Abstract
The paper presents the interaction mechanism of silver nanoparticles (AgNPs) with different antibiotics and the antibacterial efficacy of the formed conjugates. The AgNPs used in this study were synthesized from silver nitrate using sodium borohydride as a reducing agent, in the presence of PVP as a protecting agent. Two antibiotics, amikacin and vancomycin with different modes of action, were used to functionalize the synthesized PVP-capped AgNPs. The formation of antibiotic-AgNPs conjugate was confirmed by UV-vis spectroscopy, Fourier transform infrared spectroscopy (FTIR), dynamic light scattering (DLS) and X-ray diffraction (XRD) and the results suggest the conjugation of both drugs to silver nanoparticle surfaces. FTIR results indicate that intermolecular hydrogen bonding exists between PVP-coated AgNPs and antibiotics. The oxygen atom coordinated with PVP was available for interaction with either amine or amide groups of drugs. Further, the antibacterial efficacy of these PVP-capped AgNPs with selected antibiotics was evaluated against Escherichia coli and Staphylococcus aureus by agar well diffusion test. Synergetic bactericidal activity for antibiotic-AgNPs conjugate was observed against both microbes.
The paper presents the interaction mechanism of silver nanoparticles (AgNPs) with different antibiotics and the antibacterial efficacy of the formed conjugates.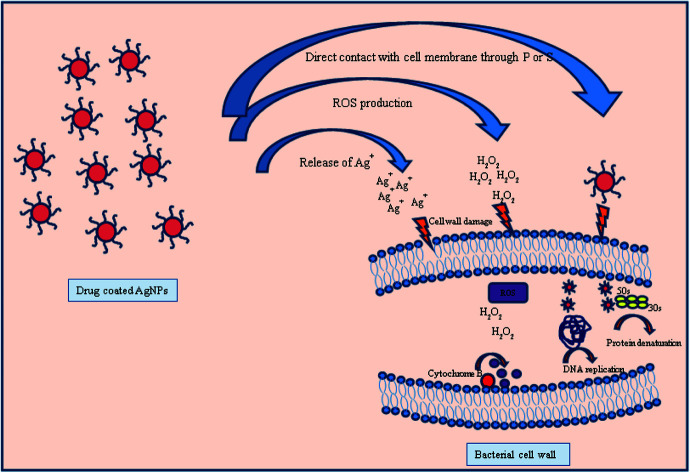
1. Introduction
In the rapidly growing field of nanotechnology, noble metallic nanomaterials are very popular among researchers due to their unique physical, chemical and biological properties. These metal nanomaterials can be used in drug delivery systems,1 bio imaging,2 biosensors3 and antimicrobial activities4 by exploiting their properties. Due to the emergence of multi-drug resistant bacterial strains, antimicrobial properties of noble nanomaterials and also in combination with different antimicrobial agents, have been explored on a large scale to make them effective against these bacteria.5–7 In the early 20's, different infectious diseases were the prime cause of death globally.8 Nowadays, the resistance to various classic antibiotics that are used in hospitals or clinics, has been reached a crucial point.9,10 By discovering new classes of antibiotics and chemically modifying the existing drugs, the resistance to bacterial strains can be improved.11 Recently, in various studies, it has been suggested that noble nanomaterials possess antimicrobial properties which can be used to control bacterial infections.12 Nowadays, nanoantibiotics are very well received among medical researchers all around the world. Nanoantibiotics are the nanomaterials, which either possess antimicrobial property by its own13 or enhance the effectiveness of conventional antibiotics,14–16 and are capable of controlling bacterial infections both in vitro and in vivo.
Various metal and metal oxide nanoparticles produce reactive oxygen species (ROS) under UV light and this increases their utilization in antimicrobial formulations and dressings.17 In particular, nanomaterials of silver, zinc, and their compounds have been reported to be effective antimicrobial agents against different microorganisms.18,19 The highly reactive surfaces of silver and zinc dioxide have been utilized on a large-scale in the bactericidal compounds that are used in filters and coatings on catheters.20–22 Recently, it was observed in different studies that antibiotics incorporated in polymer stabilized metal nanoparticles have shown the enhanced antimicrobial activities and anti-MRSA (methicillin resistance S. aureus) activities, in contrast to non-polymerized forms of penicillin and N-methylthio β-lactams.23,24 For biological applications metal nanoparticles of size up to 200 nm can be utilized but the nanoparticles with diameter up to 70 nm are highly appropriate for biosystems.25 Since ancient times, silver is the most popular antibacterial or antifungal agent, employed to treat infections or wounds.26,27 In the past decade, the notable antibacterial activity of silver nanoparticles against various class of bacteria including Salmonella, Staphylococcus, and Pseudomonas, has been observed.28–30 For biological applications, the silver nanoparticles must be stable and this stability can be achieved by using some ionic protecting agents or by some hydrophilic surfactants such as proteins,31 amino acids32 which includes polymers such as polyvinylpyrrolidone (PVP) and polyvinyl alcohol (PVA).33–37
Polyvinylpyrrolidone (PVP), a commonly used water-soluble polymer, has been extensively used for the synthesis of stable silver nanoparticles through steric stabilization mechanism. There are two highly active sites present (through N atom or –C O) on PVP surface to react with other molecules. PVP acts as a protecting agent through the coordinate bonding of N atom with the silver surface.38 Apart from this N atom, PVP has also a –C O functional group, which can be used as a bridge for drugs to interact with silver surface through hydrogen bonding. Here, in this study we have compared the antibacterial efficacy of two different drugs and their conjugates with silver nanoparticles. The silver nanoparticles (AgNPs) were prepared by a common synthetic route that is borohydride reduction of silver nitrate in the presence of stabilizing agent PVP. The nanoparticles can be stabilized by steric forces and this steric stabilization can be achieved by adsorbing PVP on silver surface. Further, the synthesized silver nanoparticles were loaded with two different drugs named as, vancomycin and amikacin. Vancomycin (C66H75Cl2N9O24) is an antibiotic of glycopeptide class whereas amikacin (C22H43N5O13) belongs to aminoglycoside class of antibiotics. Vancomycin is used to treat a number of bacterial infections caused by Gram-positive strain. It is highly recommended as a treatment for complicated skin infections, bloodstream infections, bone and joint infections, and meningitis caused by methicillin-resistant Gram-positive common bacteria Staphylococcus aureus. On the other hand, amikacin is an antibiotic of aminoglycoside class which is most often used for treating severe infections such as pneumonia, sepsis, and urinary tract infections caused by aerobic Gram-negative bacteria, especially Pseudomonas, Acinetobacter, Enterobacter, E. coli. Further, studies were extended to investigate the antibacterial effect of silver nanoparticles loaded with vancomycin and amikacin on both the Gram classes of bacteria – Staphylococcus aureus (Gram-positive) and Escherichia coli (Gram-negative).
