Abstract
Antimicrobial peptides (APs) are important components of the innate defenses of animals, plants, and microorganisms. However, some bacterial pathogens are resistant to the action of APs. For example, Proteus mirabilis is highly resistant to the action of APs, such as polymyxin B (PM), protegrin, and the synthetic protegrin analog IB-367. To better understand this resistance, a transposon mutagenesis approach was used to generate P. mirabilis mutants sensitive to APs. Four unique PM-sensitive mutants of P. mirabilis were identified (these mutants were >2 to >128 times more sensitive than the wild type). Two of these mutants were also sensitive to IB-367 (16 and 128 times more sensitive than the wild type). Lipopolysaccharide (LPS) profiles of the PM- and protegrin-sensitive mutants demonstrated marked differences in both the lipid A and O-antigen regions, while the PM-sensitive mutants appeared to have alterations of either lipid A or O antigen. Matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis of the wild-type and PM-sensitive mutant lipid A showed species with one or two aminoarabinose groups, while lipid A from the PM- and protegrin-sensitive mutants was devoid of aminoarabinose. When the mutants were streaked on an agar-containing medium, the swarming motility of the PM- and protegrin-sensitive mutants was completely inhibited and the swarming motility of the mutants sensitive to only PM was markedly decreased. DNA sequence analysis of the mutagenized loci revealed similarities to an O-acetyltransferase (PM and protegrin sensitive) and ATP synthase and sap loci (PM sensitive). These data further support the role of LPS modifications as an elaborate mechanism in the resistance of certain bacterial species to APs and suggest that LPS surface charge alterations may play a role in P. mirabilis swarming motility.
Antimicrobial peptides (APs), both natural and synthetic, are of increasing interest as antibacterial agents. These peptides are natural defense mechanisms of many plants, animals, and microorganisms. Most APs are cationic, amphipathic molecules of typically 12 to 45 amino acid residues and have a broad spectrum of activity against bacteria and fungi. In gram-negative bacteria, APs bind to the negatively charged residues of the lipopolysaccharide (LPS) of the outer membrane. These peptides can then transverse the membrane and cause the formation of pores or solubilization of the inner membrane.
In recent years, these peptides have been isolated from numerous organisms. Based on their structures, APs can be divided into five broad categories (for recent reviews, see references 1, 12, and 22). Insect cecropins and amphibian magainins are the prototypes of the most-studied group, which consist of linear peptides that form α-helices devoid of cysteine residues. APs with a high content of one or two amino acids, particularly proline and glycine, such as insect drosocin and human histatin I, constitute the second class. The most diverse and widely distributed group is the cystine-rich peptides. This group includes the cysteine-stabilized αβ (CSαβ) motif of the insect defensins and the β-sheet structures of the porcine protegrins and amphibian tachyplesins. A fourth class of APs recognized recently consists of macrocyclic peptides with tridisulfide structure, such as RTD-1 in primates, and macrocyclic peptides devoid of disulfides, such as AS-48 produced by Enterococcus faecalis and J25 from Escherichia coli. Although the peptides in this fourth group share common structures and are positively charged, they vary considerably in chain length, hydrophobicity, and distribution of charges. The last group consists of peptides with unique structure, such as polymyxin B (PM), which possesses a fatty acid attached through an amide linkage and a seven-member ring structure composed mainly of diaminobutyric acid.
Protegrins, as mentioned above, belong to the cysteine-rich class of APs. This family of APs was originally isolated from porcine leukocytes, and five native sequences (PG-1 to PG-5) have been characterized (35, 39) and found to have a broad spectrum of activity. Protegrins consist of 16 to 18 amino acids with multiple arginine residues making them highly cationic and two disulfide bonds forming a β-sheet (2, 18). Synthetic protegrins, such as IB-367, show improved bactericidal and fungicidal properties compared to those of native PG-1 under certain conditions (27).
Both gram-negative and gram-positive bacteria have developed mechanisms of resistance to these antimicrobial peptides. The sap genes, which encode proteins similar to those involved in ATP-binding cassette (ABC) transport systems and K+ transporters, have been shown to be important in AP resistance in Salmonella enterica serovar Typhimurium, Vibrio fischeri, and Erwinia chrysanthemi (8, 26, 29, 30). The outer membrane protein OmpT has been shown to proteolytically cleave APs in E. coli and S. enterica serovar Typhimurium (14, 36). Neisseria gonorrhoeae possesses an energy-dependent efflux pump (mtr), the loss of which resulted in increased susceptibility to PG-1 and the α-helical human AP LL-37 (32). Furthermore, bacteria including S. enterica serovar Typhimurium, Proteus mirabilis, Klebsiella pneumoniae, Burkholderia cepacia, Pseudomonas aeruginosa, and E. coli add covalent modifications to their LPS, which has been shown or is proposed to affect AP resistance (5, 9, 10, 19, 28). In S. enterica serovar Typhimurium, heptaacylated lipid A formed by the PhoP-PhoQ-regulated addition of palmitate leads to decreased membrane permeability and increased resistance to (mainly) α-helical peptides (16, 17). Other LPS additions include the PmrA-PmrB-regulated modifications of the phosphate groups of the LPS core and lipid A with phosphoethanolamine and modification of the 4′ (and sometimes the 1′) phosphate of lipid A with aminoarabinose (16, 20, 38, 40). The addition of phosphoethanolamine and aminoarabinose reduces the electrostatic interactions between the LPS and APs such as PM. Aminoarabinose LPS modification has been shown to be involved in PM resistance in S. enterica serovar Typhimurium, P. aeruginosa, and P. mirabilis (5, 10, 15, 24).
