Abstract
Objectives
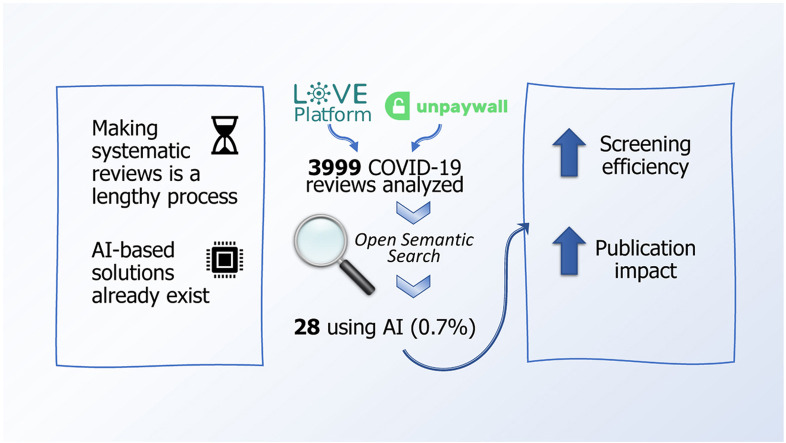
A rapidly developing scenario like a pandemic requires the prompt production of high-quality systematic reviews, which can be automated using artificial intelligence (AI) techniques. We evaluated the application of AI tools in COVID-19 evidence syntheses.
Study Design
After prospective registration of the review protocol, we automated the download of all open-access COVID-19 systematic reviews in the COVID-19 Living Overview of Evidence database, indexed them for AI-related keywords, and located those that used AI tools. We compared their journals’ JCR Impact Factor, citations per month, screening workloads, completion times (from pre-registration to preprint or submission to a journal) and AMSTAR-2 methodology assessments (maximum score 13 points) with a set of publication date matched control reviews without AI.
Results
Of the 3,999 COVID-19 reviews, 28 (0.7%, 95% CI 0.47–1.03%) made use of AI. On average, compared to controls (n = 64), AI reviews were published in journals with higher Impact Factors (median 8.9 vs. 3.5, P < 0.001), and screened more abstracts per author (302.2 vs. 140.3, P = 0.009) and per included study (189.0 vs. 365.8, P < 0.001) while inspecting less full texts per author (5.3 vs. 14.0, P = 0.005). No differences were found in citation counts (0.5 vs. 0.6, P = 0.600), inspected full texts per included study (3.8 vs. 3.4, P = 0.481), completion times (74.0 vs. 123.0, P = 0.205) or AMSTAR-2 (7.5 vs. 6.3, P = 0.119).
Conclusion
AI was an underutilized tool in COVID-19 systematic reviews. Its usage, compared to reviews without AI, was associated with more efficient screening of literature and higher publication impact. There is scope for the application of AI in automating systematic reviews.
Keywords: Artificial intelligence, Systematic review, COVID-19, Automation, Research design, Bibliometrics
Graphical abstract
What is new?
Key findings
-
•
The use of artificial intelligence (AI) in COVID-19 systematic reviews was very low.
-
•
COVID-19 reviews using AI tools showed higher publication impact and workload savings.
What this adds to what was known?
-
•
Semi-automated screening and RCT filtering are the most notable use-cases of AI tools in evidence synthesis.
-
•
There is a lack of systematic review tools cohesively integrating AI.
What is the implication and what should change now?
-
•
There is scope for the application of AI in automating systematic reviews going forward.
1. Introduction
Evidence-based medicine depends on the production of timely systematic reviews to guide and update health care practice and policies [1]. This is a resource-intensive undertaking, requiring teams of multiple reviewers to interrogate numerous repositories and databases, screen through thousands of potentially relevant citations and articles, extract the pertinent data from the selected studies, and then prepare cohesive summaries of the findings [2,3]. In the context of the SARS-CoV2/COVID-19 pandemic, methods to speed up this lengthy process were urgently needed [4,5].
Systematic evidence synthesis relies on robust and standardized procedures to achieve dependable results. However, the call to accelerate research output during the pandemic led to a decrease on reviews’ methodological quality [6,7] and the ascend of “rapid reviews” [8,9] (which shorten the usual timeframes by sacrificing on search depth, screening robustness or data extraction and at the expense of increased risk of errors). Are these unavoidable tradeoffs for timelier results?
Instead, artificial intelligence (AI) based solutions (that automate parts of the workflow by mimicking human problem-solving, comprising machine-learning, nature language processing, data mining and other subfields) [10] are now available to either complement or substitute human efforts with limited risk of bias [[11], [12], [13]], and have been previously (but scarcely) [14] employed in evidence synthesis to enhance screening [15] and data extraction [16,17]. Their aims are to shorten production times, allow for broader screenings of the literature and reduce reviewers’ workloads without compromising on methodological quality.
Here, we evaluated the use of AI techniques among COVID-19 evidence syntheses to empirically determine whether, compared to COVID-19 evidence syntheses without AI, they impacted on the production, the quality, and the publication of systematic reviews.
2. Materials and methods
This methodological study [18] is reported following PRISMA 2020 guidelines [19] (checklist provided as Supplementary material 1A), and its protocol was prospectively registered at Open Science Forum Registries (DOI 10.17605/OSF.IO/H5DAW) [20].
2.1. Search and selection of reviews
We considered for inclusion all COVID-19 related systematic reviews that could have made use of any AI tool (machine learning, deep learning, or natural language processing) to accelerate, improve or complement any aspect of the review conduct (search, screening, data extraction and synthesis). We implemented a script (available at DOI 10.5061/dryad.9kd51c5j6) [21] to process all COVID-19 bibliographic references registered in the COVID-19 Living Overview of Evidence (L·OVE) database [22], filtering articles classified as “systematic review” between December 1st, 2019 and August 15th, 2021, and then querying the “Unpaywall” database [23] for every extracted DOI to obtain a JSON record with download links. The process was repeated three times since the publication of our protocol to reduce the loss of articles due to server-side errors (last searched on August 17th, 2021).