2. Experimental materials and methods
2.1. Materials
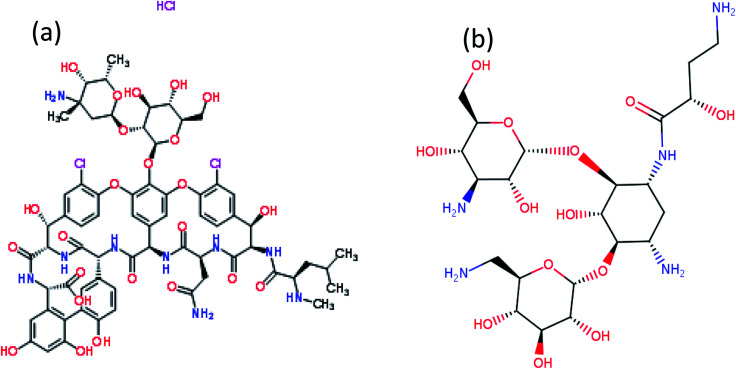
Silver nitrate (AgNO3), sodium borohydride (NaBH4) and PVP40 (C6H9NO)n were used for the synthesis of AgNPs. All chemicals were purchased from Sigma Aldrich. Two antibiotics named as vancomycin (C66H75Cl2N9O24) and amikacin (C22H43N5O13) were used for functionalization of AgNPs (Fig. 1). All samples were prepared in double-distilled water. Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) cultures were used for the antibacterial study.
Fig. 1. Chemical structure (a) vancomycin and (b) amikacin.
2.2. Synthesis of colloidal PVP-capped silver nanoparticles
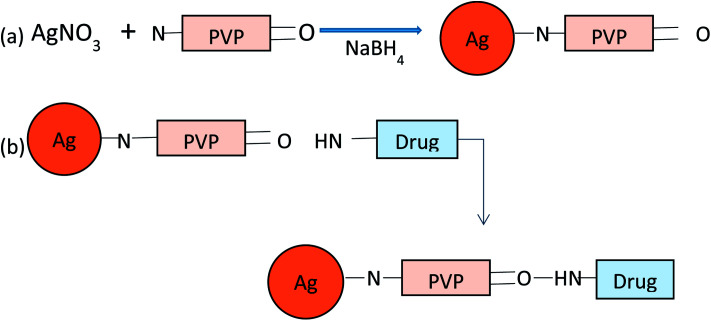
Colloidal silver nanoparticles (AgNPs) were synthesized by reduction of NaBH4. The solution of silver nanoparticles (AgNPs) was synthesised by using 1.0 mM AgNO3 as a precursor and 2.0 mM of NaBH4 in ice bath, as a reducing agent and in the presence of capping agent PVP (0.3%), by continuous stirring until the colour of solution changes to pale yellow. The solutions were kept at low temperature for further characterizations and experimentation.
2.3. Characterization of synthesized PVP-coated silver nanoparticles
The UV-vis absorption spectra of prepared PVP-AgNPs was recorded by using spectrometer between 200 to 600 nm. The FTIR measurements were done to reveal different functional groups in the range of 4000–400 cm−1 using Perkin Elmer spectrometer. The hydrodynamic diameter of AgNPs was assesses with a Zetasizer. The X-ray diffraction pattern was conducted with diffractometer using monochromatic Cu Kα radiation (λ = 1.54 Å) running at 40 kV and 30 mA. The intensity data for AgNPs deposited on a glass slide was collected over a 2θ values of range 35–80° with a scan rate of 2° min−1.
2.4. Synthesis of drug loaded silver nanoparticles
The synthesis of drug loaded AgNPs (Amk@PVP-AgNPs and Van@PVP-AgNPs) was done by preparing aqueous solution of vancomycin (5 mg mL−1) and amikacin (10 μL mL−1) separately. Keeping the concentration of AgNPs (1 mM) constant, varying concentrations (0.05 mM, 0.1 mM, 0.3 mM, 0.5 mM) of vancomycin and amikacin were used for the formation of drug–Ag complex. The solution was allowed to stir for 15–20 min on a magnetic stirrer at room temperature. After synthesis store the solution in refrigerator for further experimentation and characterizations (Fig. 2).
Fig. 2. Schematic representation of the synthesis of (a) PVP-AgNPs and (b) drug loaded AgNPs.
2.5. Bactericidal test
The agar well diffusion method was used to evaluate the antibacterial activity of drug loaded AgNPs. Two model bacterial strains Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) related to Gram-positive and Gram-negative class of bacteria, respectively, were used for the antibacterial test. S. aureus and E. coli were grown at 37 °C and maintained on Luria broth (LB) plates. Bacteria were also grown in LB media for 24 h at 37 °C. All the glassware and media used for well diffusion assay were sterilized in an autoclave at 121 °C for 15 minutes. The bacterial plaque was prepared by growing a single colony overnight in nutrient broth. The different concentrations of vancomycin and amikacin were selected for bactericidal test. The agar plate surface is inoculated by spreading a volume of microbial inoculums over the entire surface of agar plate. Then, a hole with of diameter about 5 mm was punched with a sterile cork borer and 50 μL of each of free drug, AgNPs, mixture of drug and AgNPs, and normal saline solution (as a negative control) were poured into the wells, separately. These plates were incubated at 35–37 °C for 24 hours, and then antibacterial activities were observed by measuring the zone of inhibition for each sample, which was calculated by subtracting the diameter of well from the total inhibition zone diameter.
