Abstract
Obligate intracellular pathogens in the family Chlamydiaceae infect taxonomically diverse eukaryotes ranging from amoebae to mammals. However, many fundamental aspects of chlamydial cell biology and pathogenesis remain poorly understood. Genetic dissection of chlamydial biology has historically been hampered by a lack of genetic tools. Exploitation of the ability of chlamydia to recombine genomic material by lateral gene transfer (LGT) ushered in a new era in chlamydia research. With methods to map mutations in place, genetic screens were able to assign functions and phenotypes to specific chlamydial genes. Development of an approach for stable transformation of chlamydia also provided a mechanism for gene delivery and platforms for disrupting chromosomal genes. Here, we explore how these and other tools have been used to test hypotheses concerning the functions of known chlamydial virulence factors and discover the functions of completely uncharacterized genes. Refinement and extension of the existing genetic tools to additional Chlamydia spp. will substantially advance understanding of the biology and pathogenesis of this important group of pathogens.
1. Introduction
Bacteria in the phylum Chlamydiae share a characteristic biphasic intracellular developmental cycle and reside within a parasitophorous vacuole (termed the inclusion) (Moulder 1985). This obligate intracellular lifestyle and protected intracellular niche limit opportunities for chlamydia to exchange genetic material with other microorganisms and pose barriers to their genetic manipulation in the laboratory. There are few examples of genetic exchange between chlamydiae and other organisms, and Chlamydia spp. generally have few mobile genetic elements and other horizontally acquired genes compared to free-living bacteria (Stephens et al. 1998; Kalman et al. 1999; Read et al. 2000; Horn et al. 2004; Collingro et al. 2011).
Genome sequencing and comparative genomics have played especially important roles in the investigation of chlamydial biology because methods for targeted genetic manipulation only became available recently. The first sequenced C. trachomatis genomes were highly similar, suggesting that static genomes might be another obstacle that would hinder genetic manipulation. However, subsequent comparisons of hundreds of genomes and the recapitulation of chlamydial recombination and transformation in vitro confirmed that some Chlamydia spp. readily exchange DNA, but usually only with close relatives (DeMars et al. 2007; Wang et al. 2011; Hadfield et al. 2017). Another barrier to genetic manipulation of members of Chlamydia spp. is that more than half of their pan-genome is comprised of a core set of orthologous genes that are present in all Chlamydiae (Collingro et al. 2011). Many of these core genes are predicted to mediate essential and conserved features of chlamydial development, such as maintenance of the chlamydial inclusion and the transitions between the extracellular infectious elementary body (EB) and intracellular replicative reticulate body (RB) forms, and may not be dispensable even in vitro.
Chlamydiae are estimated to have diverged from other eubacteria 2 billion years ago, and vertebrate pathogens in the family Chlamydia species diverged from environmental Chlamydia-like organisms (CLOs) 700 million years ago (Weisburg et al. 1986; Everett et al. 1999; Horn et al. 2004). Partial or full genome sequences from eight CLO families (Parachlamydieaceae, Waddliaceae, Simkaniaceae, Rhabdochlamydiaceae, Criblamydiaceae Chlavichlamydiaceae, and Parilichlamydiaceae) have been described, and there is evidence that as many as 200 more exist (Lagkouvardos et al. 2014; Taylor-Brown et al. 2015). CLO genomes contain numerous rearrangements and share little synteny with one another, which may reflect the broad taxonomic range of the hosts that these organisms parasitize (Collingro et al. 2011). Some members of Criblamydiaceae, Chlavichlamydiaceae, and Parilichlamydiaceae spp. are important fish pathogens (Draghi et al. 2004; Karlsen et al. 2008; Stride et al. 2013). Other CLOs can enter and proliferate in cultured human cells (Greub et al. 2003; Collingro et al. 2005; Casson et al. 2006). S. nevegensis, P. acanthaomoebae, W. condrophila and Rabdochlamydia spp. have been associated with disease in humans and livestock (Taylor-Brown et al. 2015). Parachlamydia can also elicit respiratory disease in experimental mouse and cattle challenge models, which is consistent with the hypothesis that they are mammalian pathogens (Casson et al. 2008; Lohr et al. 2015). The predicted expanded metabolic and recombination capacities of some CLOs, compared to Chlamydia spp., suggest that they might be excellent candidates for the development of both genetic tools and axenic culture, although no attempts to genetically modify CLOs have been described (Collingro et al. 2011; Omsland et al. 2014).
Unlike CLO genomes, Chlamydiaceae genomes share substantial synteny with one another (Read et al. 2000; Read et al. 2003; Collingro et al. 2011; Nunes and Gomes 2014). Eleven Chlamydia spp. (C. trachomatis, C. muridarum, C. pneumoniae, C. psittaci, C. pecorum, C. abortus, C. felis, C. suis, C. cavie, C. gallinacea, C. avium) are recognized, and reference genomes are available for representatives of all of these except C. avium (Stephens et al. 1998; Kalman et al. 1999; Read et al. 2000; Read et al. 2003; Thomson et al. 2005; Azuma et al. 2006; Mojica et al. 2011; Schofl et al. 2011; Donati et al. 2014; Sachse et al. 2014; Holzer et al. 2016). Extant members of the genus Chlamydia separate into two well-supported clades based on their 16S rRNA gene sequences (Stephens et al. 2009; Sachse and Laroucau 2015). One clade contains the mouse pathogen C. muridarum, the pig pathogen C. suis, and various human pathogens in the species C. trachomatis, and the other clade contains the remaining species.
Little genetic distance can separate Chlamydia spp. and strains that infect different hosts and cause distinct diseases. For example, 99% of C. muridarum genes have high similarity orthologues in C. trachomatis serovar D (Read et al. 2000). Genomes of C. trachomatis strains that cause blinding trachoma (serovars A-C, trachoma), mucosa-restricted sexually transmitted infections (serovars D-K, chlamydia), and the invasive sexually transmitted disease lymphogranuloma venereum (Lymphogranuloma venereum (LGV) serovars L1-L3) share more than 99% nucleotide sequence identity (Stephens et al. 1998; Carlson et al. 2005; Thomson et al. 2008). This phenotypic diversity in the context of little genetic distance led to the proposal that host and tissue tropism of Chlamydia spp. is determined by a small group of niche-specific genes (Read et al. 2003). These genes are attractive targets for manipulation because their variable presence in different isolates that can be cultivated in the same conditions suggests that they are dispensable (Kari et al. 2011). In this review, we describe recent advances in the development of tools for genetic manipulation of pathogenic Chlamydia, the uses and limitations of these tools, and how these might be improved for dissecting mechanisms of chlamydial cell biology and pathogenesis.
2. Chlamydial Genomes Are Malleable
Statements that chlamydiae are genetically “intractable” were fixtures in the literature until recently, but evidence that this was not so emerged earlier. For example, a study in 1983 showed that temperature-sensitive (TS) C. abortus mutants could be isolated from infected cell cultures treated with the alkylating mutagen N-methyl-N′-nitro-N-nitrosoguanidine (NTG) (Rodolakis 1983). One of these TS mutants was a potent attenuated vaccine strain that afforded strong protection against the virulent parent organism in sheep (Rodolakis and Souriau 1983, 1986). Sequencing of this mutant 30 years later confirmed that it contained multiple mutations consistent with the effects of NTG (Burall et al. 2009).
Antibiotic resistance studies provided additional evidence that chlamydial genomes are malleable. Treharne observed that C. trachomatis became rifampin resistant when passed in increasing concentrations of this antibiotic (Treharne et al. 1989). Another study identified a fluoroquinolone-resistant chlamydial gyrA allele by sequencing candidate resistance alleles in drug-resistant mutants isolated by serial passage (Dessus-Babus et al. 1998). Endogenous chlamydial resistance alleles for other antibiotics were identified using similar strategies (Dreses-Werringloer et al. 2003; McCoy et al. 2003; Misiurina et al. 2004; Binet and Maurelli 2007; Demars et al. 2007). The ease with which Chlamydia spp. developed resistance to antibiotics in vitro initially seemed inconsistent with the paucity of antibiotic-resistant C. trachomatis clinical isolates. Subsequent studies showed that laboratory selected endogenous resistance alleles have fitness costs in the absence of antibiotic explained this paradox (Binet and Maurelli 2005, 2009; Binet et al. 2010). Chlamydial metabolism has also been investigated using mutants derived by passage in toxic precursors. For example, hydroxyurea, thioguanine, trimethoprim, and sulfisoxazole-resistant C. trachomatis mutants provided insights into the mechanisms of chlamydial nucleotide metabolism (Tipples and McClarty 1991; Qin and McClarty 1992; Wylie et al. 1996).
