Abstract
Tumor initiation is driven by oncogenes that activate signaling networks for cell proliferation and survival involving protein phosphorylation. Protein kinases in these pathways have proven to be effective targets for pharmaceutical inhibitors that have progressed to the clinic to treat various cancers. Here, we offer a narrative about the development of small molecule modulators of the protein Ser/Thr phosphatase 2A (PP2A) to reduce the activation of cell proliferation and survival pathways. These novel drugs promote the assembly of select heterotrimeric forms of PP2A that act to limit cell proliferation. We discuss the potential for the near-term translation of this approach to the clinic for cancer and other human diseases.
Introduction: protein phosphorylation and cancer
A watershed of scientific discoveries approximately 50 years ago opened a new era of clinical oncology that continues to grow today. Tumor viruses were revealed as causative agents of malignancies, which led to the definition of the mechanisms that induce cellular transformation and tumorigenesis. In the early 1900s, Peyton Rous discovered a transplantable infectious agent derived from tumors in chickens that was named Rous sarcoma virus [1]. Decades later, the simple RNA genome of this virus was mapped and a single gene, named src, was shown to be necessary and sufficient for the transformation of fibroblasts grown in culture and the formation of tumors in vivo [1]. Non-infected cells were found to have an endogenous gene called sarc that is related to this viral gene [2,3]. Both the src and sarc proteins (later called v-src and c-src) were the same size and precipitated by the same antibody found in the serum of rabbits with transplanted tumors [4-7]. This implied that this gene/protein functions to regulate cell growth and proliferation. But, how? The breakthrough came in the 1970s when addition of radioactive ATP to immunoprecipitates from transformed cells resulted in 32P-labeling of the immunoglobulin heavy chain, demonstrating that there was a protein kinase activity associated with v-src [8,9]. Analysis revealed phosphorylated Tyr residues and the surprising realization was that v-src was a new and different type of kinase [9-11]. Up to this time protein phosphorylation was only known to occur on Ser/Thr residues and was thought to be primarily dedicated to control of metabolism, in particular the synthesis and degradation of glycogen for maintenance of blood sugar [12-14].
Around this same time, Cohen was pioneering work on the receptor for epidermal growth factor (EGF). Adding EGF and radioactive ATP to membrane preparations from A431 cells increased 32P-labeling, including a prominent band of 150 kDa on SDS/PAGE that proved to be the EGF receptor [15]. Finding pTyr in EGF receptor [16] linked the receptor to oncogenes such as v-src with a common biochemical mechanism. Cementing this relationship was the example of the oncogene of avian erythroblastosis virus (v-erbB) being a doppelganger of the cytoplasmic domain of the EGF receptor [17,18]. Advances in protein sequencing during the 1980s enabled discovery of protein kinase domains in multiple growth factor receptors and other viral oncogenes [19]. Thus, cellular transformation and tumorigenesis became attributed to increases in pTyr [20]. The culprits were protein Tyr kinases amplified by viruses, activated by mutations, abnormally overexpressed or propelled via autocrine loops. This posed these novel enzymes as potential targets for development of new drugs for oncology [21,22] .
Coincident with finding a common mechanism for oncogenes and mitogens was the discovery of a signaling pathway featuring mitogen-activated protein kinase (MAPK, a.k.a. ERK (extracellular signal-regulated kinase)), a Ser/Thr protein kinase [23,24]. This pathway was activated downstream of receptors for insulin or EGF, and a vexing question at the time was how pTyr signals were transduced into pSer/pThr [25-27]. Part of the answer was that the kinase activating MAPK (a.k.a. MEK (MAPK and ERK kinase kinase)) phosphorylated both Tyr and Thr residues [28]. Transduction involved docking of proteins to the pTyr residues in receptors via SRC homology domain 2 (SH2) domains first found in the src oncoprotein [29-31] and physical proximity enhanced stimulation of guanine nucleotide exchange factor (GEF) activity for activation of a prevalent oncogene, the GTPase ras. The MAPK pathway was activated by ras engaging its partner Raf (Ser/Thr kinase activated by Ras named as rapidly accelerated fibrosarcoma), yet another oncogene that encodes a Ser/Thr kinase [32,33]. MAPK was associated with activation of transcription factors leading to cell division, a proliferative response. But proliferation signals alone do not support tumorigenesis and especially metastasis. Tumor cells need to avoid apoptosis to survive and accumulate [34,35]. This function resides in yet another oncogene first found in murine leukemia virus named AKT-8, which encodes a protein with Ser/Thr kinase activity [36]. Even before many substrates for MAPK and Ser/Thr kinase encoded by murine leukemia virus (AKT) were known, the activation of these kinases (involving phosphorylation at multiple sites) served as biomarkers for cell transformation and tumor forming potential. Elevated protein phosphorylation in cells, in particular MAPK and AKT, was heralded as indicative of signaling for growth, proliferation and survival, making this a target for inhibitors [37-39]. The new age of therapeutics in oncology would be based on reduction in cellular protein phosphorylation, on Tyr as well as Ser/Thr residues.
However, prevailing prejudices thwarted development of kinase inhibitors and delayed the dawning of this new age. First was the close sequence relationship among all the 500 protein kinases in the human genome [40], predicting the near-identical 3D structures that were later visualized [41]. A common 3D structure made it difficult to imagine how small molecules would distinguish one kinase from any other. Reinforcing this concern, all these enzymes used a common substrate, ATP, with active sites that conform to this ligand. Even worse, ATP is present in millimolar concentrations in cells, preventing effective competition by a small molecule inhibitor. Nonetheless, these prejudices were overcome by persistence and inventiveness that generated new knowledge to supplant conventional wisdom. Combination of high-throughput screening and medicinal chemistry created small molecules that showed specificity for individual kinases and selectivity among protein kinases. Highly effective compounds proved to be non-competitive inhibitors, sidestepping the challenge of high ATP concentrations [42,43]. Inhibitors of the EGF receptor kinase (Tarceva, Iressa) and the MAPK pathway (Trametinib) have made their way through clinical trials and into oncology practice.
Protein kinases have become so popular as targets for drug development, they are now only second to G protein-coupled receptors (GPCRs) [44,45]. An alternative way to reduce the levels of pathological protein phosphorylation would be to target the protein phosphatases that catalyze the dephosphorylation and inactivation of protein kinases. Rather than inhibit, the intent would need to be to increase activity, a foreign concept in terms of drug development. Nevertheless, an effective way to reduce activation of both the MAPK and AKT signaling pathways would be to increase the activity of protein phosphatase 2A (PP2A). What follows is an account of the path towards developing activators of PP2A.
How enzyme activation involves relief of constraints
Unbridled catalytic activity is the antithesis of homeostasis, which requires a nuanced modulation of enzymatic action in response to physiological or environmental perturbations. Modulation of enzyme activity is often achieved by suppressing the inherent catalytic activity by occlusion or conformational distortion. Most common is the blocking of active sites by distal segments of the same polypeptide, or by separate subunits, in what is called intrasteric regulation [45]. The intracellular second messengers cyclic AMP and calcium ions can be envisioned as agents that produce activation by relieving intrasteric inhibition. The cAMP-dependent protein kinase (PKA) regulatory subunits sit as a pseudosubstrate blocking the active site of the kinase. Binding of cAMP to regulatory subunits results in dissociation from catalytic subunits, freeing the kinase to express activity [46,47]. Likewise, binding of calcium ions to calmodulin (CaM) induces conformational changes allowing CaM to bind to and displace auto-inhibitory segments in its targets, such as calcineurin, CaM-dependent kinase (CaMK), myosin light chain (LC) kinase or phosphodiesterase [48]. Interestingly both PKA R subunits and CaM have tetrameric domains for binding their respective ligands arranged as α2/β2 resembling hemoglobin. This produces a cooperative allosteric response to second messengers. The src kinase is another example where intramolecular association of the SH2 domain with a pTyr in the C-terminus holds the enzyme in a closed and inactive conformation. Dephosphorylation of this inhibitory pTyr and engagement of the SH2 and SH3 domains with ligands opens up the protein, freeing it for reaction with substrates [49]. Thus, enzyme activation arises from relief of inhibition and this perspective makes activation by pharmaceutical agents more plausible.
