Abstract
As with most organ systems that undergo continuous generation and maturation during the transition from fetal to adult life, the hematopoietic and immune systems also experience dynamic changes. Such changes lead to many unique features in blood cell function and immune responses in early childhood. The blood cells and immune cells in neonates are a mixture of fetal and adult origin due to the co-existence of both fetal and adult types of hematopoietic stem cells (HSCs) and progenitor cells (HPCs). Fetal blood and immune cells gradually diminish during maturation of the infant and are almost completely replaced by adult types of cells by 3 to 4 weeks after birth in mice. Such features in early childhood are associated with unique features of hematopoietic and immune diseases, such as leukemia, at these developmental stages. Therefore, understanding the cellular and molecular mechanisms by which hematopoietic and immune changes occur throughout ontogeny will provide useful information for the study and treatment of pediatric blood and immune diseases. In this review, we summarize the most recent studies on hematopoietic initiation during early embryonic development, the expansion of both fetal and adult types of HSCs and HPCs in the fetal liver and fetal bone marrow stages, and the shift from fetal-to-adult hematopoiesis/immunity during neonatal/infant development. We also discuss the contributions of fetal types of HSCs/HPCs to childhood leukemias.
Keywords: fetal, neonatal, infant, hematopoiesis, leukemia
Graphical Abstract
Introduction.
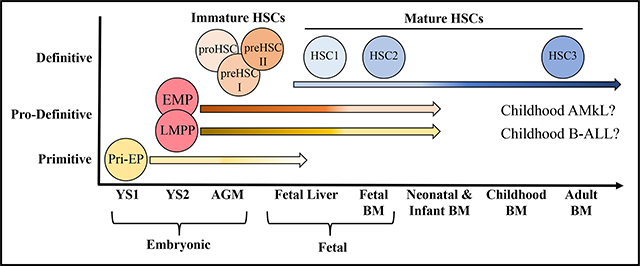
Hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) persist in a fetal phenotype until 2–3 weeks of age in mice.[1–3] Compared to adult HSCs, fetal and neonatal HSCs exhibit markedly different properties, including differences in surface markers, proliferative state, repopulation/self-renewal capabilities, gene expression, and differentiation activities.[2, 4, 5] Fetal/neonatal HSCs are highly proliferative with relatively high repopulation/self-renewal capabilities, while adult HSCs are quiescent with respect to the cell cycle (G0 phase) with reduced repopulation/self-renewal capabilities. Fetal liver (FL) HSCs display faster hematopoietic regeneration and outcompete the production of HSCs from adult bone marrow (BM) cells in irradiated recipients.[6–8] Fetal HSCs display an erythroid differentiation bias, possess a distinct potential for innate-like lymphocytes (such as B1a, Vγ3+ γδ-T and innate lymphoid cells) and produce smaller sized and lower ploidy megakaryocytes (Mks) [9, 10], while adult HSCs show more balanced differentiation, produce larger sized/higher ploidy Mks and lack potential to become innate-like lymphocytes. The erythroid bias of fetal/neonatal HSC/HPCs is due to the slower kinetics of erythropoietin (EPO)-induced differentiation.[11] In addition, fetal/neonatal T/B lymphocytes show reduced or deviant responses to antigenic stimulation compared to adult counterparts. Furthermore, fetal HSCs are resistant to many leukemic oncogenes detected in adult cases of leukemia, which might explain the different frequency of various leukemia subtypes in neonates/infants compared to adults [12–15]. Both HSC-intrinsic and environmental mechanisms have been proposed to explain such distinct properties of fetal and adult HSCs. Therefore, a better understanding of the cellular and molecular mechanisms by which the dynamic hematopoietic changes occur will provide a blueprint for the study of pediatric blood and immune disorders, as well as strategies potential for these disorders.
Most early studies consider adult HSCs/HPCs to be uniformly derived from definitive (adult-type) hematopoiesis in the para-aortic splanchnopleural/AGM region. These studies suggested a dynamic maturation theory of adult-type HSCs/HPCs during the fetal-to-adult hematopoietic transition. However, prior to definitive hematopoiesis within the AGM, at least 2 more waves of fetal type hematopoiesis are generated from the yolk sac (YS). Studies suggested that both fetal and adult types of HSCs/HPCs migrate to and expand in the FL and subsequently in the fetal BM. The hematopoietic and immune cells in the fetal stage are primarily produced by fetal types of HSCs/HPCs, which are then gradually diminished and replaced by cells produced by adult-type HSCs/HPCs during later fetal and early postnatal development. Many types of fetal hematopoietic cells, including fetal HSCs and HPCs, are present in neonatal and infant BM, which might contribute to hematopoiesis and immunity at multiple stages. Therefore, the fetal-to-adult hematopoietic transition is more likely a combination of the fetal-type to adult-type shift and gradual HSC/HPC maturation. In this review, we discuss the multiple waves of hematopoietic generation during early embryonic development and the blood/immune cell contributions of these waves to fetal, neonatal and adult stages. We specifically focus on the dynamic changes of fetal and adult types of HSCs/HPCs during the fetal-to-adult hematopoietic transition and discuss their potential roles in the pathogenesis of pediatric blood and immune diseases.
It needs to be mentioned that most of our current knowledge of fetal-to-adult hematopoietic transition is primarily drawn from the studies of mouse models. Nevertheless, hematopoietic development is conserved across vertebrates, and thus most of the results obtained from animal studies have been confirmed in humans. Therefore, we will mainly focus on studies involving mouse models and will extend our discussion to humans when there are notable differences between mouse and human.[16, 17] As there are many cell populations and developmental stages discussed, a summary of HSC and HPC phenotypes is listed in Table 1.
Table 1.
Phenotypes of HSCs and HPCs during fetal development
| Development Stage | Cell Population | Phenotype |
|---|---|---|
| E7.5–10.5 YS | Primitive Erythroid Progenitor (Pri-EP) | CD41+Kit+Sca1− |
| E8 YS | Pre-Macrophage (Mφ) | CD45+CX3CR1+ |
| E8.5 YS | Hemogenic Endothelial cells (ECs) | CD31+TIE+CD45−CD41−Runx1+ |
| E8.5–9.5 YS FL, FBM and Neonatal BM |
Erythro-Myeloid Progenitor (EMP) | CD45lowCD41+Kit+AA4.1+CD16/32+Sca1− CD45+CD34+CD44+ (H) |
| E9.5 YS FL, FBM and Neonatal BM |
Lympho-Myeloid Progenitor (LMP) | Lin−Kit+Rag1-GFP+Flt3+IL7Rα in YS, also Csf1r+ThpoR+ CD45+KIT+ FLT3+IL7Rα+ in FL |
| E9.5 YS FL, FBM and Neonatal BM |
Developmentally-Restricted HSC (drHSC) | Lin−Sca1+Kit+CD150lowFlt3+IL7Rα+ Rag1+ |
| Adult BM | Lymphoid-Primed Multipotent Progenitor (LMPP) | Lin−Sca1+Kit+CD150lowFlt3hiRag1+ |
| E8.5–9.5 YS | Common Primitive-Definitive Precursor (CPDP) | CD45−CD41+AA4.1−Kitmed |
| E8.5–9.5 YS | Multipotent Lympho-Myeloid Progenitor (MLMP) | CD45+KithighAA4.1+CD41+
|
| E8.5–9.5 YS | Multipotent Progenitors in YS | CD43+KIThighSca1+CD11A− |
| E8.5–9.5 YS | Myeloid-Restricted Progenitor | CD45+ KithighAA4.1−CD41+ |
| FL | Erythroid Progenitors (EP) | CD45−Kit+Ter119− |
| E9.5–12.5 Placenta | Placenta HSCs | CD34medc-kithi |
| AGM E9.5 E10.5 E11.5 |
imHSCs
pro-HSCs pre-HSC I pre-HSC II |
VEC+CD41loCD43−CD45− at as early VEC+ CD31+CD41loCD43+CD45− VEC+CD31+CD41lo CD43+CD45+, PROCR+ |
| E12 FL E14 FL FL |
HSCs HSCs HSCs |
Lin−Sca-1+Mac-1lowCD201+
CD45+CD150+CD48− CD201+ CD150+CD48−Sca1+CD11b+CD45+ CD41−Lin− DAPI− |
| FL and FBM | B-1 Cell Progenitor Cells (mouse) | AA4.1+CD19+B220lo/- |
| FL and Fetal BM (Human) |
Adult Type B cell Progenitors Fetal Type Early Lymphoid Progenitors (ELPs)-Mouse LMP Equivalent Pre-ProB |
CD34+CD19+CD10+ CD34+IL7R+CD19−CD10− CD34+CD19+CD10+ |
Multiple waves of hematopoietic initiation during embryonic development.
At least three consecutive overlapping waves of hematopoietic generation have been identified during embryonic development.[18, 19] The first wave, called primitive hematopoiesis, is generated directly from the mesoderm in the YS blood islands on embryonic day (E) 7.5 for mice and E17 for humans in a Runx1/Myb-independent manner. HPCs produced at this stage are called primitive erythroid progenitors (pri-EPs), defined as CD41+Kit+Sca1−, which produce red blood cells (RBCs), macrophages (Mφ) and megakaryocytes (Mks) but no lymphocytes.[20] RBCs produced by pri-EPs are large, nucleated cells that express a specific pattern of embryonic globin chains: εy- and βH1- in mice and ζ- and ε- in humans.[21] Distinct from Mφ produced by adult HPCs, pri-EPs produce Mφ without monocytic intermediates. Platelets produced by pri-EPs are larger in size compared to adult BM-derived platelets.[22]
The second wave, called pro-definitive (or transient-definitive) hematopoiesis, arises from Wnt-responsive hemogenic endothelial cells (ECs)(CD31+TIE+CD45−CD41−Runx1+) in the YS around E8.5 for mice in a Flk-1 and Scl-dependent, but Myb-independent, fashion.[19, 23–26] The HPCs generated during this wave are termed erythro-myeloid progenitors (EMPs), defined as CD45lowCD41+Kit+AA4.1+CD16/32+Sca1−. Shortly after generation, EMPs migrate to the FL and can differentiate into erythrocytes, Mks, Mφ, innate immune cells (NK cells, ILCs and mast cells), and other myeloid lineages such as neutrophils but have minimal conventional T/B lymphocyte potential. Transplantation studies suggested that EMPs lack self-renewal and long-term hematopoietic engraftment capacity,[18, 19, 23, 26–32] EMPs can contribute to the erythrocyte compartment for more than 20 days upon transplantation.[19] While these EMP-derived RBCs resemble adult RBCs, they express fetal globin βH1- (or γ-).[33] Recently, CD45+CD34+CD44+ YS-derived myeloid-biased progenitors, a type of EMP-like cell in the human with RBC, Mk, Mφ and mast cell differentiation potential, have been described.[34–36]
In addition to generating EMPs, the YS also serves as an initial source of T and B lymphoid progenitors.[24, 37, 38] Lin−Kit+Rag1-GFP+Flt3+IL7Rα+ lymphomyeloid progenitors (LMPs) have been identified in the E9.5 YS. Most LMPs also express colony stimulating factor 1 (CSF1) receptor and thrombopoietin (THPO) receptor. LMPs have both lymphoid (including canonical B2/T lymphocytes and innate B1a/dermal γδ T cells) and myeloid differentiation potential but lack megakaryocytic and erythroid potential. LMPs have been proposed to be the first progenitors to seed the developing thymus.[39] Phenotypically, LMPs might be the developmentally restricted HSCs (drHSCs) described by Forsberg,[40] which share many features of the epigenetic and transcriptional regulatory landscape with adult lymphoid-primed multipotent progenitors (LMPPs).[41] However, the exact interrelationships among EMPs, LMPs and drHSCs need to be determined. Several early studies also identified common primitive-definitive precursors, multipotent lympho-myeloid progenitors, myeloid-restricted progenitors, and multipotent progenitors in E8.5–9.5 YS, suggesting heterogeneity of the multipotent differentiation potential of YS HPCs.[25, 42–44] These studies suggested that common primitive-definitive precursors might produce multipotent lympho-myeloid, myeloid-restricted progenitors and EMPs to form a differentiation hierarchy.
The third wave of blood cell formation is called definitive hematopoiesis and emerges during E8.5-E11.5 for mice and 3–5 post-conception weeks (PCW) for humans in the major arteries in the para-aortic splanchnopleural/AGM region in a nitric oxide- and Notch1-Runx1-Gata2 axis-dependent manner.[45–52] The HPCs produced during this wave are referred to as immature HSC (imHSCs) and they can transform into multi-lineage functional HSCs in the FL environment or in proper in vitro culture conditions.[53, 54] Before colonization of the FL, imHSCs undergo quick maturation steps from pro-HSCs at as early as E9.5, through pre-HSC I at E10.5, and then pre-HSC II at E11.5.[55, 56] Endothelial protein C receptor expression could enrich all the HSCs in the E11.5 AGM region.[19] Functional transplantation study demonstrated a dramatic expansion of the imHSC pool during these maturation steps, from 5 cells at early E10 to 50 cells by late E10.5.[57] The majority (85%) of HSCs in the E11.0 and E11.5 AGM were lymphoid-biased γ-subtype and the remaining 15% were balanced β-subtype.[58] However, clonal fate-mapping experiments estimated that 600–700 HSC clones in mice and ~30 HSC clones in zebrafish contribute to life-long hematopoiesis.[59–61] This suggests that the numbers of imHSCs are significantly under-estimated from previous studies using functional assays. More effective assays are urgently needed in order to accurately examine the numbers of functional imHSCs.
In addition, colony-forming HPCs and functional HSCs have been identified within the vascular labyrinth region of mice at E9.5 and E10.5–11.0 in humans. HSCs in the placenta are expand by ~20-fold between E11.5–12.5, diminish by E13.5, and completely disappear by E15.5.[62–66] These studies suggest that the placenta serves as a major niche for HSC expansion. The emergence of HSCs in the placental vasculature can be detected in the absence of circulation, suggesting a de novo source of hematopoietic generation that is independent of YS and AGM hematopoiesis.[62, 64, 67] Placental HSCs colonize the FL on E11.5 and their diminishment in the placenta is coincides with FL HSC expansion. The emergence of placental HSCs during early embryonic development (~6 PCW) in humans suggests that this mechanism is developmentally conserved. Placental HSCs possess the classical CD34medc-kithi fetal HSC surface phenotype, similar to FL HSCs. However, it remains unknown what contribution placenta-derived HSCs make to the adult HSC pool.
The role of YS HPCs to fetal, infant and adult hematopoiesis.
Pri-EPs generated in YS at E7.5 are expanded and maintained there until E10.5. These pri-EPs produce nucleated RBCs and highly proliferative Mks for platelets that support embryonic growth up until E15.5.[53, 54, 68] In addition, pri-EPs also produce primitive Mφ progenitors starting at E7.5 in the YS. After the onset of circulation, primitive Mφ progenitors predominantly migrate into the brain for self-renewal and lifetime maintenance of microglial cells in a PU.1 and Irf8-dependent manner (Figure 1A).[18, 69, 70] Thus, before the 2nd wave of YS hematopoiesis was identified, it was believed that YS hematopoiesis was transient and only supported fetal hematopoiesis for short time in order to bridge the gap to HSC activity. However, after the identification of 2nd wave YS hematopoiesis, recent studies demonstrated that EMPs and LMPs generated in the YS can support hematopoiesis and immunity for embryonic development at least up-to birth. Fate-mapping studies demonstrated that many types of EMP- and LMP-derived cells can be sustained even in adult tissues over the long term.[4, 46, 71] Figure 1B.