3. Results and discussion
3.1. UV-vis absorption characterization of synthesized blank and drug loaded AgNPs
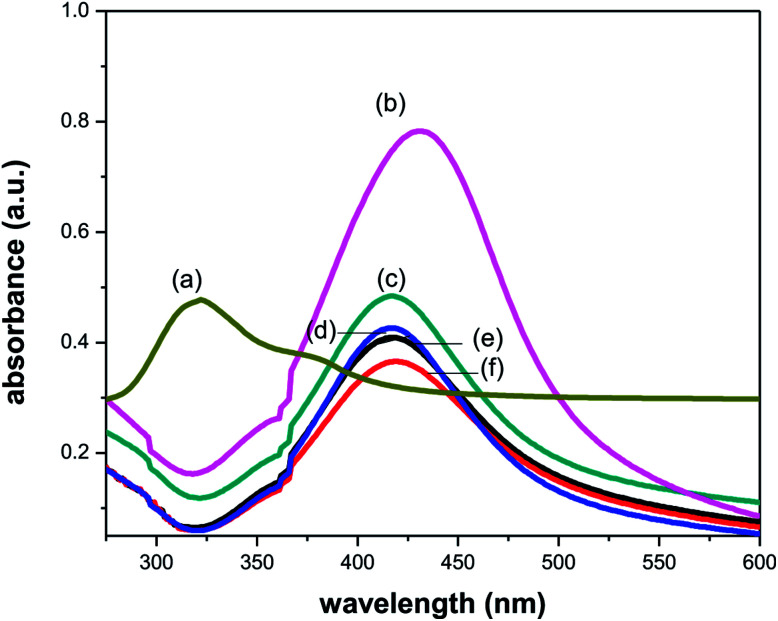
The synthesis of silver nanoparticles by the reduction of silver nitrate was followed by UV-vis spectroscopy. It is well known that silver nanoparticles exhibit a pale yellowish colour in aqueous solution due to the surface plasmon resonance (SPR) of metal nanoparticle.39 UV-vis spectra of the aqueous silver nanoparticles solution synthesized being recorded after completion of the reaction. The UV-vis spectrum of silver nanoparticles synthesized at room temperature clearly shows an intense surface plasmon resonance (SPR) band at 431 nm (visible region) which confirmed the formation of AgNPs (Fig. 3). Vancomycin and amikacin shows absorption peak around at 290 and 320 nm (UV region), respectively. After the addition of vancomycin to AgNPs solution, the absorption peak shifts to shorter wavelength (blue shift) up to 2 nm due to charge transfer between drug and PVP coated AgNPs.40 However, in case of amikacin a significant blue shift of around 15 nm in absorption peaks was observed. The cause of blue shift could be explained on the basis of either nucleophilic reaction (charge transfer) or electronic transitions between different orbitals. The nucleophilic substitution reaction takes place between lone pair of oxygen atom of PVP and hydrogen atom of amine group of drug. So, PVP acts as a nucleophile in this reaction which increases the electron density and causes hypsochromic shift in absorption peak. Further, there could also be possibility of electronic transitions occurring between bonding or non-bonding orbital to antibonding orbital. In visible region, two main electronic transitions are possible: (1) bonding to antibonding orbital (π → π*) and (2) non-bonding to antibonding orbital (n → π*). The first one occurs at low energy (higher wavelength) and second one requires high energy (lower wavelength). Due to presence of lone pair on PVP, the transition from n → π* might be more dominating than π → π*, causes the significant blue shift. Hence, PVP acts as a bridge between drug and AgNPs surface and this blue shift attributed to the attachment of drug with AgNPs surface.
Fig. 3. UV-vis absorption spectra of (a) amikacin, (b) PVP-AgNPs, (c–f) Amk@PVP-AgNPs at concentration 0.05, 0.1, 0.3 and 0.5 mM respectively.
3.2. Fourier transform infrared absorption studies of blank and drug coated AgNPs
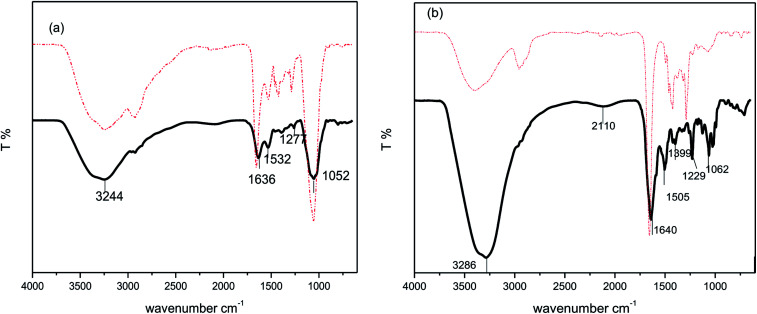
To determine the coupling of the drug to the functionalized nanoparticles FTIR spectroscopy has been used. Fig. 4 shows the typical FTIR spectra of the drugs, bare AgNPs and drug modified AgNPs. As drugs have various amino and hydroxyl groups so mainly two types of bonding takes place between drugs and metal nanoparticles one is amino bonding (through amine or amide group) and other one is thiol bonding. The free drug amikacin (Fig. 4(a) solid line) shows characteristic frequency bands around 3244, 1636, 1532, 1277 and 1052 corresponding to –OH– stretching frequency, N–H bending vibration of amines and amides, –CN– bond of amines and C–O vibrations of esters and ethers, respectively. Similarly, free vancomycin (Fig. 4(b) solid line) has vibrational bands at 3286, 1640, 1505, 1399, 1229 and 1062 cm−1corresponding to stretching –OH– bond, bending vibrations of N–H group of amines and amides, –CN– group of amines and C–O group of ethers and esters, respectively. In case of drug coated silver nanoparticles (dotted lines), there was no significant shift in frequency peaks except the peaks at 1632 and 1640 cm−1 corresponding to N–H group of secondary amines or amides, which were shifted to higher wavenumbers. Hence, the FTIR data infers that it might be the hydrogen atom of amines or amides that binds to the oxygen atom of PVP which was free for the attachment on the surface of silver.
Fig. 4. FTIR spectra of (a) Amk@PVP-AgNPs (dotted line) and amikacin (solid line) and (b) Van@PVP-AgNPs (dotted line) and vancomycin (solid line).
3.3. Particle size distribution, polydispersity and morphological studies of bare AgNPs and drug loaded AgNPs
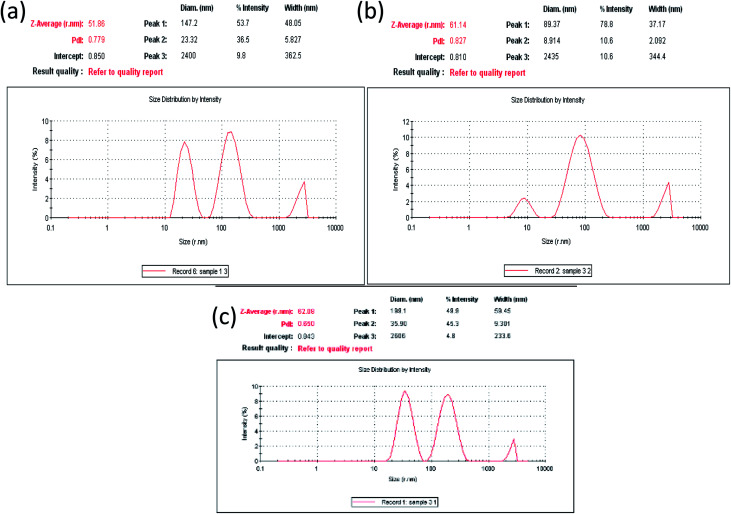
Particle size of the nanoparticles is presented as Z-average diameter, which is basically mean hydrodynamic diameter of the particles. Particle size measurement was required to confirm the production of the particles in nano-range. Attachment of drug onto the surface of AgNPs would also impact the size of the nanoparticles, and therefore should be reflected in the diameter value. The Z-average size, particle size distribution and polydispersity index (PdI) for both AgNPs and (drug@AgNPs) was recorded by using Zetasizer. The value of PdI index in Zetasizer software lies between 0 and 1. Hence, the PdI must be less than 1 for samples to be suitable for DLS analysis. Lower value of PdI attributes towards monodispersed samples (less polydisperse). The Z-average diameter of nanoparticles was calculated by using Stokes–Einstein equation:2r = kB/3πηD
In this equation, kB is the Boltzmann's constant, T is the absolute temperature, η is the viscosity of the medium and D is the diffusion coefficient. The DLS size measurement in terms of intensity distribution for blank AgNPs, Amk@AgNPs and Van@AgNPs is shown in Fig. 5. The average particle size for bare AgNPs was calculated around 51 nm with PdI 0.779 by using Stoke–Einstein equation. However, for Amk@AgNPs and Van@AgNPs, the value of Z-average was found around 61 and 62 nm with PdI 0.827 and 0.650, respectively. The decrease or increase in PdI might be due to reduction or enhancement in surface tension of AgNPs after functionalized with drugs. Calculated values of Z-average size and PdI are tabulated in Table 1. Therefore, enhanced Z-average of AgNPs with drugs (amikacin and vancomycin) also assures the loading of drugs on AgNPs surface.