Negative control of PPP family Ser/Thr phosphatases
Negative control of enzyme activity applies to various protein Ser/Thr phosphatases of the major PPP family [50]. Both PP3 (calcineurin) and PP5 are restrained in inactive conformations, by an inhibitory segment or a TPR domain, respectively. Phosphatase activation involves ligand binding (CaM for PP3 and phospholipids for PP5) to the inhibitory segments, thereby opening the protein and exposing the active site for action [51,52]. For PP1 and PP2A biochemical assays with purified subunits showed association of the catalytic subunits with regulatory subunits did not fully extinguish catalytic activity but did restrain it. Regulatory subunits for PP1 have long been assayed for their ability to reduce phosphatase activity with phosphorylase as a primary substrate, relative to other proteins such as myosin light chain or eIF2 [50]. This is attributed to formation of a different surface topology surrounding the active site, affecting substrate binding.
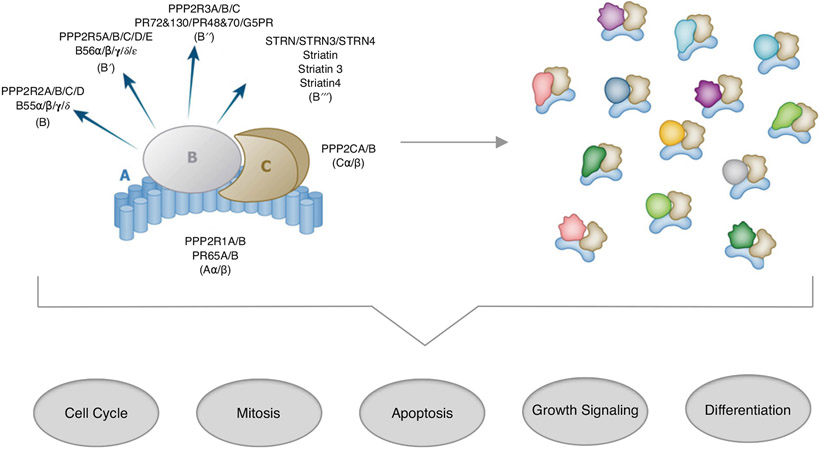
Multiple sources of evidence point to PP2A as an enzyme under negative control, in a low activity state. Phosphorylase was the traditional substrate used in the 1970–1980s to monitor the activity of phosphatases during purification from tissue extracts [50]. The activity of phosphorylase phosphatase (PP2A) preparations was stimulated by adding a heat-stable protein [53] that turned out to be histone H1 [54]. Similar PP2A phosphatase preparations were activated as much as five-fold by histone H1, or by polylysine or protamine. This phosphatase was characterized as polycation-stimulated (PCS) phosphatase [55-58] and recognized from its subunit composition and properties as PP2A. Binding of PP2A-A subunit to isolated C subunit reduces the enzyme velocity by more than 20-fold with a peptide substrate [59], and on top of that B subunit binding to the AC core dimer affected the kinetics but not velocity. Association with recombinant A subunit inhibits PP2A catalytic subunit activity to different extents with peptide and protein substrates [60]. Binding of the A subunit made the enzyme sensitive to polycation stimulation [60] showing the constraint on PP2A phosphatase activity. Most PP2A exists in cells and tissues as ABC heterotrimers. The popular concept has been that the B subunits participate as ‘regulatory’ in terms of substrate specificity and recruitment (Figure 1). Indeed, the discovery of short linear motifs (SLIMs) docking to B56 has beautifully demonstrated this principle in action [61-64]. The picture that emerges is a robustly active PP2A catalytic subunit, held in a reduced activity state by association with the A subunit, with substrate access limited by or facilitated by the various B subunits.
Figure 1. PP2A refers to a set of distinctive serine/threonine phosphatases.
The PP2A A and C subunits each have an alpha and beta isoform, while B subunits are in four protein families, each containing multiple isoforms. In a cell, this results in assembly of a heterogeneous pool of PP2A heterotrimeric holoenzymes that collectively regulate critical biological processes often associated with disease progression.
Yet another layer of negative control on PP1 and PP2A comes from multiple inhibitor proteins specific for each phosphatase [50]. These are separate genes encoding mostly low molecular weight proteins dominated by inherently unstructured regions that are phosphorylated, in most cases at multiple sites that modulate function [65]. Phosphorylation can boost inhibitory potency 1000-fold, and these inhibitors act as especially poor substrates with low turnover, preoccupying the phosphatase, described as ‘inhibition by unfair competition’ [66,67]. Once one appreciates how many ways phosphatases are controlled by inhibitory mechanisms then it becomes more plausible to embark on discovery and development of small molecule therapeutics to increase phosphatase activity and restore homeostasis. Indeed, there are efforts underway to develop drugs that prevent inhibition of PP2A by its endogenous inhibitor proteins [68,69].
PP2A as the target for novel drugs in cancer cells and tumors
Limiting proliferative signaling with antipsychotic tricyclic drugs
Early in their use, it appeared that the phenothiazine class of antipsychotics (also known as tricyclics, based on their chemical structures) may have anticancer actions. Phenothiazines were observed to antagonize CaM [70-72], that was recognized for its role in DNA synthesis and cellular proliferation. Phenothiazines emerged as a potential therapeutic approach to treat cancer cells, based on this mechanism [70,73-76]. Studies have since then identified that phenothiazines do indeed have an antiproliferative effect in cancer cells. These studies led to the identification of a number of processes and mechanisms that phenothiazines modulate as a basis for their anticancer effects [77-79]. In addition to anti-CaM activity, negative regulation of AKT, mammalian target of rapamycin (mTOR) and glycogen synthase kinase-3β (GSK3β) have been implicated, along with regulation of the cell cycle through cyclin-dependent kinases (CDKs). Inhibition of these pathways among others have suggested potential targets for phenothiazines in regulation of cell cycle, apoptosis, autophagy, epithelial–mesenchymal transition (EMT), metastasis and angiogenesis [80-84]. Lastly, phenothiazines have been found to reduce multidrug resistance (MDR) through direct inhibition of the efflux pump and by reducing its transcription [85-88]. In light of these findings, phenothiazines have been studied in cancer patients. Because these drugs are FDA approved they are ideal candidates for repurposing. On their own, the phenothiazines have been associated with dose-wlimiting toxicities, attributable to their D2 dopamine receptor antagonism. However, in addition to their use as single agents rational drug combination studies also have been proposed [77]. Because of their modulation of MDR, use of tricyclics has been proposed in combination with cytotoxic chemotherapies. In other instances, their modulation of AKT signaling has led to the hypothesis that they may be synergistic with RTK inhibitors. Notably, one pathway that emerged as a critical modulator of sensitivity to RTKi in non-small cell lung cancer is the transcription factor, forkhead box transcription factor O1 (FOXO1). Addition of the tricyclic trifluoperazine (TFP) causes FOXO1 to be mislocalized in cells, leading to aberrant regulation of proteins in the EGFR–AKT signaling axis. This TFP-dependent relocalization was shown to be synergistic in combination with EGFR inhibitor, Erlotinib [89]. However, in combination studies, particularly with chemotherapies, dose-limiting toxicities have been observed. Clinical trials with RTKs have been initiated, but are not yet completed.