Figure 1. The contribution of 2 waves of YS hematopoiesis to embryonic and postnatal blood/immune cells.
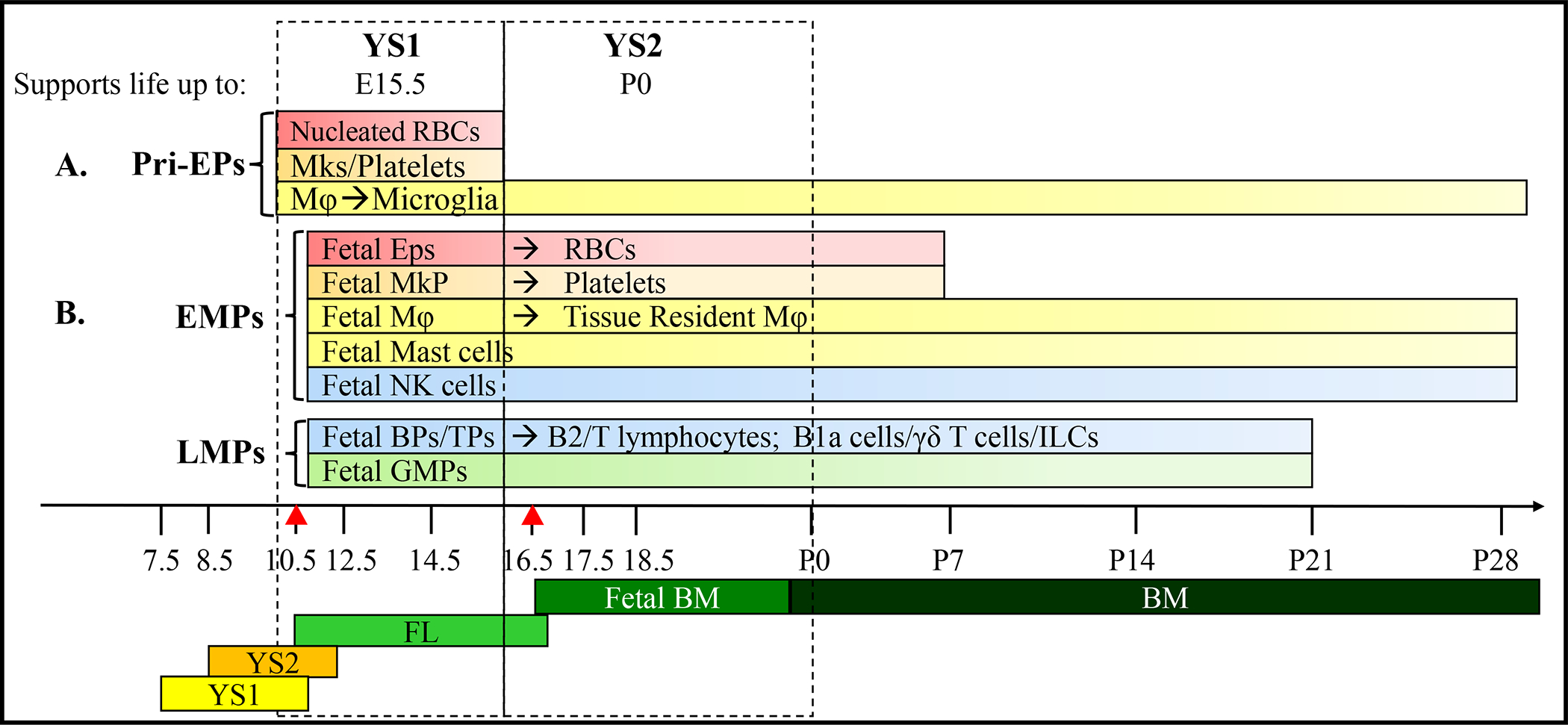
A. The 1st wave of YS hematopoiesis (YS1) is initiated on E7.5. pri-HSCs generated in this wave primarily produce nucleated RBCs and Mks/platelets to support embryonic survival up to E15.5. In addition, Mφ produced in this wave generate microglia in the brain for the remainder of life. B. The 2nd wave of YS hematopoiesis (YS2) starts on E8.5. EMPs and LMPs generated in this wave migrate to FL and then fetal BM to produce RBCs, platelets, innate immune cells and granulocytes. This wave of hematopoiesis can support the survival of the embryo up until birth. In addition, Mφ and innate immune cells infiltrate the embryo’s tissues and renew themselves, maintaining tissue resident Mφ and immune cells for the remainder of life.
Shortly after generation in YS, both EMPs and LMPs colonize the FL starting on E10.5, which is earlier than that of AGM imHSCs (starting on E11.5, see in below).[57] Therefore, it is likely that EMPs and LMPs establish the first wave of hematopoiesis in the FL.[72] EMPs are also increased 3-fold from E10.5 to E11.5, at which time they are 25 times more numerous in the FL than in E8.5 YS.[73] The early stages of FL hematopoiesis are dominated by the erythroid lineage, while lymphoid and myeloid lineages are increased at later stages.[74] At E13.5, the largest FL population is nucleated erythrocytes, but after E14.5 more mature enucleated erythroid cells are found within the vessels.[75] Most of the hematopoietic cells express the pan-hematopoietic marker CD45; however, primitive erythroid cells at this stage of FL development are CD45−.[76] A recent study demonstrated that during FL hematopoiesis, a large population of erythroid progenitors (EPs) are produced by EMPs, which comprise >70% of CD45−Ter119− cells (>10% of FL cells) at E14.5. Similar to observations in mice, EPs in human FL also do not express CD45 at stages ranging from 7 to 17 weeks after conception.[35] The FL also provides a unique microenvironment for the expansion of EMP-produced megakaryocytic progenitors (MkPs).[77] This study suggests that AGM-derived HSCs do not contribute to erythropoiesis or megakaryopoiesis up until birth, but that these processes are primarily dependent upon EMP-derived EPs and MkPs. Mechanistically, it was found that EMP-derived EPs require 10-fold lower concentrations of erythropoietin, related to the low EPO microenvironment of the FL, than AGM-derived HSC counterparts for efficient erythrocyte production. As a consequence, EMP-derived EPs efficiently outcompete AGM HSC progeny and sustain erythropoiesis throughout murine embryonic life.[78, 79]
LMPs have been identified in FL at E10.5, expand during E11.5-E14.5 in FL, and colonize fetal BM on E16.5.[24] Such cells phenotypically and functionally resemble the drHSCs defined by Forsberg’s lab. LMPs/drHSCs contribute to tissue-residual innate immune cells such as B1a cells (primarily inhabiting the peritoneal and pleural cavities), epidermal γδT cells (predominantly expressing T cell receptor with a Vγ5Vδ1 allele), NK cells, innate lymphoid cells (including ILCs 1, 2, and 3) and mast cells during later embryonic development as well as for a short time after birth. Consistently, drHSCs can still be detected in BM 2 weeks after birth. A serial transplantation study demonstrated that drHSCs are capable of self-renewal and long-term multi-lineage reconstitutive capacity with a lymphoid bias. In addition, drHSCs can produce B1a and γδT lymphocytes. The contribution of EMPs to tissue-residual innate immune cells such as B1a cells, epidermal γδT cells (predominantly express T cell receptor with a Vγ5Vδ1 allele), NK cells, ILCs and mast cells is still being debated. [1, 8, 79, 80]
Pre-Mφ are present in YS from E8.5, where they proliferate and then access to bloodstream to migrate towards the embryonic tissues, peaking around E10.5. The traffic of such cells is dramatically decreased after E12.5 and disappears after E14.5.[81] In embryonic tissues, the pre-Mφs produce an M2-like non-inflammatory phenotype of tissue-resident Mφs. Such Mφs not only help to clear up the dead cells but also produce cytokines to stimulate tissue growth. During embryonic development, YS-derived Mφs play a critical role in the emergence of AGM HSCs and the organogenesis of many embryonic organs including kidney, testis, myocardial vascular network, endocardial valvular tissues, coronary and skull.[15, 17, 20, 82–86] Genetic depletion of YS-derived Mφs in the early embryo impairs timely hematopoietic colonization and morphogenesis of many organs. Most importantly, such tissue-resident Mφs have self-renewal capacity which can persist in the adult tissues throughout life, such as Kupffer cells in liver, Langerhans cells in the epidermis, alveolar Mφ in the lungs, and microglia in the brain.[28, 53, 69, 70, 73] These tissue-resident Mφs not only mediate immune responses but are also involved in wound healing and tissue repair.
In summary, the major functions of these two early waves of YS hematopoiesis are to produce: 1) circulating RBCs for gas exchange during the early and later embryonic development;[79] 2) platelets to maintain hemostasis; 3) Mφ to clean up apoptotic cells for tissue/organ generation and remodeling; 4) inflammatory signaling for definitive HSC emergence[80, 82]; and 5) tissue-resident Mφ in adults for tissue regeneration and repair. Although pri-EPs and EMPs (as well as their progeny) can be detected in adult BM tissue (7–20%, dependent the techniques used),[71, 73, 83] future studies will need to determine if and how these varieties of HPCs produced in the YS contribute to neonatal and adult hematopoietic systems.
The role of AGM imHSCs in fetal, infant and adult hematopoiesis.
Spatiotemporal analysis and genetic fate mapping of developmental HSCs demonstrate that adult BM hematopoiesis is primarily derived from imHSCs generated within the AGM.[4, 46] imHSCs do not have long-term multi-lineage hematopoietic reconstitutive capacity.[52, 87] After generation, imHSCs first colonize the FL and then the spleen and BM, where these cells undergo several maturation changes until they become quiescent HSCs in the adult BM niche 3–4 weeks after birth. Figure 2.
Figure 2. Maturation of AGM hematopoiesis during embryonic, postnatal and adult life.
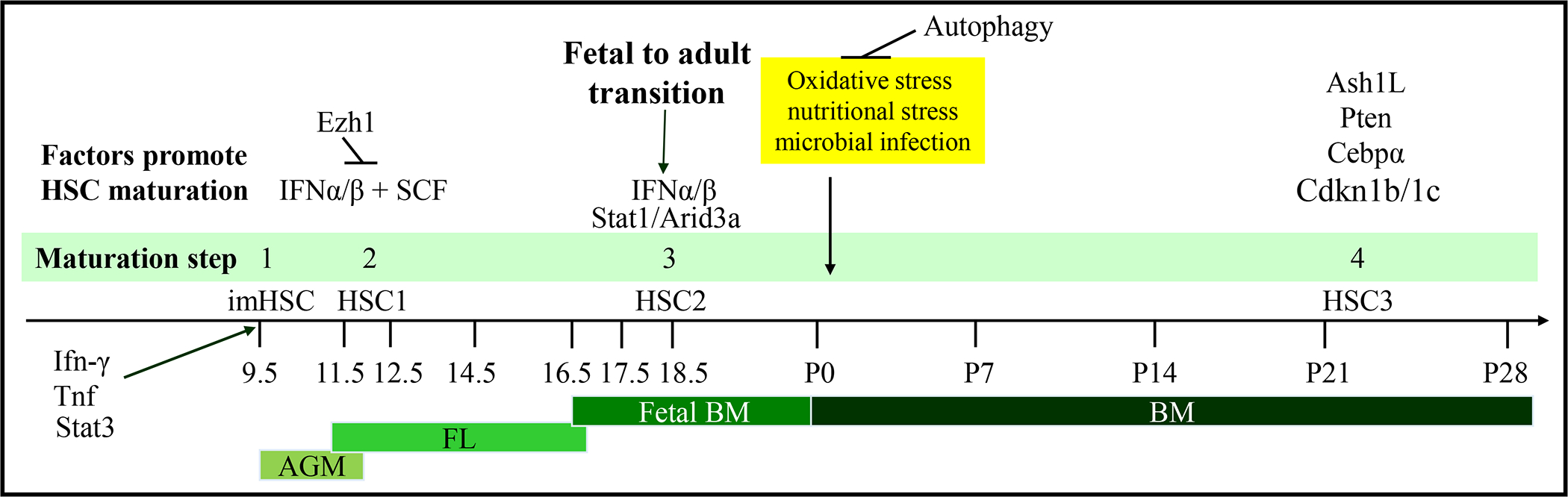
Definitive hematopoiesis is initiated in the AGM region at E9.5. imHSCs generated at this wave of hematopoiesis migrate to FL at E10.5, to fetal BM at E16.5, and finally are maintained in adult BM for lifetime hematopoiesis. During this process, imHSCs undergo multiple steps of maturation. First, inflammatory cytokines such as IFN and the hematopoietic cytokine SCF stimulate imHSCs to develop self-renewal and hematopoietic engraftment capacities to become functional HSCs (HSC1) in FL. HSC1 proliferate and expand in FL before colonizing fetal bone marrow. Starting at E18.5, stimulated by IFNs, HSC1 in BM gradually transition to adult HSCs (HSC2) by losing their fetal features and gaining adult HSC characteristics. The full fetal-to-adult maturation of HSCs is completed at approximately 3 weeks postnatal, at which time the majority of HSCs become quiescent in the cell cycle. Ash1L, Pten and Cebpα might be responsible for this quiescent state through regulation Cdkn1b and Cdkn1c expression, which are negative regulators of the cell cycle. Although the major environmental challenges of HSCs occur immediately after birth, HSCs are protected from such challenges by autophagy.
The first step of HSC maturation is the imHSC-to-functional HSC transformation which occurs in FL between E11.5-E12.5 to obtain long-term multi-lineage hematopoietic reconstitutive capacity. ImHSCs colonize FL at E11.5 (~5–6 PCW in humans). Fully functional HSCs can be detected at E12.5 (6 PCW for human) in FL.[57] Transplantation study demonstrated that the number of HSCs is expanded ~38 times from their original number (from E11.5-E14.5), peaking at around E14 for mice and E140 for humans. The phenotypes of HSCs in the FL are Lin−Sca-1+Mac-1lowCD201+ on E12 and CD45+CD150+CD48− CD201+ after E14.[8, 84][85] The 2nd step of HSC maturation is a gradual and uncoordinated epigenetic and transcriptional reprogramming to obtain most adult HSC properties which starts on E18.5, shortly after fetal BM colonization. [15] The 3rd step of HSC maturation is to acquire cell cycle quiescence 3–4 weeks after birth, which coincides with diminishment of fetal-type hematopoietic properties.[86, 88, 89]
The generation and maturation process of HSCs during embryonic and neonatal development are primarily stimulated by inflammatory cytokines, which is correlated with niche environmental changes. Interferon (IFN)-γ, interleukin (IL)-3, and tumor necrosis factor produced by Mφ are important for imHSC emergence in the AGM.[80, 82, 90–93] Together with Scf-Kit signaling, IFNα is required for the imHSC-to-HSC transformation event during early FL hematopoiesis, the 1st step in HSC maturation.[90, 92] At E18.5, IFNα and IFNβ produced by epidermal Mφs stimulate the 2nd stage of maturation by promoting the differentiation of HSCs and expansion of HPCs.[15] The 3rd step in HSC maturation might be controlled by Cebpα and Ash1-mediated transcriptional and epigenetic programs and regulated by the Pten-Akt-mTor signaling pathway.[86, 88, 89] This signaling promotes cell cycle quiescence of HSCs and may be initiated by chronic type I IFN stimulation.[94–96] Interestingly, such maturation steps do not always correlate to micro-environmental changes. For example, the most significant environmental challenge that HSCs face are oxidative stress, nutritional stress, and microbial infection immediately after birth. Such stresses induce shifts in glucose and glutamine metabolism, mitochondrial activation, and redox production in HSCs. HSCs are protected from such stresses by Atg5-dependent autophagy during the first 5–7 days after birth, but are adapted to such challenges by P7.[97, 98]
Transition of HSC niche from AGM, FL to fetal BM.