Fig. 5. Particle size distribution by intensity (a) for blank AgNPs, (b) Amk@AgNPs, and (c) Van@AgNPs.
Calculated DLS parameters for AgNPs, Amk@AgNPs and Van@AgNPs.
| Nanomaterial | Z-average (nm) | Polydispersity (PdI) |
|---|---|---|
| PVP-AgNPs | 51 | 0.779 |
| Amk@AgNPs (0.1 mM) | 61 | 0.827 |
| Van@AgNPs (0.1 mM) | 62 | 0.650 |
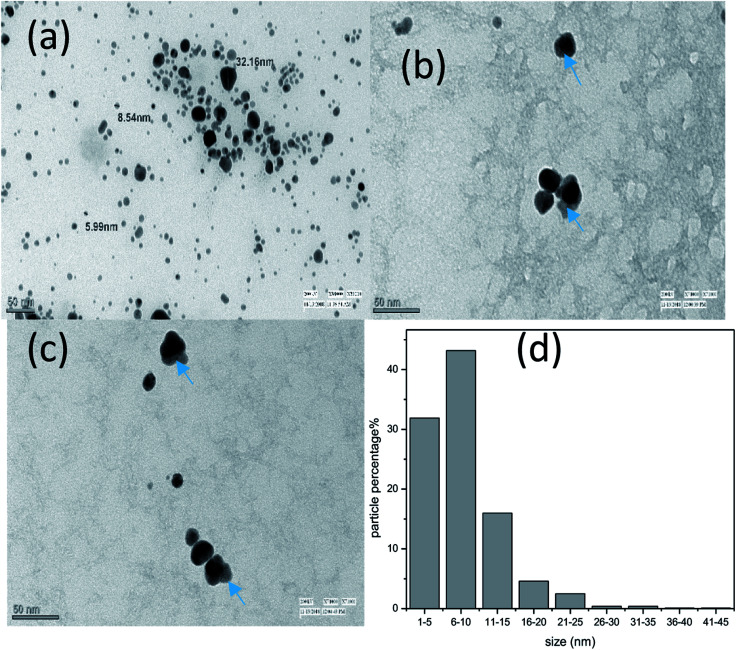
Transmission electron microscopy (TEM) has been used to study the size, shape and surface morphology of bare AgNPs, Van@AgNPs and Amk@AgNPs. The corresponding micrographs are shown in Fig. 6. The morphology of AgNPs was found spherical and the size of AgNPs lies within the range of 5–35 nm with average size of 8.3 nm. The diameter of AgNPs observed in TEM was lower as compare to DLS because by DLS wet and extended diameter of AgNPs suspension has been measured whereas TEM micrographs show the dry and shrunk configuration of AgNPs.41 Attachment of drug is shown by arrow in the TEM micrographs (Fig. 6(b) and (c)) as light and dark part corresponds to drug and nanoparticle. Hence, interaction of drugs with AgNPs also confirmed from TEM micrographs.
Fig. 6. TEM micrographs of (a) bare-AgNPs, (b) Van@AgNPs and (c) Amk@AgNPs and (d) size distribution by TEM.
3.4. X-ray diffraction analysis of AgNPs and drug coated AgNPs
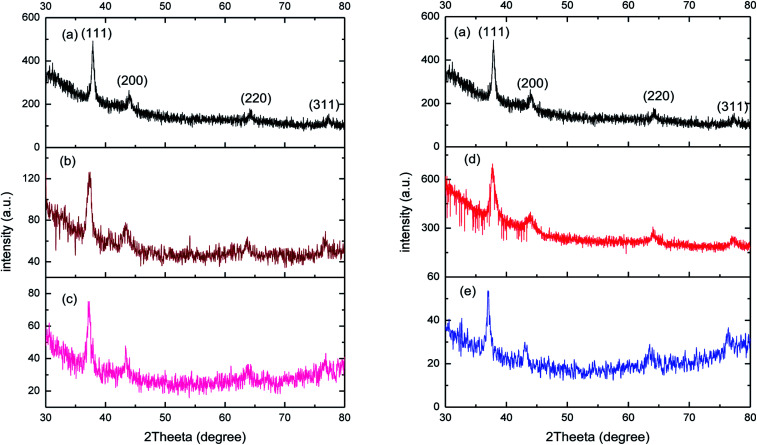
XRD spectra was recorded to confirm the crystalline nature of prepared AgNPs (Fig. 7(a)). For bare AgNPs, the XRD reflection lines were observed at 38.10°, 44.09°, 64.20° and 77.42°, ascribed to (111), (200), (220) and (311) respectively, the reflections of face-centred cubic structure of metallic silver. The estimated crystallite size for AgNPs was around 20 nm which was calculated by using Scherrer formula:D = 0.9 λ/β cos θwhere, D is the crystallite size, k is constant, λ is the wavelength of X-ray radiation, β is the line width and θ is the angle of diffraction. The lattice constant ‘a’ worked out to be 4.098 Å which is in good agreement with in standard data file JCPDS no 04. After the functionalization of nanoparticles with both drugs (amikacin and vancomycin), there was a slight shift in major peak of bare AgNPs at 38.10° to 37.38° (0.05 mM) and 37.21° (0.3 mM) for Amk@AgNPs. Similarly, for Van@AgNPs the major peak shifted to 37.75° (0.05 mM) and 37.30° (0.3 mM). There was also a minor shift in small peaks (2θ values) from 44.09°, 64.20° and 77.42° to 43.38°/43.38, 63.84°/63.84° and 76.88°/76.88° for 0.05 mM and 0.3 mM Amk@AgNPs, respectively. In case of Van@AgNPs this shift was observed at 43.90°/43.18°, 64.02°/63.64° and 77.24°/76.50° for 0.05 mM and 0.3 mM, respectively (Table 2). The different reflection peaks for drug coated nanoparticles confirmed their crystalline nature and face-centred cubic symmetry for different ratios. There was no notable change in crystallite size was observed in case of drug–AgNPs complex. The shift in XRD reflection lines is indicates the presence of lattice strain that is resulted from either compressive stress or tensile stress. Moreover, the shift in reflection lines towards lower angle implies the presence of compressive stress. Therefore, XRD results also suggest the interaction of drugs with AgNPs.