Identification of PP2A as a putative target of tricyclics in reducing cell proliferation
While some of the anticancer activity of tricyclic phenothiazines has been mapped back to direct interaction with the D2 dopamine receptor, it has long been hypothesized that the majority of anticancer activity may be due to other actions in the cell. Efforts have focused on the identification of a therapeutic target of this drug class (in addition to MDR) that may be mediating their anticancer activity. Only recently identified was PP2A identified as a target. Notably, this observation accounts for much of the research and biological consequences that have been linked to antipsychotics over the years. Through its action as a serine/threonine phosphatase, PP2A is a well-described tumor suppressor that negatively regulates (inactivates) kinases such as AKT and ERK downstream of major oncogenes like KRAS. PP2A also regulates many biological processes that align with what is known about the biological effects of phenothiazines in cellular and in vivo model systems [69].
Kim et al. [90] first proposed that antipsychotics may exert their anticancer activity through PP2A. Specifically, they demonstrated that Haloperidol (an antipsychotic but not tricyclic) inhibits MAPK signaling through PP2A, based on kinetics of phosphorylation and binding studies among PP2A, MEK and ERK. Subsequently, in 2014, Gutierrez et al. published research implicating the tricyclic perphenazine as a PP2A agonist based upon small molecule screening to identify inhibitors of MYC [91]. In addition to showing perphenazine-induced apoptosis in MYC overexpression models, they proposed PP2A as the drug target-based on data from fluorous ligand-affinity chromatography coupled with mass spectrometry. In addition to inhibition of MYC, one well-characterized substrate of PP2A, a number of other PP2A substrates were dephosphorylated upon treatment with Perphenazine [91]. With these observations in mind, there was a drive to design PP2A agonists that do not carry the dose-limiting toxicities associated with the phenothiazine class of molecules. As a result in recent years new classes of molecules that activate PP2A have been described. Different classes of these molecules are referred to as small molecule activators of PP2A (SMAPs), as well as improved heterocyclic activators of PP2A (iHAPs) [92-96]. As a whole these may also be grouped in with what has been referred to as PADs or PP2A activating drugs [97], though this term does not necessarily refer to molecules that directly bind and activate PP2A but rather act via inhibition of another protein or target such as a PP2A inhibitor.
Medicinal chemistry transformation of tricyclics into SMAPs
The evolution to both SMAPs and iHAPs was built on the observation that the tricyclic phenothiazine backbone of antipsychotics was associated with the anticancer activity of these molecules. Thus, even before PP2A was confirmed as the target, iterative versions of the molecules were made to focus on improving the anticancer activity while at the same time abrogating the neuroleptic affects associated with their interactions with the dopamine receptor [98]. This led to the generation of whole classes of novel tricyclic molecules that had robust anticancer activity in vitro and in vivo. In addition, when added to living cells these molecules induced net dephosphorylation of well-credentialed PP2A substrates such as ERK and MYC [92,94,99-101], supporting the idea that PP2A is the target of these molecules and they promoted an increase in PP2A activity. Moreover, the use of chemical inhibitors of PP2A (such as okadaic acid) abrogated the biological activity of these SMAPs [93,94,102] .
Effects of SMAPs on PP2A in biochemical assays
Despite multiple studies that demonstrated the addition of SMAP compounds to living cells reduced the phosphorylation of multiple PP2A substrates in parallel it proved elusive to demonstrate in biochemical assays direct activation of purified PP2A by SMAPs. Routine phosphatase assays utilize small molecule chromogenic substrates p-nitrophenyl phosphate (pNPP) and 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) at millimolar and micromolar concentrations, respectively. Hydrolysis of these substrates releases phosphate plus either a yellow colored or fluorescent product, readily quantified by spectroscopy. Surprisingly there was no activation of PP2A by different SMAPs over a concentration range from 30 nM to 20 μM (unpublished data). This was true for both pNPP and DiFMUP. This might be a result of the use of these artificial chemical substrates rather than proteins that exhibit lower KM values. Therefore histone H1 32P-phosphorylated at different single sites by either PKA or protein kinase C (PKC) [99,103-105] were used as substrates with purified PP2A, assaying for release of acid-soluble inorganic 32P-phosphate. However, once again there was no activation, and if anything some slight reduction in PP2A activity when SMAPs were added up to micromolar concentrations (unpublished). Different preparations of PP2A, native AC dimer purified from human red cells as well as AB56γC trimer assembled from recombinant subunits were used for these biochemical studies. No activation was seen in response to SMAPs. Finally, assay with a conjugated peptide substrate, ProFluor Ser/Thr R110 Substrate System (Promega) produced an SMAP concentration-dependent increase in approximately 20% in PP2A activity [94]. It is not understood how this one particular substrate exposes an SMAP-induced activation of purified PP2A. The PP2A catalytic subunit is restrained into a relatively low activity state by its binding to the A subunit, leaving an opportunity for allosteric effects to increase activity. However, the SMAP effect in enzyme assays is subtle and does not seem to satisfactorily account for the robust reduction in phosphorylation of PP2A substrates seen in cells. This apparent disparity stimulated a search for other effects of SMAPs on PP2A.
SMAPs alter PP2A actions by promoting B56 subunit association
Until recently, convincing biochemical and structural data confirming interaction of this SMAPs and PP2A heterotrimeric enzyme were lacking and the mechanism by which SMAPs directed tumor suppressive activity of PP2A was unknown. Additionally, it was unclear whether SMAPs broadly activated PP2A heterotrimers or whether activation was restricted to subset of distinct heterotrimers with particular regulatory B subunits. Critical experiments published by Leonard et al. honed in especially on this urgent question [106]. In cancer cell lines, a split luciferase system (NanoBret) enabled the study of dynamic conformational changes in the PP2A A-α scaffolding subunit with the luciferase protein split between the C-terminal and N-terminal ends. The addition of SMAPs to cells confirmed dynamic reduction in the intramolecular N-C terminus distance of the A scaffolding subunit, consistent with specific heterotrimer assembly. It was next observed that upon treatment with SMAPs, binding of the B56α subunit was enhanced in cells and in vivo, while the binding of other regulatory subunits was either decreased or unchanged. Furthermore, when the NanoBret system was used with a luciferase component placed on the carboxy tail of the B56α subunit, enhanced binding of this subunit to PP2A was observed. Overall, this suggested a mechanism for SMAPs involves their capacity to stabilize select PP2A holoenzymes over others. In the instance of stabilizing the B56α holoenzyme, this is particularly encouraging because B56α reportedly directs tumor suppressive activity of PP2A, primarily through targeting the oncogene MYC [106].
Additional structural and biophysical studies further demonstrated SMAPs preferentially stabilized the PP2A AB56αC holoenzyme producing an apparent increase in PP2A activity by accumulation of this specific PP2A heterotrimer in cells. Size exclusion chromatography (SEC) was used to profile holoenzyme stability in the absence and presence of SMAPs. Surface plasmon resonance (SPR) analysis confirmed that the calculated Kd for B56α to the AC core dimer was enhanced in the presence of SMAP. Notably, when SPR analysis was done with the B56γ subunit, the binding kinetics were unchanged [106]. Thus, the SMAP distinguished between different isoforms of B56.