Specific signals promoting the chemoattraction of EMPs, LMPs and imHSCs to FL are still unclear. It most likely that EMPs, LMPs and imHSCs circulate in the peripheral blood (PB) and are passively retained in FL through adhesive factor–mediated cell-extracellular matrix (ECM) and cell-cell contacts, including a number of integrins (such as β1-integrin CD29 and α2b-integrin CD41),[99, 100] VE-Cadherin/CD144, glycosylphosphatidylinositol-anchored surface protein-GPI-80, together with ItgaM-CD11b.[101–103] Seminal studies demonstrated that HPCs lacking β1-integrin were unable to colonize the FL[99, 104], while the chemokine C-X-C Motif Chemokine Ligand 12 (CXCL12) produced by FL ECs is an important factor for retaining HSCs in FL, but not for its initial colonization.[105, 106]
FL provides the first niche for HSC maturation and expansion.[2, 3, 5, 107, 108] In FL, several types of cells have been proposed to directly contact with HSCs and promote HSC maturation and expansion; these include desmin+ stellate cells, DLK1+ or CD3+DLK1+SCF+ hepatocyte progenitors.[105, 109] Such cells promote HSC expansion and maturation by producing several supportive hematopoietic cytokines such as KIT ligand (KITL), THPO, α-fetoprotein, insulin growth factor 2 (IGF2), angiopoietin-like 2 (Angptl2) and Angpt3 [105, 110–113], extracellular matrix proteins including vascular cell adhesion protein 1, fibronectin1, vitronectin1, Laminin-b1 and Laminin-c1,[114] as well as an essential fatty acid docosahexaenoic acid.[115] Detailed imaging studies suggested that EPCR+ HSCs in FL are localized around the Lyve-1+ sinusoidal network,[116] while CD150+CD48−Sca1+CD11b+CD45+ CD41−Lin− DAPI− HSCs[84] are found closely associated with the Nestin+NG2+ pericytes of portal vessels, suggesting that both the peri-sinusoidal network and peri-arterial stromal cells might be niches for FL HSCs.[117, 118] Nestin+NG2+ mesenchymal stromal cells are required for HSC expansion by producing KITL, Angptl2, and IGF2 and other HSC factors. Selective elimination of Nestin+NG2+ pericytes leads to a modest reduction in HSCs numbers and HSC proliferative.[117] In addition, DLK+ hepatoblasts produce KITL, FLT3 ligand, IGF2, Angpt3, Angptl2, Wnt family growth factors, Ephrin2a, CSF1, EPO, CXCL12, and IL-6. DLK+ hepatoblasts can support HSC expansion for ~20-fold within 3 weeks of co-culture [119]. Moreover, p75NTR+ FL hepatic stellate cells express a range of hematopoietic cytokines, such as CSF1, IGF2, THPO, KITL, EPO, IGF1, IL-1, FLT3 ligand and Oncostatin M.[110] Furthermore, bile acids, the active components of bile from the maternal and FL hepatocytes, contribute to FL HSC expansion through repressing unfolded/misfolded protein accumulation and inhibiting endoplasmic reticulum stress.[120] A most recent study demonstrated that KITL is primarily expressed by hepatic stellate and endothelial cells which create perisinusoidal vascular niche for HSCs in FL. Specific deletion of KITL from both endothelial and hepatic stellate cells in FL results in nearly complete loss of HSCs [121]. A summary of HSC niche cells at various developmental time points, as well as the factors each secretes, can be found in Table 2.
Table 2.
HSC niches during development
| Development Stage | Niche Cells | Hematopoietic Supporting Factors |
|---|---|---|
| AGM | Mesenchymal Cells Mφ |
Notch1 and Wnt, IFN-γ, IL-3, TNF, IFNα and IFNβ |
| FL | Endothelial cells[121, 217] Desmin+ Stellate Cells[114, 121] p75NTR+ Stellate Cells[110] Dlk1+ or CD3+DLK1+SCF+ Hepatocyte Progenitors[105, 109, 111, 112, 120, 121] Lyve-1+ Sinusoidal Network Nestin+NG2+ Pericytes of Portal vessels[117] Neutrophils |
CXCL12, KITL, WNT5A IL-3, IL-6, CSFs and EPO CSF1, IGF2, THCO, KITL, EPO, IGF1, IL-1, FLT3 Ligand and Oncostatin M KITL, IGF2, Angpt2, Angpt3, THPO, CXCL12, α-fetoprotein, bile acids, VCAM1, Fibronectin1, Vitronectin1, Laminin-b1 and Laminin-c1, DHA Undefined factor(s) maintain slow cycling state of HSCs KITL, Angptl2, and IGF2 MMP9 for HSCs release from FL |
| Fetal BM | Osterix+ Stromal Progenitors[128–130] | CXCL12, Osteopontin, VEGF, Slit, and SCF, Collagen, N-cadherin, VCAM, Selectins, Fibronectin, Integrins |
Fetal BM colonization of HSCs and HPCs occurs around at E16.5–17.5 for mice and 10–12 PCW for human and this becomes the dominant site of hematopoiesis going forward.[122] The HSC migration from FL to the BM is mediated by different cytokines, ECM signals and adhesion molecules (e.g., CXCL12, SCF, cadherins, integrins).[123] MMP9 produced by neutrophils promotes the release of HSCs from their FL niche by remodeling ECM.[124] And then HSCs migrate to fetal BM niche in response to the chemotaxis of several key BM environmental signals including CXCL12, VEGF, Slit, and SCF. After reaching fetal BM, HSCs are retained within the niche by several adhesive matrix proteins such as collagen, N-cadherin, VCAM, selectins and fibronectin.[106, 123, 125] The adhesive niche factors promote HSC survival by stimulating survival signaling such as the Wave-2/c-Abl pathway.[126, 127] Osterix+ primitive and definitive stromal progenitors play critical roles in BM niche formation during BM development.[128] Osx−/− fetal BM fails to generate the HSC niche due to defects in endochondral ossification, osteolineage stromal cells, and osteopontin, secreting which play important roles in the regulation of HSC quiescence and homing.[129–131]
The transition of fetal-to-adult blood cells.
The fetal-to-adult transition of HSCs/HPCs is evident by the transition of mature blood and immune cells. Significant differences are observed between fetal and adult blood and immune cells.
Fetal-to-adult B cell transition.
The major changes of B lymphopoiesis during the fetal-to-adult transition include the loss of potential of CD5+ B1a B cell production, the gain of IL-7/thymic stromal-derived lymphopoietin (TSLP)-dependent IgDhigh B2 cell growth, and fetal-to-adult B cell receptor shift.[1, 132–134] B-1a cells are innate immune cells which reside in the peritoneal and pleural cavities. B-2 cells mediate a canonical adaptive immune by producing antibodies in a T cell-dependent fashion, while B-1a cells execute important roles in the first line of defense against microbial infection by producing IgM type of stereotypic natural antibodies in a T cell-independent manner.[135–137] Such B1 to B2 type shift of the B lymphopoiesis is associated with the fetal to adult type shift of B cell progenitors.[138]
In mice, although the progenitors for B-1 B cells are still debated, it is accepted that B-1a cells are primarily produced by fetal types of HSCs/HPCs. Different from the B cell progenitor cells derived from AGM HSCs/HPCs in FL which primarily produce B-2 lymphocytes [88], the B progenitor cells derived from YS HSCs/HPCs in FL have a strong tendency toward differentiation into B-1a B lymphocytes.[9, 43, 139] It was suggested that YS LMPs produce AA4.1+CD19+B220lo/- B-1 cell progenitor cells peak in the FL. Such progenitor cells are maintained in the BM and spleen during fetal development and the first 2 weeks after birth, and then diminish in adulthood.[9, 10, 140–142] The dynamic changes in B-1 progenitor cells is closely associated with the dynamic change of drHSCs.[1, 132] The development of B-1 cells is IL-7/TSLP-independent, while the development of B2 cells absolutely requires IL-7/TSLP. This might explain the IL-7/TSLP-independent B lymphopoiesis during fetal and 2 weeks after birth, as observed in knockout mouse studies [143, 144].
In humans, just as is observed in mice, fetal HSCs/HPCs have the potential to produce B-1 B-cells, which might be associated with the fetal types of B cell progenitors. Transcriptomic data suggests that an LMP equivalent is also present in human YS.[35] In addition to adult types of B cell progenitors such as CD34+CD19+CD10+ ProB progenitors, two fetal types of B cell progenitors, CD34+IL7R+CD19−CD10− oligo-potent early lymphoid progenitors (ELPs) and CD34+CD19+CD10− uni-lineage pre-ProB, were identified in FL and fetal BM. [145–149] Biologically ELPs are potentially analogous cells of human LMPs.[148–150] Functional study demonstrated that ELPs are upstream of pro-preB cells, which then give rise to proB cells. The onset of B cell lymphopoiesis in FL was demonstrated by the detection of ELPs at 6 PCW, as well as pre-proB and proB progenitors at 7 PCW. Pre-proB and proB cells account for ~2.5% and ~8% of FL CD34+ cells, respectively, at 7 PCW. Both progenitors are significantly expanded in fetal BM. Approximately 20% and 11% of fetal BM CD34+ cells are pre-proB-progenitors and proB-progenitors, respectively, at 11 PCW. ProB-progenitors are expanded further to >30% of CD34+ cells in fetal BM during the latter part of the second trimester. In adult BM, proB- account for ~14% CD34+ cells. However, both fetal-types of B progenitors ELP and pre-proB can be detected in cord blood and early postnatal but are diminished during the postnatal-to-adult transition.[146, 148] Single cell RNAseq study suggests the presence of IL7R+ ELPs in human YS [35], however whether ELPs and pre-pro-B cells are originated from YS or AGM need to be determined.
Fetal-to-adult T cell transition.
The major change in T lymphopoiesis during the fetal-to-adult transition is the gain of adaptive immune responsiveness. Fetal and neonatal T cells exhibit innate-like functions. In contrast to adult T cells which express a higher avidity TCR (T cell receptor) and produce more conventional cytokines (IFN-γ or IL-4 and IL-13) after clonal proliferation and differentiation, naive T cells from the fetus/neonate express Toll-like receptors, complement receptors, and NK cell receptors, as well as a more peptide-promiscuous TCR (such as dermal γδTCR). Following TCR stimulation, fetal/neonatal T cells rapidly produce IL-8, which is consistent with their more broadly reactive nature. Compared to adult T lymphocytes, fetal CD4+ T cells exhibit enhanced proliferation, and preferentially become regulatory T cells (Tregs) or a Th2 phenotype in response to foreign antigen induction. As a result, fetal CD4+ T cells are immune tolerant.[151, 152] Fetal CD8+ T cells exhibit an enhanced capacity to proliferate and give rise to more short-lived effectors after infection, whereas adult CD8+ T cells respond with slower kinetics but give rise to more memory CD8+ T cells.[153] Studies demonstrated that fetal and adult T cells are distinct populations that arise from separate populations of HPCs that are present at different stages of development.[153] In addition, neonatal T cells are transcriptionally shifted to a more effector-like state early in life by developmentally regulated miRNAs.[154] Expression analyses reveal heterogeneous responsiveness of fetal lymphoid progenitors to Notch signaling [155, 156].
Transition of fetal to adult megakaryopoiesis.
Mks derived from EMPs form colonies more rapidly than those derived from adult BM, and mature more rapidly than adult-derived Mks.[22, 157] Compared to their adult counterparts, neonatal MkPs are hyperproliferative and generate 10-fold more Mks. In addition, neonatal Mks show a ten-fold higher proliferative rate compared to adult Mks. Due to the unique uncoupling of proliferation, polyploidization, and cytoplasmic maturation in neonatal Mks, large numbers of fetal/neonatal Mks are smaller and have lower ploidy than their adult counterparts, thus producing fewer pre-platelets than adult Mks.[158] Although relatively high THPO plasma levels are detected in fetuses and neonates compared to adults, the differences between neonatal and adult Mks are primarily due to the intrinsic differences of fetal and adult MkPs. Compared to adult MkPs, neonatal MkPs are more sensitive to THPO in vitro. Thus, the rapid proliferation of MkPs and Mks during fetal and neonatal phases might compensate for the lower production of MkPs from HSPCs. Interestingly, newborn mice are less responsive to THPO-receptor-agonist treatment compared to adult mice in vivo, as evidenced by the observation that neonatal Mks increase in numbers but not in size.[159] Neonatal platelets are hyporeactive to platelet agonists such as collagen, thrombin, adenosine 5′-diphosphate and thromboxane due to enhanced primary hemostasis, such as higher hematocrit values, higher von Willebrand factor (vWF) concentrations, and a predominance of ultra-long vWF polymers. [160, 161] In addition, fetal platelets have low levels of integrin-activating proteins and P-selectin (CD62P), and do not readily associate with neutrophils in vitro and in vivo.[162] The developmental alteration of MkPs and Mks from FL to early postnatal BM indicate a graded influence of ontogenic stage. Such ontogenic changes of MkPs-Mks-platelets are regulated by the dynamic alteration of Lin28b-Let-7b which negatively regulates the expression of platelet P-selectin.[163] Most importantly, such ontogenic features of neonatal HPCs and MkPs play a critical role in the pathogenesis of several fetal Mk disorders, including a transient myeloproliferative disorder (TMD) with Down Syndrome (DS) and Gata-1 mutations as well as acute megakaryocytic type (AMkL) with or without DS.
Transition of fetal to adult erythropoiesis.
Compared to adult HSCs/HPCs, FL HSCs/HPCs display erythroid bias as demonstrated by producing a significantly higher ratio of megakaryocyte-erythroid progenitors/granulocyte-monocyte progenitors and erythroid progenitors/granulocyte progenitors.[164] Such differentiation bias is primarily due to the HSC intrinsic property with minimal contribution from microenvironments. In addition, the proliferation of FL EPs is significantly higher than that of adult EPs. However, FL EPs exhibited delayed differentiation kinetics compared to adult EPs. As a consequence, fetal EPs produce increased number of both burst-forming units and colony-forming units [11]. In addition, there are 3 major differences between fetal and adult mature red blood cells (RBCs)[11]: 1) Fetal RBCs express γ-globin throughout fetal life and are silenced after birth due to the 2nd wave of globin switching (γ to β). About 98% of hemoglobin (Hb) in fetal RBCs is HbF (α2γ2 globins), while about 95% of Hb in adults is HbA (α2β2 globins); 2) Fetal PB contains a large number of nucleated RBCs, which are rare in adult PB;[165] and 3) Compared to adult RBCs, the volume of fetal and neonatal RBC was 21% larger.[166]
Distinct regulatory machinery of fetal and adult HSCs.