Other groups have harnessed spontaneous mutants to study chlamydial pathogenesis. To test if detrimental mutations accumulate in cell culture, a C. trachomatis reference isolate was passaged in a murine genital tract (GT) model, and the genomes of the resulting progeny were compared to the reference isolate (Sturdevant et al. 2010). Decreased virulence of the starting reference strain compared to progeny shed late from the murine GT mapped to a frameshift mutation in ct135. Analogous mutations accumulated in a C. muridarum ct135 orthologue (tc0412), and cell culture propagation selects against intact ct135 (Ramsey et al. 2009; Borges et al. 2013). Another study compared the virulence of a C. muridarum parent isolate and its cell culture-adapted progeny in mice. Two alleles, tc0237 and tc0668, that mediate C. muridarum attachment and pathogenicity were identified (Chen et al. 2015a; Conrad et al. 2015).
Studies of rare, naturally plasmid-negative Chlamydia isolates and later of parent and chemically cured strain pairs demonstrated that some key chlamydial virulence determinants are dispensable in vitro (Peterson et al. 1990; Farencena et al. 1997; Stothard et al. 1998; O’Connell and Nicks 2006). Most natural Chlamydia isolates, except C. pneumoniae, contain a highly conserved ~7.5 kb plasmid that encodes 8 open reading frames (ORFs) and at least two small noncoding RNAs (sRNAs) (Palmer and Falkow 1986; Thomas et al. 1997; Pickett et al. 2005; Albrecht et al. 2010; Zhong 2017). The ubiquity of plasmids in C. trachomatis reference and clinical isolates suggested that these plasmids play key roles in chlamydial biology and pathogenesis. Plasmid-free C. trachomatis (pCT−) isolates grow with near-normal kinetics in vitro, but do not accumulate glycogen and have reduced infectivity (Matsumoto et al. 1998; Russell et al. 2011). However, plasmid-free C. muridarum (pCM−) is highly attenuated, unable to elicit pathology, and unable to compete with wild type C. muridarum in the murine GT (O’Connell et al. 2007; Russell et al. 2011). Studies of pCM− and wild-type strain pairs have now shown that the plasmid plays a central role in C. muridarum GT virulence, dissemination from the murine upper GT to the gastrointestinal (GI) tract, and colonization of the murine GI tract (Carlson et al. 2008; Lei et al. 2014; Chen et al. 2015b; Shao et al. 2017; Zhong 2017). Collectively, these studies defined plasmid-associated phenotypes and laid the groundwork for contemporary studies of plasmid ORFs using transformation and plasmid genetic engineering (discussed below).
3. Moving Genes Between and into Chlamydial Genomes with Recombination and Lateral Gene Transfer
Major outer membrane protein (MOMP) gene (ompA) sequencing began to replace C. trachomatis serotyping in the early 1990s. The resulting sequences revealed that many un-typeable clinical C. trachomatis isolates contained chimeric ompA alleles (Dean et al. 1992; Fitch et al. 1993; Lampe et al. 1993; Brunham et al. 1994; Hayes et al. 1994). Hayes understood the significance of these chimeras and proposed, “efforts to manipulate the chlamydial genome in vitro by recombination should be intensified” (Hayes et al. 1994). Sequencing of the polymorphic membrane protein (pmp) genes later revealed identical pmpC sequences associated with different ompA genotypes in C. trachomatis clinical isolates, showing that recombination between Chlamydia isolates could extend beyond ompA (Gomes et al. 2004).
Recapitulation of lateral gene transfer (LGT) in vitro confirmed that chlamydiae can exchange chromosomal DNA and led to the development LGT-based mutation mapping (discussed below) (Demars et al. 2007; DeMars and Weinfurter 2008). Demars and colleagues first demonstrated that doubly antibiotic-resistant isolates arose from co-infections with various combinations of antibiotic-resistant C. trachomatis L1 parents 1000- to 10,000-fold more frequently than did spontaneous double mutants from single infections with either parent alone (Demars et al. 2007). Recombination between endogenous ofloxacin resistant C. trachomatis serovar L1 and rifampicin resistant C. trachomatis serovar D parents showed that LGT could be used to exchange DNA between dissimilar parents and that large genomic regions (123–790 kb) could be exchanged (DeMars and Weinfurter 2008). Exchange of a C. suis tetracycline resistance island between C. suis, C. muridarum, and C. trachomatis in another study showed that LGT between different chlamydial species and heterologous regions of chlamydial genomes was possible (Suchland et al. 2009). LGT does not appear to require that the donor and recipient reside in the same vacuole for C. trachomatis because wild-type and non-fusogenic incA− parents produced LGT recombinants at similar frequencies (Jeffrey et al. 2013). If LGT can occur between other non-fusogenic Chlamydia spp. is unknown. So far, analyses of genomes from dozens of LGT recombinants has not yet identified any clear recombination hotspots associated with this process (Jeffrey et al. 2010, 2013; Brothwell et al. 2016; Muramatsu et al. 2016). The mechanisms of LGT also remain mysterious, but many Chlamydia spp. encode low identity orthologues of the B. subtilis competence genes comEC and mec, so it is plausible that DNA exchange between competent RBs mediates LGT (Stephens et al. 1998; Demars et al. 2007; DeMars and Weinfurter 2008).
Two LGT-based mutation-mapping approaches have been described, positive selection LGT (LGT) and counterselection LGT (csLGT). In LGT, host cells are co-infected with a parent that has a phenotype of interest and is resistant to an antibiotic (e.g., spectinomycin, rifampicin, trimethoprim) and a second parent that does not have the phenotype but is resistant to a second antibiotic (Nguyen and Valdivia 2012). Comparison of the genomes of the resulting doubly antibiotic-resistant progeny that have either maintained or lost the phenotype of interest can identify which allele is responsible for the phenotype (Nguyen and Valdivia 2013). Importantly, reciprocal LGT of these progenies with a third antibiotic-resistant parent can permit resistance allele recycling and serial mapping attempts (Nguyen and Valdivia 2012). In csLGT, healthy recombinant progeny is generated from parents that both have conditional or nonconditional growth defects (Brothwell et al. 2016; Muramatsu et al. 2016). For example, an LGT cross of two TS parent strains performed at the non-permissive temperature will yield temperature-resistant progeny (Brothwell et al. 2016). Similar to LGT, comparing parent and recombinant progeny genomes can then identify the alleles linked to the phenotypes of the parents. The advantages of csLGT include avoidance of the fitness costs associated with some endogenous antibiotic resistance alleles and generation of progeny-parent pairs that differ only in the detrimental allele. Unlike LGT, enrichment of recombinants by csLGT is dependent upon the strength of the counter-selectable phenotypes of the parents.
Binet and Maurelli showed that homologous dsDNA sequences can also be directly introduced into C. psittaci using electroporation and recombined into the chlamydial chromosome (Binet and Maurelli 2009a, b). This clever study took advantage of the single copy rRNA operon in C. psittaci and distinct SNPs in the 16S rRNA gene that confer resistance to kasugamycin and spectinomycin. The engineered resistance alleles contained a silent mutation that destroyed a restriction site to monitor allelic exchange verses spontaneous antibiotic resistance and flanking homologous sequences that targeted the allele to the chromosome. This promising approach has not been replicated in other Chlamydia spp. or used to manipulate other genes.
4. Transformation of Chlamydia with Plasmid Shuttle Vectors
The potential of the chlamydial plasmid for genetic engineering was appreciated upon its discovery, but was not realized until almost 35 years later when C. trachomatis L2 was stably transformed with a vector constructed from its corresponding plasmid (pL2) (Wang et al. 2011). Electroporation of C. trachomatis EBs with a vector constructed from pCT and an E. coli plasmid (pBGS9) in which the pBGS9 chloramphenicol acetyltransferase (cat) gene was controlled by a Chlamydia promoter provided the first evidence that transformation was possible (Tam et al. 1994). Chloramphenicol resistant organisms and RBs containing the vector detected in the transformed cultures could survive up to four passages in chloramphenicol but were eventually lost. In hindsight, the shuttle vector may have been unstable because the E. coli plasmid elements disrupted a pCT maintenance gene (Song et al. 2013). Nonetheless, careful descriptions of positive and negative results in this study provided a roadmap for future attempts.