P. mirabilis has been identified as the causative pathogen in many different infections including meningitis in children and chronic otitis media, a disease characterized by mucopurulent otorrhea. The most common diseases caused by this organism are urinary tract infections (primarily catheter induced) and bladder and kidney stones. The high resistance of P. mirabilis to APs, such as PM and protegrins, may play a role in the virulence of this organism at mucosal surfaces. Therefore, we undertook a transposon mutagenesis approach to identify genes necessary for resistance to APs in P. mirabilis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Plasmids and bacterial strains used in this study are described in Table 1. Cultures were grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates. For P. mirabilis mutagenesis, the following antibiotics and concentrations were used: rifampin, 100 μg/ml; chloramphenicol, 175 μg/ml; and PM, 400 μg/ml. For all other work, the antibiotics ampicillin (50 μg/ml) and chloramphenicol (45 μg/ml) were used. All antibiotics (including gentamicin, nisin, novobiocin, PM, tetracycline, tobramycin, and vancomycin used in MIC assays) were purchased from Sigma (St. Louis, Mo.). IB-255, IB-256, IB-332, IB-352, IB-355, and IB-367 were synthesized by IntraBiotics Pharmaceuticals, Inc.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| P. mirabilis | ||
| BB2000 (JSG785) | Spontaneous Rifr mutant; PMr protegrin resistant | 3 |
| JSG945 | BB2000 derivative; PMs protegrin sensitive | This study |
| JSG946 | BB2000 derivative; PMs protegrin sensitive | This study |
| JSG947 | BB2000 derivative; PMs protegrin resistant | This study |
| JSG948 | BB2000 derivative; PMs protegrin resistant | This study |
| E. coli DH5α | Stratagene | |
| Plasmids | ||
| pBluescript KS+ | Ampr; cloning vector | Stratagene |
| pUT/mini-Tn5(Cam) | Ampr Camr | 3 |
| pWSK29 | Ampr; cloning vector | 11 |
| pWSK30 | Ampr; cloning vector | 11 |
| pWSK945 | Ampr; 4.5-kb clone of the WT region surrounding the JSG945 Tn5 insertion | This study |
Mini-Tn5 mutagenesis.
P. mirabilis BB2000 was mutagenized with a mini-Tn5 transposon conferring chloramphenicol resistance as previously described (3). Transconjugates were plated onto agar plates containing rifampin and chloramphenicol. Approximately 6 × 103 rifampin- and chloramphenicol-resistant P. mirabilis colonies were obtained from five separate matings. PM-sensitive mutants were identified by replica plating mini-Tn5 mutants on plates containing Mueller-Hinton broth (MHB) (10.5 g/liter), 1% agarose, and 400 μg of PM per ml. This MHB-agarose medium inhibits the swarming motility of P. mirabilis without affecting the activity of PM. Those mutants unable to grow on the plates containing PM were further analyzed.
Cloning of mutated genes.
To obtain the DNA adjacent to the mini-Tn5 insertion, chromosomal DNA from each mutant was digested with PstI. PstI does not cut within the mini-Tn5 transposon; therefore, plasmids containing the PstI fragment of interest were identified by transformation into E. coli DH5α and selection on plates containing Luria broth, ampicillin, and chloramphenicol. Approximately 400 bp of sequence was generated using primer JG258 located at the I-end of the transposon (5′-CCATTGCTGTTGACAAAGGG-3′). From the sequence information, primers were designed and used to PCR amplify a fragment of DNA specific for each mutant. The chromosomal DNA region containing the desired wild-type DNA locus was cloned from a HindIII gene bank. The correct clone was identified by PCR amplification with the primers that were used to amplify the probe.
MIC assay.
MICs were determined in a liquid medium essentially by the procedure of Steinberg et al. (34).The peptides (starting concentration 10 times the final concentration) were diluted (twofold serial dilutions) in 0.1% acetic acid and 0.02% bovine serum albumin. Eleven microliters of peptide was added to 100 μl of bacterial culture (between 104 and 105 CFU/ml), and the mixture was incubated at 37°C for 18 h. The MIC was defined as the lowest antimicrobial concentration at which no visible growth occurred. The peptides synthesized at IntraBiotics Pharmaceuticals, Inc., undergo rigorous quality control including amino acid analysis, high-performance liquid chromatography, and bioassays for potency.
Preparation and analysis of LPS.
LPS was prepared by whole-cell microextraction using proteinase K digestion (21). LPS profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 15% acrylamide gels. The gels were stained with silver by the method of Tsai and Frasch (37).
Analysis of lipid A modifications.