The different requirements of genes for the self-renewal, survival and differentiation of fetal and adult HSCs indicates that they are regulated by distinct mechanisms. Studies suggested that the Lin28b-Let7 axis and transcription factor Sox17 are selectively required for the function and identities of fetal HSCs, while Gfi-1, Tel/Etv6 and Bmi-1 are selectively required for the survival and self-renewal of adult HSCs. Lin28b and Sox17 are highly expressed in fetal HSCs and are repressed in adult HSCs. Mice with Tie2-Cre mediated Sox17 deletion in ECs die at E12.5 due to the lack of the 2nd and 3rd waves of hematopoiesis, suggesting that Sox17 is required for the generation of these 2 waves of hematopoiesis. Mice with polyI:C-induced Sox17 deletion at postnatal day 2 (P2), P4 and P6 die due to BM failure within 15 days; however, adult mice with induced deletion of Sox17 are completely normal.[167] Lin28b overexpression in adult BM HSCs induces fetal HSC identities as demonstrated by the reactivation of γ fetal globin genes in RBCs and ability to generate B1a and innate-like γδ T cells.[6] Lin28b plays such a role by repressing Let7 to de-repress an array of oncogenes including Myc, Ras, Blimp1, Arid3A and Hmga2.[6, 7, 35, 138, 168, 169] These studies suggest that Lin28b and Sox17 may act as master regulators of developmentally-timed changes in HSC programs and HSC self-renewal potential.[167] However, the knockout of many other genes selectively cause hematopoietic defects in certain stages of development. Reorganizing these genes based on developmental stages provides explanations for the gene requirements of each hematopoietic wave.
The 1st wave of hematopoiesis initiates directly from YS mesoderm. During the 2nd (YS) and 3rd waves (AGM) of hematopoiesis, HPCs are produced by specialized ECs called hemangioblasts through a program called the endothelial-to-hematopoietic transition. After generation, HPCs undergo maturation and expansion to generate blood cells and immune cells to fulfill the demands of development. The HPCs generated in the 1st and 2nd waves primarily provide blood support from E10.5-E15.5 and E15.5-P0, respectively, while HPCs generated in the 3 wave provide blood and immune cell supports for all of postnatal life. The ontogeny of HPCs produced in all 3 waves is controlled by dynamic transcriptional and epigenetic landscapes and is stimulated by microenvironment signaling.[170–172] For example, it was found that regulation of the epigenetic landscape by determination of topologically-associating domain boundaries is controlled by transcription factors TCF3 and MAFB in fetal HSCs, but by transcription factors NR4A1 and GATA3 in adult HSCs.[173] Thus, the genes required for proliferation, differentiation and survival of the 3 waves of HSCs/HPCs might not be the same. One can determine the genes that are required for each wave of hematopoiesis through various Cre-mediated knockout systems targeting different developmental stages.
Sox2Cre mediates gene deletion specifically in the epiblast; VECCre and Tie2Cre mediate gene deletion in hemogenic ECs and hemogenic precursors; VavCre mediates gene deletion in almost all HSCs/HPCs in YS, AGM and FL. Mx1Cre mediates gene deletion in HSCs and HPCs as well as hepatocytes any time after polyI:C induction, while Rosa26CreERT mediates gene deletion in all tissue cells any time after 4-hydroxytamoxifen induction. By crossing conditional knock-out animals with such Cre lines, the roles of many genes in HPC generation in YS and AGM, HPC maturation, proliferation, differentiation and survival in FL and BM have been studied. In Figure 3, mice with either germline or HEC deletion of any of the genes in group 1, including Scl1, Runx1, Ezh2, Chd1, β-catenin, Notch1, Gata2 or Sox17, failed to generate both the 2nd and 3rd waves of hematopoiesis and died before E15.5, suggesting that such genes are required for the endothelial-to-hematopoietic transition.[174] Mice with VavCre-mediated deletion of Gata2, Meis1, Myb, Myc, Pu.1, Erg, or Lis1 (Group 2) died between E12.5-E16.5 due to impaired FL hematopoiesis, suggesting that these genes are required for maturation, proliferation, differentiation survival or self-renewal/maintenance of HSCs/HPCs from both the 2nd and 3rd waves of hematopoiesis. Mice with VavCre-mediated deletion of Moz, Mof, Satb1, Rae28, Mel18, Eed, Suz12, Prdm16 or Lsd1 (Group 3) died perinatally or shortly after birth due to impaired fetal BM hematopoiesis. Although FL hematopoiesis can sufficiently support embryonic development in such genetic knock-out mice, HSCs in FL have functional defects, suggesting that these genes are required for maturation, proliferation, differentiation survival and/or self-renewal/maintenance of the HSCs that are generated from the 3rd wave of hematopoiesis. Interestingly, all Group 1 genes (except for Gata2 which also belongs to Group 2) are not required for the expansion and maintenance of either FL or adult BM HSCs and HPCs. All Group 2 genes are required for the proliferation, self-renewal, and maintenance of fetal HPCs and adult HSCs, while all group 3 genes are required for the self-renewal and maintenance of adult HSCs. In addition, only Atg5 and Sox17 have been assessed by inducing deletion immediately after birth. Although neither of these genes is required in adult HSCs, induced deletion of either Atg5 or Sox17 between P0-P7 causes severe defects in HSCs. It was suggested that Atg5-mediated autophagy is required to protect HSCs from stresses during early postnatal life, including oxidative stress, nutritional stress, and microbial infection. Future study will be needed to determine whether Sox17 is also required for the proliferation, self-renewal, and maintenance of fetal HPCs by using the VavCre -mediated deletion method.
Figure 3. Genes required for the generation, maturation and maintenance of HSCs as demonstrated by knock-out mouse studies.
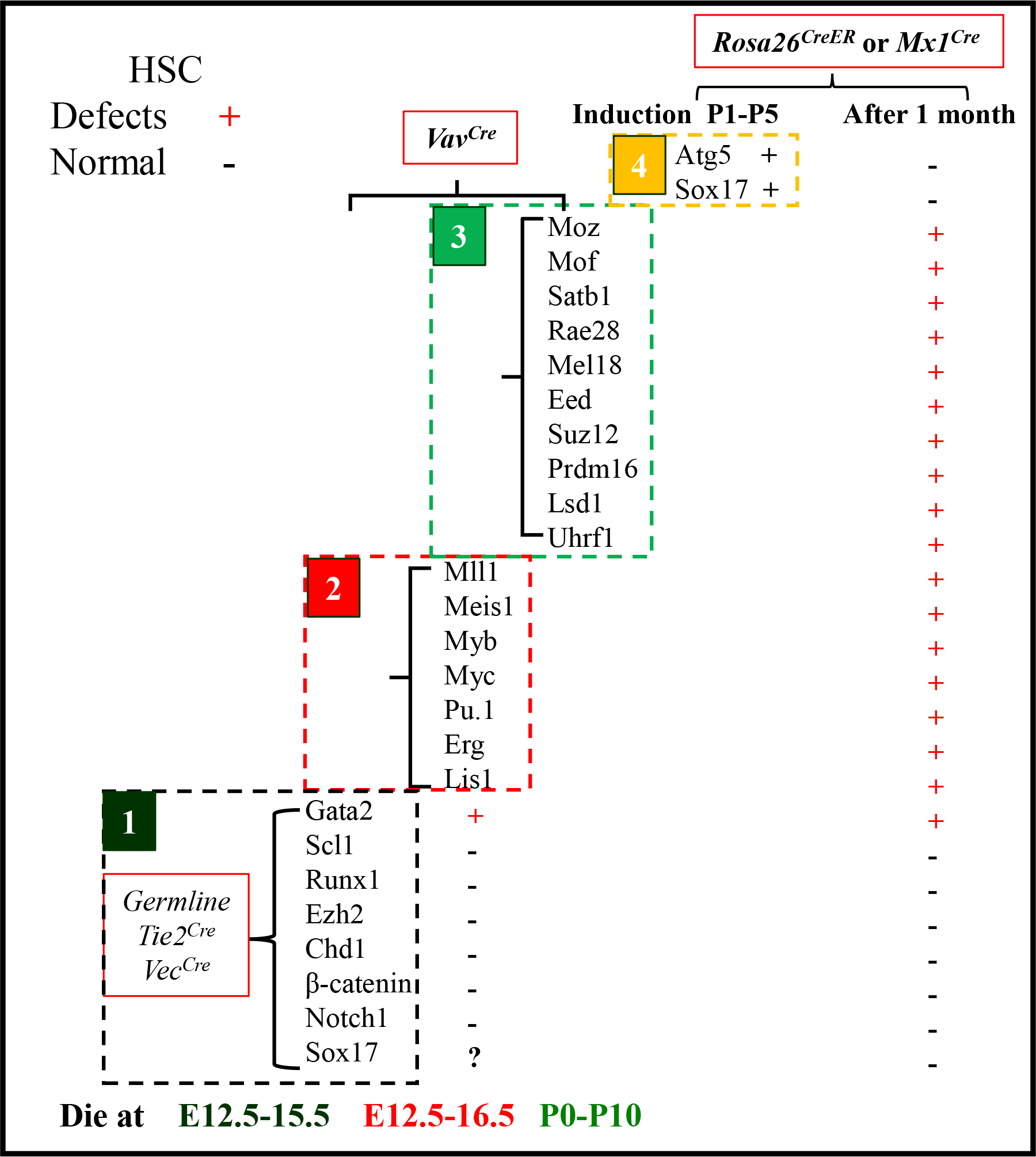
VECCre-mediated gene deletion in hemogenic ECs. Tie2Cre mediates gene deletion in hemogenic precursors of hematopoietic and endothelial lineages including the majority of ECs and a subset of HCs. VavCre mediates gene deletion in FL HSCs/HPCs. Mx1Cre mediates gene deletion in HSCs/HPCs and hepatocytes and time post polyI:C induction. Rosa26Cre-ERT mediates gene deletion in all types of tissue cells after 4-hydroxytamoxifen induction. Genes in Group 1 are not necessary for the generation of the 1st wave of YS hematopoiesis but are required for the generation of the 2nd wave of YS hematopoiesis and AGM hematopoiesis. Genes in Group 2 are not necessary for hematopoietic initiation but are required for maturation, proliferation, differentiation survival and/or self-renewal/maintenance of HSCs/HPCs from both the 2nd and 3rd waves of hematopoiesis. Genes in Group 3 are only required for the maturation, proliferation, differentiation, survival, and/or self-renewal/maintenance of the functional HSCs that are generated from the 3rd wave of hematopoiesis. (+) indicates severe defects in HSCs/HPCs after deletion of the indicated gene. (−) indicates normal hematopoiesis after deletion of the indicated gene.
5. Fetal origin of pediatric leukemia.
Pediatric leukemia presents before 14 years of age. Based on age of onset, pediatric leukemia can be further divided into neonatal leukemia (< 1 month) infant leukemia (1 month to 1 year), and childhood leukemia (1 to 14 years). Studies suggests that the leukemic subtypes and characteristics among neonates, infants and adults are associated with the ontogenic changes of HSCs and HPCs.[13, 14, 175] For example, the high incidence of AMkL and B-ALL in fetal stages might be associated with the transient existence of platelet-biased HSCs and lymphoid-biased drHSCs, respectively. Convincing evidence demonstrated that most pediatric leukemias originate before birth. However, it has not been determined from which of the 3 waves of hematopoiesis these leukemic cells originate. The unique features of pediatric leukemia suggest that at least some may originate from the earlier YS hematopoiesis rather than the 3rd wave of AGM hematopoiesis.[149, 176–185] Understanding the ontology of fetal hematopoiesis is of particular interest in understanding the pathogenesis of childhood blood disorders.
Distinct genetic abnormalities of pediatric and adult leukemia due to the different response of fetal/neonatal HSCs/HPCs and adult HSCs/HPCs to leukemic oncogenes.
As opposed to leukemia in adults and elderly patients, which are dominated by acute myeloid leukemia (AML), childhood leukemias are primarily of the acute lymphoblastic type (ALL > 85%) and AMkL with fewer AML cases.[186] Among ALL cases, ~85% are B cell ALL (B-ALL) and 15% are T cell ALL (T-ALL). The peak incidence of B-ALL is 2–6 years of age, while T-ALL and AML show a relatively constant incidence throughout childhood.
The genetic abnormalities in pediatric leukemia are different from those in adult leukemia. In ALL, Bcr-Abl and Ph-like mutations are most commonly detected in adults while hyperdiploidy and several chromosomal translocations, including t(12;21), t(1;19) and t(17;19), are most common in childhood. Such translocations lead to fusions of ETV6-RUNX1, TCF3-PBX1 and TCF3-HLF genes respectively. In AML, mixed lineage leukemia (MLL) gene rearrangements (MLL-r) and GATA1 mutations (such as GATA1s mutation) are common in infant and early childhood but rare in adults[187–189], while mutations in FLT3, NPM1, DNMT3A, TET2 and IDH1 are all common in adults but rare in infancy and early childhood.[190, 191] Such differences in genetic abnormalities between pediatric and adult leukemia are primarily attributable to the different responses of fetal/neonatal and adult HSCs/HPCs to stimulation of leukemic oncogenes. [12–15]
Transgenic overexpression and animal model studies demonstrated that fetal/neonatal HSCs/HPCs are resistant to genetic transcriptions induced by some adult types of leukemic oncogenes, while adult HSCs/HPCs are resistant to genetic transcriptions induced by some pediatric types of leukemic oncogenes [12–15]. Consequently, some of the pediatric types of oncogenes preferentially induce leukemia when expressed in fetal/neonatal stages while some of the adult types of oncogenes preferentially induce leukemia when expressed in adult HSCs/HPCs. For example: 1) FLT3-ITD induces the expression of a significant panel of genes in adult HSCs/HPCs but does not do so in fetal and neonatal HSCs/HPCs. FLT3-ITD can only induce leukemia development when expressed in adult HSCs/HPCs, while fetal and neonatal HSCs/HPCs are resistant to FLT3-ITD-induced leukemic transformation. A recent study suggests that IFNα-induced HSC maturation and HPC expansion are required for FLT3-ITD-induced leukemia development. Fetal and neonatal HSCs/HPCs lack of such IFNα-induced maturation and expansion.[14, 15] 2) Gata1s induces the expression of megakaryocytic genes and represses erythroid genes in fetal HSCs/HPCs but fails to do so in adult HSCs/HPCs. As a consequence, Gata1s induces a TMD in mice only when overexpressed in fetal/neonatal HSCs/HPCs. Gata1s does so by stimulating fetal HSCs/HPCs to produce significantly more CD41loAChE− immature megakaryoblasts and CD71+Ter119lo immature pro-erythroblasts. However, Gata1s failed to do so in adult BM HSCs/HPCs[12]; 3) MLL-ENL preferentially induces a significant panel of gene expression in neonatal HSCs/HPCs but does not do so in adult HSCs/HPCs. Thus, MLL-ENL overexpression drives highly efficient leukemic initiation in neonatal HSCs/HPCs but is poorly efficient in doing so in adult HPSCs.[13] Interestingly, some oncogenes induce different types of leukemia in fetal and adult HSCs/HPCs. For example, ETO2-GLIS2 induces the activities of key transcription factors, including ERG, SPI1, GATA1, and CEBPα, in neonatal HSCs/HPCs that are different in adult HSCs/HPCs. ETO2-GLIS2 in fetal HSCs/HPCs induces rapid AMkL development whereas expression in adult BM HSCs/HPCs results in no AMkL types of AML with long latency.[175]
Fetal origin of pediatric leukemia.