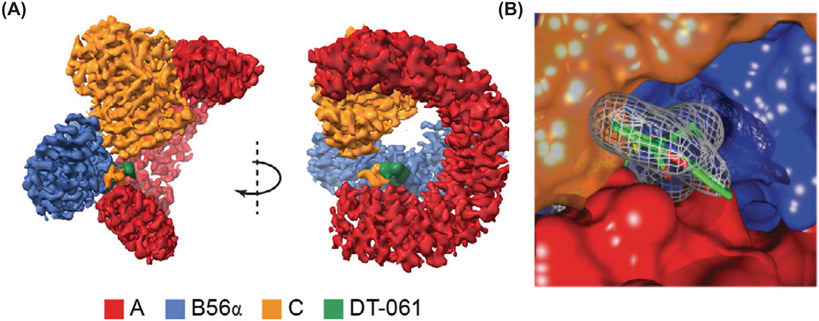
Critically, perhaps the most conclusive evidence for binding of SMAPs to a specific PP2A heterotrimer came from the first ever high resolution 3D structure of the B56α holoenzyme bound to the SMAP DT-061 (Figure 2A,B). This work confirmed how SMAPs interacted with PP2A, provided structural insight into PP2A holoenzyme stability and identified a unique interfacial drug binding pocket. This structure confirmed previous results from X-ray crystallography that the C subunit binds to the C-terminal portion of the A subunit while the B subunit binds near the N-terminal end of the scaffolding subunit. Additionally, this structure provided new high resolution data placing the C-terminal tail of the C subunit at the site of contact between the A and B subunits resulting in an unique three-way intersubunit interaction. It is within a pocket at this subunit–subunit junction that SMAP DT-061 interacts with the holoenzyme. The trifluoromethoxy end of the molecule interacts with two residues on the A subunit (E100 and E101) and one side chain on the B subunit (K316) while the hydrophobic benzene ring of the molecule is buried within a pocket of B subunit residues (I237, Y238 and F317) and a residue in the C-terminal tail of the C subunit. Lastly, phenoxazine moiety contacts were observed between Y307 and P305 of the C subunit and T102 and V103 of the A subunit. The C-terminal tail of the C subunit has been shown to be inherently flexible but this interaction with DT-061 at Y307 effectively pins the tail of the C subunit in space between the A and B subunits. It is thought that this stabilization of the C-terminal of the catalytic subunit contributes to the overall stability of the holoenzyme in the presence of DT-061. The minimal effects of SMAPs on canonical phosphatase activity originally employed (vide infra) compared with the stabilizing effects on subunit interactions highlights the complexity of drug development for a multisubunit holoenzyme. The activity assays originally employed focused primarily on the AC dimer. Only upon inclusion of the appropriate B subunit in additional biochemical assays could the binding and mechanism of action of SMAPs be elucidated, further emphasizing the importance of B subunits in modulating and directing the activity of the enzyme [106].
Figure 2. The 3D molecular structure of PP2A bound to the SMAP DT-061.
(A) Binding of DT-061 to the PP2A AB56αC holoenzyme determined at 2.2 Å resolution by cryo-E-M. Color code for PP2A subunits and DT-061 shown. (B) Close-up of DT-061 at interface of A, B56α and C subunits.
Lastly, in the structure with SMAP DT-061, it was observed that the C-terminal Leu of the C subunit is methylated(methyl esterified). Methylation of the C-terminal of the catalytic subunit is critical for holoenzyme biogenesis and this post-translational modification directs the selection of certain B subunits over others [107-109]. Critically, it was observed in vitro and in vivo that methylation occurs upon treatment of cells with DT-061. Specifically, methylation increases in the first 3 h after DT-061 treatment, and by 12 h post-treatment it returns to basal levels. The observation of the post-translational modification can support future research as a biomarker for stabilization of the holoenzyme and also inform development of future generations of SMAPs as a marker for holoenzyme engagement [106].
At the same time, the structural and mechanistic basis for SMAP binding to PP2A was identified, the concept of biased subunit availability was reinforced independently with emerging research about the iHAP class of PP2A activators as well. In the case of iHAPs, the molecules are proposed to selectively stabilize holoenzymes containing the B56ε regulatory subunit. Morita et al. identified the subunit responsible with a combination of binding studies and CRISPR-Cas 9 deletion of different subunits to disrupt holoenzyme formation. Additionally, through their work they identified MYBL2 as a substrate of the PP2A B56ε holoenzyme. They further demonstrated that the PP2A-mediated dephosphorylation of MYBL2 at S241 is critical for mediating the anticancer activity of iHAPs in T-ALL [96].
Research from both SMAPs and iHAPs demonstrates an emerging and exciting new chapter in small molecule design towards the stabilization of select PP2A holoenzymes (Figure 3). These molecules can first be used as tools to help us better understand the environmental factors and mechanisms guiding PP2A holoenzyme formation. Further, these discoveries can help influence the evolution of the small molecules such that they are increasingly refined and optimized as they make their way through preclinical development and into clinical translation.
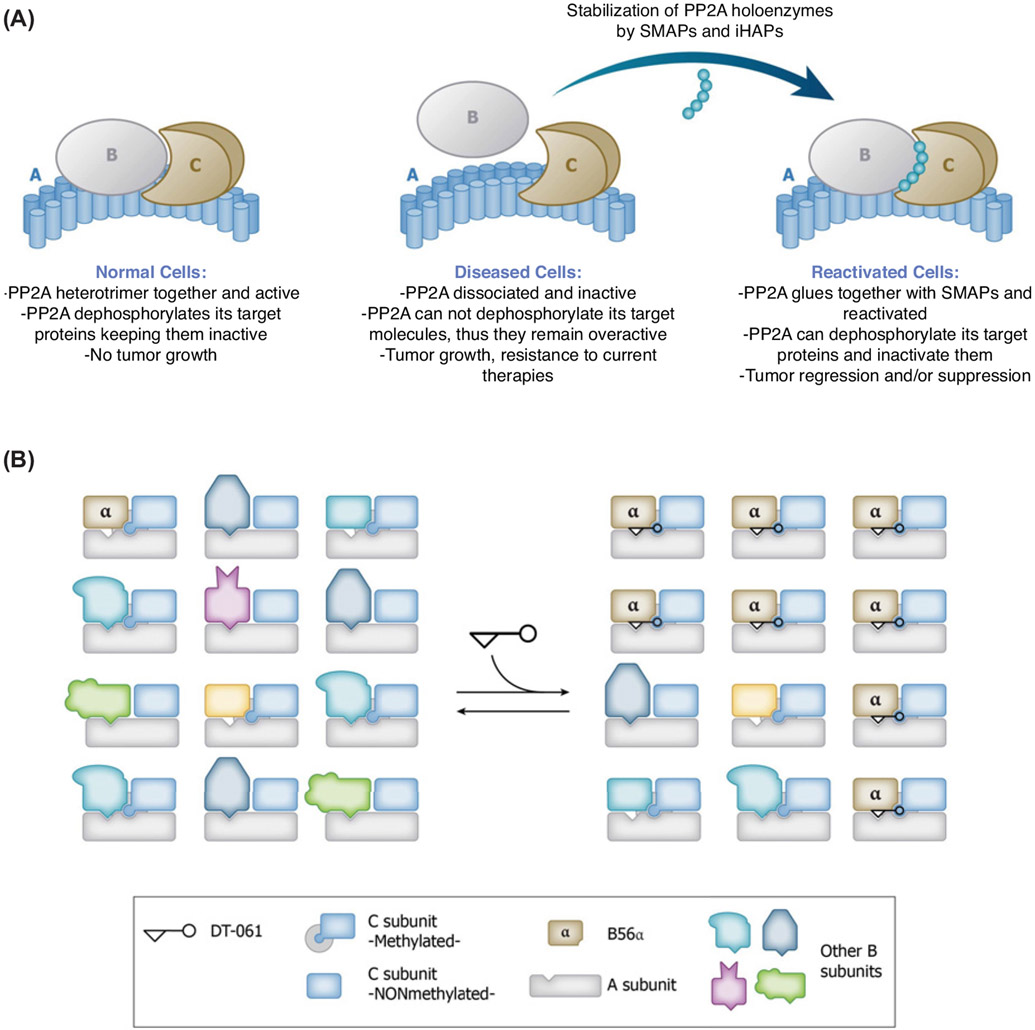
Figure 3. Novel Therapeutics Shift Populations of PP2A Holoenzymes.
(A) Schematic representation of how SMAPs and iHAPs act to stabilize a particular PP2A holoenzyme in disease settings where PP2A is inactivated. (B) Illustration of biased subunit assembly. DT-061 acts to shift the overall population of PP2A by selectively stabilizing the B56α containing holoenzyme with methylated C subunit.