An approach developed by the Clarke group is the basis for most contemporary transformation protocols (Wang et al. 2011). Their initial vector consisted of a portion of pBR325 encoding β-lactamase (bla) and pBR oriC ligated into the pL2 plasmid (pBR325::L2). The shuttle vector and EBs were incubated in CaCl2-Tris buffer, and McCoy cells infected with this mixture yielded stable penicillin-resistant transformants. Transformation of the glycogen-deficient pCT− C. trachomatis isolate 25667R with pBR325::L2 restored glycogen accumulation. This confirmed that plasmid loss–not background mutations—caused the glycogen deficiency of pCT− strains and was the first example of genetic complementation in Chlamydia. Finally, a similar vector containing a green fluorescent protein (GFP) gene under a Neisseria promoter was introduced into the backbone of the new Swedish variant plasmid (pSW2), which contains a deletion in CDS1 and a duplication in CDS3 (Seth-Smith et al. 2009). The transformants stably expressed GFP. This demonstrated that pCT-derived vectors can express foreign transgenes and that some regions of chlamydial plasmids are dispensable for their in vitro maintenance.
5. Applications of the Plasmid as a Shuttle Vector
The ability to deliver and express transgenes facilitated the development of many new approaches for investigating chlamydial biology. For example, Agaisse and Derré developed vectors that express GFP, mCherry, and cyan fluorescent protein genes under the control of chlamydial promoters (Agaisse and Derre 2013). This permitted real-time visualization of developing inclusions and their interactions with host cell organelles. Introduction of a tetracycline promoter and operator sequences upstream of a multiple cloning site in pASK permitted anhydrotetracycline-regulated expression of transgenes in C. trachomatis (Wickstrum et al. 2013). Further tuning of a tetracycline-regulated vector permitted comprehensive interrogation of the secretion and localization of known and putative inclusion membrane proteins (Incs) (Bauler and Hackstadt 2014; Weber et al. 2015). New chlamydial expression and shuttle vectors and are evolving rapidly, and we refer readers to a more extensive review of this topic (Bastidas and Valdivia 2016).
Despite remarkable progress and recent demonstration of transformation in additional Chlamydia spp., chlamydial transformation still has many limitations (Ding et al. 2013, Song et al. 2014). Inability to transform specific constructs, achieve expression of specific products from vectors and/or complement chromosomal defects is common (Nelson DE, unpublished) (Weber et al. 2015, 2017). We and others have observed subtle—and sometimes not so subtle—growth defects in transformants (Nelson and Zhong, unpublished; Wang et al. 2011). Possible explanations for these vector effects could include inappropriate timing of gene expression during the developmental cycle and/or improper levels of gene expression from multi-copy vectors (Wickstrum et al. 2013). Recently identified roles of the plasmid in the regulation of chlamydial virulence, chromosomal gene expression, and lytic exit suggest multiple reasons why manipulation of the chlamydial plasmid could elicit unanticipated phenotypes (discussed below). Considering these limitations, it is essential that vector controls be included in complementation experiments when feasible. Alternately, complementation results can sometimes be validated using phenotypic complementation and/or ectopic expression of the chlamydial gene in host cells (Clifton et al. 2004; Nelson et al. 2007).
6. Reverse Genetic Dissection of Chlamydia Plasmids
Development of transformation permitted reverse genetic dissection of the functions of chlamydial plasmids. The inability to transform non-LGV strains with pBR325::L2 or pGFP::SW2 suggested that plasmid or chromosomal determinants dictated plasmid tropism. C. trachomatis serovar A and C. muridarum could only be stably transformed with shuttle vectors constructed from their corresponding plasmids, indicating that plasmid sequences mediate stable transformation (Song et al. 2014). Forcing recombination between C. muridarum and LGV plasmids in another study revealed that CDS2 is a key determinant of plasmid tropism (Wang et al. 2014). Inactivation of specific plasmids ORFs revealed that Pgp1, 2 and 6 proteins and pgp8, but not Pgp8, are also essential for plasmid maintenance and identified additional cis-acting plasmid regulatory regions (Gong et al. 2013; Song et al. 2013; Liu et al. 2014; Zhong 2017). Pgp4 is dispensable for plasmid maintenance, but regulates multiple chromosomal genes including glgA (Carlson et al. 2008; Song et al. 2013). Loss of pgp4 is also sufficient to elicit glycogen deficiency (Song et al. 2013). Pgp4 also plays an essential and type III secretion-dependent role in the lytic exit of C. trachomatis (Yang et al. 2015). Collectively, these studies revealed that chlamydial plasmids play central roles in multiple aspects of chlamydial metabolism and pathogenesis.
7. Genetic Manipulation of Chlamydial Chromosomes
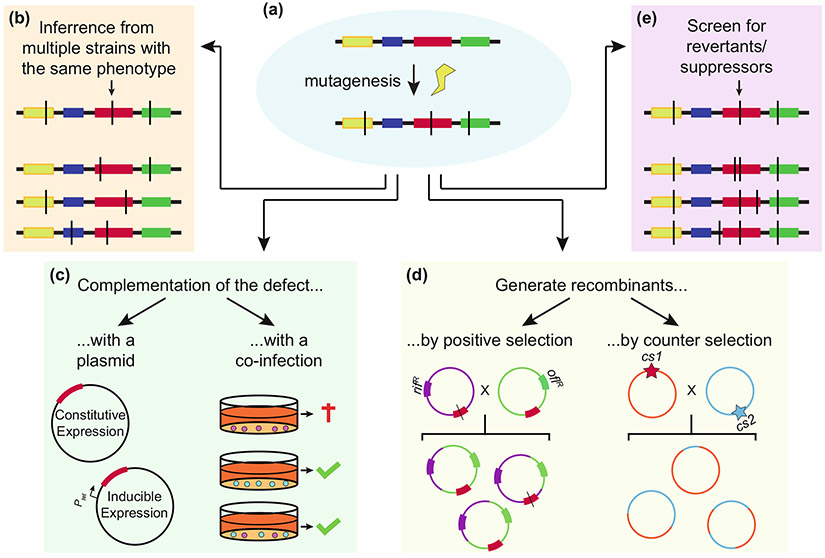
Four approaches for genetic analysis of chlamydial chromosomal gene functions have been reported (Table 1): Targeting Induced Local Lesions in Genomes (TILLING), TILLING by sequencing, TargeTron®, and Fluorescence-Reported Allelic Exchange Mutagenesis (FRAEM) (Kari et al. 2011; Johnson and Fisher 2013; Kokes et al. 2015; Mueller et al. 2016). Chemical mutagens are used to generate libraries of random mutants in the TILLING approaches, whereas TargeTron and FRAEM permit targeted mutagenesis (Fig. 1).
Table 1.