To capture reviews which deployed AI, we constructed a list of keywords with high probability of appearing in papers with AI tools (Supplementary Material 1B). We indexed every downloaded file with the OpenSemanticSearch search engine, running on a local Linux virtual machine. Every file that matched any of our keywords was manually inspected independently by two authors (JRTH and RFL). Pre-prints and non-English articles were included. The only exclusion criterion applied was non-open access status, due to the need to evaluate the methods section of each included review. To create a comparison group with sufficient statistical power of reviews without AI, for each included review we used the obtained records to randomly select three controls with the same publication date (within a 1-day margin if not enough articles were available for a given date). In addition, we located and included for analysis all previous versions of reviews labeled as living or “updated”.
2.2. Data extraction
The following data were manually extracted independently by two authors (JRTH and RFL) from each review: type of review (as described by its authors: standard, rapid/scoping, living, or update of a prior version); disclosed funding and conflicts of interest information; publication status, 2020 Journal Citation Reports (JCR) Impact Factor of the publishing journal and number of citations received (up to August 17th, 2021); number of abstracts screened, full texts reviewed and included studies; number of authors and of reviewers participating in the screening; and dates of protocol registration (if available) and of the review's earliest version. For living and updated reviews, we computed the increase in records screened and included between each of their versions and attributed their citation count to the newest one (to avoid double counting). Excel was used to record all variables.
Three authors (JRTH and RFL, assisted by CAP) graded all reviews with the AMSTAR-2 quality appraisal and risk of bias rating [24]. We excluded items 11-12 and 15, which apply to meta-analyses (as pre-specified by our protocol) and gave 0.5 points for “partial YES” answers when applicable, making for a maximum score of 13 points. For living and updated reviews, we only evaluated their most recent version (to avoid double counting). For reviews that included both randomized controlled trials and observational studies, question 9 (assessment of the risk of bias of individual studies) was graded separately for each study type. The list of the quality items evaluated is provided as Supplementary material 1C.
2.3. Data synthesis
We calculated the ratios of abstracts screened and full texts inspected per author (as workload measurement) and per included study (screening precision). The number of reviewers participating in the screening was reported inconsistently between studies and was therefore not used in the calculations. We calculated the completion time of the pre-registered reviews as the difference between their protocol's date and the first pre-print's date of publication (or reception date at the journal, for published articles with no pre-prints available). Living and updated reviews' completion times were calculated as the difference between the publication dates of each of their versions. We excluded non pre-registered reviews from this metric due to heterogeneity in the reporting of their starting dates. We used Pearson's chi-square test to compare the percentage of rapid, living, funded, and published reviews between groups. Publishing journals' JCR Impact Factor, citation counts, screening workloads, completion times and AMSTAR-2 ratings were presented as medians with interquartile ranges (IQR), represented using box-and-whisker diagrams and compared using the Wilcoxon–Mann–Whitney test. R version 4.0.5 was used for statistical computing, and GraphPad Prism 9.2.0 for graphing. We also provided a narrative description of reviews using artificial intelligence, detailing which parts of the review process were automated and what software they used, how the AMSTAR-2 ratings differed among them, and how authors justified or what impact they attributed to the use of AI tools.
3. Results
3.1. Search and selection of reviews
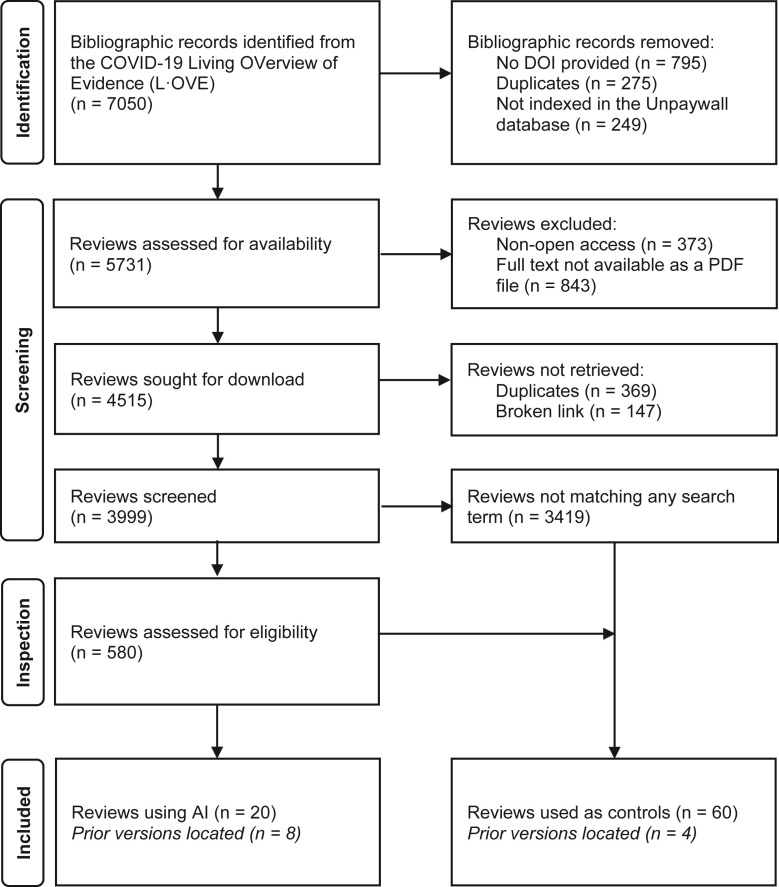
As outlined in Figure 1 , we identified 7,050 bibliographic records of COVID-19 systematic reviews, successfully downloaded 3,999, and manually inspected 580 that matched some of our keywords. We selected 20 reviews, of which there were 8 prior versions, making a total of 28 reviews (0.7% of the total, 95% CI 0.47–1.03%) with use of AI. Of the 60 articles selected as publication-date-matched controls, we located another 4 prior versions, making a total of 64 articles without use of AI. The complete list of selected articles is provided as an Excel document (Supplementary Material 2, sheet “Included reviews”) with all the extracted variables and the AMSTAR-2 quality appraisal's breakdown for each question. The full list of manually inspected and finally discarded articles is also provided (sheet “Excluded reviews”).
Fig. 1.
Flowchart of included reviews: Flowchart of records obtained, screened, assessed for eligibility, and included in our study.
