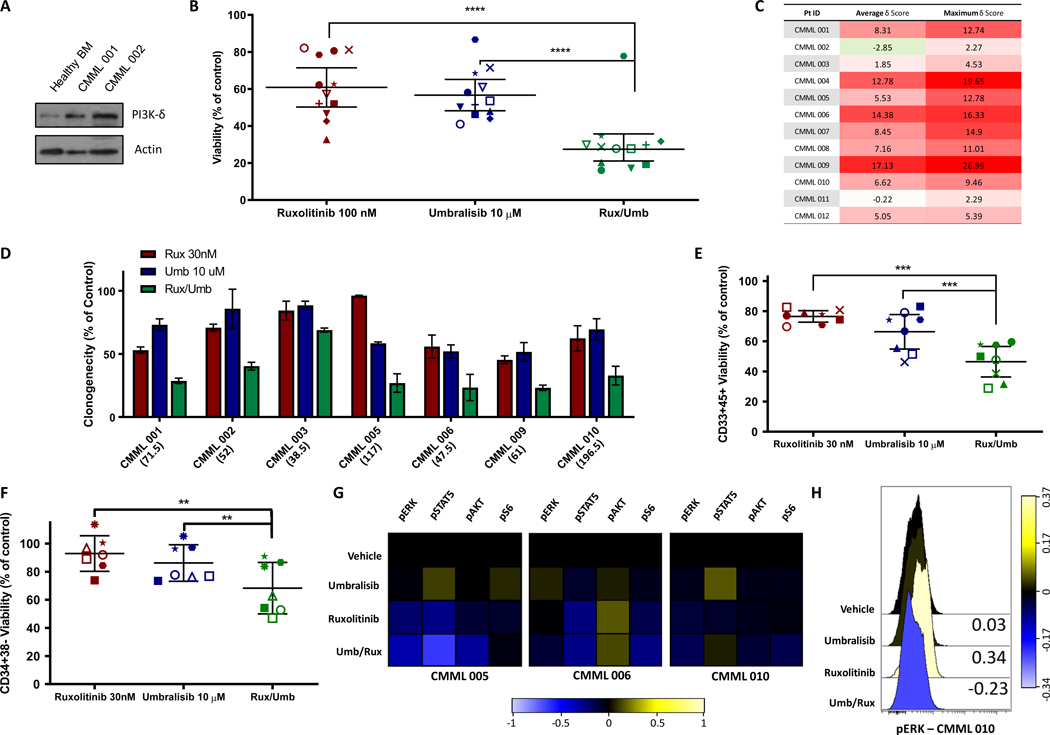
CMML samples contained significant amounts of the PI3K-δ isoform protein as examined by western blot (Figure 3A). The combination of 100 nM ruxolitinib and 10 μM umbralisib decreased viability in most samples tested beyond the monotherapy treatments and resulted in ZIP synergy scores ranging from 5 to 17, indicating an additive to synergistic effect, in a majority of patients tested (Figure 3 B-C, S4). The strongest synergy was found in the CMML samples 004, 006, and 009. All three harbor mutations in pro-growth signaling pathways, including the only 2 samples with JAK2 mutations and a third containing a NRAS mutation. Colony forming unit (CFU) assays further demonstrated the ability of ruxolitinib + umbralisib to reduce clonogenicity to a greater extent than either agent alone (Figure 3D). Finally, the combination also decreased the viability, defined using flow cytometry as Annexin V−/DAPI− cells, of myeloid CD33+/CD45+ and hematopoietic stem cell CD34+CD38− cell subpopulations (Figure 3E-F). Representative dot plots and gating strategy for these experiments can be found in the supplementary materials (Figure S5).
Figure 3. Ruxolitinib/Umbralisib combination ex vivo treatment of CMML patient samples.