Genetic analysis of chlamydial chromosomal gene functions
| Method | Strain | Gene(s) | Advantage | Caveat | References |
|---|---|---|---|---|---|
|
Serial passage with increasing selection Non-mutagenized organisms were serially passaged in increasing concentrations of a selection agent |
C. trachomatis L2 | Various | Mutants with one or a few mutations per genome | Mutants may not arise; isolation of mutants highly dependent on selection conditions | Tipples and McClarty (1991) |
|
In vitro serial passage Organisms were passaged 10Xin between embryonating hen’s eggs and 17 X in HeLa 229 cells. Virulence selection in mice |
C. trachomatis D/UW-3/CX | ct135 | Isolate phenotypic mutants with one or a few mutations per genome | Mutants may not arise; isolation of mutants highly dependent on selection conditions | Sturdevant et al. (2010) |
|
In vitro serial passage and selection C. muridarum was passaged for 28 generation in vitro using unassisted and assisted attachment infection models |
C. muridarum | tc0237, tc0412, tc0668 | Isolate phenotypic mutants with one or a few mutations per genome | Mutants may not arise; isolation of mutants highly dependent on selection conditions | Chen et al. (2015a, b) |
|
Chemical mutagenesis NTG mutagenesis and screening for temperature-sensitive mutants |
C. psittaci AB7 | TS genes | Identify multiple phenotypically related mutants; mutate essential genes | Multiple mutations requiring mapping to determine the causative TS allele(s) | Rodolakis (1983) |
|
Chemical mutagenesis & antibiotic LGT EMS mutagenesis and screening for diverse plaque morphologies Antibiotic recombination to map causative alleles |
C. trachomatis L2 | Glycogen genes | Identify multiple phenotypically related mutants; map causative alleles | Labor intensive library creation, screening, and mapping; compare multiple recombinants to identify causative allele; antibiotic recombinants may have growth defects | Nguyen and Valdivia (2012) |
|
Chemical mutagenesis & WGS EMS or ENU mutagenesis and screening for mutants that failed to promote F-actin assembly |
C. trachomatis L2 | inaC | Identify multiple phenotypically related mutants; rapidly map causative alleles | WGS of a large mutant library may be cost prohibitive; mutants may not share the same mutation related to phenotype of interest | Kokes et al. (2015) |
|
Chemical mutagenesis & csLGT EMS mutagenesis of a GFP-expressing parent and screening for mutants. Counter selection recombination to map causative alleles |
C. trachomatis L2 | TS & IFN-γ persistance genes | Identify multiple phenotypically related mutants; map multiple causative alleles at the same time and isolate isogenic recombinants | Labor intensive library creation, screening, and mapping; selection of recombinants limited by strength of phenotype | Brothwell et al. (2016), Muramatsu et al. (2016) |
|
Allelic exchange Homologous recombination of linear and circular suicide plasmids |
C. psittaci 6BC | 16S rRNA | Modify genes with specific mutations | Electroporation equipment; suicide vector may be maintained episomally after a number of passages | Binet and Maurelli (2009a, b) |
|
TILLING for null mutant EMS mutagenesis and screening for null mutants within a subset of mutants found to have mismatched sequence in a region of interest |
C. trachomatis D/UW-3/CX | trpB | Knock out any nonessential gene; mutate essential genes | Labor-intensive library creation, screening, and mapping; potential for multiple chromosomal mutations that need to be mapped and complemented | Kari et al. (2011) |
|
TargeTron—gene disruption Homologous Recombination of a group II Intron on a suicide plasmid |
C. trachomatis L2 | incA | Disrupt any nonessential gene | Proprietary algorithm to determine appropriate targeting sites; potential lack of insertion sites in 5′ sequence of the gene of interest; transformation efficiency | Johnson and Fisher (2013) |
|
FRAEM—gene knockout Homologous recombination of an insertional cassette using a conditional suicide plasmid |
C. trachomatis L2 | trpA; ctl063-065 | Knock out any nonessential gene(s); transient suicide plasmid with fluorescent reporter | Polar effects; transformation efficiency | Mueller et al. (2016) |
Abbreviations used: NTG N-methyl-N′-nitro-N-nitrosoguanidine; EMS ethyl methanesulfonate; ENU N-ethylnitrosourea; WGS whole genome sequencing; TILLING targeting induced local lesions in genomes; FRAEM fluorescence reported allelic exchange mutagenesis; LGT lateral gene transfer; cs counterselction; TS temperature sensitive
Fig. 1.
Mutation mapping in chlamydia. a Chemical mutagenesis strategies can result in multiple mutations (denoted by black lines). Multiple approaches can resolve which mutation(s) (shown in red) are linked to a phenotype. b Additional mutants with the same phenotype that have different mutant alleles of the same gene can be isolated. c A plasmid that expresses the wild-type allele of the gene can be used to complement the phenotype. Alternately, co-infection with wild-type chlamydia and/or ectopic expression of the gene in host cells can be used to complement phenotypes associated with secreted effectors, inclusion membrane transporters, or enzymes that detoxify the inclusion. d LGT can generate recombinants using endogenous antibiotic resistance or counter-selectable alleles. e Continued passage of a mutant in detrimental screening conditions can also generate revertants and intragenic suppressors that can identify genes linked to detrimental phenotypes (Nelson, unpublished). The original mutation is indicated by the arrow.
Two methods to identify naturally occurring or mutagen-induced mutations in a gene of interest have been described: TILLING and TILLING by sequencing. TILLING was adapted from plant genetics for application in Chlamydia in 2011 (McCallum et al. 2000; Kari et al. 2011). Mutants that contained less than one mutation per genome on average were generated by ethyl methanesulfonate (EMS) mutagenesis and were expanded in pools in HeLa cells. The tryptophan synthase operon (trpBA) was PCR amplified from genomic DNA from each of these pools. Cleavage of the PCR products with a mismatch-specific endonuclease (CEL-I) then identified pools that contained both wild-type and mutant trpBA amplicons. Multiple trpBA mutants were identified, including an isogenic trpB null (trpB) mutant. Unlike the library parent, growth of the isogenic trpB− mutant was not rescued from tryptophan starvation by indole, which is consistent with the predicted function of TrpB (Fehlner-Gardiner et al. 2002; Caldwell et al. 2003; Carlson et al. 2005). A similar approach was used to isolate a near-isogenic C. trachomatis pmpD− mutant (Kari et al. 2014). Although the pmpD− mutant behaved similarly to its parent in the murine GT, it was highly attenuated in a macaque trachoma model, again demonstrating that key chlamydial virulence factors can be dispensable in vitro.
Other TILLING studies have used higher mutagen doses and different Chlamydia species. TILLING of a highly mutagenized C. trachomatis serovar D library identified three chxR− mutants and five ChxR regulated genes (Yang et al. 2017). The chxR− mutants were attenuated in mice, suggesting that ChxR is a conserved chlamydial virulence factor. TILLING also identified a collection of C. muridarum plasticity zone (PZ) ORF nonsense mutants in another study (Rajaram et al. 2015). These PZ mutants were similarly virulent to their parent in a murine GT model, but some were profoundly attenuated in a mouse GI tract model (Nelson, unpublished).
Kokes et al. coupled chemical mutagenesis with genome sequencing to produce a mutant library for simultaneous forward (discussed below) and reverse genetic analyses using a strategy that we refer to as “TILLING by sequencing” (Kokes et al. 2015). A rifampicin resistant C. trachomatis L2 isolate was mutagenized with either EMS or N-ethylnitrosourea (ENU) and the resulting mutants were plaque-cloned. Sequencing of the mutants in pools identified over 8,000 different mutations, including nonsense mutations in 84 C. trachomatis ORFs. Detection of nonsense mutations in genes that play roles in carbon metabolism, DNA damage repair, and virulence revealed that a variety of cellular processes are dispensable for chlamydial survival in vitro. Use of a rifampicin-resistant parent in this study also facilitated downstream LGT mapping efforts used to link phenotypes to specific mutations.
Major limitations of TILLING by sequencing compared to TILLING include the higher costs of library construction, genome sequencing, analysis, and the general requirement for more sophisticated technology. However, since TILLING by sequencing libraries are clonal, multiple isolates with distinct mutant alleles of a gene of interest can be identified by inspection of the corresponding genome sequences (Kokes et al. 2015). These libraries are also amenable to phenotypic screening. For example, Snavely et al. identified chlamydial protease-like activity factor (CPAF) null mutants by screening for isolates that failed to stain with the anti-CPAF antibody (Snavely et al. 2014). Theoretically, any Chlamydia spp. that can be propagated in vitro, mutagenized, and clonally isolated is amenable to TILLING. The most important limitation of TILLING is mapping phenotypes to genotypes. These approaches invariably suffer from the trade-off between the overall size of the mutant library and difficulty of mapping mutations of interest in the library isolates. Fewer heavily mutagenized isolates need to be screened to identify a mutant with a given phenotype of interest, but linking phenotypes to genotypes become more difficult as numbers of background mutations increase.
The Fisher group successfully adapted an insertional mutagenesis approach (TargeTron®, Sigma Inc.) to target type II introns to specific locations on the chlamydial chromosome (Johnson and Fisher 2013). A type II intron carrying a bla cassette and incA targeting sequences was transformed into C. trachomatis L2 using a suicide vector. Integrants (incA::bla) were selected with ampicillin and confirmed by PCR and Southern blot. Similar to naturally occurring incA mutant strains, inclusions of the incA::bla mutant were non-fusogenic (Suchland et al. 2000; Johnson and Fisher 2013). Subsequently, a spectinomycin resistant intron was developed to create an incA::aadA rsbV1::bla double mutant (Lowden et al. 2015). TargeTron has now been used to disrupt multiple genes including the chaperones groEL2 and groEL3, cpoS, ct813/inaC, and various inc genes in C. trachomatis L2 (Illingworth et al. 2017; Sixt et al. 2017; Weber et al. 2017, Wesolowski et al. 2017). The applicability of the TargeTron approach is limited by the presence of optimal intron targeting sequences within the gene of interest (Hooppaw and Fisher 2016; Key and Fisher 2017). Although insertional mutagenesis approaches can disrupt specific genes, they can also induce polar effects and are less suitable than chemical mutagenesis for generation of partial loss of function alleles.