Perspectives of PP2A targeting by anticancer drugs
It is now well appreciated that PP2A is not a single entity but exists as a collection of trimeric holoenzymes with different B subunits, and in this way separate PP2A affect a variety of intracellular processes. The assembly of holoenzymes is a dynamic equilibrium involving a pool of AC dimers and individual B subunits of different structural types. The capacity to selectively stabilize individual PP2A holoenzymes with small molecule ligands holds much potential within the field of cancer. PP2A is dysregulated in different cancers and a number of other human diseases, including Alzheimer’s and cardiovascular diseases. While in some cases there is a genetic underpinning to the observed dysregulation of PP2A, such as point mutations in the A subunit in cancers, the majority of other instances can be traced back to dysregulation of specific heterotrimers or overexpression of PP2A endogenous inhibitors. The studies outlined in this review describe some steps along the way in the development of a series of small molecules directed at PP2A holoenzymes. To this end, we are now able to imagine pharmaceutical interventions that one day may selectively target individual PP2A holoenzymes in a rational, context-dependent manner. This is dramatically illustrated in the work described above by the isoform differences in B56 regulatory heterotrimerization directed by SMAPs and iHAPs. In work recently published online [110], there was potentiation of CDK9 inhibition by SMAP DBK-1154, attributed to activation of PP2A in the Integrator complex, which was present as a trimer with INTS6 as a subunit instead of a canonical B subunit. The action of this SMAP might involve stabilization of INTS6 binding to PP2A core dimer because DBK-1154 did enhance recruitment of PP2A to chromatin sites of active transcription. Our conclusion is that SMAPs have a common mode of action, albeit involving different PP2A regulatory subunits.
The structural work identifying how one specific SMAP DT-061 stabilizes the B56α holoenzyme does more than shed light on this specific holoenzyme. It provides insight into the stabilization of one PP2A holoenzyme that can potentially be applied to other B56 holoenzymes, extended to trimers with different B regulatory subunits and even to non-canonical PP2A heterotrimers. Compounds that shift the PP2A equilibrium among different heterotrimers should facilitate deciphering substrates and pathways under control of different PP2A heterotrimers. This should lead to a better understanding of the actions of these compounds and their possible side effects as they are developed as therapeutics. Collectively, we expect the latest findings about PP2A will spur future drug development for the treatment of cancer as well as other human diseases.
Abbreviations
- AKT
Ser/Thr kinase encoded by murine leukemia virus
- CaM
calmodulin
- CDK
cyclin-dependent kinase
- DiFMUP
6,8-difluoro-4-methylumbelliferyl phosphate
- eIF2
eucaryotic initiation factor 2
- ERK
extracellular signal-regulated kinase, a.k.a. MAPK
- FOXO1
forkhead box transcription factor O1
- iHAP
improved heterocyclic activators of PP2A
- MAPK,
mitogen-activated protein kinase, a.k.a. ERK
- MDR
multidrug resistance
- MEK
MAPK and ERK kinase kinase
- pNPP
p-nitrophenyl phosphate
- PAD
phosphatase activating drug
- PP2A
protein phosphatase 2A
- PPP
phospho protein phosphatase family
- RTK
receptor tyrosine kinase
- RTKi
receptor tyrosine kinase inhibitor
- SH2,
SRC homology domain 2
- SMAP
small molecule activators of PP2A
- SPR
surface plasmon resonance
- T-ALL
T-cell acute lymphoblastic leukemia
- TFP
trifluoperazine
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Van Epps HL (2005) Peyton Rous: father of the tumor virus. J. Exp. Med 201, 320, 10.1084/jem.2013fta [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbs CP, Tanaka A, Anderson SK, Radul J, Baar J, Ridgway A et al. (1985) Isolation and structural mapping of a human c-src gene homologous to the transforming gene (v-src) of Rous sarcoma virus. J. Virol 53, 19–24, 10.1128/jvi.53.1.19-24.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanstrom R, Parker RC, Varmus HE and Bishop JM (1983) Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc. Natl. Acad. Sci. U.S.A 80, 2519–2523, 10.1073/pnas.80.9.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collett MS, Erikson E, Purchio AF, Brugge JS and Erikson RL (1979) A normal cell protein similar in structure and function to the avian sarcoma virus transforming gene product. Proc. Natl. Acad. Sci. U.S.A 76, 3159–3163, 10.1073/pnas.76.7.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jay G, Shiu RP, Jay FT, Levine AS and Pastan I (1978) Identification of a transformation-specific protein induced by a Rous sarcoma virus. Cell 13, 527–534, 10.1016/0092-8674(78)90326-4 [DOI] [PubMed] [Google Scholar]
- 6.Karess RE and Hanafusa H (1981) Viral and cellular src genes contribute to the structure of recovered avian sarcoma virus transforming protein. Cell 24, 155–164, 10.1016/0092-8674(81)90511-0 [DOI] [PubMed] [Google Scholar]
- 7.Purchio AF, Erikson E, Brugge JS and Erikson RL (1978) Identification of a polypeptide encoded by the avian sarcoma virus src gene. Proc. Natl. Acad. Sci. U.S.A 75, 1567–1571, 10.1073/pnas.75.3.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collett MS and Erikson RL (1978) Protein kinase activity associated with the avian sarcoma virus src gene product. Proc. Natl. Acad, Sci, U.S.A 75, 2021–2024, 10.1073/pnas.75.4.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levinson AD, Oppermann H, Levintow L, Varmus HE and Bishop JM (1978) Evidence that the transforming gene of avian sarcoma virus encodes a protein kinase associated with a phosphoprotein. Cell 15, 561–572, 10.1016/0092-8674(78)90024-7 [DOI] [PubMed] [Google Scholar]
- 10.Erikson RL, Purchio AF, Erikson E, Collett MS and Brugge JS (1980) Molecular events in cells transformed by Rous Sarcoma virus. J. Cell Biol 87, 319–325, 10.1083/jcb.87.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter T (2015) Discovering the first tyrosine kinase. Proc. Natl. Acad. Sci. U.S.A 112, 7877–7882, 10.1073/pnas.1508223112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin CS and Rosen OM (1975) Protein phosphorylation. Annu. Rev. Biochem 44, 831–887, 10.1146/annurev.bi.44.070175.004151 [DOI] [PubMed] [Google Scholar]
- 13.Krebs EG and Beavo JA (1979) Phosphorylation-dephosphorylation of enzymes. Annu. Rev. Biochem 48, 923–959, 10.1146/annurev.bi.48.070179.004423 [DOI] [PubMed] [Google Scholar]
- 14.Larner J, Roach PJ, Huang LC, Brooker G, Murad F and Hazen R (1979) Hormonal control of glycogen metabolism. Adv. Exp. Med. Biol 111, 103–123, 10.1007/978-1-4757-0734-2_6 [DOI] [PubMed] [Google Scholar]
- 15.Cohen S, Carpenter G and King L Jr (1980) Epidermal growth factor-receptor-protein kinase interactions. Co-purification of receptor and epidermal growth factor-enhanced phosphorylation activity. J. Biol. Chem 255, 4834–4842, 10.1016/S0021-9258(19)85573-4 [DOI] [PubMed] [Google Scholar]
- 16.Ushiro H and Cohen S (1980) Identification of phosphotyrosine as a product of epidermal growth factor-activated protein kinase in A-431 cell membranes. J. Biol. Chem 255, 8363–8365, 10.1016/S0021-9258(18)43497-7 [DOI] [PubMed] [Google Scholar]