3.2. Description of the included reviews
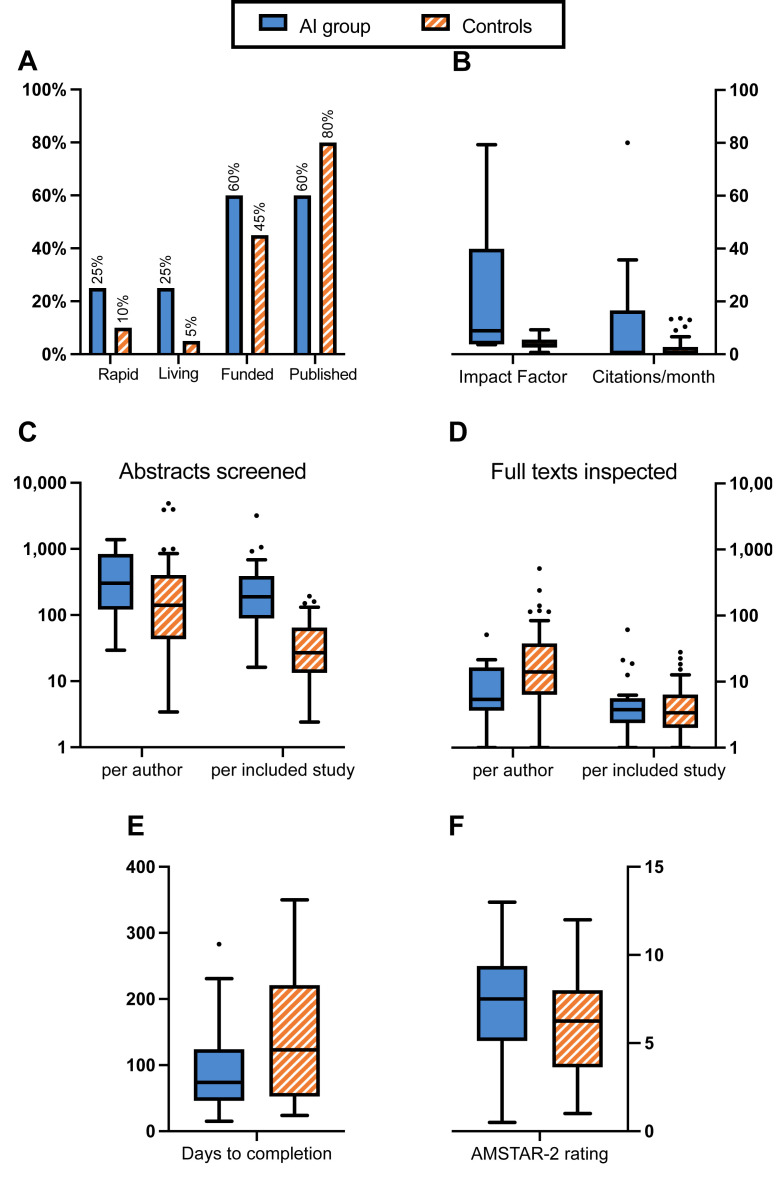
Extracted variables are summarized in Table 1 and can be visualized in Figure 2 . Of the 20 reviews selected for using AI, there were five rapid reviews (25%, with one scoping review and one rapid evidence map) and five living reviews (25%). Fifteen reviews provided a conflicts of interest statement, of which 12 (60%) declared having received external funding; 12 (60%) were published. Of the 60 control reviews, there were 6 rapid reviews (10%, with one scoping review) and three living reviews (5%). Fifty-seven reviews provided a conflicts of interest statement, of which 27 (45%) declared having received external funding; 48 (80%) were published. JCR Impact Factors and citation counts showed high variability in the AI group, mainly due to the inclusion of three BMJ [[25], [26], [27]], two Cochrane [28,29] and one Lancet [30] reviews. Furthermore, only 10 reviews in the AI group (50%) and 22 in the controls (36%) pre-registered a protocol, making for a total of 44 data points for the completion times’ calculation.
Table 1.
Extracted variables for artificial intelligence (AI) and control reviews: We used Pearson's chi-square test to compare the proportions of rapid, living, funded, and published reviews, and the Wilcoxon–Mann–Whitney test for the rest of the comparisons. Medians and IQR (Q1-Q3) are rounded to the nearest integer
| Characteristics | AI group (n = 20) |
Controls (n = 60) |
Δ | χ2 | P-value | ||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | ||||
| Rapid reviews | 5 | (25%) | 6 | (10%) | 15% | 2.846 | 0.092 |
| Living reviews | 5 | (25%) | 3 | (5%) | 20% | 6.667 | 0.010 |
| Received funding | 12 | (60%) | 27 | (45%) | 15% | 1.351 | 0.245 |
| Published |
12 |
(60%) |
48 |
(80%) |
−20% |
3.200 |
0.074 |
|
Median |
IQR |
Median |
IQR |
Wilcoxon W |
P-value |
||
| Journals' JCR Impact Factor | 9 | (4–40) | 3 | (3–6) | 409.0 | <0.001 | |
| Citations per month | 1 | (0–13) | 1 | (0–3) | 647.0 | 0.600 | |
| Abstracts screened | |||||||
| Per author | 302 | (127–804) | 140 | (44–378) | 1,126.0 | 0.009 | |
| Per included study | 189 | (94–366) | 27 | (14–64) | 1,443.0 | <0.001 | |
| Full texts inspected | |||||||
| Per author | 5 | (4–16) | 14 | (7–37) | 504.5 | 0.005 | |
| Per included study | 4 | (2–5) | 3 | (2–6) | 883.5 | 0.481 | |
| Days to completion | 74 | (48–118) | 123 | (53–221) | 183.5 | 0.205 | |
| AMSTAR-2 rating | 8 | (5–9) | 6 | (4–8) | 740.5 | 0.119 | |
Δ, absolute differences in percentage points between AI and control reviews; χ2, test statistic for Pearson's chi-square test; Wilcoxon W, test statistic for the Wilcoxon–Mann–Whitney rank sum test.
Fig. 2.