(A) Western blot analysis of 2 CMML patient bone marrow samples and 1 healthy bone marrow sample. Actin was used as a loading control. (B) The viability of treated cells compared with DMSO control using the CellTiter-Glo assay was plotted for the indicated concentrations. Individual patients are represented by specific shapes. (two-tailed t-test; n.s. not significant, *p < 0.05, **p <0 .01, ***p<0.005, ****p<0.001) (C) The overall ZIP δ synergy score was based on the entire range of doses tested, and the most synergistic area score was based on the specific concentrations that had the highest synergy for each CMML sample tested. Red shading indicates increasing synergy values. (D) Inhibitory effect of ruxolitinib (Rux), umbralisib (Umb) and the combination (Rux/Umb) on CFU-GM growth from CMML samples stimulated with GM-CSF. Colony growth was assessed after 14 d. Results are expressed as % CFU-GM growth relative to cultures without drug. Multiple regression analysis confirmed that the combination treatment reduced clonogenicity by more than either individual drug treatment (ps < 0.001). Parenthetical below horizontal labels represents the average number of colonies counted in each experiments respective control condition. (E-F) Viability was determined by Annexin V/DAPI staining using flow cytometry with viable cell defined as AnnexinV−DAPI− for both early myeloid (CD45+/CD33+) at 24 hours and hematopoietic stem cells (CD34+CD38−) at 48 hours. Each symbol represents a single experimental replicate for each patient sample. (G) Heatmap showing phosphorylated signaling protein changes measured using phospho-specific flow cytometry. Samples were incubated with DMSO, umbralisib, ruxolitinib, or the combination for 30 minutes and subsequently incubated with GM-CSF for 15 minutes before fixing and staining. Color shading represents arcsinh values of the median fold change from DMSO treated cells. (H) Histograms of pERK protein levels in unstimulated culture conditions (without addition of GM-CSF) in response to ruxolitinib, umbralisib or their combination in a single NRAS-mutant patient sample. Values represent arcsinh of the median fold change from DMSO treated cells.
Just as was done with cell lines above, a small number of available samples were subsequently investigated for cell signaling changes. As previously reported, ruxolitinib alone decreased pSTAT5 levels across 3 samples tested in response to stimulation with GM-CSF (Figure 3G). Assessment of pAKT signaling was limited by a weak response of this signaling pathway to GM-CSF stimulation in our patient samples, however, umbralisib has previously been shown to dose-dependently inhibit AKT phosphorylation across a range of cell lines.25 Solo agent treatment with umbralisib resulted in significant increases in pSTAT5 in 2/3 samples, consistent with studies demonstrating STAT5 mediated resistance to solo agent PI3K-AKT pathway inhibition.7 The combination treatment resulted in a unique pattern of signaling changes, including consistent pS6 and pERK down regulation, as was seen in both cell lines. Although not observed consistently across samples tested, a unique basal pERK signaling pattern was observed in a single NRAS-mutant sample in the absence of GM-CSF stimulation, CMML 010, which showed an increase in pERK in the presence of ruxolitinib alone but a decrease when combined with umbralisib, just as was seen in the THP-1 cell line (Figure 3H).
Discussion
CMML is associated with significant morbidity and mortality, and predominately affects patients of advanced age, limiting the utility of HSCT. Current treatment options centered around hydroxyurea and DNA methyltransferase inhibitors (DNMTi) do not significantly alter the natural disease course in most patients.26,27 Early clinical trial data has shown that ruxolitinib may improve the inflammatory symptoms that burden CMML patients, but no evidence yet exists that ruxolitinib alone can improve survival in CMML.3 Pre-clinical studies have identified complex hyperactivated signaling networks that sustain clonal growth of myeloid cells in the presence of ruxolitinib, suggesting that treatment with a combination of signal transduction inhibitors may be necessary to effectively treat the disease in some cases.26
In this study, we have identified a pattern of PI3K catalytic subunit expression, namely increased PI3KCD and decreased PIK3CA, which correlates with a monocytic histological phenotype, including both M5 AML and CMML. While a mechanistic explanation for this gene expression pattern was not sought by this study, we propose a likely mechanism based on recently published CMML single-cell RNA sequencing data.28 PI3K-δ is regulated primarily at the transcription level and transcription factor (TF) binding cluster analysis has identified a handful of key TFs that regulate PIK3CD expression, including IRF and NFAT family members.20 IRF has recently been shown to be upregulated in CMML hematopoietic stem cells and both IRF and NFAT TFs are known to play a key role in expression of monocyte specific genes.28–30 It is possible that the common phenotype of monocytic predominance shared by the otherwise heterogeneous disease of CMML is driven by a pro-monocytic differentiation and pro-inflammatory TF networks and these same TFs preferentially promote the expression of PIK3CD over the other PI3K catalytic subunit genes.