FRAEM insertional mutagenesis utilizes DNA homology to target genomic regions of interest and introduce a selection cassette and dual reporter genes to facilitate identification of recombinants (Mueller et al. 2016). The FRAEM vector is a conditional suicide vector that places the plasmid maintenance gene (pgp6) under control of tetracycline-regulated operator sequence. This permits expansion of shuttle vector transformants within the population to increase the possibility of recombination between homologous sequences on the chromosome and vector, while still maintaining the ability to cure the FRAEM vector. The selection cassette contains bla for penicillin selection of recombinants, and GFP to monitor transformation and integration of the selection cassette into the chromosome. Finally, visual screening of single and double crossover (allelic exchange) events is facilitated by a mCherry gene located outside of the selection cassette and flanking chromosomal homology regions. C. trachomatis L2 trpA was deleted and replaced with the shuttle-vector selection cassette in a proof of principle experiment. The limitations of FRAEM include its relative complexity, the risk of deletion of unknown cis-acting chromosomal sequences, and polar effects (Mueller et al. 2017). Nonetheless, FRAEM is clearly a powerful tool for targeted manipulation of chlamydial genomes.
8. Forward Genetic Analysis of Chlamydial Genomes
Forward genetic screens have identified genes that mediate multiple aspects of chlamydial development, virulence, and immune evasion. These screens have relied on chemically mutagenized libraries because the TargeTron and FRAEM approaches are low-throughput (Table 1). In addition to the relative ease of mutagenesis, chemically mutagenized libraries contain a mixture of isolates with total and partial loss of function alleles, which permits interrogation of essential gene functions.
Nguyen and Valdivia compared the plaque morphologies of a collection of EMS mutagenized isolates derived from a rifampicin resistant C. trachomatis L2 parent (Nguyen and Valdivia 2012). The mutations causing altered plaque phenotypes were mapped using LGT and phenotypic linkage analysis. Three mutants that produced granular plaques had distinct missense mutations in glgB, a glycogen-branching enzyme. One of these missense mutations segregated with the granular plaque phenotypes when the mutant was crossed by LGT with spectinomycin and trimethoprim resistant C. trachomatis parents. Missense mutations in the type II secretion gene, gspE, were also linked to granular and small plaque phenotypes.
The same library of mutagenized C. trachomatis L2 isolates was screened for mutants that failed to promote F-actin assembly at the inclusion (Kokes et al. 2015). LGT with a spectinomycin parent linked this phenotype to a nonsense mutation in an uncharacterized inc gene, inaC/ctl0184. InaC expression from a plasmid complemented the inaC mutant, confirming the results of the LGT mapping. Another screen identified mutants that elicited increased lactose dehydrogenase (LDH) release from HeLa cells, which is an indicator of host cell death (Sixt et al. 2017). A nonsense mutation in the Inc gene ctl0481 (renamed cpoS for chlamydia promoter of survival) was linked to the early cell death phenotype.
Two screens from the Nelson laboratory utilized an EMS-mutagenized library derived from a wild-type C. trachomatis L2 isolate transformed with pGFP::SW2 to avoid fitness costs associated with chromosomal antibiotic resistance alleles (Brothwell et al. 2016; Muramatsu et al. 2016). The use of pGFP::SW2 (L2-GFP) facilitated visual screens of mutant phenotypes and GFP expression served as a reporter of chlamydial metabolic activity. TS mutants that produced a reduced ratio of inclusions at 32°C versus 37°C (cold sensitive, CS) or 40°C versus. 37°C (heat sensitive, HS), compared to the library parent, were identified in the first screen (Brothwell et al. 2016). Thirty-one unique mutants with CS, TS or CS/TS phenotypes were identified and the mutations in 14 of these were mapped by csLGT. TS alleles of genes that play roles in protein synthesis, DNA replication, fatty acid biosynthesis, and carbohydrate metabolism in other bacteria as well as conserved chlamydial genes of unknown function were identified. A second screen identified six mutants that were sensitive to interferon (IFN)-γ-mediated persistence (Sip) (Muramatsu et al. 2016). Three sip alleles were mapped using csLGT. One Sip mutant contained a missense mutation in trpB, which is consistent with the known role of trpB in C. trachomatis during indole-rescue from IFN-γ-mediated tryptophan starvation (Fehlner-Gardiner et al. 2002; Kari et al. 2011). The other two mutants had missense mutations in a predicted oxidative stress protein gene cysJ and a predicted small neutral amino acid transporter. These mutants suggested that the chlamydial response to IFN-γ involves more than tryptophan metabolism.
Successful genetic screens in chlamydiae hinge upon both library diversity and the ability to map specific mutations to the phenotype of interest. Multiple approaches can link phenotypes to genotypes (Fig. 1). As with TILLING, fewer isolates need to be screened to achieve saturation when a higher concentration of mutagen is employed, but parsing out the relevant mutation becomes more difficult downstream. Additionally, high mutation burdens increase the occurrence of both directly and synthetically lethal mutations. Despite the initial challenges of library construction, forward genetic screens are powerful tools for identifying the functions of chlamydial genes, which can then be validated using the reverse genetics approaches discussed above.
9. Overview and Future Perspectives
When the field of molecular microbial pathogenesis began to expand in the 1980s Stanley Falkow proposed a guide that was later deemed Koch’s postulate of molecular pathogenesis (Falkow 1988). Paraphrasing these postulates: (1) the phenotype under investigation should be associated with a pathogen; (2) specific inactivation of the gene associated with the virulence phenotype should reduce pathogenicity; and (3) reversion or replacement of the mutant gene should restore pathogenicity. Falkow also stressed that genetic approaches for testing these postulates were not rigid, and that a lack of relevant in vivo models was a serious obstacle for identifying virulence factors of many genetically tractable pathogens. The latter points may be especially pertinent to the current state of chlamydial genetics where our perspective is that the excitement about the latest tools has sometimes led to incomplete consideration of their limitations as well as the best model systems in which the mutation associated phenotypes would be apparent.
There is no single or perfect approach for genetic manipulation of chlamydiae for all applications and choosing the best method for a given study requires consideration of the strengths and weaknesses of the existing tools. For example, forward genetic screens that employ TILLING approaches benefit from the ability of mutagens to introduce desirable point mutations that are less likely to cause cis- and trans-acting effects, but are limited by downstream mutation mapping. A report describing two C. trachomatis Himar transposon mutants suggests that transposon mutant libraries that could help circumvent this limitation may soon be available (Fischer et al. 2017). TargeTron and FRAEM can alter specific genes, but these approaches introduce disruptions and/or deletions that could disrupt cis- and trans-acting sequences. These limitations have been circumvented in other bacteria using CRISPR/Cas9 gene editing since inactivation of specific targets is achieved without altering the larger genomic context, and adaptation of this approach for chlamydiae could be valuable (Luo et al. 2016; Zhang et al. 2017). Looking more long-term, adaptation of methods pioneered in E. coli for markerless allelic exchange and single copy gene expression from neutral loci seem like promising approaches for inactivating chlamydial genes while avoiding cis-and trans-acting effects and for expressing chlamydial proteins at physiological levels (Datsenko and Wanner 2000). Multiple tools in the chlamydial genetic toolbox remain limited by transformation efficiency, and further optimization of transformation techniques including electroporation and dendrimer-based approaches could be fruitful (Tam et al. 1994; Binet and Maurelli 2009a, b; Mishra et al. 2012; Gerard et al. 2013; Kannan et al. 2013). Probably the most significant impediment in the genetic dissection of chlamydial pathogenesis remains that these organisms need to be propagated in host cells. Genetic manipulation of other “obligate” intracellular organisms has clearly benefited from the development of anexic growth media, and promising findings that might be on the horizon for chlamydiae too (Omsland et al. 2009, 2012; Haider et al. 2010).