LPS from the wild type and the PM-sensitive mutants (JSG945 to JSG948) was isolated by a modified phenol-water technique (23). Lipid A was extracted from LPS samples with detergent and mild acid by the method of Caroff et al. (7). For analysis of lipid A by paper chromatography, 1 μg of purified sample was treated with 0.5 N HCl for 18 h at 37°C, lyophilized, and spotted onto Whatman 3MM filter paper in a 10-μl volume. Samples were run in a descending fashion in a solvent system of isopropanol-ethyl acetate-water (7:1:2). Aminoarabinose was detected as an orange spot after staining with ninhydrin.
Mass spectrometry analysis of lipid A.
Lipid A samples were analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. The data were acquired using Perceptive Biosystems' Voyager DE mass spectrometer. The accelerating voltage was set at 20,000, with a grid voltage of 93%, and a guide wire voltage of 0.001. The laser was fired in the negative-ion mode with delayed extraction of 75 ns. Twenty-five to 50 scans were averaged for each acquisition. A CMBT (5-chloro-2-mercaptobenzothiazole) matrix was prepared at 20 mg/ml in chloroform-methanol (1:1). Lipid A and matrix solutions were added to the MALDI sample plate and allowed to dry. The machine was calibrated with an external standard (angiotensin I) before and after the lipid A sample was run to ensure no drift had occurred.
Nucleotide sequence accession number.
The P. mirabilis JSG945 chromosomal region containing the transposon-disrupted gene has been submitted to GenBank data bank under accession number AY029614.
RESULTS
Isolation of PM-sensitive P. mirabilis mutants.
In order to identify loci involved in the innate resistance of P. mirabilis to APs, P. mirabilis BB2000 was mutagenized with a mini-Tn5 transposon. Mini-Tn5 transposons have been used previously to mutagenize P. mirabilis, and the insertions in the P. mirabilis chromosomal DNA were shown to be both random and stable (3). From replica platings of 6,000 Rifr Camr mini-Tn5 mutants from five independent matings, four PM-sensitive strains were identified (JSG945, JSG946, JSG947, and JSG948). All four strains were shown by Southern blot analysis to contain a single mini-Tn5 on the chromosome (data not shown).
The MICs of polymyxin and numerous other APs and antibiotics for wild-type P. mirabilis and the four mutants were determined (Table 2). The MICs of polymyxin for the four mutants showed that the mutants were >2 to >128 times more sensitive than the wild type. Interestingly, JSG947 and JSG948 are more sensitive to PM on solid agar than in liquid, as these strains do not grow on plates containing 400 μg of PM per ml, but their MICs are 1,600 μg/ml (JSG947) and 3,200 μg/ml (JSG948) in liquid media. Two of the four PM-sensitive mutants (JSG945 and JSG946) were also sensitive to protegrin analogs (IB-332, -352, -355, and -367) and polyphemusin II (IB-256). These two mutants were 16 (JSG945) and 128 (JSG946) times more sensitive to the action of the protegrin analog IB-367 than the parental P. mirabilis strain (JSG785 [BB2000]). IB-367, containing two fewer residues between both the amino- and carboxy-terminal cystines, was the most-active peptide tested. There was no significant effect of any of the peptides tested on the PMs mutants JSG947 and JSG948. In addition, there was little increased sensitivity of the four PMs mutants to the antibiotics tobramycin, gentamicin, nisin, and tetracycline, but all mutants were more sensitive to vancomycin and novobiocin. Therefore, in general, JSG945 and JSG946 were sensitive to PM and protegrin analogs, while JSG947 and JSG948 had reduced resistance only to PM.
TABLE 2.
MICs of the parent P. mirabilis strain and the four AP-sensitive mutants
| Antimicrobial compound | MIC (μg/ml)a for strain:
|
||||
|---|---|---|---|---|---|
| JSG785 | JSG945 | JSG946 | JSG947 | JSG948 | |
| PM | >6,400 | 20 | 5 | 1,600 | 3,200 |
| IB-367b | 100 | 6.25 | 0.78 | 100 | 100 |
| IB-255c | >420 | >420 | >420 | >420 | >420 |
| IB-256d | 640 | 80 | 6.7 | 640 | 640 |
| IB-332e | >440 | 440 | 110 | >440 | >440 |
| IB-352f | >600 | 150 | 37.5 | 600 | 600 |
| IB-355g | 600 | 38 | 18.8 | 150 | 300 |
| Novobiocin | >1,000 | 250 | 63 | 250 | 125 |
| Tetracycline | 31 | 15 | 63 | 31 | 31 |
| Vancomycin | >1,000 | 250 | 250 | 500 | 500 |
| Gentamicin | 1 | 0.5 | 0.5 | 0.75 | 0.75 |
| Tobramycin | 1 | 0.5 | 0.5 | 0.5 | 0.75 |
| Nisin | >2,000 | >2,000 | >2,000 | >2,000 | >2,000 |
MIC is defined as the lowest concentration of antimicrobial compound that completely inhibited visually observable bacterial growth.
IB-367 is a protegrin analog (NH2-RGGLCYCRGRFCVCVGR-CONH2).
IB-255 is a synthetic analog of polyphemusin (NH2-RRWCYRKCYKGYCYRKCR-CONH2).
IB-256 is polyphemusin II (NH2-RRWCRFVCYKGFCYRKCR-CONH2).