Our investigation of primary CMML patient samples reveals a promising relationship between JAK1/2 inhibition and PI3K-δ inhibition. Studies of signaling protein changes offer a hypothesis based on the unique ability of this combination to prevent activation of the S6, STAT5 and ERK pathways simultaneously. ERK has previously been identified as a compensatory mechanism to JAK inhibitors in MPNs and has been shown to play a key role in the GM-CSF hypersensitivity observed in an oncogenic NRAS CMML model.31,32 PI3K/AKT signaling has been shown to regulate ERK activation in JAK2V617F mutant myelofibrosis patient samples and, notably, the two CMML samples we studied that harbored JAK2 mutations had the two highest synergy scores.33 PI3K-δ, specifically, was necessary for GM-CSF mediated ERK activation in a juvenile myelomonocytic leukemia model.34 While this pattern was observed, it was not consistent across our already limited number of patient samples. It is unsurprising a clear and consistent signaling pattern was not generated with CMML patient samples given the heterogeneity of this disease and these signaling changes should not be expected to be observed across all CMML patients.
Among the PI3K subunit isoforms, the PI3K-δ protein has unique relevance for the treatment of bone marrow neoplasms.10 PI3K-δ targeting therapies have been frequently associated with autoimmune phenomena which limit their tolerability, attributed to their suppressive effects on T-regulatory cells, but this appears to be less true of umbralisib.9,35,36 In this context, these preclinical data demonstrate novel synergistic activity in CMML by co-inhibition of PI3K-δ and JAK1/2 with two well-tolerated pharmaceuticals at clinically relevant doses and support the use of ruxolitinib/umbralisib combination therapy in CMML under active clinical investigation (NCT02493530).
Supplementary Material
Acknowledgements
This work was generously supported by the Vanderbilt-Ingram Cancer Center (NCI P30 CA68485), the Biff Rittenberg Foundation, and the Leukemia and Lymphoma Society for which, MRS is a Clinical Scholar. The authors also thank Peter Sportelli and Hari Miskin of TG Therapeutics who provided insight from ongoing umbralisib studies. TG Therapeutic also provided umbralisib and support for this work. Patient samples were garnered from the Vanderbilt-Ingram Cancer Center (VICC) Hematologic Malignancy Tumor Bank. The REDCap database tool is supported by grant UL1 TR000445 from NCATS/NIH. Flow Cytometry experiments were performed in the VUMC Flow Cytometry Shared Resource. The VUMC Flow Cytometry Shared Resource is supported by the Vanderbilt-Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). The authors were aided by the Cancer Cell Line Encyclopedia, generously provided to the public by the Broad Institute.
Footnotes
Conflicts of interest
M.R.S. receives research funding from Astex, Incyte, Millennium, TG Therapeutics; serves on consultancy/advisory board/monitoring committees for AbbVie, Astex, BMS, Celgene, Geron, Karyopharm, Millennium, Ryvu, Sunesis, and TG Therapeutics; has equity in Karyopharm; and has patents and royalties with Boehringer-Ingelheim. The remaining authors have no competing financial interests.