Finally, the overwhelming focus on the development of tools for a single model strain, C. trachomatis L2 434/Bu, has limited our ability to use genetic approaches to dissect crucial aspects of chlamydial pathogenesis. Despite the lack of a good in vivo model for C. trachomatis L2, the advent of genetic approaches has greatly enhanced our understanding of chlamydial biology. The challenge is now to extend these approaches to other Chlamydia spp.—especially those for which relevant animal models exist. LGV biology differs from the more common and medically important trachoma biovar strains in many ways. Trachoma biovar strains primarily infect columnar epithelial cells that protect the conjunctiva, urethra, GI, lung, cervix, and upper female reproductive tract. In contrast, LGV biovar strains infect via epithelial micro-abrasions where they enter into and proliferate in lymphatic cells. Innate immunity also rapidly clears C. trachomatis strains in mice (Morrison and Caldwell 2002). In contrast, C. muridarum can cause long-lasting infections in mice that are cleared by adaptive immunity (Barron et al. 1981; Morrison and Caldwell 2002). Existing murine C. muridarum pulmonary and GI infection models also mimic aspects of these infections with C. trachomatis STI strains in humans and the latter model was crucial to the discovery of the virulence role of chlamydial plasmids (Morrison and Caldwell 2002; Ramsey et al. 2009; Yeruva et al. 2013; Rank and Yeruva 2014). Virulence factors initially identified in the mouse GT were more important for C. muridarum GI colonization, showing that the tissue choice can also profoundly impact relevance of specific chlamydial virulence factors in the same animals (Shao et al. 2017). Beyond C. muridarum, C. caviae Guinea pig models can investigate urethral infections and sexual transmission (Rank et al. 2003; Wang et al. 2010). Primate models of trachoma and cervicitis exist that use the same isolates that cause these diseases in humans (Wolner-Hanssen et al. 1991; Kari et al. 2008). Understanding the animal and tissue-specific context in which chlamydial virulence genes are required during infection will shed light on both chlamydial biology and the limits of a given infection model. Looking forward, adapting the existing genetic tools to additional species and strains could enhance the utility of genetic approaches for dissecting mechanisms of chlamydial pathogenesis.
Acknowledgements
DE Nelson was supported by grants AI099278 and AI116706, and G Zhong was supported by grants AI121989, AI105712, AI047997, from the United States National Institutes of Health, Division of Allery and Infectious Diseases. We would like Drs. Harlan Caldwell and Derek Fisher for discussion and insights regarding aspects of this manuscript. Finally, any oversights of relevant studies were not intentional and the authors would like to apologize for any instance of this in advance.
Contributor Information
Julie A. Brothwell, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA
Matthew K. Muramatsu, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA
Guangming Zhong, Department of Microbiology, Immunology and Molecular Genetics, University of Texas Health Science Center at San Antonio, San Antonio, TX, USA.
David E. Nelson, Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA
References
- Agaisse H, Derre I (2013) A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS ONE 8(2):e57090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T (2010) Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38(3):868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Hirakawa H, Yamashita A, Cai Y, Rahman MA, Suzuki H, Mitaku S, Toh H, Goto S, Murakami T, Sugi K, Hayashi H, Fukushi H, Hattori M, Kuhara S, Shirai M (2006) Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res 13(1):15–23 [DOI] [PubMed] [Google Scholar]
- Barron AL, White HJ, Rank RG, Soloff BL, Moses EB (1981) A new animal model for the study of Chlamydia trachomatis genital infections: infection of mice with the agent of mouse pneumonitis. J Infect Dis 143(1):63–66 [DOI] [PubMed] [Google Scholar]
- Bastidas RJ, Valdivia RH (2016) Emancipating Chlamydia: advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol Mol Biol Rev 80(2):411–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauler LD, Hackstadt T (2014) Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196(7):1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT (2005) Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrob Agents Chemother 49 (11):4455–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT (2007) Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob Agents Chemother 51(12):4267–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT (2009a) The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. Bmc Microbiology 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT (2009b) Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci USA 106(1):292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Bowlin AK, Maurelli AT, Rank RG (2010) Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob Agents Chemother 54(3):1094–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Ferreira R, Nunes A, Sousa-Uva M, Abreu M, Borrego MJ, Gomes JP (2013) Effect of long-term laboratory propagation on Chlamydia trachomatis genome dynamics. Infect Genet Evol 17:23–32 [DOI] [PubMed] [Google Scholar]
- Brothwell JA, Muramatsu MK, Toh E, Rockey DD, Putman TE, Barta ML, Hefty PS, Suchland RJ, Nelson DE (2016) Interrogating genes that mediate Chlamydia trachomatis survival in cell culture using conditional mutants and recombination. J Bacteriol 198 (15):2131–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F (1994) Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Invest 94(1):458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burall LS, Rodolakis A, Rekiki A, Myers GS, Bavoil PM (2009) Genomic analysis of an attenuated Chlamydia abortus live vaccine strain reveals defects in central metabolism and surface proteins. Infect Immun 77(9):4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G (2003) Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111(11):1757–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Porcella SF, McClarty G, Caldwell HD (2005) Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 73(10):6407–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ 3rd, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD (2008) The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76(6):2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson N, Medico N, Bille J, Greub G (2006) Parachlamydia acanthamoebae enters and multiplies within pneumocytes and lung fibroblasts. Microbes Infect 8(5):1294–1300 [DOI] [PubMed] [Google Scholar]
- Casson N, Entenza JM, Borel N, Pospischil A, Greub G (2008) Murine model of pneumonia caused by Parachlamydia acanthamoebae. Microb Pathog 45(2):92–97 [DOI] [PubMed] [Google Scholar]
- Chen C, Zhou Z, Conrad T, Yang Z, Dai J, Li Z, Wu Y, Zhong G (2015a) In vitro passage selects for Chlamydia muridarum with enhanced infectivity in cultured cells but attenuated pathogenicity in mouse upper genital tract. Infect Immun 83(5):1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang Z, Sun X, Tang L, Ding Y, Xue M, Zhou Z, Baseman J, Zhong G (2015b) Intrauterine infection with plasmid-free Chlamydia muridarum reveals a critical role of the plasmid in chlamydial ascension and establishes a model for evaluating plasmid-independent pathogenicity. Infect Immun 83(6):2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T (2004) A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101(27):10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Poppert S, Heinz E, Schmitz-Esser S, Essig A, Schweikert M, Wagner M, Horn M (2005) Recovery of an environmental Chlamydia strain from activated sludge by co-cultivation with Acanthamoeba sp. Microbiology 151(Pt 1):301–309 [DOI] [PubMed] [Google Scholar]
- Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GS, Horn M (2011) Unity in variety–the pan-genome of the Chlamydiae. Mol Biol Evol 28(12):3253–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TA, Gong S, Yang Z, Matulich P, Keck J, Beltrami N, Chen C, Zhou Z, Dai J, Zhong G (2015) The chromosome-encoded hypothetical protein TC0668 Is an upper genital tract pathogenicity factor of Chlamydia muridarum. Infect Immun 84(2):467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97(12):6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D, Schachter J, Dawson CR, Stephens RS (1992) Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J Infect Dis 166(2):383–392 [DOI] [PubMed] [Google Scholar]
- DeMars R, Weinfurter J (2008) Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol 190(5):1605–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMars R, Weinfurter J, Guex E, Lin J, Potucek Y (2007) Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol 189(3):991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessus-Babus S, Bebear CM, Charron A, Bebear C, de Barbeyrac B (1998) Sequencing of gyrase and topoisomerase IV quinolone-resistance-determining regions of Chlamydia trachomatis and characterization of quinolone-resistant mutants obtained In vitro. Antimicrob Agents Chemother 42(10):2474–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Gong S, Tian Y, Yang Z, Brunham R, Zhong G (2013) Transformation of sexually transmitted infection-causing serovars of Chlamydia trachomatis using Blasticidin for selection. PLoS ONE 8(11):e80534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati M, Huot-Creasy H, Humphrys M, Di Paolo M, Di Francesco A, Myers GS (2014) Genome Sequence of Chlamydia suis MD56, isolated from the conjunctiva of a weaned piglet. Genome Announc 2(3):e00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghi A 2nd, Popov VL, Kahl MM, Stanton JB, Brown CC, Tsongalis GJ, West AB, Frasca S Jr (2004) Characterization of “Candidatus piscichlamydia salmonis” (order Chlamydiales), a chlamydia-like bacterium associated with epitheliocystis in farmed Atlantic salmon (Salmo salar). J Clin Microbiol 42(11):5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Padubrin I, Kohler L, Hudson AP (2003) Detection of nucleotide variability in rpoB in both rifampin-sensitive and rifampin-resistant strains of Chlamydia trachomatis. Antimicrob Agents Chemother 47(7):2316–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett KD, Bush RM, Andersen AA (1999) Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol 49(Pt 2):415–440 [DOI] [PubMed] [Google Scholar]