Approximately 70–80% of infant ALL are associated with MLL-r. The most common genetic abnormality in B-ALL in early childhood (<5 years) is the ETV6-RUNX1 fusion. Other common types of genetic abnormalities in childhood B-ALL include hyperdiploidy, TCF3-PBX1, TCF3-HLF, and BCR-ABL1. B-ALL with these genetic abnormalities occurs relatively constantly throughout childhood. See Figure 4. All of these chromosomal translocations and alterations in chromosome numbers detected in pediatric leukemias are the primary genetic events. Studies of leukemia patients in monozygotic twins and detection of genetic abnormalities in newborns and their cord blood suggested that most of these genetic abnormalities are intra-utero in origin. The prenatal origin of pediatric leukemias has been demonstrated in ALL with MLL-r, ETV6-RUNX1, TCF3-PBX1, BCR-ABL1 and hyperdiploidy. [192]
Figure 4. Almost all infant/neonatal leukemias and most of childhood leukemias originate from infant.
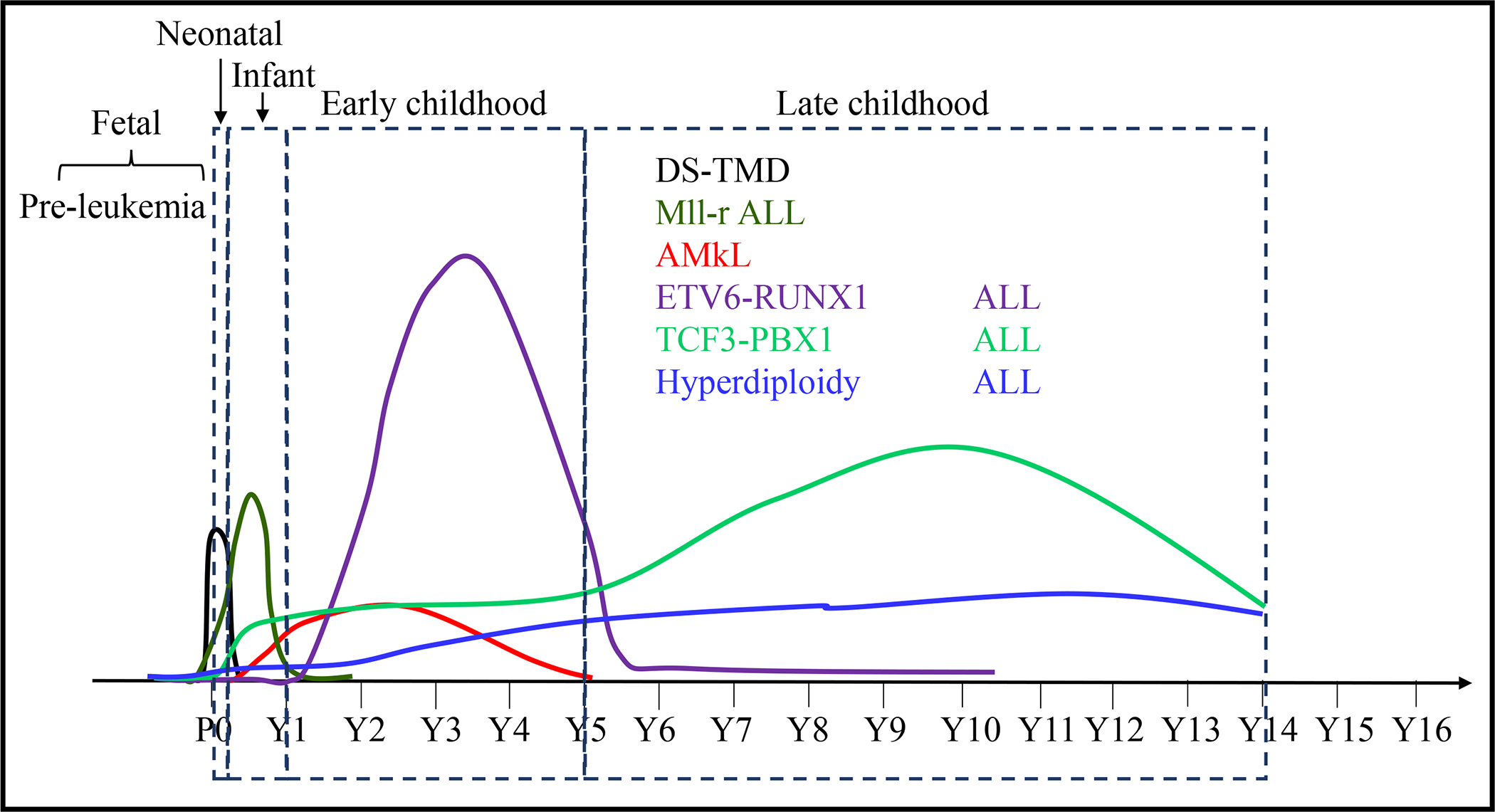
Almost all primary genetic abnormalities can be detected in fetal HSCs/HPCs, suggesting a fetal stage of pre-leukemia development. The different peaks of leukemia onsets induced by distinct genetic abnormalities are determined by: 1) efficiency of leukemia induction capacity of the primary genetic abnormalities; 2) the occurrence time of the secondary genetic abnormalities. For examples, Gata1s or MLL-r alone is sufficient to induce TMD or ALL development respectively, explaining the early onset peaks of DS-TMD and ALL in neonatal and infant. Secondary genetic mutations are required for the leukemia onsets induced by other primary genetic abnormalities, explaining the latter onset peaks of childhood AMkL and ALL.
AMkL is characterized by expression of megakaryocytic lineage markers (e.g., CD41, CD42, and CD61).[193] The majority of AMkL cases are diagnosed in early childhood (<4 years) (Figure 4), and most are DS-associated, related trisomy 21.[194, 195] Approximately 10–30% of DS neonates are born with a TMD. Among them, 20–30% progress to AMkL with a few cases developing into ALL by 4 years of age. Nonsense mutations in exon 2 of GATA1 lead to production of a short form of GATA1 (GATA1s) and loss of the long form of GATA1; these occur in approximately 90% of DS-TMD and DS-AMkL cases. Such mutations are the only somatic mutations seen in TMD in most instances. During progression of DS-TMD to DS-AMkL, 2–5 additional mutations are found to have occurred. The remaining AMkL develop without DS and are called non-DS AMkL. The most frequently observed chromosomal translocations in non-DS-AMkL are RBM15-MKL1, ETO2-GLIS2, KAT6A-CREBBP and NUP98-KDM5A.[196–198] GATA1s mutations and chromosomal translocations can be detected in newborns, also suggesting that these originate in fetal life.
MLL-r in infant ALL can always be detected before birth. In most infant ALL cases, MLL-r is the only driver mutation, indicating that MLL-r alone is sufficient to induce leukemia in infants.[187] To further support this notion, it was found almost all neonates harboring MLL-r developed leukemia within one year. In some cases, ALL is diagnosed immediately after birth. For monozygotic twins, if one develops MLL-r leukemia the other twin always also develops leukemia. There are no healthy MLL-r carrier cases reported. However, with ETV6-RUNX1 and TCF3-PBX1, significantly higher frequencies of such fusions can be detected in the PB and/or cord blood of healthy newborns. Depending on the sensitivity of the assays used, ETV6-RUNX1 fusions can be detected in 0.01–5% of newborns, hundreds of times higher than patients with ETV6-RUNX1 ALL.[177–184] TCF3-PBX1 fusion can be detected in ~0.6% of healthy newborns, also hundreds of times higher than the incidence of TCF3-PBX1 ALL cases.[185] This suggests that either ETV6-RUNX1 or TCF3-PBX1 alone induces only a pre-leukemic clone and is therefore insufficient to induce leukemia development.[199, 200] Secondary genetic events are absolutely required for full-blown ALL in such cases. The fact that a majority of healthy carriers of ETV6-RUNX1 or TCF3-PBX1 do not develop leukemia during their lifetimes suggests that ETV6-RUNX1+ or TCF3-PBX1+ clones are diminished early in life, just as numbers of drHSCs diminish during the early neonatal period. Such might be the same for hyperdiploidy and some cases of BCR-ABL1. Recent studies suggest that all of these above mentioned genetic abnormalities are primary events. Certain stress conditions such as infections or abnormal T cell/NK cell immune responses are required to induce the pre-leukemic lesions by promoting the clonal evolution of mutant HSCs/HPCs.[200–204] Further additional genetic events are absolutely required for full-blown leukemia to develop.[177, 178, 184, 205–207] Currently, it is unknown whether all these primary genetic abnormalities originate in utero or whether some of them can be acquired after birth, specifically for these later onset cases.
Spontaneous regression of leukemia in neonates and infants.
A distinctive feature of neonatal and infant leukemia is the occurrence of spontaneous regression in some cases. The TMD in DS often resolves spontaneously within the first 3 months of life.[208] Spontaneous regression is also frequently observed in neonatal and infant leukemia cases associated with t(8;16)/KAT6A-CREBBP, especially leukemia cutis. In addition, spontaneous regression has been reported in juvenile chronic myelomonocytic leukemia cases involving RAS signaling pathway mutations, such as active mutations of PTPN11 gene.[209] PTPN11 gene mutations are the cause of Noonan syndrome, which increases the risk of developing juvenile chronic myelomonocytic in early childhood. However, the propensity for leukemia development in those with Noonan syndrome declines to baseline later in life despite the persistence of the congenital genetic abnormalities. Two mechanisms have been proposed to explain the spontaneous regression of neonatal/infant leukemia: 1) the nature of the mutant cells. Many of the pediatric leukemias are initiated and propagated in certain types of HPCs that have limited self-renewal capacity such as drHSCs. Such mutant HPCs subsequently differentiate or die in the postnatal microenvironment if the oncogenes failed to induce self-renewal. 2) The changes in the hematopoietic microenvironment from the FL to postnatal BM. The postnatal BM cannot support mutant fetal cell growth due to the absence of certain factors, which remain to be determined.
6. Prospective
Primitive hematopoiesis in YS and definitive hematopoiesis in the AGM region have been well-documented by many earlier studies. Based on these studies, it was suggested that primitive hematopoiesis only supports a short period of blood cell production during early embryonic development and is then replaced by definitive hematopoiesis. Thus, it was believed that all adult hematopoietic cells and immune cells and all hematopoietic disorders are initiated from definitive hematopoiesis. However, since the identification of pre-definitive hematopoiesis in YS (the 2nd wave of hematopoiesis), such a concept must be questioned. As with the 1st and 3rd waves of hematopoiesis, the 2nd wave of hematopoiesis is conserved in all mammals. In mice, the 2nd wave of hematopoiesis provides blood and immune support for fetal development at least up to birth. HSCs/HPCs derived from the 2nd wave of hematopoiesis can still be detected in BM during the first 2 weeks of postnatal life and are completely replaced by HSCs/HPCs derived from the 3rd wave by 3–4 weeks of age in mice. The contribution of HSCs/HPCs derived from the 2nd wave of hematopoiesis to tissue resident Mφ and innate immune cells is well-documented. However, the contribution of this wave of hematopoiesis to other types of blood cells and immune cells in adult is largely unknown. In addition, although HPCs that biologically and functionally similar to EMPs and LMPs have been identified in human FL and fetal BM, [35] the contribution of such HPCs to pediatric leukemia has been suggested, whether such HPCs are true analogues of EMPs and LMPs need to be determined.
Compared to adult ALL, the outcomes for pediatric ALL patients have significantly improved over the last 3 decades owing to the combination of improved chemotherapy and supportive therapy.[210, 211] Currently, 5 year overall survival (OS) in children with ALL is >90%, and the cure rate is approaching 90%.[212] However, ALL in infants and neonates are associated with extremely poor treatment outcomes with only ~60% 5 year OS and <40% cure rate, mostly due to primary chemotherapy refractory disease and/or early relapse.[213, 214] Such disparate outcomes have been primarily attributed to the differences in genetic abnormalities. [215] However, there is increased evidence suggesting that pediatric leukemias outcomes are associated with the developmental origins of leukemic cells.[216]
Several subtypes of childhood B-ALL have been shown to arise in utero including those characterized by KMT2A-r, ETV6-RUNX1, BCR-ABL, TCF3-PBX1 or TCF3-ZNF384 gene fusions and hyperdiploid ALL. MLL-r infant ALL blasts share many of the biologic properties with fetal type of B cell progenitors, pre-proB, as demonstrated by studies of immunophenotypic profiles as well as transcriptomic and IgH rearrangement patterns. As is true for as pre-proB progenitors, MLL-r infant ALL blasts typically carry immunoglobulin loci with rearranged D-JH but not VH-to-DJH segments, fail to express CD10, and express myeloid and stem cell gene programs.[148] In contrast, blast cells from most childhood-ALL share many biological properties with more mature CD19+CD10+ adult types of B-progenitors such as proB-progenitors. However, less is known about the “pre-leukemic” cells within which first-hit events occur. Human modeling data showed that a small portion of CD34+CD38–CD19+ cells present in both infant and childhood ALL patients which cells have leukemia stem cell properties. However the primary rearrangements probably occur at the earliest stage in CD34+CD19−IL-7R+ ELPs [176], a kind of cell population shares the biological phenotype with fetal LMPs/drHSCs in mice [149]. The disappearance of pre-leukemic clones observed in most individuals during early infant development suggests that they may be derived from developmentally restricted types of cells [177–185]. Confirmation of the leukemic cell of origin is fundamental to identification and understanding of the cellular and molecular mechanisms that underlie leukemogenesis. Future studies need to determine whether pediatric ALL originates from YS LMPs/drHSCs, while infant/neonatal AMkL originate from YS EMPs.
Key points:
Hematopoiesis undergoes dynamic alterations during fetal-to-neonatal and neonatal-to-adult transitions.
Both fetal and adult types of HSCs and HPCs exist in fetuses and neonates.
The fetal types of HSCs and HPCs are expanded in fetal liver and fetal bone marrow, and diminish during neonatal-to-adult transition.
Adult types of HSCs and HPCs undergo dynamic and gradual maturation in bone marrow during perinatal-to-adult transition.
Almost all neonatal and infant leukemias originate from the fetal stage and have a poor prognosis.
Most subtypes of leukemia in childhood are also of fetal origin but have a more favorable prognosis.
Funding
This work was supported by NIH grants R01 HL133560–01 and R01 CA223194–01 through Loyola University Chicago, as well as Loyola program development funds to Jiwang Zhang.
Abbreviations
- AGM
Aorta-mesonephros-gonad
- Angptl2
Angiopoietin-like 2
- AMkL
Acute megakaryocytic type
- AML
Acute myeloid leukemia
- ALL
Acute lymphoblastic type
- BM
Bone marrow
- CSF1
Colony-stimulating factor 1
- CXCL12
C-X-C Motif Chemokine Ligand 12
- drHSCs
Developmentally-restricted HSCs
- DS
Down Syndrome
- E
Embryonic day
- ECs
Endothelial cells
- EPO
Erythropoietin
- EMPs
Erythro-myeloid progenitors
- ECM
Extracellular matrix
- FL
Fetal liver
- HSCs
Hematopoietic stem cells
- HPCs
Hematopoietic progenitor cells
- Hb
Hemoglobin
- imHSCs
Immature HSC
- IGF2
Insulin-like growth factor 2
- IL
Interleukin
- IFN
Interferon
- KITL
KIT ligand
- LMPs
Lympho-myeloid progenitors
- Mφ
Macrophages
- Mks
Megakaryocytes
- MLL
Mixed lineage leukemia
- ELPs
Early lymphoid progenitors
- PB
Peripheral blood
- PCW
Post-conception weeks
- pri-Eps
Primitive erythroid progenitors
- RBCs
Red blood cells
- THPO
Thrombopoietin
- TSLP
Thymic stromal-derived lymphopoietin
- TMD
Transient myeloproliferative disorder
- vWF
von Willebrand factor
- YS
Yolk Sac
Footnotes
Conflicts of interest
The authors declare that they have no competing financial or professional interests.