Characteristics of the included reviews: Box-and-whisker diagram (the boxes enclose the Q1-Q3 quartiles, their middle lines represent the median, and whiskers extend to the furthest data points within 1.5 IQR). Panel A compares the proportion of rapid, living, funded, and published reviews between groups; Panel B presents the journals' 2020 JCR Impact Factors and citation counts of each group; Panels C and D show authors' workload measurements: abstracts screened and full-texts inspected, per author and per included study; Panel E exhibits the average times to completion (in days) of the reviews in each group; and Panel F represents their measured AMSTAR-2 ratings.
3.3. Comparison of AI reviews with controls
The AI group included a higher proportion of living reviews than the controls (5/20 vs. 3/60, 95% CI absolute difference 0.2–39.8%, P = 0.010), while showing no differences in rapid reviews (5/20 vs. 6/60, 95% CI −5.4 to 35.4%, P = 0.092), funding (12/20 vs. 27/60, 95% CI −9.9 to 39.9%, P = 0.245) or publication status (12/20 vs. 48/60, 95% CI −43.7 to 3.7%, P = 0.074). JCR impact factors among published reviews in the AI group were significantly higher than the controls (median [IQR]: 8.9 [3.9–39.9] vs. 3.5 [2.6–5.5], P < 0.001); citation counts showed no differences (0.5 [0.0–13.5] vs. 0.6 [0.0–2.8], P = 0.600).
Concerning the workload measurements, the AI group screened more abstracts per author (302.2 [126.7–804.3] vs. 140.3 [43.8–378.2], P = 0.009) and per included study (189.0 [94.1–365.8] vs. 26.9 [13.7–64.1], P < 0.001), while inspecting less full texts per author (5.3 [3.7–16.1] vs. 14.0 [6.5–37.2], P = 0.005) and as many per included study (3.8 [2.4–5.3] vs. 3.4 [2.0–6.2], P = 0.481).
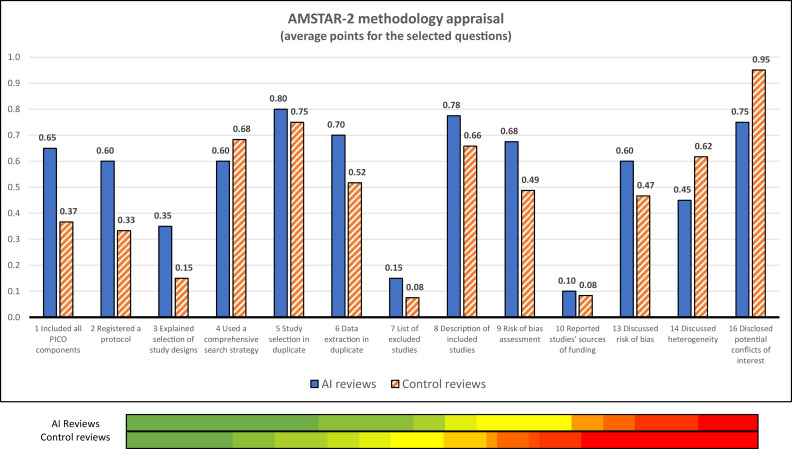
We observed no differences in the pre-registered reviews’ times to completion (74.0 [47.5–117.5] vs. 123.0 [53.0–221.0], P = 0.205). The average scores obtained in the AMSTAR-2 risk of bias rating were not significantly higher in the AI group (7.5 [5.3–9.1] vs. 6.3 [3.9–8.0] points out of 13, P = 0.119), with both groups showing high heterogeneity of results as shown in Figure 3 . Measured against the controls, the AI reviews scored worse on question 4 (literature search strategy, −12%) and better on question 6 (data extraction in duplicate, 35%), while showing minimal differences on question 5 (duplicate screening, 7%). Both groups scored the lowest on questions 7 (providing a list of excluded studies) and 10 (reporting on the sources of funding of the included studies).
Fig. 3.
AMSTAR-2 methodology appraisals' summary: The graph on the top shows the average ratings obtained in each of the evaluated questions by the reviews using Artificial Intelligence (AI) techniques (blue bars) and by the control group (orange bars). The colored bars on the bottom provide a visual representation of the quality appraisal's heterogeneity in both groups (a gradient was used to represent the obtained scores: red = 4; yellow = 6.5; green = 9).
3.4. Narrative description of the uses of AI in the included reviews
According to the step of the review process where AI was used, we can classify the 20 reviews in the AI group in three categories, as shown in Table 2 .
Table 2.
AI tools used in COVID-19 reviews: Table showing the different artificial intelligence (AI) tools that have been used in the elaboration of COVID-19 systematic reviews, according to their area of application: search assistance, randomized controlled trials (RCT) filtering and screening automation
| Ref. | Title | Authors | Journal | AI used in… | Software used | Is open source? |
|---|---|---|---|---|---|---|
| [31] | Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients with Coronavirus Disease 2019 | Parasa et al. | JAMA Network Open | Search | CORD-19 | Partially |
| [32] | The influence of comorbidity on the severity of COVID-19 disease: systematic review and analysis | Zaki et al. | Pre-print | Search | CORD-19 + Okapi BM25 | Yes |
| [33] | The Estimations of the COVID-19 Incubation Period: A Scoping Reviews of the Literature | Zaki et al. | Journal of Infection and Public Health | Search | CORD-19 + BioBERT | Yes |
| [34] | Ocular toxicity and Hydroxychloroquine: A Rapid Meta-Analysis | Michelson et al. | Pre-print | Search | GenesisAI (formerly Evid Science) | No |
| [35] | A Systematic Review of the Incubation Period of SARS-CoV-2: The Effects of Age, Biological Sex, and Location on Incubation Period | Daley et al. | Pre-print | Search | Not reported | No |
| [36] | Impact of remdesivir on 28 day mortality in hospitalized patients with COVID-19: February 2021 Meta-analysis | Robinson et al. | Pre-print | RCT filtering | RobotSearch | Yes |
| [37] | Impact of systemic corticosteroids on hospitalized patients with COVID-19: January 2021 Meta-analysis of randomized controlled trials | Robinson et al. | Pre-print | RCT filtering | RobotSearch | Yes |
| [25] | Prophylaxis against COVID-19: living systematic review and network meta-analysis | Bartoszko et al. | BMJ | RCT filtering | RobotSearch | Yes |
| [26] | Drug treatments for COVID-19: living systematic review and network meta-analysis | Siemieniuk et al. | BMJ | RCT filtering | RobotSearch | Yes |
| [38] | Adverse effects of remdesivir, hydroxychloroquine, and lopinavir/ritonavir when used for COVID-19: systematic review and meta-analysis of randomized trials | Izcovich et al. | Pre-print | RCT filtering | RobotSearch | Yes |
| [39] | Tocilizumab and sarilumab alone or in combination with corticosteroids for COVID-19: A systematic review and network meta-analysis | Zeraatkar et al. | Pre-print | RCT filtering | RobotSearch | Yes |
| [40] | Clinical trials in COVID-19 management & prevention: A meta-epidemiological study examining methodological quality | Honarmand et al. | Journal of Clinical Epidemiology | RCT filtering | RobotSearch | Yes |
| [41] | Impacts of school closures on physical and mental health of children and young people: a systematic review | Viner et al. | Pre-print | Screening | EPPI-Reviewer | No |