- Falkow S (1988) Molecular Koch’s postulates applied to microbial pathogenicity. Rev Infect Dis 10(Suppl 2):S274–276 [DOI] [PubMed] [Google Scholar]
- Farencena A, Comanducci M, Donati M, Ratti G, Cevenini R (1997) Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun 65(7):2965–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G (2002) Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem 277(30):26893–26903 [DOI] [PubMed] [Google Scholar]
- Fischer A, Harrison KS, Ramirez Y, Auer D, Chowdhury BK, Prusty BK, Sauer F, Dimond Z, Kisker C, Scott Hefty P, Rudel T (2017) Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. Elife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Peterson EM, de la Maza LM (1993) Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol Biol Evol 10(4):892–913 [DOI] [PubMed] [Google Scholar]
- Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Kannan RM, Hudson AP (2013) Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9(7):996–1008 [DOI] [PubMed] [Google Scholar]
- Gomes JP, Bruno WJ, Borrego MJ, Dean D (2004) Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol 186(13):4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yang Z, Lei L, Shen L, Zhong G (2013) Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195(17):3819–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Berger P, Papazian L, Raoult D (2003) Parachlamydiaceae as rare agents of pneumonia. Emerg Infect Dis 9(6):755–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J, Harris SR, Seth-Smith HMB, Parmar S, Andersson P, Giffard PM, Schachter J, Moncada J, Ellison L, Vaulet MLG, Fermepin MR, Radebe F, Mendoza S, Ouburg S, Morre SA, Sachse K, Puolakkainen M, Korhonen SJ, Sonnex C, Wiggins R, Jalal H, Brunelli T, Casprini P, Pitt R, Ison C, Savicheva A, Shipitsyna E, Hadad R, Kari L, Burton MJ, Mabey D, Solomon AW, Lewis D, Marsh P, Unemo M, Clarke IN, Parkhill J, Thomson NR (2017) Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Genome Res 27(7):1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S, Wagner M, Schmid MC, Sixt BS, Christian JG, Hacker G, Pichler P, Mechtler K, Muller A, Baranyi C, Toenshoff ER, Montanaro J, Horn M (2010) Raman microspectroscopy reveals long-term extracellular activity of Chlamydiae. Mol Microbiol 77(3):687–700 [DOI] [PubMed] [Google Scholar]
- Hayes LJ, Yearsley P, Treharne JD, Ballard RA, Fehler GH, Ward ME (1994) Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect Immun 62(12):5659–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer M, Laroucau K, Creasy HH, Ott S, Vorimore F, Bavoil PM, Marz M, Sachse K (2016) Whole-genome sequence of Chlamydia gallinacea type strain 08-1274/3. Genome Announc 4 (4):e00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooppaw AJ, Fisher DJ (2016) A coming of age story: Chlamydia in the Post-Genetic Era. Infect Immun 84(3):612–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M (2004) Illuminating the evolutionary history of chlamydiae. Science 304(5671):728–730 [DOI] [PubMed] [Google Scholar]
- Illingworth M, Hooppaw AJ, Ruan L, Fisher DJ, Chen L (2017) Biochemical and genetic analysis of the Chlamydia GroEL Chaperonins. J Bacteriol 199(12):e00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD (2010) Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78(6):2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD (2013) Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol 13:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ (2013) Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS ONE 8(12):e83989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS (1999) Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 21(4):385–389 [DOI] [PubMed] [Google Scholar]
- Kannan RM, Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Hudson AP (2013) Dendrimer-enabled transformation of Chlamydia trachomatis. Microb Pathog 65:29–35 [DOI] [PubMed] [Google Scholar]
- Kari L, Whitmire WM, Carlson JH, Crane DD, Reveneau N, Nelson DE, Mabey DC, Bailey RL, Holland MJ, McClarty G, Caldwell HD (2008) Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis 197(3):449–456 [DOI] [PubMed] [Google Scholar]
- Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD (2011) Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci USA 108(17):7189–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari L, Southern TR, Downey CJ, Watkins HS, Randall LB, Taylor LD, Sturdevant GL, Whitmire WM, Caldwell HD (2014) Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect Immun 82(7):2756–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen M, Nylund A, Watanabe K, Helvik JV, Nylund S, Plarre H (2008) Characterization of ‘Candidatus Clavochlamydia salmonicola’: an intracellular bacterium infecting salmonid fish. Environ Microbiol 10(1):208–218 [DOI] [PubMed] [Google Scholar]
- Key CE, Fisher DJ (2017) Use of group II intron technology for targeted mutagenesis in Chlamydia trachomatis. Methods Mol Biol 1498:163–177 [DOI] [PubMed] [Google Scholar]
- Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ (2015) Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe 17(5):716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Jehl MA, Rattei T, Horn M (2014) Signature protein of the PVC superphylum. Appl Environ Microbiol 80(2):440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe MF, Suchland RJ, Stamm WE (1993) Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun 61(1):213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G (2014) Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect Immun 82(3):983–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, Baseman J, Zhong G (2014) Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J Bacteriol 196(5):989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr M, Prohl A, Ostermann C, Liebler-Tenorio E, Schroedl W, Aeby S, Greub G, Reinhold P (2015) A bovine model of a respiratory Parachlamydia acanthamoebae infection. Pathog Dis 73(1):1–14 [DOI] [PubMed] [Google Scholar]
- Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ (2015) Use of aminoglycoside 3’ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes 8:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Leenay RT, Beisel CL (2016) Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng 113(5):930–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Izutsu H, Miyashita N, Ohuchi M (1998) Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol 36(10):3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18(4):455–457 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Sandlin RC, Maurelli AT (2003) In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J Bacteriol 185(4):1218–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM (2012) Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol Pharm 9(3):413–421 [DOI] [PubMed] [Google Scholar]
- Misiurina O, Shipitsina EV, Finashutina Iu P, Lazarev VN, Akopian TA, Savicheva AM, Govorun VM (2004) Analysis of point mutations in the ygeD, gyrA and parC genes in fluoroquinolones resistant clinical isolates of Chlamydia trachomatis. Mol Gen Mikrobiol Virusol 3:3–7 [PubMed] [Google Scholar]
- Mojica S, Huot Creasy H, Daugherty S, Read TD, Kim T, Kaltenboeck B, Bavoil P, Myers GS (2011) Genome sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J Bacteriol 193(14):3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison RP, Caldwell HD (2002) Immunity to murine chlamydial genital infection. Infect Immun 70(6):2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW (1985) Comparative biology of intracellular parasitism. Microbiol Rev 49(3):298–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Wolf K, Fields KA (2016) Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. MBio 7(1):e01817–01815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Wolf K, Fields KA (2017) Chlamydia trachomatis Transformation and Allelic Exchange Mutagenesis. Curr Protoc Microbiol 45: 11A 13 11–11A 13 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu MK, Brothwell JA, Stein BD, Putman TE, Rockey DD, Nelson DE (2016) Beyond tryptophan synthase: identification of genes that contribute to Chlamydia trachomatis survival during IFN-gamma induced persistence and reactivation. Infect Immun [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Taylor LD, Shannon JG, Whitmire WM, Crane DD, McClarty G, Su H, Kari L, Caldwell HD (2007) Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cell Microbiol 9(9):2289–2298 [DOI] [PubMed] [Google Scholar]
- Nguyen BD, Valdivia RH (2012) Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci USA 109(4):1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Valdivia RH (2013) Forward genetic approaches in Chlamydia trachomatis. J Vis Exp 80:e50636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes A, Gomes JP (2014) Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect Genet Evol 23:49–64 [DOI] [PubMed] [Google Scholar]
- O’Connell CM, Nicks KM (2006) A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152(Pt 6):1601–1607 [DOI] [PubMed] [Google Scholar]
- O’Connell CM, Ingalls RR, Andrews CW Jr, Scurlock AM, Darville T (2007) Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179(6):4027–4034 [DOI] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA (2009) Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA 106(11):4430–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T (2012) Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci USA 109(48):19781–19785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Sixt BS, Horn M, Hackstadt T (2014) Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol Rev 38(4):779–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer L, Falkow S (1986) A common plasmid of Chlamydia trachomatis. Plasmid 16(1):52–62 [DOI] [PubMed] [Google Scholar]