Fig. 7. XRD pattern for (a) bare PVP-AgNPs, (b) 0.05 mM Amk@PVP-AgNPs, (c) 0.3 mM Amk@PVP-AgNPs, (d) 0.05 mM Van@PVP-AgNPs and (e) 0.3 mM Van@PVP-AgNPs.
2θ (in degree) values for Amk@AgNPs and Van@AgNPs corresponding to different reflections lines.
| Material | (111) | (200) | (220) | (311) |
|---|---|---|---|---|
| AgNPs | 38.10° | 44.09° | 64.20° | 77.42° |
| Amk@AgNPs (0.05 mM) | 37.38° | 43.38° | 63.85° | 76.88° |
| Amk@AgNPs (0.3 mM) | 37.21° | 43.38° | 63.85° | 76.88° |
| Van@AgNPs (0.05 mM) | 37.75° | 43.90° | 64.02° | 77.24° |
| Van@AgNPs (0.3 mM) | 37.30° | 43.18° | 63.64° | 76.50° |
3.5. Antimicrobial efficacies of Amk@AgNPs and Van@AgNPs
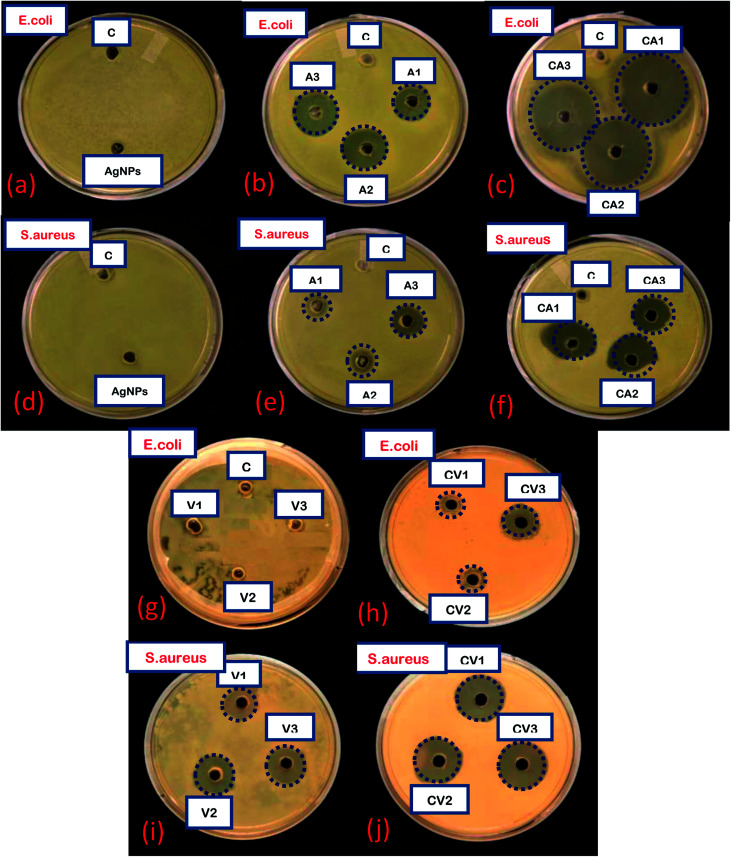
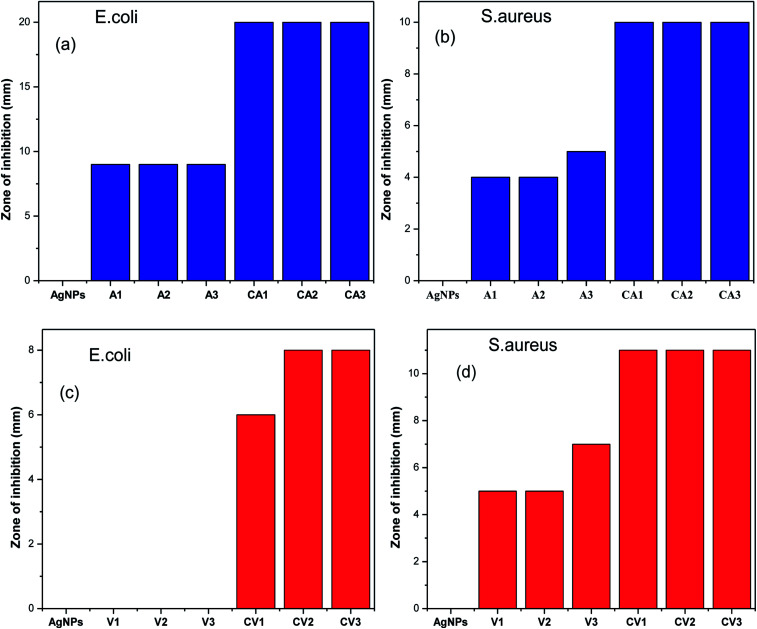
To determine the antibacterial efficacy of Van@AgNPs and Amk@AgNPs, antibacterial test has been conducted against panel of bacterial strains. Two common bacterial model strains i.e. S. aureus (Gram-positive) and E. coli (Gram-negative) were used in this investigation. Antibacterial efficacy of AgNPs with or without conjugation with drugs was expressed in terms of zone of inhibition (ZOI). For each strain, ZOI for pure drugs and drug@AgNPs was calculated by agar-well diffusion method at different concentrations. Bare-AgNPs did not show any antibacterial effect at evaluated concentration (60 μg mL−1) against both strains that might be due to low concentration. But due to its intrinsic bactericidal action, antibacterial efficacy enhanced upon conjugation with drugs. The evaluated amount of Ag in AgNPs, for different concentrations of drug@AgNPs i.e. 0.1 mM, 0.3 mM and 0.5 mM, are approximately 288, 276 and 264 μg in 50 μL solution (used for antibacterial assay), respectively. Amikacin is an aminoglycoside antibiotic, mainly effective against aerobic Gram-negative bacteria. There are only Gram-positive bacteria that are strongly affected by amikacin that are Staphylococcus and Nocardia.42 Therefore, both E. coli and S. aureus are amikacin sensitive bacteria. On the other hand, vancomycin is glycopeptide drug which only treats the infection caused by Gram-positive bacteria such as Staphylococcus, Streptococcus, and Enterococcus etc. Hence, E. coli and S. aureus are vancomycin resistant and sensitive bacteria, respectively. It was observed that E. coli also became vancomycin sensitive bacteria after treated with Van@AgNPs. In all other cases, enhanced antibacterial activity was observed with Van@AgNPs and Amk@AgNPs as compare to vancomycin and amikacin, respectively. The images of agar plates with ZOI for blank AgNPs, Van, Amk, Van@AgNPs and Amk@AgNPs at different concentrations 0.1 mM (1), 0.3 mM (2) and 0.5 mM (3) has been shown in Fig. 8 against E. coli and S. aureus.