Declarations
Ethics approval
This is not applicable for this review.
Consent to participate
This is not applicable for this review.
Consent for publication
This is not applicable for this review.
Code availability
This is not applicable for this review.
Availability of data and material
This is not applicable for this review.
References
- 1.Kikuchi K, Kondo M: Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci U S A 2006, 103(47):17852–17857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie MB, Kent DG, Dykstra B, McKnight KD, McCaffrey L, Hoodless PA, Eaves CJ: Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci U S A 2007, 104(14):5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ: Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest 2006, 116(10):2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gothert JR, Gustin SE, Hall MA, Green AR, Gottgens B, Izon DJ, Begley CG: In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 2005, 105(7):2724–2732. [DOI] [PubMed] [Google Scholar]
- 5.Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL: The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A 1995, 92(22):10302–10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA: Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 2012, 335(6073):1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K et al. : The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 2013, 15(8):916–925. [DOI] [PubMed] [Google Scholar]
- 8.Benz C, Copley MR, Kent DG, Wohrer S, Cortes A, Aghaeepour N, Ma E, Mader H, Rowe K, Day C et al. : Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 2012, 10(3):273–283. [DOI] [PubMed] [Google Scholar]
- 9.Ghosn EE, Yamamoto R, Hamanaka S, Yang Y, Herzenberg LA, Nakauchi H, Herzenberg LA: Distinct B-cell lineage commitment distinguishes adult bone marrow hematopoietic stem cells. Proc Natl Acad Sci U S A 2012, 109(14):5394–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barber CL, Montecino-Rodriguez E, Dorshkind K: Reduced production of B-1-specified common lymphoid progenitors results in diminished potential of adult marrow to generate B-1 cells. Proc Natl Acad Sci U S A 2011, 108(33):13700–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan H, Hale J, Jaffray J, Li J, Wang Y, Huang Y, An X, Hillyer C, Wang N, Kinet S et al. : Developmental differences between neonatal and adult human erythropoiesis. Am J Hematol 2018, 93(4):494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birger Y, Goldberg L, Chlon TM, Goldenson B, Muler I, Schiby G, Jacob-Hirsch J, Rechavi G, Crispino JD, Izraeli S: Perturbation of fetal hematopoiesis in a mouse model of Down syndrome’s transient myeloproliferative disorder. Blood 2013, 122(6):988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okeyo-Owuor T, Li Y, Patel RM, Yang W, Casey EB, Cluster AS, Porter SN, Bryder D, Magee JA: The efficiency of murine MLL-ENL-driven leukemia initiation changes with age and peaks during neonatal development. Blood advances 2019, 3(15):2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter SN, Cluster AS, Yang W, Busken KA, Patel RM, Ryoo J, Magee JA: Fetal and neonatal hematopoietic progenitors are functionally and transcriptionally resistant to Flt3-ITD mutations. eLife 2016, 5:e18882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Kong W, Yang W, Patel RM, Casey EB, Okeyo-Owuor T, White JM, Porter SN, Morris SA, Magee JA: Single-Cell Analysis of Neonatal HSC Ontogeny Reveals Gradual and Uncoordinated Transcriptional Reprogramming that Begins before Birth. Cell Stem Cell 2020, 27(5):732–747 e737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parekh C, Crooks GM: Critical differences in hematopoiesis and lymphoid development between humans and mice. J Clin Immunol 2013, 33(4):711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, Medvinsky A: Human haematopoietic stem cell development: from the embryo to the dish. Development 2017, 144(13):2323–2337. [DOI] [PubMed] [Google Scholar]
- 18.Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A, Godin I: Three pathways to mature macrophages in the early mouse yolk sac. Blood 2005, 106(9):3004–3011. [DOI] [PubMed] [Google Scholar]
- 19.McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, Palis J: Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell reports 2015, 11(12):1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palis J, Robertson S, Kennedy M, Wall C, Keller G: Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999, 126(22):5073–5084. [DOI] [PubMed] [Google Scholar]
- 21.Kingsley PD, Malik J, Emerson RL, Bushnell TP, McGrath KE, Bloedorn LA, Bulger M, Palis J: “Maturational” globin switching in primary primitive erythroid cells. Blood 2006, 107(4):1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, Waugh R, Palis J: The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 2007, 109(4):1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frame JM, Fegan KH, Conway SJ, McGrath KE, Palis J: Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells 2016, 34(2):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boiers C, Carrelha J, Lutteropp M, Luc S, Green JC, Azzoni E, Woll PS, Mead AJ, Hultquist A, Swiers G et al. : Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 2013, 13(5):535–548. [DOI] [PubMed] [Google Scholar]
- 25.Inlay MA, Serwold T, Mosley A, Fathman JW, Dimov IK, Seita J, Weissman IL: Identification of multipotent progenitors that emerge prior to hematopoietic stem cells in embryonic development. Stem cell reports 2014, 2(4):457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasaai B, Caolo V, Peacock HM, Lehoux S, Gomez-Perdiguero E, Luttun A, Jones EA: Erythro-myeloid progenitors can differentiate from endothelial cells and modulate embryonic vascular remodeling. Scientific reports 2017, 7:43817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGrath KE, Frame JM, Palis J: Early hematopoiesis and macrophage development. Semin Immunol 2015, 27(6):379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, Launay P, Chen J, Ginhoux F, Bajenoff M: Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 2018, 48(6):1160–1171 e1165. [DOI] [PubMed] [Google Scholar]
- 29.Dege C, Fegan KH, Creamer JP, Berrien-Elliott MM, Luff SA, Kim D, Wagner JA, Kingsley PD, McGrath KE, Fehniger TA et al. : Potently Cytotoxic Natural Killer Cells Initially Emerge from Erythro-Myeloid Progenitors during Mammalian Development. Dev Cell 2020, 53(2):229–239 e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Liu S, Xu J, Zhang X, Han D, Liu J, Xia M, Yi L, Shen Q, Xu S et al. : Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity 2018, 49(4):640–653 e645. [DOI] [PubMed] [Google Scholar]
- 31.Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, Locksley RM: Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 2019, 50(6):1425–1438 e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentek R, Ghigo C, Hoeffel G, Jorquera A, Msallam R, Wienert S, Klauschen F, Ginhoux F, Bajenoff M: Epidermal gammadelta T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J Exp Med 2018, 215(12):2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath KE, Frame JM, Fromm GJ, Koniski AD, Kingsley PD, Little J, Bulger M, Palis J: A transient definitive erythroid lineage with unique regulation of the beta-globin locus in the mammalian embryo. Blood 2011, 117(17):4600–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bian Z, Gong Y, Huang T, Lee CZW, Bian L, Bai Z, Shi H, Zeng Y, Liu C, He J et al. : Deciphering human macrophage development at single-cell resolution. Nature 2020, 582(7813):571–576. [DOI] [PubMed] [Google Scholar]
- 35.Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M et al. : Decoding human fetal liver haematopoiesis. Nature 2019, 574(7778):365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner Y, Mass E, Ashok Kumar P, Ulas T, Handler K, Horne A, Klee K, Lupp A, Schutz D, Saaber F et al. : Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat Neurosci 2020, 23(3):351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimoto M, Porayette P, Glosson NL, Conway SJ, Carlesso N, Cardoso AA, Kaplan MH, Yoder MC: Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood 2012, 119(24):5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Y, Kobayashi M, Azevedo Portilho N, Mishra A, Gao H, Liu Y, Wenzel P, Davis B, Yoder MC, Yoshimoto M: Long-Term Engraftment of ESC-Derived B-1 Progenitor Cells Supports HSC-Independent Lymphopoiesis. Stem cell reports 2019, 12(3):572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luis TC, Luc S, Mizukami T, Boukarabila H, Thongjuea S, Woll PS, Azzoni E, Giustacchini A, Lutteropp M, Bouriez-Jones T et al. : Initial seeding of the embryonic thymus by immune-restricted lympho-myeloid progenitors. Nat Immunol 2016, 17(12):1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaudin AE, Boyer SW, Perez-Cunningham J, Hernandez GE, Derderian SC, Jujjavarapu C, Aaserude E, MacKenzie T, Forsberg EC: A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell 2016, 19(6):768–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA et al. : Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 2005, 121(2):295–306. [DOI] [PubMed] [Google Scholar]
- 42.Yamane T, Hosen N, Yamazaki H, Weissman IL: Expression of AA4.1 marks lymphohematopoietic progenitors in early mouse development. Proc Natl Acad Sci U S A 2009, 106(22):8953–8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito C, Yamazaki H, Yamane T: Earliest hematopoietic progenitors at embryonic day 9 preferentially generate B-1 B cells rather than follicular B or marginal zone B cells. Biochem Biophys Res Commun 2013, 437(2):307–313. [DOI] [PubMed] [Google Scholar]
- 44.Yamane T, Ito C, Washino A, Isono K, Yamazaki H: Repression of Primitive Erythroid Program Is Critical for the Initiation of Multi-Lineage Hematopoiesis in Mouse Development. J Cell Physiol 2017, 232(2):323–330. [DOI] [PubMed] [Google Scholar]
- 45.Medvinsky A, Dzierzak E: Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 1996, 86(6):897–906. [DOI] [PubMed] [Google Scholar]
- 46.North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA: Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 2002, 16(5):661–672. [DOI] [PubMed] [Google Scholar]
- 47.Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D: Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010, 464(7285):108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C: In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010, 464(7285):116–120. [DOI] [PubMed] [Google Scholar]
- 49.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA: Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 2009, 457(7231):887–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eilken HM, Nishikawa S, Schroeder T: Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 2009, 457(7231):896–900. [DOI] [PubMed] [Google Scholar]
- 51.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G: The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 2009, 457(7231):892–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC et al. : Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 2008, 3(6):625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW et al. : A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336(6077):86–90. [DOI] [PubMed] [Google Scholar]
- 54.Tober J, McGrath KE, Palis J: Primitive erythropoiesis and megakaryopoiesis in the yolk sac are independent of c-myb. Blood 2008, 111(5):2636–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rybtsov S, Sobiesiak M, Taoudi S, Souilhol C, Senserrich J, Liakhovitskaia A, Ivanovs A, Frampton J, Zhao S, Medvinsky A: Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med 2011, 208(6):1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A: Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43(−) embryonic precursor. Stem cell reports 2014, 3(3):489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rybtsov S, Ivanovs A, Zhao S, Medvinsky A: Concealed expansion of immature precursors underpins acute burst of adult HSC activity in foetal liver. Development 2016, 143(8):1284–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye H, Wang X, Li Z, Zhou F, Li X, Ni Y, Zhang W, Tang F, Liu B, Lan Y: Clonal analysis reveals remarkable functional heterogeneity during hematopoietic stem cell emergence. Cell Res 2017, 27(8):1065–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganuza M, Hall T, Finkelstein D, Chabot A, Kang G, McKinney-Freeman S: Lifelong haematopoiesis is established by hundreds of precursors throughout mammalian ontogeny. Nat Cell Biol 2017, 19(10):1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henninger J, Santoso B, Hans S, Durand E, Moore J, Mosimann C, Brand M, Traver D, Zon L: Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nat Cell Biol 2017, 19(1):17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D: Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007, 1(4):428–442. [DOI] [PubMed] [Google Scholar]
- 62.Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK: The placenta is a niche for hematopoietic stem cells. Dev Cell 2005, 8:365–375. [DOI] [PubMed] [Google Scholar]
- 63.Robin C, Bollerot K, Mendes S, Haak E, Crisan M, Cerisoli F, Lauw I, Kaimakis P, Jorna R, Vermeulen M et al. : Human placenta is a potent hematopoietic niche containing hematopoietic stem and progenitor cells throughout development. Cell Stem Cell 2009, 5(4):385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ottersbach K, Dzierzak E: The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell 2005, 8(3):377–387. [DOI] [PubMed] [Google Scholar]
- 65.Barcena A, Muench MO, Kapidzic M, Fisher SJ: A new role for the human placenta as a hematopoietic site throughout gestation. Reproductive sciences 2009, 16(2):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barcena A, Kapidzic M, Muench MO, Gormley M, Scott MA, Weier JF, Ferlatte C, Fisher SJ: The human placenta is a hematopoietic organ during the embryonic and fetal periods of development. Dev Biol 2009, 327(1):24–33. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HK: The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell 2008, 2(3):252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr., Potter SS: A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 1991, 65(4):677–689. [DOI] [PubMed] [Google Scholar]
- 69.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER et al. : Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330(6005):841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheng J, Ruedl C, Karjalainen K: Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity 2015, 43(2):382–393. [DOI] [PubMed] [Google Scholar]
- 71.Samokhvalov IM, Samokhvalova NI, Nishikawa S: Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 2007, 446(7139):1056–1061. [DOI] [PubMed] [Google Scholar]
- 72.Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC: All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 2008, 111(7):3435–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F et al. : Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518(7540):547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kashem SW, Haniffa M, Kaplan DH: Antigen-Presenting Cells in the Skin. Annu Rev Immunol 2017, 35:469–499. [DOI] [PubMed] [Google Scholar]
- 75.Ayres-Silva Jde P, Manso PP, Madeira MR, Pelajo-Machado M, Lenzi HL: Sequential morphological characteristics of murine fetal liver hematopoietic microenvironment in Swiss Webster mice. Cell Tissue Res 2011, 344(3):455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miles C, Sanchez MJ, Sinclair A, Dzierzak E: Expression of the Ly-6E.1 (Sca-1) transgene in adult hematopoietic stem cells and the developing mouse embryo. Development 1997, 124(2):537–547. [DOI] [PubMed] [Google Scholar]