- 17.Gilmore T, DeClue JE and Martin GS (1985) Protein phosphorylation at tyrosine is induced by the v-erbB gene product in vivo and in vitro. Cell 40, 609–618, 10.1016/0092-8674(85)90209-0 [DOI] [PubMed] [Google Scholar]
- 18.Kris RM, Lax I, Gullick W, Waterfield MD, Ullrich A, Fridkin M et al. (1985) Antibodies against a synthetic peptide as a probe for the kinase activity of the avian EGF receptor and v-erbB protein. Cell 40, 619–625, 10.1016/0092-8674(85)90210-7 [DOI] [PubMed] [Google Scholar]
- 19.Berthois Y and Martin PM (1989) Growth factors and oncogenes. Biomed. Pharmacother 43, 635–639, 10.1016/0753-3322(89)90081-4 [DOI] [PubMed] [Google Scholar]
- 20.Lipsick J (2019) A history of cancer research: tyrosine kinases. Cold Spring Harb. Perspect. Biol 11, 10.1101/cshperspect.a035592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levitzki A and Gazit A (1995) Tyrosine kinase inhibition: an approach to drug development. Science 267, 1782–1788, 10.1126/science.7892601 [DOI] [PubMed] [Google Scholar]
- 22.Zwick E, Bange J and Ullrich A (2002) Receptor tyrosine kinases as targets for anticancer drugs. Trends Mol. Med 8, 17–23, 10.1016/S1471-4914(01)02217-1 [DOI] [PubMed] [Google Scholar]
- 23.Ray LB and Sturgill TW (1987) Rapid stimulation by insulin of a serine/threonine kinase in 3T3-L1 adipocytes that phosphorylates microtubule-associated protein 2 in vitro. Proc. Natl. Acad. Sci. U.S.A 84, 1502–1506, 10.1073/pnas.84.6.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seger R and Krebs EG (1995) The MAPK signaling cascade. FASEB J. 9, 726–735, 10.1096/fasebj.9.9.7601337 [DOI] [PubMed] [Google Scholar]
- 25.Rossomando AJ, Payne DM, Weber MJ and Sturgill TW (1989) Evidence that pp42, a major tyrosine kinase target protein, is a mitogen-activated serine/threonine protein kinase. Proc. Natl. Acad. Sci. U.S.A 86, 6940–6943, 10.1073/pnas.86.18.6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson NG, Maller JL, Tonks NK and Sturgill TW (1990) Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature 343, 651–653, 10.1038/343651a0 [DOI] [PubMed] [Google Scholar]
- 27.Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD et al. (1991) ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65, 663–675, 10.1016/0092-8674(91)90098-J [DOI] [PubMed] [Google Scholar]
- 28.Nakielny S, Cohen P, Wu J and Sturgill T (1992) MAP kinase activator from insulin-stimulated skeletal muscle is a protein threonine/tyrosine kinase. EMBO J. 11, 2123–2129, 10.1002/j.1460-2075.1992.tb05271.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter G (1992) Receptor tyrosine kinase substrates: src homology domains and signal transduction. FASEB J. 6, 3283–3289, 10.1096/fasebj.6.14.1385243 [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Olivier P, Rozakis-Adcock M, McGlade J and Henkemeyer M (1993) Proteins with SH2 and SH3 domains couple receptor tyrosine kinases to intracellular signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci 340, 279–285, 10.1098/rstb.1993.0069 [DOI] [PubMed] [Google Scholar]
- 31.Marengere LE and Pawson T (1994) Structure and function of SH2 domains. J. Cell Sci. Suppl 18, 97–104, 10.1242/jcs.1994.Supplement_18.14 [DOI] [PubMed] [Google Scholar]
- 32.Johnson GL and Vaillancourt RR (1994) Sequential protein kinase reactions controlling cell growth and differentiation. Curr. Opin. Cell Biol 6, 230–238, 10.1016/0955-0674(94)90141-4 [DOI] [PubMed] [Google Scholar]
- 33.Marshall MS (1995) Ras target proteins in eukaryotic cells. FASEB J. 9, 1311–1318, 10.1096/fasebj.9.13.7557021 [DOI] [PubMed] [Google Scholar]
- 34.McGill G and Fisher DE (1997) Apoptosis in tumorigenesis and cancer therapy. Front. Biosci 2, d353–d379, 10.2741/A197 [DOI] [PubMed] [Google Scholar]
- 35.Lopez J and Tait SW (2015) Mitochondrial apoptosis: killing cancer using the enemy within. Br. J. Cancer 112, 957–962, 10.1038/bjc.2015.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellacosa A, Testa JR, Staal SP and Tsichlis PN (1991) A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science 254, 274–277, 10.1126/science.1833819 [DOI] [PubMed] [Google Scholar]
- 37.De Luca A, Maiello MR, D’Alessio A, Pergameno M and Normanno N (2012) The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther Targets 16, S17–S27, 10.1517/14728222.2011.639361 [DOI] [PubMed] [Google Scholar]
- 38.Asati V, Mahapatra DK and Bharti SK (2016) PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur. J. Med. Chem 109, 314–341, 10.1016/j.ejmech.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 39.Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA et al. (2011) Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget 2, 135–164, 10.18632/oncotarget.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning G, Whyte DB, Martinez R, Hunter T and Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298, 1912–1934, 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 41.Westbrook JD, Soskind R, Hudson BP and Burley SK (2020) Impact of the Protein Data Bank on antineoplastic approvals. Drug Discov. Today 25, 837–850, 10.1016/j.drudis.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cozza G, Bortolato A, Menta E, Cavalletti E, Spinelli S and Moro S (2009) ATP non-competitive Ser/Thr kinase inhibitors as potential anticancer agents. Anticancer Agents Med. Chem 9, 778–786, 10.2174/187152009789056930 [DOI] [PubMed] [Google Scholar]
- 43.Kimura S, Ashihara E and Maekawa T (2006) New tyrosine kinase inhibitors in the treatment of chronic myeloid leukemia. Curr. Pharm. Biotechnol 7, 371–379, 10.2174/138920106778521532 [DOI] [PubMed] [Google Scholar]
- 44.Zhao Y, Bilal M, Raza A, Khan MI, Mehmood S, Hayat U et al. (2021) Tyrosine kinase inhibitors and their unique therapeutic potentialities to combat cancer. Int. J. Biol. Macromol 168, 22–37, 10.1016/j.ijbiomac.2020.12.009 [DOI] [PubMed] [Google Scholar]
- 45.Roskoski Jr, R. (2021) Properties of FDA-approved small molecule protein kinase inhibitors: a 2021 update. Pharmacol. Res 165, 105463, 10.1016/j.phrs.2021.105463 [DOI] [PubMed] [Google Scholar]
- 46.Scott JD (1991) Cyclic nucleotide-dependent protein kinases. Pharmacol. Ther. 50, 123–145, 10.1016/0163-7258(91)90075-W [DOI] [PubMed] [Google Scholar]
- 47.Francis SH and Corbin JD (1999) Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit. Rev. Clin. Lab. Sci 36, 275–328, 10.1080/10408369991239213 [DOI] [PubMed] [Google Scholar]
- 48.Crivici A and Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct 24, 85–116, 10.1146/annurev.bb.24.060195.000505 [DOI] [PubMed] [Google Scholar]
- 49.Fukami Y, Nagao T, Iwasaki T and Sato K (2002) Inhibition and activation of c-Src: the head and tail of a coin. Pharmacol. Ther 93, 263–270, 10.1016/S0163-7258(02)00195-X [DOI] [PubMed] [Google Scholar]
- 50.Brautigan DL and Shenolikar S (2018) Protein serine/threonine phosphatases: keys to unlocking regulators and substrates. Annu. Rev. Biochem 87, 921–964, 10.1146/annurev-biochem-062917-012332 [DOI] [PubMed] [Google Scholar]
- 51.Rusnak F and Mertz P (2000) Calcineurin: form and function. Physiol. Rev 80, 1483–1521, 10.1152/physrev.2000.80.4.1483 [DOI] [PubMed] [Google Scholar]