References
- 1.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic Myelomonocytic Leukemia. J Clin Oncol. 2013;31(19):2428–2436. doi: 10.1200/JCO.2012.47.3314 [DOI] [PubMed] [Google Scholar]
- 2.Padron E, Painter JS, Kunigal S, et al. GM-CSF-dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood. 2013;121(25):5068–5077. doi: 10.1182/blood-2012-10-460170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padron E, Dezern A, Andrade-Campos M, et al. A multi-institution phase I trial of ruxolitinib in patients with chronic myelomonocytic leukemia (CMML). Clin Cancer Res. 2016;22(15):3746–3754. doi: 10.1158/1078-0432.CCR-15-2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geissler K, Ohler L, Födinger M, et al. Interleukin 10 inhibits growth and granulocyte/macrophage colony-stimulating factor production in chronic myelomonocytic leukemia cells. J Exp Med. 1996;184(4):1377–1384. doi: 10.1084/JEM.184.4.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britschgi A, Andraos R, Brinkhaus H, et al. JAK2/STAT5 Inhibition Circumvents Resistance to PI3K/mTOR Blockade: A Rationale for Cotargeting These Pathways in Metastatic Breast Cancer. Cancer Cell. 2012;22(6):796–811. doi: 10.1016/J.CCR.2012.10.023 [DOI] [PubMed] [Google Scholar]
- 6.Bartalucci N, Tozzi L, Bogani C, et al. Co-targeting the PI3K/mTOR and JAK2 signalling pathways produces synergistic activity against myeloproliferative neoplasms. J Cell Mol Med. 2013;17(11):1385–1396. doi: 10.1111/jcmm.12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choong ML, Pecquet C, Pendharkar V, et al. Combination treatment for myeloproliferative neoplasms using JAK and pan-class I PI3K inhibitors. J Cell Mol Med. 2013;17(11):1397–1409. doi: 10.1111/jcmm.12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtz SE, Eide CA, Kaempf A, et al. Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies. Proc Natl Acad Sci U S A. 2017;114(36):E7554–E7563. doi: 10.1073/pnas.1703094114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burris HA, Flinn IW, Patel MR, et al. Umbralisib, a novel PI3Kδ and casein kinase-1ε inhibitor, in relapsed or refractory chronic lymphocytic leukaemia and lymphoma: an open-label, phase 1, dose-escalation, first-in-human study. Lancet Oncol. 2018;19(4):486–496. doi: 10.1016/S1470-2045(18)30082-2 [DOI] [PubMed] [Google Scholar]
- 10.Sujobert P, Bardet V, Cornillet-Lefebvre P, et al. Essential role for the p110δ isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood. 2005;106(3):1063–1066. doi: 10.1182/blood-2004-08-3225 [DOI] [PubMed] [Google Scholar]
- 11.Billottet C, Grandage VL, Gale RE, et al. A selective inhibitor of the p110δ isoform of PI 3-kinase inhibits AML cell proliferation and survival and increases the cytotoxic effects of VP16. Oncogene. 2006;25(50):6648–6659. doi: 10.1038/sj.onc.1209670 [DOI] [PubMed] [Google Scholar]
- 12.Moyo TK, Palmer J, Huang Y, et al. Resurrecting Response to Ruxolitinib: a Phase I Study Testing The Combination of Ruxolitinib and the PI3K Delta Inhibitor Umbralisib in Ruxolitinib-Experienced Myelofibrosis. Hemasphere. 2018;2:19–20. [Google Scholar]
- 13.Pölönen P, Mehtonen J, Lin J, et al. HEMap: An interactive online resource for characterizing molecular phenotypes across hematologic malignancies. Cancer Res. 2019;79(10):2466–2479. doi: 10.1158/0008-5472.CAN-18-2970 [DOI] [PubMed] [Google Scholar]
- 14.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012, 483(7391):603–607. doi: 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babicki S, Arndt D, Marcu A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for Drug Synergy in Complex Dose–Response Landscapes Using an Interaction Potency Model. Comput Struct Biotechnol J. 2015;13:504–513. doi: 10.1016/J.CSBJ.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: Towards single-cell proteomics. Nat Rev Cancer. 2006;6(2):146–155. [DOI] [PubMed] [Google Scholar]