IB-332 is a tachytegrin (NH2-RGGCRLYCRRRFCVVGCR-CONH2).
IB-352 is a tachytegrin (NH2-RGRVCLRYCRGRFCVRLCFR-CONH2).
IB-355 is a tachytegrin (NH2-RGGRVCLRYCRGKFCVRLCLR-CONH2).
The P. mirabilis AP-sensitive mutants have altered LPS profiles.
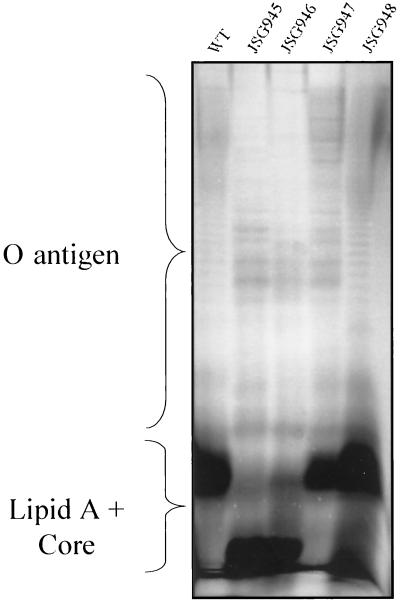
Because LPS defects have been shown to result in AP sensitivity for a number of bacteria, we examined our AP-sensitive mutants for such defects. Silver-stained SDS-PAGE profiles (Fig. 1) showed that each mutant contained an LPS having an O-antigen ladder. Therefore, the PM sensitivity of these mutants is not due to incomplete formation of the LPS (rough phenotype). The migration patterns of lipid A-core and O-antigen regions from strains JSG945 and JSG946 (PM and protegrin sensitive) were different from those of the parent strain BB2000, whereas either the O-antigen region of JSG947 or the lipid A-core region of JSG948 appear to be altered. From this analysis, it is not clear whether the aberrant migrations are due to sugar alterations, lipid A alterations, or different LPS modifications.
FIG. 1.
LPS SDS-PAGE profiles of the wild-type (WT) P. mirabilis strain and the AP-sensitive mutants. O-antigen and lipid A-core regions are marked. Note the alterations in the lipid A-core regions of JSG945, JSG946, and JSG948 and the altered O-antigen banding patterns of JSG945, JSG946, and JSG947.
Mass spectrometry analysis of lipid A from P. mirabilis AP-sensitive mutants.
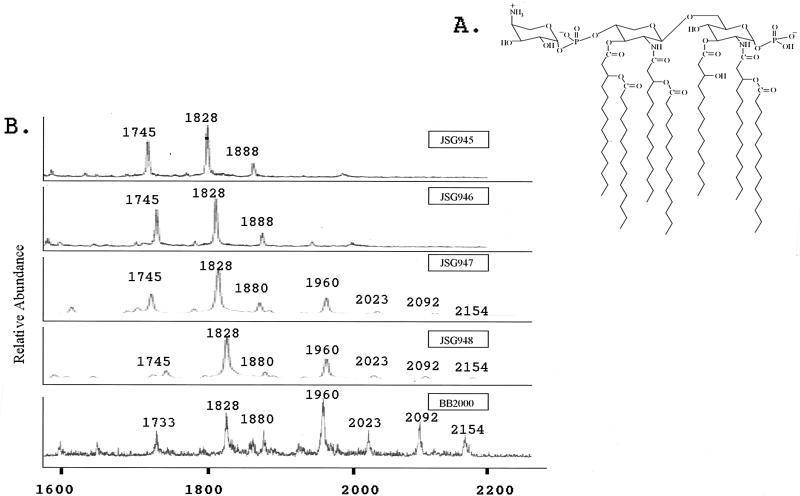
We further examined the lipid A region of the LPS from each mutant by mass spectrometry. Mass spectrometry has been used previously to identify the presence of aminoarabinose in both the core and lipid A of P. mirabilis and the lipid A of organisms such as S. enterica serovar Typhimurium (16, 20, 33, 38). Figure 2A shows the previously proposed structure of P. mirabilis lipid A (33), which has a molecular weight of 1,960. In the wild-type lipid A spectrum, three main peaks are observed (Fig. 2B). Peak m/z 1960 is representative of the proposed lipid A structure, which has aminoarabinose on the 4′ phosphate. The peak at m/z 1828 corresponds to the loss of aminoarabinose (m/z 131) from lipid A, while the addition of a second aminoarabinose moiety to the structure results in the peak at m/z 2092. Lipid A from strains JSG947 and JSG948 (PM sensitive) also contains one or two aminoarabinose moieties (m/z 1960 and 2092), but there may be less aminoarabinose on the lipid A from these two mutant strains than in the wild-type strain. Lipid A from JSG945 and JSG946 (PM and protegrin sensitive) is devoid of aminoarabinose (loss of peaks at m/z 1960 and 2092). Besides the peak at m/z 1880 (which probably reflects the loss of myristate from lipid A at m/z 2092), the chemical compositions of the other observed peaks are unknown. Paper chromatography was also performed to examine aminoarabinose addition. These studies confirmed what was observed for aminoarabinose by mass spectrometry (data not shown).