Fig. 8. Images of agar plates showing ZOI of: (a–c) AgNPs, Amk, Amk@AgNPs at different concentration against E. coli, (d–f) AgNPs, Amk, Amk@AgNPs at different concentration against S. aureus, (g and h) Van and Van@AgNPs at different concentration against E. coli and (i and j) Van and Van@AgNPs at different concentration against S. aureus. C: control, A1, A2, A3: Amk1, Amk2, Amk3 CA1, CA2, CA3: Amk@AgNPs1, Amk@AgNPs2, Amk@AgNPs3 V1, V2, V3: Van1, Van2, Van3 CV1, CV2, CV3: Van@AgNPs1, Van@AgNPs2, Van@AgNPs3.
The calculated ZOI of Amk for E. coli and S. aureus was approximately around 9 and 5 mm, respectively for three different concentrations whereas ZOI for Amk@AgNPs against E. coli and S. aureus was increased up to 20 and 10 mm, respectively. As already mentioned amikacin is a kind of antibiotic which treats infections caused by Gram-negative bacteria and rarely caused by Gram-positive bacteria (Staphylococcus). Hence, Amk@AgNPs show more enhanced bactericidal effect for E. coli than S. aureus but against S. aureus effect of drug–AgNPs complex was also significant rather than pure amikacin. On the other hand, E. coli was totally resistant to vancomycin as it is Gram-negative bacteria and hence, no ZOI was observed. But it was observed that when E. coli was treated with Van@AgNPs at different concentrations, the bacterial growth was inhibited to some extent. So, the ZOI with Van@AgNPs for E. coli increases from 0 to 8 mm and made the bacteria also sensitive to this conjugate. In case of S. aureus, bacterial growth inhibition also enhances from 7 to 11 mm. The zone of inhibition with Amk@AgNPs and Van@AgNPs for both test strains has been listed in Table 3 and 4, and their corresponding histograms are shown in Fig. 9. The synergetic effect is due to the efficient delivery of drug attached to AgNPs to bacterial cell membrane which is hydrophobic in nature. As vancomycin and amikacin are hydrophilic in nature and AgNPs are hydrophobic. Therefore, AgNPs can easily interact with cell membrane and the drug attached to AgNPs delivered to the bacterial cell more efficiently.43 Subsequently, the surface charge on AgNPs also contributes to the synergy as negatively charged AgNPs disrupt the permeability of cell membrane which in turn affects the cellular respiration severely. Due to disrupted permeability the drug can easily damage the bacterial cell and inhibit the cell wall and protein biosynthesis.44,45 It has also been reported that as compared to bare AgNPs, the drug–AgNPs conjugates will enhance the release of silver ion which also attribute towards the synergetic effect.46–50
Antibacterial activity of AgNPs, Amk and Amk@AgNPs and their zone of inhibition (mm) against E. coli and S. aureus.
| Microorganism | AgNPs | Amk1 (0.1 mM) | Amk2 (0.3 mM) | Amk3 (0.5 mM) | Amk@AgNPs1 (0.1 mM) | Amk@AgNPs2 (0.3 mM) | Amk@AgNPs3 (0.5 mM) |
|---|---|---|---|---|---|---|---|
| E. coli | 0 | 9 | 9 | 9 | 20 | 20 | 20 |
| S. aureus | 0 | 4 | 4 | 5 | 10 | 10 | 10 |
Antibacterial activity of AgNPs, Van and Van@AgNPs and their zone of inhibition (mm) against E. coli and S. aureus.
| Microorganism | AgNPs | Van1 (0.1 mM) | Van2 (0.3 mM) | Van3 (0.5 mM) | Van@AgNPs1 (0.1 mM) | Van@AgNPs2 (0.3 mM) | Van@AgNPs3 (0.5 mM) |
|---|---|---|---|---|---|---|---|
| E. coli | 0 | 0 | 0 | 0 | 6 | 8 | 8 |
| S. aureus | 0 | 5 | 5 | 7 | 11 | 11 | 11 |
Fig. 9. Enhancement in ZOI of drug coated nanoparticles (a and b) with Amk@AgNPs against E. coli and S. aureus and (c and d) with Van@AgNPs against E. coli and S. aureus.
The calculated density of AgNPs is around 2.0 × 1012/mL. For this calculated density of AgNPs, the loaded amount of drug is around 40 μg mL−1 and 20 μg mL−1 in case of Amk@AgNPs and Van@AgNPs, respectively. Less loading of vancomycin to AgNPs is due to its larger molecular mass (size) than amikacin. This concentration is almost same as 0.1 mM in case of pure drug. The antibacterial activity for this concentration (0.1 mM) of pure drug is shown in Fig. 8(b), (e) and (i) for amikacin and vancomycin, respectively. From the collected data it was concluded that Amk@AgNPs are more active against E. coli whereas Van@AgNPs against S. aureus. Hence, our data demonstrated that AgNPs act synergistically with both amikacin and vancomycin against E. coli and S. aureus.