- 77.Brouard N, Jost C, Matthias N, Albrecht C, Egard S, Gandhi P, Strassel C, Inoue T, Sugiyama D, Simmons PJ et al. : A unique microenvironment in the developing liver supports the expansion of megakaryocyte progenitors. Blood advances 2017, 1(21):1854–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soares-da-Silva F, Freyer L, Elsaid R, Burlen-Defranoux O, Iturri L, Sismeiro O, Pinto-do OP, Gomez-Perdiguero E, Cumano A: Yolk sac, but not hematopoietic stem cell-derived progenitors, sustain erythropoiesis throughout murine embryonic life. J Exp Med 2021, 218(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, Vink CS, Bhandoola A, Dzierzak E, Speck NA: Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 2011, 9(6):541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Travnickova J, Tran Chau V, Julien E, Mateos-Langerak J, Gonzalez C, Lelievre E, Lutfalla G, Tavian M, Kissa K: Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat Commun 2015, 6:6227. [DOI] [PubMed] [Google Scholar]
- 81.Stremmel C, Schuchert R, Wagner F, Thaler R, Weinberger T, Pick R, Mass E, Ishikawa-Ankerhold HC, Margraf A, Hutter S et al. : Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat Commun 2018, 9(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Espin-Palazon R, Stachura DL, Campbell CA, Garcia-Moreno D, Del Cid N, Kim AD, Candel S, Meseguer J, Mulero V, Traver D: Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 2014, 159(5):1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee LK, Ghorbanian Y, Wang W, Wang Y, Kim YJ, Weissman IL, Inlay MA, Mikkola HKA: LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage. Cell reports 2016, 17(9):2286–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ: Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 2006, 108(2):737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, Cheyne J, Zhao Y, Bowie MB, Zhao Y et al. : Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 2009, 113(25):6342–6350. [DOI] [PubMed] [Google Scholar]
- 86.Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, Levantini E, Alberich-Jorda M, Zhang J, Kawasaki A et al. : C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol 2013, 15(4):385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Z, Zhou F, Chen D, He W, Ni Y, Luo L, Liu B: Generation of hematopoietic stem cells from purified embryonic endothelial cells by a simple and efficient strategy. Journal of genetics and genomics = Yi chuan xue bao 2013, 40(11):557–563. [DOI] [PubMed] [Google Scholar]
- 88.Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J, Friedman A, Malek S, Grembecka J, Cierpicki T et al. : Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest 2015, 125(5):2007–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ: Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell 2012, 11(3):415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Esain V, Teng L, Xu J, Kwan W, Frost IM, Yzaguirre AD, Cai X, Cortes M, Maijenburg MW et al. : Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev 2014, 28(23):2597–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sawamiphak S, Kontarakis Z, Stainier DY: Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev Cell 2014, 31(5):640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim PG, Canver MC, Rhee C, Ross SJ, Harriss JV, Tu HC, Orkin SH, Tucker HO, Daley GQ: Interferon-alpha signaling promotes embryonic HSC maturation. Blood 2016, 128(2):204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robin C, Ottersbach K, Durand C, Peeters M, Vanes L, Tybulewicz V, Dzierzak E: An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev Cell 2006, 11(2):171–180. [DOI] [PubMed] [Google Scholar]
- 94.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A: IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009, 458(7240):904–908. [DOI] [PubMed] [Google Scholar]
- 95.Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, Wurzer S, Prendergast AM, Schnell A, Hexel K et al. : Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell 2015, 17(4):422–434. [DOI] [PubMed] [Google Scholar]
- 96.Pietras EM, Lakshminarasimhan R, Techner JM, Fong S, Flach J, Binnewies M, Passegue E: Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med 2014, 211(2):245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nomura N, Ito C, Ooshio T, Tadokoro Y, Kohno S, Ueno M, Kobayashi M, Kasahara A, Takase Y, Kurayoshi K et al. : Essential role of autophagy in protecting neonatal haematopoietic stem cells from oxidative stress in a p62-independent manner. Scientific reports 2021, 11(1):1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung HE, Shim YR, Oh JE, Oh DS, Lee HK: The autophagy Protein Atg5 Plays a Crucial Role in the Maintenance and Reconstitution Ability of Hematopoietic Stem Cells. Immune network 2019, 19(2):e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hirsch E, Iglesias A, Potocnik AJ, Hartmann U, Fassler R: Impaired migration but not differentiation of haematopoietic stem cells in the absence of beta1 integrins. Nature 1996, 380(6570):171–175. [DOI] [PubMed] [Google Scholar]
- 100.Potocnik AJ, Brakebusch C, Fassler R: Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity 2000, 12(6):653–663. [DOI] [PubMed] [Google Scholar]
- 101.Mazo IB, Massberg S, von Andrian UH: Hematopoietic stem and progenitor cell trafficking. Trends Immunol 2011, 32(10):493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prashad SL, Calvanese V, Yao CY, Kaiser J, Wang Y, Sasidharan R, Crooks G, Magnusson M, Mikkola HK: GPI-80 defines self-renewal ability in hematopoietic stem cells during human development. Cell Stem Cell 2015, 16(1):80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ueda T, Yokota T, Okuzaki D, Uno Y, Mashimo T, Kubota Y, Sudo T, Ishibashi T, Shingai Y, Doi Y et al. : Endothelial Cell-Selective Adhesion Molecule Contributes to the Development of Definitive Hematopoiesis in the Fetal Liver. Stem cell reports 2019, 13(6):992–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Emambokus NR, Frampton J: The glycoprotein IIb molecule is expressed on early murine hematopoietic progenitors and regulates their numbers in sites of hematopoiesis. Immunity 2003, 19(1):33–45. [DOI] [PubMed] [Google Scholar]
- 105.Chou S, Lodish HF: Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc Natl Acad Sci U S A 2010, 107(17):7799–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T: Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity 2003, 19(2):257–267. [DOI] [PubMed] [Google Scholar]
- 107.Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R: Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp Hematol 2014, 42(8):669–683. [DOI] [PubMed] [Google Scholar]
- 108.Ema H, Nakauchi H: Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 2000, 95(7):2284–2288. [PubMed] [Google Scholar]
- 109.Chou S, Flygare J, Lodish HF: Fetal hepatic progenitors support long-term expansion of hematopoietic stem cells. Exp Hematol 2013, 41(5):479–490 e474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tan KS, Kulkeaw K, Nakanishi Y, Sugiyama D: Expression of cytokine and extracellular matrix mRNAs in fetal hepatic stellate cells. Genes Cells 2017, 22(9):836–844. [DOI] [PubMed] [Google Scholar]
- 111.Zhang CC, Lodish HF: Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood 2004, 103(7):2513–2521. [DOI] [PubMed] [Google Scholar]
- 112.Zheng J, Huynh H, Umikawa M, Silvany R, Zhang CC: Angiopoietin-like protein 3 supports the activity of hematopoietic stem cells in the bone marrow niche. Blood 2011, 117(2):470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sugiyama D, Kulkeaw K, Mizuochi C, Horio Y, Okayama S: Hepatoblasts comprise a niche for fetal liver erythropoiesis through cytokine production. Biochem Biophys Res Commun 2011, 410(2):301–306. [DOI] [PubMed] [Google Scholar]
- 114.Kordes C, Sawitza I, Gotze S, Haussinger D: Hepatic stellate cells support hematopoiesis and are liver-resident mesenchymal stem cells. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 2013, 31(2–3):290–304. [DOI] [PubMed] [Google Scholar]
- 115.Liu C, Han T, Stachura DL, Wang H, Vaisman BL, Kim J, Klemke RL, Remaley AT, Rana TM, Traver D et al. : Lipoprotein lipase regulates hematopoietic stem progenitor cell maintenance through DHA supply. Nat Commun 2018, 9(1):1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iwasaki H, Arai F, Kubota Y, Dahl M, Suda T: Endothelial protein C receptor-expressing hematopoietic stem cells reside in the perisinusoidal niche in fetal liver. Blood 2010, 116(4):544–553. [DOI] [PubMed] [Google Scholar]
- 117.Khan JA, Mendelson A, Kunisaki Y, Birbrair A, Kou Y, Arnal-Estape A, Pinho S, Ciero P, Nakahara F, Ma’ayan A et al. : Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 2016, 351(6269):176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, Yzaguirre AD, Speck NA, Zon LI: Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 2015, 160(1–2):241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gerlach JC, Thompson RL, Gridelli B, Schmelzer E: Effects of Delta-Like Noncanonical Notch Ligand 1 Expression of Human Fetal Liver Hepatoblasts on Hematopoietic Progenitors. Stem cells international 2019, 2019:7916275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sigurdsson V, Takei H, Soboleva S, Radulovic V, Galeev R, Siva K, Leeb-Lundberg LM, Iida T, Nittono H, Miharada K: Bile Acids Protect Expanding Hematopoietic Stem Cells from Unfolded Protein Stress in Fetal Liver. Cell Stem Cell 2016, 18(4):522-532. [DOI] [PubMed] [Google Scholar]
- 121.Lee Y, Leslie J, Yang Y, Ding L: Hepatic stellate and endothelial cells maintain hematopoietic stem cells in the developing liver. J Exp Med 2021, 218(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Christensen JL, Wright DE, Wagers AJ, Weissman IL: Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol 2004, 2(3):E75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ciriza J, Thompson H, Petrosian R, Manilay JO, Garcia-Ojeda ME: The migration of hematopoietic progenitors from the fetal liver to the fetal bone marrow: lessons learned and possible clinical applications. Exp Hematol 2013, 41(5):411–423. [DOI] [PubMed] [Google Scholar]
- 124.Theodore LN, Hagedorn EJ, Cortes M, Natsuhara K, Liu SY, Perlin JR, Yang S, Daily ML, Zon LI, North TE: Distinct Roles for Matrix Metalloproteinases 2 and 9 in Embryonic Hematopoietic Stem Cell Emergence, Migration, and Niche Colonization. Stem cell reports 2017, 8(5):1226–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Papayannopoulou T, Priestley GV, Nakamoto B, Zafiropoulos V, Scott LM: Molecular pathways in bone marrow homing: dominant role of alpha(4)beta(1) over beta(2)-integrins and selectins. Blood 2001, 98(8):2403–2411. [DOI] [PubMed] [Google Scholar]
- 126.Shao L, Chang J, Feng W, Wang X, Williamson EA, Li Y, Schajnovitz A, Scadden D, Mortensen LJ, Lin CP et al. : The Wave2 scaffold Hem-1 is required for transition of fetal liver hematopoiesis to bone marrow. Nat Commun 2018, 9(1):2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK: Structure and control of the actin regulatory WAVE complex. Nature 2010, 468(7323):533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mizoguchi T, Pinho S, Ahmed J, Kunisaki Y, Hanoun M, Mendelson A, Ono N, Kronenberg HM, Frenette PS: Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell 2014, 29(3):340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coskun S, Chao H, Vasavada H, Heydari K, Gonzales N, Zhou X, de Crombrugghe B, Hirschi KK: Development of the fetal bone marrow niche and regulation of HSC quiescence and homing ability by emerging osteolineage cells. Cell reports 2014, 9(2):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cao H, Cao B, Heazlewood CK, Domingues M, Sun X, Debele E, McGregor NE, Sims NA, Heazlewood SY, Nilsson SK: Osteopontin is An Important Regulative Component of the Fetal Bone Marrow Hematopoietic Stem Cell Niche. Cells 2019, 8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boulais PE, Frenette PS: Making sense of hematopoietic stem cell niches. Blood 2015, 125(17):2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Montecino-Rodriguez E, Dorshkind K: B-1 B cell development in the fetus and adult. Immunity 2012, 36(1):13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Souto-Carneiro MM, Sims GP, Girschik H, Lee J, Lipsky PE: Developmental changes in the human heavy chain CDR3. J Immunol 2005, 175(11):7425–7436. [DOI] [PubMed] [Google Scholar]
- 134.Roy A, Bystry V, Bohn G, Goudevenou K, Reigl T, Papaioannou M, Krejci A, O’Byrne S, Chaidos A, Grioni A et al. : High resolution IgH repertoire analysis reveals fetal liver as the likely origin of life-long, innate B lymphopoiesis in humans. Clinical immunology 2017, 183:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gronwall C, Vas J, Silverman GJ: Protective Roles of Natural IgM Antibodies. Frontiers in immunology 2012, 3:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shaw PX, Horkko S, Chang MK, Curtiss LK, Palinski W, Silverman GJ, Witztum JL: Natural antibodies with the T15 idiotype may act in atherosclerosis, apoptotic clearance, and protective immunity. J Clin Invest 2000, 105(12):1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moins-Teisserenc H, Busson M, Herda A, Apete S, Peffault de Latour R, Robin M, Xhaard A, Toubert A, Socie G: CD19+CD5+ B cells and B1-like cells following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2013, 19(6):988–991. [DOI] [PubMed] [Google Scholar]
- 138.Xu X, Deobagkar-Lele M, Bull KR, Crockford TL, Mead AJ, Cribbs AP, Sims D, Anzilotti C, Cornall RJ: An ontogenetic switch drives the positive and negative selection of B cells. Proc Natl Acad Sci U S A 2020, 117(7):3718–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, Yoder MC: Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A 2011, 108(4):1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yoshimoto M: The first wave of B lymphopoiesis develops independently of stem cells in the murine embryo. Ann N Y Acad Sci 2015, 1362:16–22. [DOI] [PubMed] [Google Scholar]
- 141.Montecino-Rodriguez E, Leathers H, Dorshkind K: Identification of a B-1 B cell-specified progenitor. Nat Immunol 2006, 7(3):293–301. [DOI] [PubMed] [Google Scholar]
- 142.Kobayashi M, Shelley WC, Seo W, Vemula S, Lin Y, Liu Y, Kapur R, Taniuchi I, Yoshimoto M: Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfbeta for their development. Proc Natl Acad Sci U S A 2014, 111(33):12151–12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vosshenrich CA, Cumano A, Muller W, Di Santo JP, Vieira P: Pre-B cell receptor expression is necessary for thymic stromal lymphopoietin responsiveness in the bone marrow but not in the liver environment. Proc Natl Acad Sci U S A 2004, 101(30):11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carpino N, Thierfelder WE, Chang MS, Saris C, Turner SJ, Ziegler SF, Ihle JN: Absence of an essential role for thymic stromal lymphopoietin receptor in murine B-cell development. Mol Cell Biol 2004, 24(6):2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alhaj Hussen K, Vu Manh TP, Guimiot F, Nelson E, Chabaane E, Delord M, Barbier M, Berthault C, Dulphy N, Alberdi AJ et al. : Molecular and Functional Characterization of Lymphoid Progenitor Subsets Reveals a Bipartite Architecture of Human Lymphopoiesis. Immunity 2017, 47(4):680–696 e688. [DOI] [PubMed] [Google Scholar]
- 146.Sanz E, Munoz AN, Monserrat J, Van-Den-Rym A, Escoll P, Ranz I, Alvarez-Mon M, de-la-Hera A: Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways. Proc Natl Acad Sci U S A 2010, 107(13):5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sanz E, Alvarez-Mon M, Martinez AC, de la Hera A: Human cord blood CD34+Pax-5+ B-cell progenitors: single-cell analyses of their gene expression profiles. Blood 2003, 101(9):3424–3430. [DOI] [PubMed] [Google Scholar]
- 148.O’Byrne S, Elliott N, Rice S, Buck G, Fordham N, Garnett C, Godfrey L, Crump NT, Wright G, Inglott S et al. : Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood 2019, 134(13):1059–1071. [DOI] [PubMed] [Google Scholar]