FIG. 2.
Mass spectrometry analysis of lipid A from the P. mirabilis PMS mutants. (A) Structure of the wild-type P. mirabilis lipid A (33). (B) MALDI-TOF analysis of lipid A. Peaks at m/z 1828, 1960, and 2092 have been identified as a heptaacylated lipid A with zero, one, and two aminoarabinose moieties, respectively. Peaks at m/z 1880 correspond to the loss of a fatty acid chain from m/z 2092. All other peaks have yet to be identified. Note the loss of the peaks representing aminoarabinose-containing lipid A in JSG945 and JSG946.
DNA sequence analysis of P. mirabilis AP-sensitive mutants.
Chromosomal DNA from each of the four mutants was isolated to clone the DNA flanking the mini-Tn5. The DNA was digested with PstI (which does not digest within the transposon) to produce fragments containing the mini-Tn5(Cam) transposon and flanking P. mirabilis chromosomal DNA. Upon identification of chloramphenicol-resistant clones, a primer specific to the I-end of the transposon was used to sequence each of the four plasmid clones. Approximately 400 to 600 bp of sequence was obtained and used to search the protein and DNA sequence database.
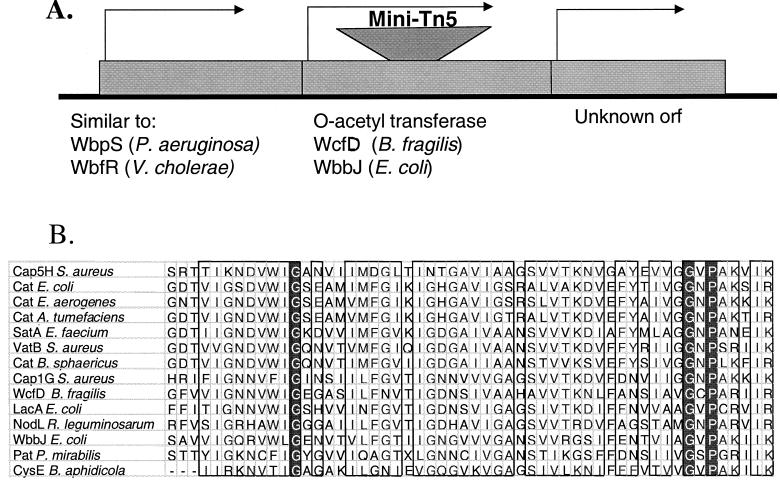
P. mirabilis JSG945 contains a transposon integration in a gene whose product is similar to a putative acetyltransferase in Bacteroides fragilis and the LacA galactoside-O-acetyltransferase in E. coli. These acetyltransferases belong to the CysE-LacA-NodL family of acetyltransferases, which acetylate a variety of substrates including antibiotics, sugar moieties, and serine residues (25). The P. mirabilis O-acetyltransferase identified in this study and other acetyltransferases of this family contain highly conserved regions at their carboxy termini (Fig. 3B).
FIG. 3.
Analysis of the Tn5-disrupted locus of P. mirabilis JSG945. (A) Approximately 1.5 kb of sequence surrounding the Tn5 insert in JSG945 was obtained from a 4.5-kb fragment. GenBank comparison identified three loci, including the O-acetyltransferase that was disrupted by the Tn5 transposon. WbpS and WbfR are thought to be involved in O-antigen synthesis. orf, open reading frame. (B) PatA, the putative O-acetyltransferase, is a member of the CysE-LacA-NodL family of acetyltransferases. This and other acetyltransferases of this family are aligned in one highly conserved amino-terminal region to show protein sequence homology. Groups of amino acids with strong similarity are boxed, and amino acids that are 100% conserved within the aligned sequences are indicated by white lettering on a black background.
P. mirabilis JSG947 has a transposon integration in a gene whose product is similar to ATP synthases. The gene containing the transposon integration in JSG948 encodes a product homologous to the SapD proteins of bacteria including Salmonella and Erwinia spp. (26, 30). sapD is part of an operon whose products exhibit similarity to the ABC family of transporters shown to be important in resistance to APs such as melittin and protamine. Unfortunately, the gene product disrupted by the transposon insertion in JSG946 could not be determined, as the entire pUT delivery plasmid carrying the transposon recombined onto the chromosome. However, available data including MICs, LPS profiles, and Southern blot analysis (data not shown) suggest that JSG945 and JSG946 contain insertions in the same chromosomal region.
A 4.5-kb wild-type clone was identified for the region surrounding the mini-Tn5 in P. mirabilis JSG945. DNA sequence analysis showed this clone to contain the entire disrupted open reading frame. The region immediately upstream contained an open reading frame homologous to the P. aeruginosa WbpS and Vibrio cholerae WbfR gene products, which are thought to be involved in LPS O-antigen synthesis. The downstream gene shows no significant similarity to sequences in GenBank. Attempts to use this cloned fragment in complementation experiments were unsuccessful due to the multiple antibiotic resistance of JSG945, resulting in the inability to select for the transformed plasmid and the relatively high frequency of reversion of the PM-sensitive phenotype when attempting to select directly for the complementing phenotype (increased PM resistance). Although we were unable to perform plasmid complementation experiments, upon examination of the spontaneous revertants, many had lost the chloramphenicol resistance (associated with the transposon) and had regained the wild-type P. mirabilis lipid A band as assayed by SDS-PAGE (data not shown). These data suggest that the defect in both PM resistance and LPS was due to the mini-Tn5 insertion.
