Abstract
The purpose of this review is to explore self-organizing mechanisms that pattern microtubules (MTs) and spatially organize animal cell cytoplasm, inspired by recent experiments in frog egg extract. We start by reviewing conceptual distinctions between self-organizing and templating mechanisms for subcellular organization. We then discuss self-organizing mechanisms that generate radial MT arrays and cell centers in the absence of centrosomes. These include autocatalytic MT nucleation, transport of minus ends, and nucleation from organelles such as melanosomes and Golgi vesicles that are also dynein cargoes. We then discuss mechanisms that partition the cytoplasm in syncytia, in which multiple nuclei share a common cytoplasm, starting with cytokinesis, when all metazoan cells are transiently syncytial. The cytoplasm of frog eggs is partitioned prior to cytokinesis by two self-organizing modules, protein regulator of cytokinesis 1 (PRC1)-kinesin family member 4A (KIF4A) and chromosome passenger complex (CPC)-KIF20A. Similar modules may partition longer-lasting syncytia, such as early Drosophila embryos. We end by discussing shared mechanisms and principles for the MT-based self-organization of cellular units.
Keywords: microtubules, centrosome, syncytium, self-organization, cytoskeleton, nucleation
1. INTRODUCTION
In a paper that surprised many, Cheng & Ferrell (2019) reported that cytoplasm from frog eggs could self-organize into regularly spaced cell-like units reminiscent of syncytial cells (Figure 1a). The reaction depended on microtubules (MTs) but not on nuclei, centrosomes, actin, or plasma membranes. We wrote a short perspective to accompany that report (Mitchison & Field 2019). This article is an extension of that discussion in which we address the principles by which cellular units self-organize and some history of the research into this topic. Our main focus is on MT-based systems. The self-organization of plasma membrane domains and cortical actin cytoskeletons is a fascinating topic, but it is beyond the scope of this review. For an additional perspective on self-organization of embryo cytoplasm, see Shamipour et al. (2021).
Figure 1.
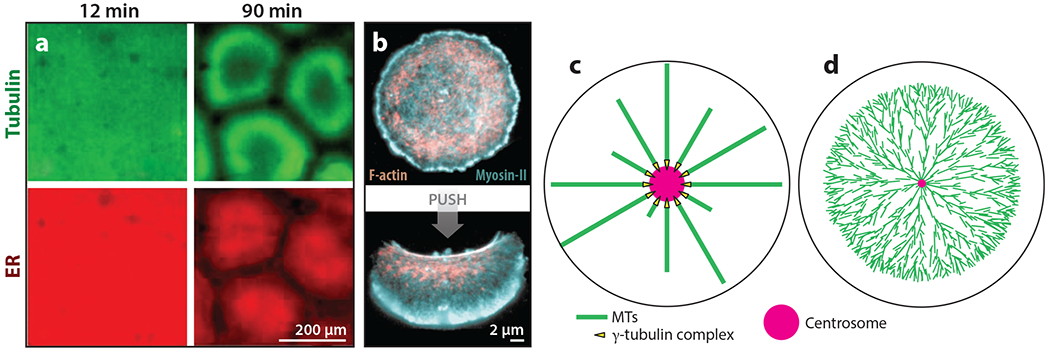
Examples of self-organization and templating in cellular organization. (a) The self-organization of cellular units in interphase frog egg extract. Panel a adapted with permission from Cheng & Ferrell (2019). (b) An example of bistability in actomyosin-based cellular organization. Keratocyte fragments were stable in two organizational states, (top) circular/immobile and (bottom) polarized/migrating. Circular nonmotile fragments were converted into polarized motile fragments by pushing with a pipette. Panel b adapted from Verkhovsky et al. (1999). (c) The templating of MT organization by the centrosome in a small cell. (d) The self-organization of a MT aster in a frog egg, in which the aster grows as a traveling wave by MT-stimulated MT nucleation. The cell radius is ~600 μm and the average MT length is ~;15 μm. Panel d adapted from Ishihara et al. (2016). Abbreviations: ER, endoplasmic reticulum; MT, microtubule.