- 52.Chinkers M (2001) Protein phosphatase 5 in signal transduction. Trends Endocrinol. Metab 12, 28–32, 10.1016/S1043-2760(00)00335-0 [DOI] [PubMed] [Google Scholar]
- 53.Wilson SE, Mellgren RL and Schlender KK (1982) Isolation of a heat-stable protein activator of phosphorylase phosphatase. FEBS Lett. 146, 331–334, 10.1016/0014-5793(82)80946-0 [DOI] [PubMed] [Google Scholar]
- 54.Wilson SE, Mellgren RL and Schlender KK (1983) Evidence that the heat-stable protein activator of phosphorylase phosphatase is histone H1. Biochem. Biophys. Res. Commun 116, 581–586, 10.1016/0006-291X(83)90563-6 [DOI] [PubMed] [Google Scholar]
- 55.Bollen M, Vandenheede JR, Goris J and Stalmans W (1988) Characterization of glycogen-synthase phosphatase and phosphorylase phosphatase in subcellular liver fractions. Biochim. Biophys. Acta 969, 66–77, 10.1016/0167-4889(88)90089-4 [DOI] [PubMed] [Google Scholar]
- 56.Waelkens E, Goris J and Merlevede W (1987) Purification and properties of polycation-stimulated phosphorylase phosphatases from rabbit skeletal muscle. J. Biol. Chem 262, 1049–1059, 10.1016/S0021-9258(19)75748-2 [DOI] [PubMed] [Google Scholar]
- 57.Schlender KK, Wilson SE and Mellgren RL (1986) Purification and characterization of the polycation-stimulated protein phosphatase catalytic subunit from porcine renal cortex. Biochim. Biophys. Acta 872, 1–10, 10.1016/0167-4838(86)90140-8 [DOI] [PubMed] [Google Scholar]
- 58.Jakes S, Mellgren RL and Schlender KK (1986) Isolation and characterization of an inhibitor-sensitive and a polycation-stimulated protein phosphatase from rat liver nuclei. Biochim. Biophys. Acta 888, 135–142, 10.1016/0167-4889(86)90079-0 [DOI] [PubMed] [Google Scholar]
- 59.Price NE and Mumby MC (2000) Effects of regulatory subunits on the kinetics of protein phosphatase 2A. Biochemistry 39, 11312–11318, 10.1021/bi0008478 [DOI] [PubMed] [Google Scholar]
- 60.Turowski P, Favre B, Campbell KS, Lamb NJ and Hemmings BA (1997) Modulation of the enzymatic properties of protein phosphatase 2A catalytic subunit by the recombinant 65-kDa regulatory subunit PR65alpha. Eur. J. Biochem 248, 200–208, 10.1111/j.1432-1033.1997.t01-1-00200.x [DOI] [PubMed] [Google Scholar]
- 61.Keating L, Touati SA and Wassmann K (2020) A PP2A-B56-centered view on metaphase-to-anaphase transition in mouse oocyte meiosis I. Cells 9, 390, 10.3390/cells9020390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hertz EPT, Kruse T, Davey NE, Lopez-Mendez B, Sigurethsson JO, Montoya G et al. (2016) A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell 63, 686–695, 10.1016/j.molcel.2016.06.024 [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Wang Z, Yu T, Yang H, Virshup DM, Kops GJ et al. (2016) Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein Cell 7, 516–526, 10.1007/s13238-016-0283-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Bajaj R, Bollen M, Peti W and Page R (2016) Expanding the PP2A interactome by defining a B56-specific SLiM. Structure 24, 2174–2181, 10.1016/j.str.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eto M and Brautigan DL (2012) Endogenous inhibitor proteins that connect Ser/Thr kinases and phosphatases in cell signaling. IUBMB Life 64, 732–739, 10.1002/iub.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams BC, Filter JJ, Blake-Hodek KA, Wadzinski BE, Fuda NJ, Shalloway D et al. (2014) Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A-B55 heterotrimers. Elife 3, e01695, 10.7554/eLife.01695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filter JJ, Williams BC, Eto M, Shalloway D and Goldberg ML (2017) Unfair competition governs the interaction of pCPI-17 with myosin phosphatase (PP1-MYPT1). Elife 6, e24665, 10.7554/eLife.24665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kauko O and Westermarck J (2018) Non-genomic mechanisms of protein phosphatase 2A (PP2A) regulation in cancer. Int. J. Biochem. Cell Biol 96, 157–164, 10.1016/j.biocel.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 69.Sangodkar J, Farrington CC, McClinch K, Galsky MD, Kastrinsky DB and Narla G (2016) All roads lead to PP2A: exploiting the therapeutic potential of this phosphatase. FEBS J 283, 1004–1024, 10.1111/febs.13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hait WN and Lazo JS (1986) Calmodulin: a potential target for cancer chemotherapeutic agents. J. Clin. Oncol 4, 994–1012, 10.1200/JCO.1986.4.6.994 [DOI] [PubMed] [Google Scholar]
- 71.Levin RM and Weiss B (1977) Binding of trifluoperazine to the calcium-dependent activator of cyclic nucleotide phosphodiesterase. Mol. Pharmacol 13, 690–697 [PubMed] [Google Scholar]
- 72.Levin RM and Weiss B (1978) Specificity of the binding of trifluoperazine to the calcium-dependent activator of phosphodiesterase and to a series of other calcium-binding proteins. Biochim. Biophys. Acta 540, 197–204, 10.1016/0304-4165(78)90132-0 [DOI] [PubMed] [Google Scholar]
- 73.Hait WN, Grais L, Benz C and Cadman EC (1985) Inhibition of growth of leukemic cells by inhibitors of calmodulin: phenothiazines and melittin. Cancer Chemother. Pharmacol 14, 202–205, 10.1007/BF00258116 [DOI] [PubMed] [Google Scholar]
- 74.Hait WN and Lee GL (1985) Characteristics of the cytotoxic effects of the phenothiazine class of calmodulin antagonists. Biochem. Pharmacol 34, 3973–3978, 10.1016/0006-2952(85)90374-0 [DOI] [PubMed] [Google Scholar]
- 75.Lee GL and Hait WN (1985) Inhibition of growth of C6 astrocytoma cells by inhibitors of calmodulin. Life Sci. 36, 347–354, 10.1016/0024-3205(85)90120-1 [DOI] [PubMed] [Google Scholar]
- 76.Hait WN (1987) Targeting calmodulin for the development of novel cancer chemotherapeutic agents. Anticancer Drug Des. 2, 139–149 [PubMed] [Google Scholar]
- 77.Tzadok S, Beery E, Israeli M, Uziel 0, Lahav M, Fenig E et al. (2010) In vitro novel combinations of psychotropics and anti-cancer modalities in U87 human glioblastoma cells. Int. J. Oncol 37, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 78.Varga B, Csonka A, Csonka A, Molnar J, Amaral L and Spengler G (2017) Possible biological and clinical applications of phenothiazines. Anticancer Res. 37, 5983–5993 [DOI] [PubMed] [Google Scholar]
- 79.Zhelev Z, Ohba H, Bakalova R, Hadjimitova V, Ishikawa M, Shinohara Y et al. (2004) Phenothiazines suppress proliferation and induce apoptosis in cultured leukemic cells without any influence on the viability of normal lymphocytes. Phenothiazines and leukemia. Cancer Chemother. Pharmacol 53, 267–275, 10.1007/s00280-003-0738-1 [DOI] [PubMed] [Google Scholar]
- 80.Choi JH, Yang YR, Lee SK, Kim SH, Kim YH, Cha JY et al. (2008) Potential inhibition of PDK1/Akt signaling by phenothiazines suppresses cancer cell proliferation and survival. Ann. N.Y. Acad. Sci 1138, 393–403, 10.1196/annals.1414.041 [DOI] [PubMed] [Google Scholar]
- 81.Eriksson A, Yachnin J, Lewensohn R and Nilsson A (2001) DNA-dependent protein kinase is inhibited by trifluoperazine. Biochem. Biophys. Res. Commun 283, 726–731, 10.1006/bbrc.2001.4830 [DOI] [PubMed] [Google Scholar]