P. mirabilis AP-sensitive mutants are defective in swarming motility.
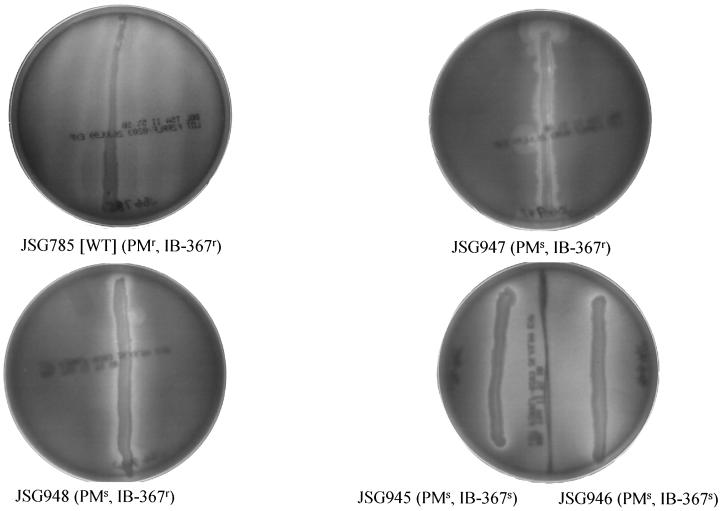
P. mirabilis displays a form of multicellular behavior called swarming, in which typical vegetative rods differentiate into long hyperflagellate swarm cells that undergo rapid and coordinated population migration across surfaces. It was observed that PM- and protegrin-sensitive mutants (JSG945 and JSG946) were completely unable to swarm, while the PM-sensitive, protegrin-resistant mutants (JSG947 and JSG948) exhibited only weak swarming ability (Fig. 4). Therefore, the LPS defects or other unknown phenotypic effects of the transposon insertions in these mutants abolish or dramatically reduce Proteus swarming.
FIG. 4.
Swarming motility of the parental P. mirabilis strain and the AP-sensitive mutants. Agar plates inoculated in a single vertical line with P. mirabilis wild-type (WT) strain (JSG785 [BB2000]) or the AP-sensitive mutants. While the wild-type strain swarmed across the agar surface in a typical wavelike pattern, the mutants were either completely defective (JSG945 and JSG946) or severely defective (JSG947 and JSG948; note the blebs of swarming from the inoculum) in swarming ability.
DISCUSSION
In recent years, much interest has been focused on APs as therapeutic agents, as they are often the first line of defense for animals, plants, and microorganisms. Both gram-positive and -negative bacteria possess mechanisms of resistance to the action of many APs, and one important mechanism of resistance to APs in gram-negative bacteria involves the modification of LPS. These modifications include the addition of charged moieties to the core and lipid A. Alteration of the acylation status of lipid A has also been shown to affect resistance to APs. Until now, LPS modifications have not been identified as playing a role in resistance to β-sheet APs. In fact, although several pathogenic bacteria are relatively resistant to the effects of β-sheet APs, such as the defensins, until this study, a β-sheet AP resistance mechanism had not been identified in any microorganism.
We have identified three P. mirabilis loci that are necessary for resistance to PM with one also necessary for resistance to protegrin (IB-367), a β-sheet AP. P. mirabilis JSG948, whose resistance was decreased only to PM, contained a transposon insertion in a gene homologous to the sapD locus. In S. enterica serovar Typhimurium, the sapABCDF operon was found to be necessary for resistance to some APs, including melittin and protamine, but strains with mutations in this locus remained resistant to defensins (β-sheet APs) (13, 29). Similar to what was observed in Salmonella, the P. mirabilis sap mutant was involved in resistance to PM but not the β-sheet AP protegrin. Thus, this family of transporters appears to be specifically involved in the resistance to α-helical and other non-β-sheet peptides. It is not clear why LPS defects (O antigen and possibly lipid A) were observed in this mutant based on the predicted roles of these proteins as an ABC transport system. A possible explanation is that these Sap proteins indirectly affect the biosynthesis or modification of LPS through interactions with LPS precursors or other proteins involved in constructing LPS in the inner membrane.
The gene containing the transposon insertion in P. mirabilis JSG947 produces a product with similarity to a member of the A subunit of the F1F0 ATPase. With a deficiency in the ability to produce ATP by oxidative phosphorylation, this strain may have less energy reserved to repair cell damage caused by the APs. However, this does not explain why the AP sensitivity was selective, because JSG947 is still resistant to the action of protegrins. Also not clearly explained is why an ATPase mutant would possess defects in LPS, as the O-antigen banding pattern of this strain was different from that of the wild-type strain. Interestingly, in two-dimensional gel experiments examining Salmonella proteins that increased or decreased in abundance by the addition of APs, one of the affected proteins was also shown to be a subunit of the F1F0 ATPase (31). Therefore, it appears that proper maintenance of ATP levels in the cell may be important in resistance to these APs.