- 149.Boiers C, Richardson SE, Laycock E, Zriwil A, Turati VA, Brown J, Wray JP, Wang D, James C, Herrero J et al. : A Human IPS Model Implicates Embryonic B-Myeloid Fate Restriction as Developmental Susceptibility to B Acute Lymphoblastic Leukemia-Associated ETV6-RUNX1. Dev Cell 2018, 44(3):362–377 e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roy A, Cowan G, Mead AJ, Filippi S, Bohn G, Chaidos A, Tunstall O, Chan JK, Choolani M, Bennett P et al. : Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci U S A 2012, 109(43):17579–17584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michaelsson J, Mold JE, McCune JM, Nixon DF: Regulation of T cell responses in the developing human fetus. J Immunol 2006, 176(10):5741–5748. [DOI] [PubMed] [Google Scholar]
- 152.Takahata Y, Nomura A, Takada H, Ohga S, Furuno K, Hikino S, Nakayama H, Sakaguchi S, Hara T: CD25+CD4+ T cells in human cord blood: an immunoregulatory subset with naive phenotype and specific expression of forkhead box p3 (Foxp3) gene. Exp Hematol 2004, 32(7):622–629. [DOI] [PubMed] [Google Scholar]
- 153.Mold JE, Venkatasubrahmanyam S, Burt TD, Michaelsson J, Rivera JM, Galkina SA, Weinberg K, Stoddart CA, McCune JM: Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 2010, 330(6011):1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rudd BD: Neonatal T Cells: A Reinterpretation. Annu Rev Immunol 2020, 38:229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chea S, Schmutz S, Berthault C, Perchet T, Petit M, Burlen-Defranoux O, Goldrath AW, Rodewald HR, Cumano A, Golub R: Single-Cell Gene Expression Analyses Reveal Heterogeneous Responsiveness of Fetal Innate Lymphoid Progenitors to Notch Signaling. Cell reports 2016, 14(6):1500–1516. [DOI] [PubMed] [Google Scholar]
- 156.Rackaityte E, Halkias J: Mechanisms of Fetal T Cell Tolerance and Immune Regulation. Frontiers in immunology 2020, 11:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu MJ, Matsuoka S, Yang FC, Ebihara Y, Manabe A, Tanaka R, Eguchi M, Asano S, Nakahata T, Tsuji K: Evidence for the presence of murine primitive megakaryocytopoiesis in the early yolk sac. Blood 2001, 97(7):2016–2022. [DOI] [PubMed] [Google Scholar]
- 158.Ma DC, Sun YH, Chang KZ, Zuo W: Developmental change of megakaryocyte maturation and DNA ploidy in human fetus. Eur J Haematol 1996, 57(2):121–127. [DOI] [PubMed] [Google Scholar]
- 159.Sparger KA, Ramsey H, Lorenz V, Liu ZJ, Feldman HA, Li N, Laforest T, Sola-Visner MC: Developmental differences between newborn and adult mice in response to romiplostim. Platelets 2018, 29(4):365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Israels SJ, Odaibo FS, Robertson C, McMillan EM, McNicol A: Deficient thromboxane synthesis and response in platelets from premature infants. Pediatr Res 1997, 41(2):218–223. [DOI] [PubMed] [Google Scholar]
- 161.Davizon-Castillo P, Allawzi A, Sorrells M, Fisher S, Baltrunaite K, Neeves K, Nozik-Grayck E, DiPaola J, Delaney C: Platelet activation in experimental murine neonatal pulmonary hypertension. Physiological reports 2020, 8(5):e14386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Margraf A, Nussbaum C, Rohwedder I, Klapproth S, Kurz ARM, Florian A, Wiebking V, Pircher J, Pruenster M, Immler R et al. : Maturation of Platelet Function During Murine Fetal Development In Vivo. Arterioscler Thromb Vasc Biol 2017, 37(6):1076–1086. [DOI] [PubMed] [Google Scholar]
- 163.Stolla MC, Catherman SC, Kingsley PD, Rowe RG, Koniski AD, Fegan K, Vit L, McGrath KE, Daley GQ, Palis J: Lin28b regulates age-dependent differences in murine platelet function. Blood advances 2019, 3(1):72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rowe RG, Wang LD, Coma S, Han A, Mathieu R, Pearson DS, Ross S, Sousa P, Nguyen PT, Rodriguez A et al. : Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis. J Exp Med 2016, 213(8):1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dzierzak E, Philipsen S: Erythropoiesis: development and differentiation. Cold Spring Harbor perspectives in medicine 2013, 3(4):a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Linderkamp O, Wu PY, Meiselman HJ: Geometry of neonatal and adult red blood cells. Pediatr Res 1983, 17(4):250–253. [DOI] [PubMed] [Google Scholar]
- 167.Kim I, Saunders TL, Morrison SJ: Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 2007, 130(3):470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oliveira-Mateos C, Sanchez-Castillo A, Soler M, Obiols-Guardia A, Pineyro D, Boque-Sastre R, Calleja-Cervantes ME, Castro de Moura M, Martinez-Cardus A, Rubio T et al. : The transcribed pseudogene RPSAP52 enhances the oncofetal HMGA2-IGF2BP2-RAS axis through LIN28B-dependent and independent let-7 inhibition. Nat Commun 2019, 10(1):3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li YS, Zhou Y, Tang L, Shinton SA, Hayakawa K, Hardy RR: A developmental switch between fetal and adult B lymphopoiesis. Ann N Y Acad Sci 2015, 1362:8–15. [DOI] [PubMed] [Google Scholar]
- 170.McKinney-Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, Loewer S, Naveiras O, Kathrein KL, Konantz M et al. : The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell 2012, 11(5):701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HK, Yuan GC et al. : Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell 2012, 23(4):796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC et al. : Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell 2016, 36(1):9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen C, Yu W, Tober J, Gao P, He B, Lee K, Trieu T, Blobel GA, Speck NA, Tan K: Spatial Genome Re-organization between Fetal and Adult Hematopoietic Stem Cells. Cell reports 2019, 29(12):4200–4211 e4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gomes AM, Kurochkin I, Chang B, Daniel M, Law K, Satija N, Lachmann A, Wang Z, Ferreira L, Ma’ayan A et al. : Cooperative Transcription Factor Induction Mediates Hemogenic Reprogramming. Cell reports 2018, 25(10):2821–2835 e2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lopez CK, Noguera E, Stavropoulou V, Robert E, Aid Z, Ballerini P, Bilhou-Nabera C, Lapillonne H, Boudia F, Thirant C et al. : Ontogenic Changes in Hematopoietic Hierarchy Determine Pediatric Specificity and Disease Phenotype in Fusion Oncogene-Driven Myeloid Leukemia. Cancer discovery 2019, 9(12):1736–1753. [DOI] [PubMed] [Google Scholar]
- 176.Hotfilder M, Rottgers S, Rosemann A, Schrauder A, Schrappe M, Pieters R, Jurgens H, Harbott J, Vormoor J: Leukemic stem cells in childhood high-risk ALL/t(9;22) and t(4;11) are present in primitive lymphoid-restricted CD34+CD19- cells. Cancer Res 2005, 65(4):1442–1449. [DOI] [PubMed] [Google Scholar]
- 177.Ford AM, Bennett CA, Price CM, Bruin MC, Van Wering ER, Greaves M: Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc Natl Acad Sci U S A 1998, 95(8):4584–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, Saha V, Biondi A, Greaves MF: Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 1999, 354(9189):1499–1503. [DOI] [PubMed] [Google Scholar]
- 179.Zuna J, Madzo J, Krejci O, Zemanova Z, Kalinova M, Muzikova K, Zapotocky M, Starkova J, Hrusak O, Horak J et al. : ETV6/RUNX1 (TEL/AML1) is a frequent prenatal first hit in childhood leukemia. Blood 2011, 117(1):368–369; author reply 370–361. [DOI] [PubMed] [Google Scholar]
- 180.Lausten-Thomsen U, Madsen HO, Vestergaard TR, Hjalgrim H, Nersting J, Schmiegelow K: Prevalence of t(12;21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood 2011, 117(1):186–189. [DOI] [PubMed] [Google Scholar]
- 181.Fueller E, Schaefer D, Fischer U, Krell PF, Stanulla M, Borkhardt A, Slany RK: Genomic inverse PCR for exploration of ligated breakpoints (GIPFEL), a new method to detect translocations in leukemia. PLoS One 2014, 9(8):e104419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kosik P, Skorvaga M, Durdik M, Jakl L, Nikitina E, Markova E, Kozics K, Horvathova E, Belyaev I: Low numbers of pre-leukemic fusion genes are frequently present in umbilical cord blood without affecting DNA damage response. Oncotarget 2017, 8(22):35824–35834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Skorvaga M, Nikitina E, Kubes M, Kosik P, Gajdosechova B, Leitnerova M, Copakova L, Belyaev I: Incidence of common preleukemic gene fusions in umbilical cord blood in Slovak population. PLoS One 2014, 9(3):e91116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schafer D, Olsen M, Lahnemann D, Stanulla M, Slany R, Schmiegelow K, Borkhardt A, Fischer U: Five percent of healthy newborns have an ETV6-RUNX1 fusion as revealed by DNA-based GIPFEL screening. Blood 2018, 131(7):821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hein D, Dreisig K, Metzler M, Izraeli S, Schmiegelow K, Borkhardt A, Fischer U: The preleukemic TCF3-PBX1 gene fusion can be generated in utero and is present in approximately 0.6% of healthy newborns. Blood 2019, 134(16):1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Puumala SE, Ross JA, Aplenc R, Spector LG: Epidemiology of childhood acute myeloid leukemia. Pediatr Blood Cancer 2013, 60(5):728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J et al. : The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 2015, 47(4):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pine SR, Guo Q, Yin C, Jayabose S, Druschel CM, Sandoval C: Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood 2007, 110(6):2128–2131. [DOI] [PubMed] [Google Scholar]
- 189.Horton SJ, Jaques J, Woolthuis C, van Dijk J, Mesuraca M, Huls G, Morrone G, Vellenga E, Schuringa JJ: MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia 2013, 27(5):1116–1126. [DOI] [PubMed] [Google Scholar]
- 190.Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, Chen SH, Hou JY, Huang YJ, Shih YS et al. : Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 2013, 121(15):2988–2995. [DOI] [PubMed] [Google Scholar]
- 191.Cancer Genome Atlas Research N: Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013, 368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bateman CM, Colman SM, Chaplin T, Young BD, Eden TO, Bhakta M, Gratias EJ, van Wering ER, Cazzaniga G, Harrison CJ et al. : Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood 2010, 115(17):3553–3558. [DOI] [PubMed] [Google Scholar]
- 193.Pagano L, Pulsoni A, Vignetti M, Mele L, Fianchi L, Petti MC, Mirto S, Falcucci P, Fazi P, Broccia G et al. : Acute megakaryoblastic leukemia: experience of GIMEMA trials. Leukemia 2002, 16(9):1622–1626. [DOI] [PubMed] [Google Scholar]
- 194.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S: Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet 2004, 5(10):725–738. [DOI] [PubMed] [Google Scholar]
- 195.Henry E, Walker D, Wiedmeier SE, Christensen RD: Hematological abnormalities during the first week of life among neonates with Down syndrome: data from a multihospital healthcare system. Am J Med Genet A 2007, 143A(1):42–50. [DOI] [PubMed] [Google Scholar]
- 196.de Rooij JD, Branstetter C, Ma J, Li Y, Walsh MP, Cheng J, Obulkasim A, Dang J, Easton J, Verboon LJ et al. : Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet 2017, 49(3):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gruber TA, Downing JR: The biology of pediatric acute megakaryoblastic leukemia. Blood 2015, 126(8):943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, Ta HQ, Chen SC, Su X, Ogden SK, Dang J et al. : An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012, 22(5):683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Morrow M, Horton S, Kioussis D, Brady HJ, Williams O: TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood 2004, 103(10):3890–3896. [DOI] [PubMed] [Google Scholar]
- 200.Torrano V, Procter J, Cardus P, Greaves M, Ford AM: ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood 2011, 118(18):4910–4918. [DOI] [PubMed] [Google Scholar]
- 201.Martin-Lorenzo A, Hauer J, Vicente-Duenas C, Auer F, Gonzalez-Herrero I, Garcia-Ramirez I, Ginzel S, Thiele R, Constantinescu SN, Bartenhagen C et al. : Infection Exposure is a Causal Factor in B-cell Precursor Acute Lymphoblastic Leukemia as a Result of Pax5-Inherited Susceptibility. Cancer discovery 2015, 5(12):1328–1343. [DOI] [PubMed] [Google Scholar]
- 202.Rodriguez-Hernandez G, Hauer J, Martin-Lorenzo A, Schafer D, Bartenhagen C, Garcia-Ramirez I, Auer F, Gonzalez-Herrero I, Ruiz-Roca L, Gombert M et al. : Infection Exposure Promotes ETV6-RUNX1 Precursor B-cell Leukemia via Impaired H3K4 Demethylases. Cancer Res 2017, 77(16):4365–4377. [DOI] [PubMed] [Google Scholar]
- 203.Burgler S, Nadal D: Pediatric precursor B acute lymphoblastic leukemia: are T helper cells the missing link in the infectious etiology theory? Molecular and cellular pediatrics 2017, 4(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sullivan EM, Jeha S, Kang G, Cheng C, Rooney B, Holladay M, Bari R, Schell S, Tuggle M, Pui CH et al. : NK cell genotype and phenotype at diagnosis of acute lymphoblastic leukemia correlate with postinduction residual disease. Clin Cancer Res 2014, 20(23):5986–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cazzaniga G, van Delft FW, Lo Nigro L, Ford AM, Score J, Iacobucci I, Mirabile E, Taj M, Colman SM, Biondi A et al. : Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood 2011, 118(20):5559–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M: Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood 1999, 94(3):1057–1062. [PubMed] [Google Scholar]
- 207.Maia AT, van der Velden VH, Harrison CJ, Szczepanski T, Williams MD, Griffiths MJ, van Dongen JJ, Greaves MF: Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia 2003, 17(11):2202–2206. [DOI] [PubMed] [Google Scholar]
- 208.Gamis AS, Smith FO: Transient myeloproliferative disorder in children with Down syndrome: clarity to this enigmatic disorder. Br J Haematol 2012, 159(3):277–287. [DOI] [PubMed] [Google Scholar]
- 209.Matsuda K, Shimada A, Yoshida N, Ogawa A, Watanabe A, Yajima S, Iizuka S, Koike K, Yanai F, Kawasaki K et al. : Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood 2007, 109(12):5477–5480. [DOI] [PubMed] [Google Scholar]
- 210.Pieters R, De Lorenzo P, Ancliffe P, Aversa LA, Brethon B, Biondi A, Campbell M, Escherich G, Ferster A, Gardner RA et al. : Outcome of Infants Younger Than 1 Year With Acute Lymphoblastic Leukemia Treated With the Interfant-06 Protocol: Results From an International Phase III Randomized Study. J Clin Oncol 2019, 37(25):2246–2256. [DOI] [PubMed] [Google Scholar]
- 211.Tomizawa D, Miyamura T, Imamura T, Watanabe T, Moriya Saito A, Ogawa A, Takahashi Y, Hirayama M, Taki T, Deguchi T et al. : A risk-stratified therapy for infants with acute lymphoblastic leukemia: a report from the JPLSG MLL-10 trial. Blood 2020, 136(16):1813–1823. [DOI] [PubMed] [Google Scholar]
- 212.Pui CH, Carroll WL, Meshinchi S, Arceci RJ: Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol 2011, 29(5):551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Salzer WL, Devidas M, Carroll WL, Winick N, Pullen J, Hunger SP, Camitta BA: Long-term results of the pediatric oncology group studies for childhood acute lymphoblastic leukemia 1984–2001: a report from the children’s oncology group. Leukemia 2010, 24(2):355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Jackson TR, Ling RE, Roy A: The Origin of B-cells: Human Fetal B Cell Development and Implications for the Pathogenesis of Childhood Acute Lymphoblastic Leukemia. Frontiers in immunology 2021, 12:637975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Greaves M: A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer 2018, 18(8):471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Montecino-Rodriguez E, Li K, Fice M, Dorshkind K: Murine B-1 B cell progenitors initiate B-acute lymphoblastic leukemia with features of high-risk disease. J Immunol 2014, 192(11):5171–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Choi YJ, Heck AM, Hayes BJ, Lih D, Rayner SG, Hadland B, Zheng Y: WNT5A from the fetal liver vascular niche supports human fetal liver hematopoiesis. Stem cell research & therapy 2021, 12(1):321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is not applicable for this review.