- 82.Kang S, Dong SM, Kim BR, Park MS, Trink B, Byun HJ et al. (2012) Thioridazine induces apoptosis by targeting the PI3K/Akt/mTOR pathway in cervical and endometrial cancer cells. Apoptosis 17, 989–997, 10.1007/s10495-012-0717-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park MS, Dong SM, Kim BR, Seo SH, Kang S, Lee EJ et al. (2014) Thioridazine inhibits angiogenesis and tumor growth by targeting the VEGFR-2/PI3K/mTOR pathway in ovarian cancer xenografts. Oncotarget 5, 4929–4934, 10.18632/oncotarget.2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ronald S, Awate S, Rath A, Carroll J, Galiano F, Dwyer D et al. (2013) Phenothiazine inhibitors of TLKs affect double-strand break repair and DNA damage response recovery and potentiate tumor killing with radiomimetic therapy. Genes Cancer 4, 39–53, 10.1177/1947601913479020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donmez Y, Akhmetova L, Iseri OD, Kars MD and Gunduz U (2011) Effect of MDR modulators verapamil and promethazine on gene expression levels of MDR1 and MRP1 in doxorubicin-resistant MCF-7 cells. Cancer Chemother. Pharmacol 67, 823–828, 10.1007/s00280-010-1385-y [DOI] [PubMed] [Google Scholar]
- 86.Ford JM, Prozialeck WC and Hait WN (1989) Structural features determining activity of phenothiazines and related drugs for inhibition of cell growth and reversal of multidrug resistance. Mol. Pharmacol 35, 105–115 [PubMed] [Google Scholar]
- 87.Tsakovska I and Pajeva I (2006) Phenothiazines and structurally related compounds as modulators of cancer multidrug resistance. Curr. Drug Targets 7, 1123–1134, 10.2174/138945006778226660 [DOI] [PubMed] [Google Scholar]
- 88.Yde CW, Clausen MP, Bennetzen MV, Lykkesfeldt AE, Mouritsen OG and Guerra B (2009) The antipsychotic drug chlorpromazine enhances the cytotoxic effect of tamoxifen in tamoxifen-sensitive and tamoxifen-resistant human breast cancer cells. Anticancer Drugs 20, 723–735, 10.1097/CAD.0b013e32832ec041 [DOI] [PubMed] [Google Scholar]
- 89.Sangodkar J, Dhawan NS, Melville H, Singh VJ, Yuan E, Rana H et al. (2012) Targeting the FOXO1/KLF6 axis regulates EGFR signaling and treatment response. J. Clin. Invest 122, 2637–2651, 10.1172/JCI62058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim SH, Seo MS, Jeon WJ, Yu HS, Park HG, Jung GA et al. (2008) Haloperidol regulates the phosphorylation level of the MEK-ERK-p90RSK signal pathway via protein phosphatase 2A in the rat frontal cortex. Int. J. Neuropsychopharmacol 11, 509–517, 10.1017/S1461145707008292 [DOI] [PubMed] [Google Scholar]
- 91.Gutierrez A, Pan L, Groen RW, Baleydier F, Kentsis A, Marineau J et al. (2014) Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J. Clin. Invest 124, 644–655, 10.1172/JCI65093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farrington CC, Yuan E, Mazhar S, Izadmehr S, Hurst L, Allen-Petersen BL et al. (2020) Protein phosphatase 2A activation as a therapeutic strategy for managing MYC-driven cancers. J. Biol. Chem 295, 757–770, 10.1016/S0021-9258(17)49933-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McClinch K, Avelar RA, Callejas D, Izadmehr S, Wiredja D, Perl A et al. (2018) Small-molecule activators of protein phosphatase 2A for the treatment of castration-resistant prostate cancer. Cancer Res. 78, 2065–2080, 10.1158/0008-5472.CAN-17-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N et al. (2017) Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J. Clin, Invest 127, 2081–2090, 10.1172/JCI89548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tohme R, Izadmehr S, Gandhe S, Tabaro G, Vallabhaneni S, Thomas A et al. (2019) Direct activation of PP2A for the treatment of tyrosine kinase inhibitor-resistant lung adenocarcinoma. JCI Insight 4, 10.1172/jci.insight.125693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morita K, He S, Nowak RP, Wang J, Zimmerman MW, Fu C et al. (2020) Allosteric activators of protein phosphatase 2A display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell 181, 702.e720–715.e720, 10.1016/j.cell.2020.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Ciccone M, Calin GA and Perrotti D (2015) From the biology of PP2A to the PADs for therapy of hematologic malignancies. Front. Oncol 5, 21, 10.3389/fonc.2015.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kastrinsky DB, Sangodkar J, Zaware N, Izadmehr S, Dhawan NS, Narla G et al. (2015) Reengineered tricyclic anti-cancer agents. Bioorg. Med. Chem 23, 6528–6534, 10.1016/j.bmc.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Allen-Petersen BL, Risom T, Feng Z, Wang Z, Jenny ZP, Thoma MC et al. (2019) Activation of PP2A and inhibition of mTOR synergistically reduce MYC signaling and decrease tumor growth in pancreatic ductal adenocarcinoma. Cancer Res. 79, 209–219, 10.1158/0008-5472.CAN-18-0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kauko O, O’Connor CM, Kulesskiy E, Sangodkar J, Aakula A, Izadmehr S et al. (2018) PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci. Transl. Med 10, eaaq1093, 10.1126/scitranslmed.aaq1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Connor CM, Perl A, Leonard D, Sangodkar J and Narla G (2018) Therapeutic targeting of PP2A. Int. J. Biochem. Cell Biol 96, 182–193, 10.1016/j.biocel.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Perl AL, O’Connor CM, Fa P, Mayca Pozo F, Zhang J, Zhang Y et al. (2019) Protein phosphatase 2A controls ongoing DNA replication by binding to and regulating cell division cycle 45 (CDC45). J. Biol. Chem 294, 17043–17059, 10.1074/jbc.RA119.010432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chu CS, Hsu PH, Lo PW, Scheer E, Tora L, Tsai HJ et al. (2011) Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome. J. Biol. Chem 286, 35843–35851, 10.1074/jbc.M111.228064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jakes S and Schlender KK (1988) Histone H1 phosphorylated by protein kinase C is a selective substrate for the assay of protein phosphatase 2A in the presence of phosphatase 1. Biochim. Biophys. Acta 967, 11–16, 10.1016/0304-4165(88)90182-1 [DOI] [PubMed] [Google Scholar]
- 105.Jakes S, Hastings TG, Reimann EM and Schlender KK (1988) Identification of the phosphoserine residue in histone H1 phosphorylated by protein kinase C. FEBS Lett. 234, 31–34, 10.1016/0014-5793(88)81296-1 [DOI] [PubMed] [Google Scholar]
- 106.Leonard D, Huang W, Izadmehr S, O’Connor CM, Wiredja DD, Wang Z et al. (2020) Selective PP2A enhancement through biased heterotrimer stabilization. Cell 181, 688.e616–701.e616, 10.1016/j.cell.2020.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tolstykh T, Lee J, Vafai S and Stock JB (2000) Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. EMBO J. 19, 5682–5691, 10.1093/emboj/19.21.5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei H, Ashby DG, Moreno CS, Ogris E, Yeong FM, Corbett AH et al. (2001) Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J. Biol. Chem 276, 1570–1577, 10.1074/jbc.M008694200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Longin S, Zwaenepoel K, Louis JV, Dilworth S, Goris J and Janssens V (2007) Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J. Biol. Chem 282, 26971–26980, 10.1074/jbc.M704059200 [DOI] [PubMed] [Google Scholar]
- 110.Vervoort SJ, Welsh SA, Devlin JR, Barbieri E, Knight DA, Offley S et al. (2021) The PP2A-Integrator-CDK9 axis fine tunes transcription and can be targeted therapeutically in cancer. Cell, 10.1016/j.cell.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]