| [27] | Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal | Wynants et al. | BMJ | Screening | EPPI-Reviewer | No |
| [28] | Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection (Review) | Dinnes et al. | Cochrane Database of Systematic Reviews | Screening | EPPI-Reviewer | No |
| [29] | Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 | Struyf et al. | Cochrane Database of Systematic Reviews | Screening | EPPI-Reviewer | No |
| [42] | Are medical procedures that induce coughing or involve respiratory suctioning associated with increased generation of aerosols and risk of SARS-CoV-2 infection? A rapid systematic review | Wilson et al. | Journal of Hospital Infection | Screening | EPPI-Reviewer | No |
| [43] | Risk and Protective Factors in the COVID-19 Pandemic: A Rapid Evidence Map | Elmore et al. | Frontiers in Public Health | Screening | SWIFT-Active Screener | No |
| [44] | Tocilizumab and Systemic Corticosteroids in the Management of COVID-19 Patients: A Systematic Review and Meta-Analysis | Alkofide et al. | International Journal of Infectious Diseases | Screening | Abstrackr | Yes |
| [30] | Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis | Chu et al. | The Lancet | Screening | Evidence Prime | No |
3.4.1. Search process
Three reviews [[31], [32], [33]] complemented their search procedures with open-ended question queries on CORD-19 [45], an open dataset of COVID-19 related articles structured to facilitate the use of text mining and machine learning systems: Zaki et al. [32] used a GitHub repository based on the Okapi BM25 search algorithm; Zaki et al. [33] employed BioBERT, a peer-reviewed [46] and open-source text mining system pre-trained for biomedical content analysis; and Parasa et al. [31] provided no details on the search engine employed. Additionally, Michelson et al. [34] used proprietary software from the “GenesisAI” company to produce a “rapid meta-analysis” as proof-of-concept of their product. Daley et al. [35] disclosed no information on the software employed. Only two reviews in this sub-group were published, and none registered a protocol. The average AMSTAR-2 score was 3.7/13.
3.4.2. Filtering of randomized controlled trials
Seven articles [25,26,[36], [37], [38], [39], [40]] employed RobotSearch, a peer-reviewed [47] and open-source software to identify randomized controlled trials (RCT) from a citations list. It is based on a neural network trained with data from Cochrane's reviews and stands out for its ease of use (no installation is required) and flexibility (as it allows for different levels of sensitivity, including one developed specifically for systematic reviews, as well as integration with other scripts).
In our sample, RobotSearch was often incorporated in the workflows of living or partially automated reviews. Two of the reviews that made use of RobotSearch were Bartoszko et al. [25], a network meta-analysis of the evidence for COVID-19 prophylaxis, and Siemieniuk et al. [26], a living meta-analysis of randomized trials to inform World Health Organization (WHO) Living Guidelines on drugs for treatment of COVID-19, of which Izcovich et al. [38] and Zeraatkar et al. [39] are separate sub-studies. Both are part of the “BMJ Rapid Recommendations” project and maintain a website where summaries of the evidence available and interim analyses are published. The average AMSTAR-2 score was 7.5/13.
3.4.3. Screening of titles and abstracts
We found eight articles [[27], [28], [29], [30],[41], [42], [43], [44]] that made use of AI-powered screening procedures. Five of them [[27], [28], [29],41,42] used EPPI-Reviewer, a web-based tool (distributed as shareware) to assist in the elaboration of all kinds of literature reviews. It offers a wide variety of features, from bibliographic management to collaborative working, as well as study identification capabilities, automatic clustering of articles, and text mining. In particular, the included reviews used its “SGCClassifier” module to prioritize the screening of articles more likely to be included. As a result, both Wynants et al. [27] and two Cochrane reviews [28,29] quoted a 80% reduction in the screening burden due to this tool.
Similar screening automation techniques from systematic reviews' elaboration platforms were used by other two articles: SWIFT-Active Screener [48] by Elmore et al. [43], which was set to achieve a certain study recall objective as the screening's stopping criterion; and Evidence Prime by Chu et al. [30], to double-check the screening process. Finally, Alkofide et al. [44] used Abstrackr, the only open-source software in this category, which uses feedback from previously selected and rejected articles to guide the screening process. Evaluations of this tool published in the literature [49] suggest high workload savings in the production of systematic reviews at the cost of 0.1% false negative rates.
Among the reviews analyzed in this study, this subgroup presented the highest scores in the AMSTAR-2 appraisal tool (9.1/13), with the notable mentions of two Cochrane reviews [28,29] (12 points) and a rapid meta-analysis [30] published in the Lancet (10.5 points). Contrary to reviews in the other categories that prioritized search depth, the use of AI-powered tools in this subgroup was motivated by the screening burden faced by the reviewers: quoting Dinnes et al. [28], “a more efficient approach [was needed] to keep up with the rapidly increasing volume of COVID-19 literature”.
4. Discussion
We evaluated if the potential benefits of deploying AI in evidence syntheses have been realized in COVID-19 reviews. We found that AI was rarely utilized, appearing in only 0.7% of the studied reviews, but that it was significantly associated with reductions in authors’ screening workload and publication in journals with higher Impact Factor. Being a living review was associated with using AI, with the most common use cases being the optimization of screening (prioritizing studies with high likelihood of being relevant) and the selection of randomized controlled trials.