- 18.Earl DC, Ferrell PB, Leelatian N, et al. Discovery of human cell selective effector molecules using single cell multiplexed activity metabolomics. Nat Commun. 2018;9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3(5):361–368. [DOI] [PubMed] [Google Scholar]
- 20.Kok K, Nock GE, Verrall EAG, et al. Regulation of p110δ PI 3-kinase gene expression. PLoS One. 2009;4(4):e5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelsi-Boyer V, Cervera N, Bertucci F, et al. Gene expression profiling separates chronic myelomonocytic leukemia in two molecular subtypes. Leukemia. 2007, 21(11):2359–2362. [DOI] [PubMed] [Google Scholar]
- 22.Iyengar S, Clear A, Maharaj L, et al. P110a-mediated constitutive PI3K signaling limits the efficacy of p110d-selective inhibition in mantle cell lymphoma, particularly with multiple relapse. Blood. 2013;121(12):2274–2284. doi: 10.1182/blood [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locatelli SL, Careddu G, Inghirami G, et al. The novel PI3K-δ inhibitor TGR-1202 enhances Brentuximab Vedotin-induced Hodgkin lymphoma cell death via mitotic arrest. Leukemia. 2016;30(12):2402–2405. doi: 10.1038/leu.2016.224 [DOI] [PubMed] [Google Scholar]
- 24.Burris HA, Flinn IW, Patel MR, et al. Umbralisib, a novel PI3Kδ and casein kinase-1ε inhibitor, in relapsed or refractory chronic lymphocytic leukaemia and lymphoma: an open-label, phase 1, dose-escalation, first-in-human study. Lancet Oncol. 2018, 19(4):486–496. doi: 10.1016/S1470-2045(18)30082-2 [DOI] [PubMed] [Google Scholar]
- 25.Deng C, Lipstein MR, Scotto L, et al. Silencing c-Myc translation as a therapeutic strategy through targeting PI3Kδ and CK1ε in hematological malignancies. Blood. 2017;129(1):88–99. doi: 10.1182/blood-2016-08-731240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moyo TK, Savona MR. Therapy for Chronic Myelomonocytic Leukemia in a New Era. Curr Hematol Malig Rep. 2017;12(5):468–477. doi: 10.1007/s11899-017-0408-8 [DOI] [PubMed] [Google Scholar]
- 27.Merlevede J, Droin N, Qin T, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7(1):10767. doi: 10.1038/ncomms10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiseman DH, Baker SM, Dongre A V., et al. Chronic myelomonocytic leukaemia stem cell transcriptomes anticipate disease morphology and outcome. EBioMedicine. 2020;58:102904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J Biol Chem. 2007;282:20065–20069. [DOI] [PubMed] [Google Scholar]
- 30.Rao A (1994) NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol Today 1994;15:274–281. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Liu Y, Li Z, et al. Endogenous oncogenic NRAS mutation promotes aberrant GM-CSF signaling in granulocytic/monocytic precursors in a murine model of chronic myelomonocytic leukemia. Blood. 2010;116(26):5991–6002. doi: 10.1182/blood-2010-04-281527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stivala S, Codilupi T, Brkic S, et al. Targeting compensatory MEK/ERK activation increases JAK inhibitor efficacy in myeloproliferative neoplasms. J Clin Invest. 2019;129(4):1596–1611. doi: 10.1172/JCI98785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf A, Eulenfeld R, Gäbler K, et al. JAK2-V617F-induced MAPK activity is regulated by PI3K and acts synergistically with PI3K on the proliferation of JAK2-V617F-positive cells. JAK-STAT. 2013;2(3):e24574. doi: 10.4161/jkst.24574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin CB, Li XJ, Mali RS, et al. PI3K p110δ uniquely promotes gain-of-function Shp2-induced GM-CSF hypersensitivity in a model of JMML. Blood. 2014;123(18):2838–2842. doi: 10.1182/blood-2013-10-535104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chellappa S, Kushekhar K, Munthe LA, et al. The PI3K p110δ Isoform Inhibitor Idelalisib Preferentially Inhibits Human Regulatory T Cell Function. J Immunol. 2019;202(5):1397–1405. doi: 10.4049/jimmunol.1701703 [DOI] [PubMed] [Google Scholar]
- 36.Maharaj K, Powers JJ, Achille A, et al. The dual PI3Kδ/CK1ε inhibitor umbralisib exhibits unique immunomodulatory effects on CLL T cells. Blood Adv. 2020;4(13):3072–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.