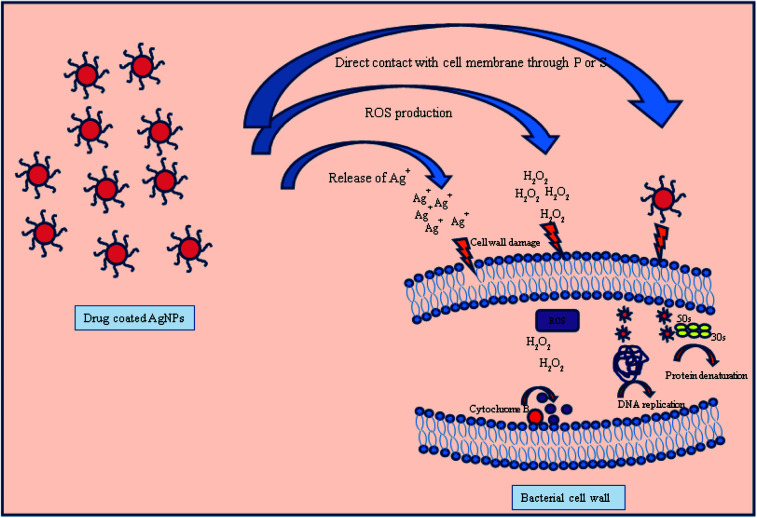
The antibacterial mechanism of drug coated silver nanoparticles is still not much explored but the different mechanisms could be possible through (1) as silver is a soft acid so it has a superior tendency to react with soft bases such as sulphur and phosphorous containing proteins and DNA present in bacterial cell wall and cause bacterial death due to change in permeability.51,52 (2) The other possible mechanism is through vancomycin and amikacin. Both drugs have different way to interact with bacterial cell wall and cause death of bacteria. Vancomycin inhibit the cell wall growth by inhibiting peptidoglycan synthesis by binding to amino acids (particularly to acyl-d-alanyl-d-alanine) within cell wall which prevents the addition of new units to peptidoglycan. On the other hand amikacin due to its cationic nature tends to bind with anionic compound found in bacterial surface and inhibits the protein synthesis by further binding with 30S ribosome unit.53,54 (3) If bacteria resists the antibacterial action of either AgNPs or antibiotic, then the combined action of drug–AgNPs complex disrupt the bacterial cell wall and inhibit the growth of bacteria by increasing ROS generation either agent alone Fig. 10.55 Therefore, combining PVP-AgNPs with both antibiotics at a lower concentration has the potential to become an excellent antibacterial treatment.
Fig. 10. Different possible modes of action of drug coated silver nanoparticles with bacterial cell wall.
Hence, this study shows that Amk@PVP-AgNPs have excellent antibacterial efficacy against E. coli whereas Van@PVP-AgNPs have higher effect against S. aureus. However, further studies on bactericidal effect of these drug–AgNPs conjugates on other Gram-positive and Gram-negative bacteria are necessary to evaluate its use as a new biocidal system.
4. Conclusions
Amikacin and vancomycin has been encapsulated on PVP-capped AgNPs through hydrogen bonding for improving in vitro antibacterial property. Interaction of drugs with AgNPs surface was confirmed from the shift in UV-vis absorption peak and XRD reflection lines. The calculated values of Z-average after drug incorporation also infer the successful loading of drugs on AgNPs surface. Also, FTIR studies attributed towards the hydrogen bonding between amino group of drug and oxygen atom of PVP. Further, it was shown that the drug–AgNPs complex has a profound synergetic antibacterial efficacy against S. aureus and E. coli test strains and this synergetic effect was augmented. The results showed that Amk@PVP-AgNPs conjugates were more effective against E. coli whereas Van@PVP-AgNPs conjugates against S. aureus. As PVP-AgNPs shows significant shift with amikacin as compared to vancomycin in UV-vis spectra, it can be concluded that PVP is a better linker for amikacin than vancomycin. Hence, PVP-AgNPs acts as carrier for both vancomycin and amikacin and can be used as a system to enhance the antibacterial activity against multi-drug resistant bacteria.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
One of the authors (Amritpal Kaur) are obliged to the University Grants Commission (UGC) for providing financial support under MANF scheme (201415-MANF-2014-15-SIK-PUN-36453). The authors would like to thank Dr Simran Preet, Department of Biophysics, Panjab University, Chandigarh for antibacterial test. Authors are also thankful to Central Instrumental Laboratory, Panjab University, Chandigarh for FTIR studies.
References
- Keun S. O. Ree S. K. Jinho L. Dongmin K. Sun H. C. Soon H. Y. J. Appl. Polym. Sci. 2008;108:3239–3244. doi: 10.1002/app.27767. [DOI] [Google Scholar]
- Hongwei L. Colleen L. N. Jason H. H. Nanomedicine. 2006;1:201–208. doi: 10.2217/17435889.1.2.201. [DOI] [PubMed] [Google Scholar]
- Amanda J. H. Lei C. William L. K. Richard P. V. D. J. Am. Chem. Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- Fayaz A. M. Balaji K. Girilal M. Ruchi Y. Kalaichelvan P. T. Venketesan R. Nanomedicine. 2009;6:103–109. doi: 10.1016/j.nano.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Rai M. Yadav A. Gade A. Biotechnol. Adv. 2009;27:76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Goodman C. M. McCusker C. D. Yilmaz T. Rotello V. M. Bioconjugate Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- Schaller M. Laude J. Bodewaldt H. Hamm G. Korting H. C. Skin Pharmacol. Physiol. 2004;17:31–36. doi: 10.1159/000074060. [DOI] [PubMed] [Google Scholar]
- Cohen M. L. Nature. 2000;406:762–767. doi: 10.1038/35021206. [DOI] [PubMed] [Google Scholar]
- Gold H. S. Moellering R. C. N. Engl. J. Med. 1996;335:1445–1453. doi: 10.1056/NEJM199611073351907. [DOI] [PubMed] [Google Scholar]
- Walsh C. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- Boucher H. W. Talbot G. H. Bradley J. S. Edwards Jr J. E. Gilbert D. Rice L. B. Scheld M. Spellberg B. Bartlett J. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Huh A. J. Kwon Y. J. J. Controlled Release. 2011;156:128–145. doi: 10.1016/j.jconrel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Kim J. S. Kuk E. Yu K. N. Kim J. H. Park S. J. Lee H. J. Kim S. H. Park Y. K. Park Y. H. Hwang C. Y. Kim Y. K. Lee Y. S. Jeong D. H. Cho M. H. Nanomedicine. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Shan W. Q. Jian J. JinHong F. JiaCong S. Sci. China, Ser. B: Chem. 2007;50:418–424. doi: 10.1007/s11426-007-0028-6. [DOI] [Google Scholar]