As a limitation of our study, we would highlight its low statistical power due to the small number of reviews using AI. Anticipating the limited availability of reviews with AI, we adopted a highly sensitive screening procedure, processing more than 7,000 bibliographic references of COVID-19 systematic reviews (combining expert advice in the selection of keywords and a fully-featured search engine), and chose a 3:1 control group size to minimize the risk of type II statistical errors. Using L-OVE as our primary database allowed access to all relevant and updated sources in a systematic and machine-readable way; however, our search strategy might show a reduced sensitivity to institutional reports and whitepapers, often not indexed by traditional databases. The impact of download errors and excluding non-open-access reviews from our study is uncertain; its influence on generalizing our results should be interpreted in light of the diversity of secondary sources reachable through L-OVE and the high accessibility of COVID-19 research during the pandemic. Furthermore, the use of publication dates as a matching variable allowed for a bias-minimizing (script-driven) selection of controls but it prevented the use of other desirable controlling variables such as review sizes or goals.
We also note that reporting workloads “per author” instead of “per reviewer participating in the screening” may underestimate workload measurements for large teams (when not all their authors participate in the screening). A higher author count might also be related to resource availability, and thus access to expert advice regarding AI. Likewise, better-resourced groups with AI expert support might have greater access to well-indexed journals, potentially biasing Impact Factor analyses in favor of AI. The AMSTAR-2 tool was inevitably applied without blinding the reviewers to use or non-use of AI, which, given the subjectiveness of certain aspects of the methodology assessment, might have influenced this evaluation. Finally, the use of citation counts to measure reviews’ impact has known deficiencies such as being influenced by citation bias or the authority of the authors [50], and this approach may underestimate the impact of recently published reports.
On average, it takes 15 months for teams of five reviewers to complete a traditional systematic review [51], with estimated screening error rates of around 10% [52]. Facing the COVID-19 pandemic demanded robust evidence summaries with urgency as delays incurred cost in terms of lost lives and economic damage. However, despite the explosive growth that the AI and machine learning fields have experienced during the last years, they played a surprisingly limited role in COVID-19 evidence synthesis. Our findings are consistent with previous reports [14] that the benefits AI can provide in the conduct of systematic reviews are unknown to most review authors, while the relative unorthodoxy of its methods might initially hinder their acceptance by the research community. Open-source software, more prone to community adoption, will be essential in this aspect. Hopefully, our article will raise the profile of AI in evidence syntheses.
Our narrative description of the reviews included in this study showed that none made use of more than one AI-tool. A more cohesive approach, seamlessly merging AI into every step of the review process, would save reviewers’ time trying to interconnect different tools with sometimes incompatible formats. Semi-automated screening procedures were one of the areas where AI showed more adoption, and the variety of software options (such as EPPI-Reviewer, already adopted as a Cochrane Review Production Tool) was higher. On the contrary, full automation was only employed by RobotSearch (an extensively appraised randomized trials identifier), suggesting that the adoption of increasingly automated solutions may be hindered by the need to further assess their potential cost on recall and risk-of-bias against their productivity contributions.
5. Conclusion
The need for automated solutions in research synthesis is obvious, as reviewers' workload is growing with the rapidly expanding biomedical field. Adoption of new technologies can take time, but realizing AI's potential in evidence synthesis should be a priority. Going forward, AI must be incorporated to systematic reviews as the next step toward timely, better, and more responsive decision-making.
CRediT authorship contribution statement
Juan R. Tercero-Hidalgo: Methodology, Software, Investigation, Writing – Original Draft. Khalid S. Khan: Conceptualization, Methodology, Writing – Review & Editing, Supervision. Aurora Bueno-Cavanillas: Conceptualization, Methodology, Supervision, Project administration. Rodrigo Fernández-López: Investigation, Data Curation, Writing – Review & Editing. Juan F. Huete: Software, Validation, Resources, Writing – Review & Editing. Carmen Amezcua-Prieto: Validation, Visualization, Writing – Review & Editing. Javier Zamora: Methodology, Formal analysis, Resources, Writing – Review & Editing. Juan M. Fernández-Luna: Conceptualization, Methodology, Software, Validation, Writing – Review & Editing, Supervision. All authors contributed to the editing of the paper and approved its final version to be submitted.
Footnotes
Funding and conflicts of interest information: JRTH thanks the University of Granada for supporting his work through a Research Initiation Grant for Undergraduate Students. There are no conflicts of interest to declare.
Data statement: The data that support the findings of this study are available in Dryad (DOI 10.5061/dryad.9kd51c5j6). These data were derived from the following resources available in the public domain: Living Overview of the Evidence, Unpaywall, 2020 Journal Citation Reports. The protocol of this study was pre-registered at Open Science Forum Registries (DOI 10.17605/OSF.IO/H5DAW).
CRediT author statement: Juan R. Tercero-Hidalgo: Methodology, Software, Investigation, Writing – Original Draft. Khalid S. Khan: Conceptualization, Methodology, Writing – Review & Editing, Supervision. Aurora Bueno-Cavanillas: Conceptualization, Methodology, Supervision, Project administration. Rodrigo Fernández-López: Investigation, Data Curation, Writing – Review & Editing. Juan F. Huete: Software, Validation, Resources, Writing – Review & Editing. Carmen Amezcua-Prieto: Validation, Visualization, Writing – Review & Editing. Javier Zamora: Methodology, Formal analysis, Resources, Writing – Review & Editing. Juan M. Fernández-Luna: Conceptualization, Methodology, Software, Validation, Writing – Review & Editing, Supervision. All authors contributed to the editing of the paper and approved its final version to be submitted.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jclinepi.2022.04.027.