It was found that the mutation in P. mirabilis JSG946 was not formed by just the mini-Tn5 but by the entire delivery plasmid and transposon. This had been shown to occur at a relatively high frequency in previous P. mirabilis work with this transposon delivery system (3). Based on the phenotypes of the mutants and the results of Southern blot experiments, JSG945 and JSG946 appear to contain insertions in the same chromosomal region. Because there are slight differences in the MICs of some APs for JSG945 and JSG946, it is possible that the insertions are in adjacent genes that have slightly different effects on AP resistance. It is also possible that the insertions are in different locations in the same gene, which may also have differential effects on the MICs. Because of the inherent difficulties in analyzing the insertion site in JSG946 and the similarity between the phenotypes of these two strains, only JSG945 was further characterized.
The transposon insertion in strain JSG945 (which resulted in a 320- and 16-fold decrease in resistance to polymyxin and protegrin, respectively) was found to be located in a gene producing a putative O-acetyltransferase. This putative O-acetyltransferase belongs to the CysE-LacA-NodL family of acetylases, which act upon a wide variety of substrates (25). The putative acetyltransferase identified in this study is necessary for the addition of aminoarabinose to lipid A in P. mirabilis, as JSG954 LPS was shown by mass spectrometry and paper chromatography to be devoid of aminoarabinose. Several genes have been isolated in S. enterica serovar Typhimurium that are necessary for the addition of aminoarabinose to lipid A (15), but none of these genes appear to be acetyltransferases. The mechanism by which this acetylation is needed for aminoarabinose addition is unknown, but it is possible that (i) this insertion has caused a polar effect on the transcription of downstream genes that are necessary for the biosynthesis or addition of aminoarabinose, (ii) acetylation of the O antigen is necessary before aminoarabinose can be added to the LPS, or (iii) this gene product is not an O-antigen acetylase but is involved in the biosynthesis or addition of aminoarabinose to LPS. S. enterica serovar Typhimurium mutants devoid of aminoarabinose on lipid A remain resistant to β-sheet APs, but the loss of aminoarabinose (and likely O acetylation) from the LPS of P. mirabilis renders it sensitive to the β-sheet AP protegrin. Thus, O acetylation may be the LPS modification necessary for resistance to β-sheet APs in P. mirabilis. These data represent the first identification of a nonregulatory bacterial gene and a potential mechanism necessary for resistance to a β-sheet AP.
It was observed that P. mirabilis JSG945 and JSG946 (PM and protegrin sensitive) were unable to swarm on agar surfaces. In addition, the swarming abilities of JSG947 and JSG948 (PM and protegrin sensitive) were severely defective. It had been previously observed in P. mirabilis that mutations in the O antigen of the LPS, creating rough mutants, altered swarming motility (4). The O antigen was also shown to be necessary for Myxococcus xanthus S motility (6). Because all of our mutants display alterations in lipid A or O antigen of the LPS, we hypothesize that a smooth, properly modified LPS is necessary for swarming motility in P. mirabilis. Because aminoarabinose is positively charged, alterations in LPS modification with this molecule may affect the surface charge necessary for cell-cell or cell surface interactions important in swarming motility.
Because APs represent a unique class of antibiotics, they are currently being tested as such in humans for a number of indications. However, before this type of treatment becomes common, it is important to understand how microorganisms resist the action of APs. This study adds to the growing list of resistance mechanisms identified in various gram-negative organisms implicating LPS modifications as a major focal point.
ACKNOWLEDGMENTS
This work was supported in part by IntraBiotics Pharmaceuticals, Inc.
We thank Bob Belas for providing strains and for helpful discussions and Evgeny Vinogradov for reviewing the mass spectrometry results. We also thank Steve Mouton and Lynda Bonewald of the mass spectrometry core facility at the University of Texas Health Science Center at San Antonio for technical assistance.