- Peterson EM, Markoff BA, Schachter J, de la Maza LM (1990) The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23 (2):144–148 [DOI] [PubMed] [Google Scholar]
- Pickett MA, Everson JS, Pead PJ, Clarke IN (2005) The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology-Sgm 151:893–903 [DOI] [PubMed] [Google Scholar]
- Qin B, McClarty G (1992) Effect of 6-thioguanine on Chlamydia trachomatis growth in wild-type and hypoxanthine-guanine phosphoribosyltransferase-deficient cells. J Bacteriol 174(9):2865–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram K, Giebel AM, Toh E, Hu S, Newman JH, Morrison SG, Kari L, Morrison RP, Nelson DE (2015) Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun 83(7):2870–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG (2009) Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect Immun 77 (8):3284–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank RG, Yeruva L (2014) Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect Immun 82(4):1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank RG, Bowlin AK, Reed RL, Darville T (2003) Characterization of chlamydial genital infection resulting from sexual transmission from male to female guinea pigs and determination of infectious dose. Infect Immun 71(11):6148–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28(6):1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM (2003) Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31(8):2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis A (1983) In vitro and in vivo properties of chemically induced temperature-sensitive mutants of Chlamydia psittaci var. ovis: screening in a murine model. Infect Immun 42 (2):525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis A, Souriau A (1983) Response of ewes to temperature-sensitive mutants of Chlamydia psittaci (var ovis) obtained by NTG mutagenesis. Ann Rech Vet 14(2):155–161 [PubMed] [Google Scholar]
- Rodolakis A, Souriau A (1986) Response of goats to vaccination with temperature-sensitive mutants of Chlamydia psittaci obtained by nitrosoguanidine mutagenesis. Am J Vet Res 47 (12):2627–2631 [PubMed] [Google Scholar]
- Russell M, Darville T, Chandra-Kuntal K, Smith B, Andrews CW Jr, O’Connell CM (2011) Infectivity acts as in vivo selection for maintenance of the chlamydial cryptic plasmid. Infect Immun 79(1):98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse K, Laroucau K (2015) Two more species of Chlamydia-does it make a difference? Pathog Dis 73(1):1–3 [DOI] [PubMed] [Google Scholar]
- Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, Weidmann M, Myers G, Vorimore F, Vicari N, Magnino S, Liebler-Tenorio E, Ruettger A, Bavoil PM, Hufert FT, Rossello-Mora R, Marz M (2014) Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol 37(2):79–88 [DOI] [PubMed] [Google Scholar]
- Schofl G, Voigt A, Litsche K, Sachse K, Saluz HP (2011) Complete genome sequences of four mammalian isolates of Chlamydophila psittaci. J Bacteriol 193(16):4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN (2009) Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genom 10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Melero J, Zhang N, Arulanandam B, Baseman J, Liu Q, Zhong G (2017) The cryptic plasmid is more important for Chlamydia muridarum to colonize the mouse gastrointestinal tract than to infect the genital tract. PLoS ONE 12(5):e0177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixt BS, Bastidas RJ, Finethy R, Baxter RM, Carpenter VK, Kroemer G, Coers J, Valdivia RH (2017) The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21(1):113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH (2014) Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71(3):336–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD (2013) Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81(3):636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Carlson JH, Zhou B, Virtaneva K, Whitmire WM, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD (2014) Plasmid-mediated transformation tropism of chlamydial biovars. Pathog Dis 70(2):189–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282(5389):754–759 [DOI] [PubMed] [Google Scholar]
- Stephens RS, Myers G, Eppinger M, Bavoil PM (2009) Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol 55 (2):115–119 [DOI] [PubMed] [Google Scholar]
- Stothard DR, Williams JA, Van Der Pol B, Jones RB (1998) Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect Immun 66 (12):6010–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stride MC, Polkinghorne A, Miller TL, Groff JM, Lapatra SE, Nowak BF (2013) Molecular characterization of “Candidatus Parilichlamydia carangidicola,” a novel Chlamydia-like epitheliocystis agent in yellowtail kingfish, Seriola lalandi (Valenciennes), and the proposal of a new family, “Candidatus Parilichlamydiaceae” fam. nov. (order Chlamydiales). Appl Environ Microbiol 79(5):1590–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturdevant GL, Kari L, Gardner DJ, Olivares-Zavaleta N, Randall LB, Whitmire WM, Carlson JH, Goheen MM, Selleck EM, Martens C, Caldwell HD (2010) Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect Immun 78(9):3660–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Rockey DD, Bannantine JP, Stamm WE (2000) Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun 68(1):360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD (2009) Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother 53 (11):4604–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JE, Davis CH, Wyrick PB (1994) Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol 40(7):583–591 [DOI] [PubMed] [Google Scholar]
- Taylor-Brown A, Vaughan L, Greub G, Timms P, Polkinghorne A (2015) Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog Dis 73(1):1–15 [DOI] [PubMed] [Google Scholar]
- Thomas NS, Lusher M, Storey CC, Clarke IN (1997) Plasmid diversity in Chlamydia. Microbiology 143(Pt 6):1847–1854 [DOI] [PubMed] [Google Scholar]
- Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, Cerdeno-Tarraga AM, Harris B, Doggett J, Ormond D, Mungall K, Clarke K, Feltwell T, Hance Z, Sanders M, Quail MA, Price C, Barrell BG, Parkhill J, Longbottom D (2005) The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res 15(5):629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O’Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN (2008) Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18(1):161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipples G, McClarty G (1991) Isolation and initial characterization of a series of Chlamydia trachomatis isolates selected for hydroxyurea resistance by a stepwise procedure. J Bacteriol 173(16):4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treharne JD, Yearsley PJ, Ballard RC (1989) In vitro studies of Chlamydia trachomatis susceptibility and resistance to rifampin and rifabutin. Antimicrob Agents Chemother 33 (8):1393–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nagarajan U, Hennings L, Bowlin AK, Rank RG (2010) Local host response to chlamydial urethral infection in male guinea pigs. Infect Immun 78(4):1670–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN (2011) Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7(9):e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YB, Cutcliffe LT, Skilton RJ, Ramsey KH, Thomson NR, Clarke IN (2014) The genetic basis of plasmid tropism between Chlamydia trachomatis and Chlamydia muridarum. Pathogens and Disease 72(1):19–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, Bauler LD, Lam J, Hackstadt T (2015) Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83(12):4710–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, Lam JL, Dooley CA, Noriea NF, Hansen BT, Hoyt FH, Carmody AB, Sturdevant GL, Hackstadt T (2017) Absence of specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep 19(7):1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Hatch TP, Woese CR (1986) Eubacterial origin of chlamydiae. J Bacteriol 167 (2):570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowski J, Weber MM, Nawrotek A, Dooley CA, Calderon M, St Croix CM, Hackstadt T, Cherfils J, Paumet F (2017) Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and golgi complex positioning. MBio 8(3):e02280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrum J, Sammons LR, Restivo KN, Hefty PS (2013) Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS ONE 8(10):e76743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner-Hanssen P, Patton DL, Holmes KK (1991) Protective immunity in pig-tailed macaques after cervical infection with Chlamydia trachomatis. Sex Transm Dis 18(1):21–25 [DOI] [PubMed] [Google Scholar]
- Wylie JL, Wang LL, Tipples G, McClarty G (1996) A single point mutation in CTP synthetase of Chlamydia trachomatis confers resistance to cyclopentenyl cytosine. J Biol Chem 271 (26):15393–15400 [DOI] [PubMed] [Google Scholar]
- Yang C, Starr T, Song L, Carlson JH, Sturdevant GL, Beare PA, Whitmire WM, Caldwell HD (2015) Chlamydial lytic exit from host cells is plasmid regulated. MBio 6(6):e01648–01615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Kari L, Sturdevant GL, Song L, Patton MJ, Couch CE, Ilgenfritz JM, Southern TR, Whitmire WM, Briones M, Bonner C, Grant C, Hu P, McClarty G, Caldwell HD (2017) Chlamydia trachomatis ChxR is a transcriptional regulator of virulence factors that function in in vivo host-pathogen interactions. Pathog Dis 75(3):ftx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeruva L, Spencer N, Bowlin AK, Wang Y, Rank RG (2013) Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 68(3):88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cheng QX, Liu AM, Zhao GP, Wang J (2017) A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front Microbiol 8:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G (2017) Chlamydial plasmid-dependent pathogenicity. Trends Microbiol 25(2):141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]