Appendix A. Supplementary Data
References
- 1.Lasserson T.J., Thomas J., Higgins J.P.T. Cochrane handbook for systematic reviews of interventions. Cochrane; 2022. Chapter 1: Starting a review. 6.3 (updated February 2022). Available at: www.training.cochrane.org/handbook. [Google Scholar]
- 2.Tsafnat G., Glasziou P., Choong M.K., Dunn A., Galgani F., Coiera E. Systematic review automation technologies. Syst Rev. 2014;3:1–15. doi: 10.1186/2046-4053-3-74. 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussbaumer-Streit B., Ellen M., Klerings I., Sfetcu R., Riva N., Mahmić-Kaknjo M., et al. Resource use during systematic review production varies widely: a scoping review. J Clin Epidemiol. 2021;139:287–296. doi: 10.1016/j.jclinepi.2021.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Gill D., Baker E.H., Hitchings A.W. We need clinical guidelines fit for a pandemic. BMJ. 2021;373:n1093. doi: 10.1136/bmj.n1093. [DOI] [PubMed] [Google Scholar]
- 5.Knottnerus J.A., Tugwell P. Methodological challenges in studying the COVID-19 pandemic crisis. J Clin Epidemiol. 2020;121:A5–A7. doi: 10.1016/j.jclinepi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Cao L., Zhang Z., Hou L., Qin Y., Hui X., et al. Reporting and methodological quality of COVID-19 systematic reviews needs to be improved: an evidence mapping. J Clin Epidemiol. 2021;135:17–28. doi: 10.1016/j.jclinepi.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung R.G., Di Santo P., Clifford C., Prosperi-Porta G., Skanes S., Hung A., et al. Methodological quality of COVID-19 clinical research. Nat Commun. 2021;12:1–10. doi: 10.1038/s41467-021-21220-5. 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tricco A.C., Garritty C.M., Boulos L., Lockwood C., Wilson M., McGowan J., et al. Rapid review methods more challenging during COVID-19: commentary with a focus on 8 knowledge synthesis steps. J Clin Epidemiol. 2020;126:177–183. doi: 10.1016/j.jclinepi.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesty L., Meskell P., Glenton C., Delaney H., Smalle M., Booth A., et al. A QuESt for speed: rapid qualitative evidence syntheses as a response to the COVID-19 pandemic. Syst Rev. 2020;9:1–6. doi: 10.1186/s13643-020-01512-5. 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amezcua-Prieto C., Fernández-Luna J.M., Huete-Guadix J.F., Bueno-Cavanillas A., Khan K.S. Artificial intelligence and automation of systematic reviews in women’s health. Curr Opin Obstet Gynecol. 2020;32:335–341. doi: 10.1097/GCO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 11.O’Mara-Eves A., Thomas J., McNaught J., Miwa M., Ananiadou S. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev. 2015;4:1–22. doi: 10.1186/2046-4053-4-5. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall I.J., Wallace B.C. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst Rev. 2019;8:1–10. doi: 10.1186/s13643-019-1074-9. 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas J., Noel-Storr A., Marshall I., Wallace B., McDonald S., Mavergames C., et al. Living systematic reviews: 2. Combining human and machine effort. J Clin Epidemiol. 2017;91:31–37. doi: 10.1016/j.jclinepi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Scott A.M., Forbes C., Clark J., Carter M., Glasziou P., Munn Z. Systematic review automation tools improve efficiency but lack of knowledge impedes their adoption: a survey. J Clin Epidemiol. 2021;138:80–94. doi: 10.1016/j.jclinepi.2021.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Thomas J., McDonald S., Noel-Storr A., Shemilt I., Elliott J., Mavergames C., et al. Machine learning reduced workload with minimal risk of missing studies: development and evaluation of a randomized controlled trial classifier for Cochrane Reviews. J Clin Epidemiol. 2021;133:140–151. doi: 10.1016/j.jclinepi.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt L., Olorisade B.K., McGuinness L.A., Thomas J., Higgins J.P.T. Data extraction methods for systematic review (semi)automation: a living systematic review [version 1; peer review: 3 approved] F1000Res. 2021;10:1–35. doi: 10.12688/f1000research.51117.1. 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonnalagadda S.R., Goyal P., Huffman M.D. Automating data extraction in systematic reviews: a systematic review. Syst Rev. 2015;4:1–16. doi: 10.1186/s13643-015-0066-7. 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mbuagbaw L., Lawson D.O., Puljak L., Allison D.B., Thabane L. A tutorial on methodological studies: the what, when, how and why. BMC Med Res Methodol. 2020;20:1–12. doi: 10.1186/s12874-020-01107-7. 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tercero-Hidalgo J.R., Khan K.S., Bueno-Cavanillas A., Fernández-López R., Huete J.F., Amezcua-Prieto C., et al. Covid-19 systematic evidence synthesis with artificial intelligence: a review of reviews. Open Sci Forum Regist. 2021 doi: 10.17605/OSF.IO/H5DAW. [DOI] [Google Scholar]
- 21.Tercero-Hidalgo J.R., Khan K.S., Bueno-Cavanillas A., Fernández-López R., Huete J.F., Amezcua-Prieto C., et al. COVID-19 evidence syntheses with artificial intelligence: an empirical study of systematic reviews. Dryad Dataset. 2021 doi: 10.5061/dryad.9kd51c5j6. [DOI] [Google Scholar]
- 22.Rada G., Verdugo-Paiva F., Ávila C., Morel-Marambio M., Bravo-Jeria R., Pesce F., et al. Evidence synthesis relevant to COVID-19: a protocol for multiple systematic reviews and overviews of systematic reviews. Medwave. 2020;20:e7868. doi: 10.5867/medwave.2020.03.7867. [DOI] [PubMed] [Google Scholar]
- 23.Dhakal K. Unpaywall. J Med Libr Assoc. 2019;107:286–288. [Google Scholar]