REFERENCES
- 1.Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 2.Aumelas A, Mangoni M, Roumestand C, Chiche L, Despaux E, Grassy G, Calas B, Chavanieu A. Synthesis and solution structure of the antimicrobial peptide protegrin-1. Eur J Biochem. 1996;237:575–583. doi: 10.1111/j.1432-1033.1996.0575p.x. [DOI] [PubMed] [Google Scholar]
- 3.Belas R, Erskine D, Flaherty D. Transposon mutagenesis in Proteus mirabilis. J Bacteriol. 1991;173:6289–6293. doi: 10.1128/jb.173.19.6289-6293.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belas R, Goldman M, Ashliman K. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J Bacteriol. 1995;177:823–828. doi: 10.1128/jb.177.3.823-828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boll M, Radziejewska-Lebrecht J, Warth C, Krajewska-Pietrasik D, Mayer H. 4-Amino-4-deoxy-L-arabinose in LPS of enterobacterial R-mutants and its possible role for their polymyxin reactivity. FEMS Immunol Med Microbiol. 1994;8:329–341. doi: 10.1111/j.1574-695X.1994.tb00460.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowden M G, Kaplan H B. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 7.Caroff M, Tacken A, Szabo L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr Res. 1988;175:273–282. doi: 10.1016/0008-6215(88)84149-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen H Y, Weng S F, Lin J W. Identification and analysis of the sap genes from Vibrio fischeri belonging to the ATP-binding cassette gene family required for peptide transport and resistance to antimicrobial peptides. Biochem Biophys Res Commun. 2000;269:743–748. doi: 10.1006/bbrc.1999.1506. [DOI] [PubMed] [Google Scholar]
- 9.Cox A D, Wilkinson S G. Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol Microbiol. 1991;5:641–646. doi: 10.1111/j.1365-2958.1991.tb00735.x. [DOI] [PubMed] [Google Scholar]
- 10.Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 11.Fu R, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 12.Ganz T, Lehrer R I. Antibiotic peptides from higher eukaryotes: biology and applications. Mol Med Today. 1999;5:292–297. doi: 10.1016/s1357-4310(99)01490-2. [DOI] [PubMed] [Google Scholar]
- 13.Groisman E A, Parra-Lopez C, Salcedo M, Lipps C J, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guina T, Yi E C, Wang H, Hackett M, Miller S I. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 16.Guo L, Lim K, Gunn J S, Bainbridge B, Darveau R, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 18.Harwig S S, Swiderek K M, Lee T D, Lehrer R I. Determination of disulphide bridges in PG-2, an antimicrobial peptide from porcine leukocytes. J Pept Sci. 1995;1:207–215. doi: 10.1002/psc.310010308. [DOI] [PubMed] [Google Scholar]
- 19.Helander I M, Kato Y, Kilpelainen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem. 1996;237:272–278. doi: 10.1111/j.1432-1033.1996.0272n.x. [DOI] [PubMed] [Google Scholar]
- 20.Helander I M, Kilpelainen I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol. 1994;11:481–487. doi: 10.1111/j.1365-2958.1994.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttner K M, Bevins C L. Antimicrobial peptides as mediators of epithelial host defense. Pediatr Res. 1999;45:785–794. doi: 10.1203/00006450-199906000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Johnson K G, Perry M B. Improved techniques for the preparation of bacterial lipopolysaccharides. Can J Microbiol. 1976;22:29–34. doi: 10.1139/m76-004. [DOI] [PubMed] [Google Scholar]
- 24.Kaca W, Radziejewska-Lebrecht J, Bhat U R. Effect of polymyxins on the lipopolysaccharide-defective mutants of Proteus mirabilis. Microbios. 1990;61:23–32. [PubMed] [Google Scholar]
- 25.Lewendon A, Ellis J, Shaw W V. Structural and mechanistic studies of galactoside acetyltransferase, the Escherichia coli LacA gene product. J Biol Chem. 1995;270:26326–26331. doi: 10.1074/jbc.270.44.26326. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Solanilla E, Garcia-Olmedo F, Rodriguez-Palenzuela P. Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell. 1998;10:917–924. [PMC free article] [PubMed] [Google Scholar]
- 27.Mosca D A, Hurst M A, So W, Viajar B S C, Fujii C A, Falla T J. IB-367, a protegrin peptide with in vitro and in vivo activities against the microflora associated with oral mucositis. Antimicrob Agents Chemother. 2000;44:1803–1808. doi: 10.1128/aac.44.7.1803-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nummila K, Kilpelainen I, Zahringer U, Vaara M, Helander I M. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol. 1995;16:271–278. doi: 10.1111/j.1365-2958.1995.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 29.Parra-Lopez C, Baer M T, Groisman E A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra-Lopez C, Lin R, Aspedon A, Groisman E A. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 1994;13:3964–3972. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi S Y, Li Y, Szyroki A, Giles I G, Moir A, O'Connor C D. Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol Microbiol. 1995;17:523–531. doi: 10.1111/j.1365-2958.1995.mmi_17030523.x. [DOI] [PubMed] [Google Scholar]
- 32.Shafer W M, Qu X, Waring A J, Lehrer R I. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidorczyk Z, Zahringer U, Reitschel E T. Chemical structure of the lipid A component of lipopolysaccharide from a Proteus mirabilis Re-mutant. Eur J Biochem. 1983;137:15–22. doi: 10.1111/j.1432-1033.1983.tb07789.x. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg D A, Hurst M A, Fujii C A, Kung A H C, Ho J F, Cheng F-C, Loury D J, Fiddes J C. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storici P, Zanetti M. A novel cDNA sequence encoding a pig leukocyte antimicrobial peptide with a cathelin-like pro-sequence. Biochem Biophys Res Commun. 1993;196:1363–1368. doi: 10.1006/bbrc.1993.2403. [DOI] [PubMed] [Google Scholar]
- 36.Stumpe S, Schmid R, Stephens D L, Georgiou G, Bakker E P. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. 1998;180:4002–4006. doi: 10.1128/jb.180.15.4002-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 38.Vaara M. Increased outer membrane resistance to ethylenediaminetetraacetate and cations in novel lipid A mutants. J Bacteriol. 1981;148:426–434. doi: 10.1128/jb.148.2.426-434.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao C, Liu L, Lehrer R I. Identification of a new member of the protegrin family by cDNA cloning. FEBS Lett. 1994;346:285–288. doi: 10.1016/0014-5793(94)00493-5. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z, Lin S, Cotter R J, Raetz C R. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]