- Oloffs A. Crosse-Siestrup C. Bisson S. Rinck M. Rudolvh R. Gross U. Biomaterials. 1994;15:753–758. doi: 10.1016/0142-9612(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Turos E. Shim J. Y. Wang Y. Greenhalgh K. K Reddy G. S. Dickey S. Lim D. V. Bioorg. Med. Chem. Lett. 2007;17:53–56. doi: 10.1016/j.bmcl.2006.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaker R. P. Ren G. Trans. R. Soc. Trop. Med. Hyg. 2008;102:1–2. doi: 10.1016/j.trstmh.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turos E. Reddy G. S. Greenhalgh K. Ramaraju P. Abeylath S. C. Jang S. Dickey S. Lim D. V. Bioorg. Med. Chem. Lett. 2007;17:3468–3472. doi: 10.1016/j.bmcl.2007.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühling M. Bradford A. Readman J. W. Somerfiel P. J. Handy R. D. Mar. Environ. Res. 2009;68:278–283. doi: 10.1016/j.marenvres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Huang Z. Zheng X. Yan D. Yin G. Liao X. Kang Y. Yao Y. Huang D. Hao B. Langmuir. 2008;24:4140–4144. doi: 10.1021/la7035949. [DOI] [PubMed] [Google Scholar]
- Maness P. C. Smolinski S. Blake D. M. Huang Z. Wolfrum E. J. Jacoby W. A. Appl. Environ. Microbiol. 1999;65:4094–4098. doi: 10.1128/aem.65.9.4094-4098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Mahendra S. Lyon D. Y. Brunet L. Liga M. V. Li D. Alvarez P. J. Water Res. 2008;42:4591–4602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Paknikar K., US Pat., US2007/0218555A1, 2007
- Dibrov P. Dzioba J. Khoosheh K. Hase C. Antimicrob. Agents Chemother. 2002;46:2668–2670. doi: 10.1128/AAC.46.8.2668-2670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahverdi A. R. Fakhimi A. Shahverdi H. R. Minaian S. Nanomedicine. 2007;3:168–171. doi: 10.1016/j.nano.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Suvorova E. I. Klechkovskaya V. V. Kopeikin V. V. Buffat P. A. J. Cryst. Growth. 2005;275:2351–2356. doi: 10.1016/j.jcrysgro.2004.11.333. [DOI] [Google Scholar]
- Sergeev B. M. Kiryukhin M. V. Rubtsova M. Y. Prusov A. N. Colloid J. 2003;65:636–638. doi: 10.1023/A:1026192327440. [DOI] [Google Scholar]
- Yakutik I. M. Shevchenko P. G. Surf. Sci. 2004;566:414–418. doi: 10.1016/j.susc.2004.06.122. [DOI] [Google Scholar]
- Kasuga N. C. Sato M. Amano A. Hara A. Tsuruta S. Sugie A. Nomiya K. Inorg. Chim. Acta. 2008;361:1267–1273. doi: 10.1016/j.ica.2007.08.020. [DOI] [Google Scholar]
- Khanna P. K. Singh N. Kulkarni D. Deshmukh S. Charan S. Adhyapak P. Mater. Lett. 2007;61:3366–3370. doi: 10.1016/j.matlet.2006.11.064. [DOI] [Google Scholar]
- Shin H. S. Yang H. J. Kim S. B. Lee M. S. J. Colloid Interface Sci. 2004;274:89–94. doi: 10.1016/j.jcis.2004.02.084. [DOI] [PubMed] [Google Scholar]
- Temgire M. K. Joshi S. S. Radiat. Phys. Chem. 2004;71:1039–1044. doi: 10.1016/j.radphyschem.2003.10.016. [DOI] [Google Scholar]
- Khanna P. K. Singh N. Charan S. Subbarao V. V. V. S. Gokhale R. Mulik U. P. Mater. Chem. Phys. 2005;93:117–121. doi: 10.1016/j.matchemphys.2005.02.029. [DOI] [Google Scholar]
- Khanna P. K. Gokhale R. Subbarao V. V. V. S. J. Mater. Sci. 2004;39:3773–3776. doi: 10.1023/B:JMSC.0000030735.08903.a9. [DOI] [Google Scholar]
- Lim P. Liu R. She P. Hung C. Shih H. Chem. Phys. Lett. 2006;420:304–308. doi: 10.1016/j.cplett.2005.12.075. [DOI] [Google Scholar]
- Kvítek L. Panacek A. Soukupova J. Kolar M. Vecerova R. Prucek R. Holecova M. Zboril R. J. Phys. Chem. C. 2008;112:5825–5834. doi: 10.1021/jp711616v. [DOI] [Google Scholar]
- Patil S. S. Dhumal R. S. Varghese M. V. Paradkar A. R. Khanna P. K. Synth. React. Inorg. Met.-Org. Chem. 2009;39:265–272. [Google Scholar]
- Link S. El-Sayed M. A. Annu. Rev. Phys. Chem. 2003;54:331–366. doi: 10.1146/annurev.physchem.54.011002.103759. [DOI] [PubMed] [Google Scholar]
- Chaodong H. Lanlan L. Zeguo F. Jia L. Jinbao G. Jie W. Ultrason. Sonochem. 2014;21:542–548. doi: 10.1016/j.ultsonch.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Kaur A. Goyal D. Kumar R. Appl. Surf. Sci. 2018;449:23–30. doi: 10.1016/j.apsusc.2017.12.066. [DOI] [Google Scholar]
- Wang J. Zhu R. Sun X. Zhu Y. Liu H. Wang S. L. Int. J. Nanomed. 2014;9:3987–3998. doi: 10.2147/IJN.S64103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M. I. J. Clin. Pharmacol. 1975;15:246–251. doi: 10.1002/j.1552-4604.1975.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Jamaran S. Zarif B. R. Open Ecol. J. 2016;6:452–459. doi: 10.4236/oje.2016.67043. [DOI] [Google Scholar]
- Basiuk V. A. and Basiuk E. V., Green Processes for Nanotechnology, Springer, New York, 2015, vol. 446 [Google Scholar]
- Marambio-Jones C. Hoek E. M. J. Nanopart. Res. 2010;12:1531–1551. doi: 10.1007/s11051-010-9900-y. [DOI] [Google Scholar]
- Dixit A. Das S. Jyoti A. Kaushik S. J. Pharm. Sci. Res. 2017;9:14–17. [Google Scholar]
- Kumar N. Das S. Jyoti A. Kaushik S. Int. J. Pharm. Sci. 2016;8:183–186. [Google Scholar]
- Rajora N. Kaushik S. Jyoti A. Kothari S. L. IET Nanobiotechnol. 2016;10:367–373. doi: 10.1049/iet-nbt.2015.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. Kaushik S. Jyoti A. J. Pharm. Sci. Res. 2016;8:313–316. [Google Scholar]
- Allahverdiyev A. M. Kon K. V. Abamor E. S. Bagirova M. Rafailovich M. Expert Rev. Anti-Infect. Ther. 2011;9:1035–1052. doi: 10.1586/eri.11.121. [DOI] [PubMed] [Google Scholar]
- Palza H. Int. J. Mol. Sci. 2015;16:2099–2116. doi: 10.3390/ijms16012099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S. Poulose E. K. Int. Nano Lett. 2012;2:1–10. doi: 10.1186/2228-5326-2-1. [DOI] [Google Scholar]
- Foster C. Champney W. S. Arch. Microbiol. 2008;189:441–449. doi: 10.1007/s00203-007-0334-6. [DOI] [PubMed] [Google Scholar]
- Mehta R. Champney W. S. Curr. Microbiol. 2003;47:237–243. doi: 10.1007/s00284-002-3945-9. [DOI] [PubMed] [Google Scholar]
- Morones-Ramirez J. R. Winkler J. A. Spina C. S. Collins J. J. Sci. Transl. Med. 2013;5:190ra81. doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]