- 24.Shea B.J., Reeves B.C., Wells G., Thuku M., Hamel C., Moran J., et al. Amstar 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ. 2017;358:j4008. doi: 10.1136/bmj.j4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartoszko J.J., Siemieniuk R.A.C., Kum E., Qasim A., Zeraatkar D., Ge L., et al. Prophylaxis against covid-19: living systematic review and network meta-analysis. BMJ. 2021;373:n949. doi: 10.1136/bmj.n949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemieniuk R.A.C., Bartoszko J.J., Ge L., Zeraatkar D., Izcovich A., Kum E., et al. Drug treatments for covid-19: living systematic review and network meta-analysis. BMJ. 2020;370:m2980. doi: 10.1136/bmj.m2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinnes J., Deeks J.J., Berhane S., Taylor M., Adriano A., Davenport C., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2021;3:CD013705. doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struyf T., Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Leeflang M.M.G., et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19. Cochrane Database Syst Rev. 2021;2:CD013665. doi: 10.1002/14651858.CD013665.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schünemann H.J., et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., et al. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw Open. 2020;3:e2011335. doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaki N., Mohamed E.A., Ibrahim S., Khan G. The influence of comorbidity on the severity of COVID-19 disease: a systematic review and analysis. medRxiv. 2020:1–17. doi: 10.1101/2020.06.18.20134478. 2020.06.18.20134478. [DOI] [Google Scholar]
- 33.Zaki N., Mohamed E.A. The estimations of the COVID-19 incubation period: a scoping reviews of the literature. J Infect Public Health. 2021;14:638–646. doi: 10.1016/j.jiph.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michelson M., Chow T., Martin N., Ross M., Tee A., Minton S. Ocular toxicity and hydroxychloroquine: a rapid meta-analysis. MedRxiv. 2020;22:e20007. doi: 10.2196/20007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daley C., Fydenkevez M., Ackerman-Morris S. A systematic review of the incubation period of SARS-CoV-2: the effects of age, biological sex, and location on incubation period. MedRxiv. 2020:1–19. doi: 10.1101/2020.12.23.20248790. 2020.12.23.20248790. [DOI] [Google Scholar]
- 36.Robinson R., Prakash V., Tamimi R Al, Albast N., Al-Bast B., Wieland E., et al. Impact of remdesivir on 28 day mortality in hospitalized patients with COVID-19: February 2021 Meta-analysis. MedRxiv. 2021:1–15. doi: 10.1101/2021.03.04.21252903. 2021.03.04.21252903. [DOI] [Google Scholar]
- 37.Robinson R., Prakash V., Tamimi R Al, Albast N., Al-Bast B. Impact of systemic corticosteroids on hospitalized patients with COVID-19: January 2021 Meta-analysis of randomized controlled trials. MedRxiv. 2021:1–15. doi: 10.1101/2021.02.03.21251065. medRxiv 2021.02.03.21251065. [DOI] [Google Scholar]
- 38.Izcovich A., Siemieniuk R.A.C., Bartoszko J.J., Ge L., Zeraatkar D., Kum E., et al. Adverse effects of remdesivir, hydroxychloroquine, and lopinavir/ritonavir when used for COVID-19: systematic review and meta-analysis of randomized trials. BMJ Open. 2022;12(3):1–12. doi: 10.1136/bmjopen-2020-048502. e048502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeraatkar D., Cusano E., Martinez J.P.D., Qasim A., Mangala S.O., Kum E., et al. Use of tocilizumab and sarilumab alone or in combination with corticosteroids for covid-19: systematic review and network meta-analysis. BMJ Medicine. 2022;1(1):1–14. doi: 10.1101/2021.07.05.21259867. e000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honarmand K., Penn J., Agarwal A., Siemieniuk R., Brignardello-Petersen R., Bartoszko J.J., et al. Clinical trials in COVID-19 management & prevention: a meta-epidemiological study examining methodological quality. J Clin Epidemiol. 2021;139:68–79. doi: 10.1016/j.jclinepi.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viner R., Russell S., Saulle R., Croker H., Stansfeld C., Packer J., et al. Impacts of school closures on physical and mental health of children and young people: a systematic review. MedRxiv. 2020:1–31. doi: 10.1101/2021.02.10.21251526. 2021.02.10.21251526. [DOI] [Google Scholar]
- 42.Wilson J., Carson G., Fitzgerald S., Llewelyn M.J., Jenkins D., Parker S., et al. Are medical procedures that induce coughing or involve respiratory suctioning associated with increased generation of aerosols and risk of SARS-CoV-2 infection? A rapid systematic review. J Hosp Infect. 2021;116:37–46. doi: 10.1016/j.jhin.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmore R., Schmidt L., Lam J., Howard B.E., Tandon A., Norman C., et al. Risk and protective factors in the COVID-19 pandemic: a rapid evidence map. Front Public Heal. 2020;8:582205. doi: 10.3389/fpubh.2020.582205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alkofide H., Almohaizeie A., Almuhaini S., Alotaibi B., Alkharfy K.M. Tocilizumab and systemic corticosteroids in the management of patients with COVID-19: a systematic review and meta-analysis. Int J Infect Dis. 2021;110:320–329. doi: 10.1016/j.ijid.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Wang L., Lo K., Chandrasekhar Y., Reas R., Yang J., Eide D., et al. CORD-19: the COVID-19 Open Research Dataset. ArXiv. 2020:1–11. doi: 10.48550/arXiv.2004.10706. 2004.10706v4. [DOI] [Google Scholar]
- 46.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall I.J., Noel-Storr A., Kuiper J., Thomas J., Wallace B.C. Machine learning for identifying randomized controlled trials: an evaluation and practitioner’s guide. Res Synth Methods. 2018;9:602–614. doi: 10.1002/jrsm.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard B.E., Phillips J., Tandon A., Maharana A., Elmore R., Mav D., et al. SWIFT-Active Screener: accelerated document screening through active learning and integrated recall estimation. Environ Int. 2020;138:105623. doi: 10.1016/j.envint.2020.105623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gates A., Johnson C., Hartling L. Technology-assisted title and abstract screening for systematic reviews: a retrospective evaluation of the Abstrackr machine learning tool. Syst Rev. 2018;7:1–9. doi: 10.1186/s13643-018-0707-8. 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urlings M.J.E., Duyx B., Swaen G.M.H., Bouter L.M., Zeegers M.P. Citation bias and other determinants of citation in biomedical research: findings from six citation networks. J Clin Epidemiol. 2021;132:71–78. doi: 10.1016/j.jclinepi.2020.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Borah R., Brown A.W., Capers P.L., Kaiser K.A. Analysis of the time and workers needed to conduct systematic reviews of medical interventions using data from the PROSPERO registry. BMJ Open. 2017;7:e012545. doi: 10.1136/bmjopen-2016-012545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z., Nayfeh T., Tetzlaff J., O’Blenis P., Murad M.H. Error rates of human reviewers during abstract screening in systematic reviews. PLoS One. 2020;15:e0227742. doi: 10.1371/journal.pone.0227742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.