2. PRINCIPLES OF SELF-ORGANIZATION
2.1. Self-Organization in Biology
Self-organization can be defined as a process in which some form of overall order arises from local interactions between parts of an initially disordered system. This process is related to the concept of self-assembly, in which a disordered system of preexisting components forms an organized structure or pattern as a consequence of specific, local interactions among the components themselves without external direction (Wikipedia 2021). The term self-organization is conventionally applied to dissipative systems, while the term self-assembly is most often used for systems that proceed to thermodynamic equilibrium.
Self-assembly is ubiquitous in protein biochemistry, in which it leads to stable oligomeric assemblies that are sometimes called quaternary structures. Self-organization is much more complicated. Assembly converges on a stable point in a multidimensional organizational landscape that is highly dynamic and can perform mechanical work. Classic examples include Benard convection cells in physics and reaction–diffusion systems in developmental biology [reviewed in Landge et al. (2020)]. Self-organizing systems can break symmetry and organize spontaneously. This requires positive feedback mechanisms to amplify random fluctuations. These systems can also be bistable such that an outside stimulus is required to break symmetry. A very clear example of bistability in cell organization is provided by circular fragments of fish keratocyte lamellipodia, which are stably circular until nudged with a pipette, whereupon they become stably polarized and migrate in a straight line (Figure 1b).
2.2. Alternatives to Self-Organization
To critically discuss self-organization, we need to define biologically relevant alternative organizing principles: candidates include instructed, molded, templated, and nucleated. The concept of “instructed” is defined here as a situation in which a preexisting structure directly determines the size or shape of a larger assembly. For example, the length of bacteriophage tails is instructed by the length of a tape measure protein (Xu et al. 2014). Defined this way, instructed is limited to systems of molecular size. The concept of “molded” is not common in molecular assembly processes, though it is one way to conceptualize the central dogma processes of DNA replication, transcription, and translation in which a polynucleotide molds its complement by base pairing. The concept of “templated” is similar to molded, in that we apply it to situations in which an assembly initiates in a mold and then grows out to become much larger. A good example is prion propagation, which constitutes an epigenetic mechanism in fungi (Benkemoun & Saupe 2006).
The concept of “nucleated” is a subset of templating mechanisms and is highly relevant to MT biology. Nucleation must be carefully parsed, because it can apply in different ways to templated and self-organizing systems. The hypothetical centrosome in Figure 1c templates a fixed number of MTs, and the system is not self-organized. Nucleation can also describe an external stimulus that breaks symmetry and triggers self-organization. The centrosome in Figure 1d nucleates MTs that nucleate more MTs by an autocatalytic branching process, as observed in frog eggs (Ishihara et al. 2016). The resulting aster is largely self-organized. The centrosome determines where it initiates, but the density and direction of MT growth are determined locally. This mechanism can also initiate spontaneously with slower kinetics (Ishihara et al. 2014). The spontaneous initiation of autocatalytic nucleation likely explains the formation of centrosome-free units in frog egg extract (Figure 1a), based on the inspection of recent live imaging of this process (Afanzar et al. 2020). The spontaneous units must possess some radial order based on their ability to transport organelles inward. How this is achieved in the absence of centrosomes is an unsolved problem.
2.3. Self-Organization versus Templating in the Growth of Cylindrical Cells
Self-organization is ubiquitous in biology. DNA encodes biology but does not template it. Preexisting cells are needed as factories to convert DNA sequences and metabolites into the macromolecular subunits and chemical energy needed to support self-organization, but these cells do not directly template the structure of new cells. In cylindrical cells, as exemplified by rod-shaped bacteria, the existing cellular architecture might appear to template new cell growth (Figure 2a). However, experiments in which the cylindrical shape reemerges from experimental perturbation with the correct radius show that it actually self-organizes (Figure 2b,c). The self-organization of cylindrical geometry in bacteria depends on the guidance of cell wall polymerase complexes along the line of maximal curvature by intrinsically curved MreB polymers (Hussain et al. 2018). The radial length scale is determined, at least in Bacillus subtilis, by the ratio between cell wall polymerases that move on MreB-guided versus random paths (Dion et al. 2019).
Figure 2.
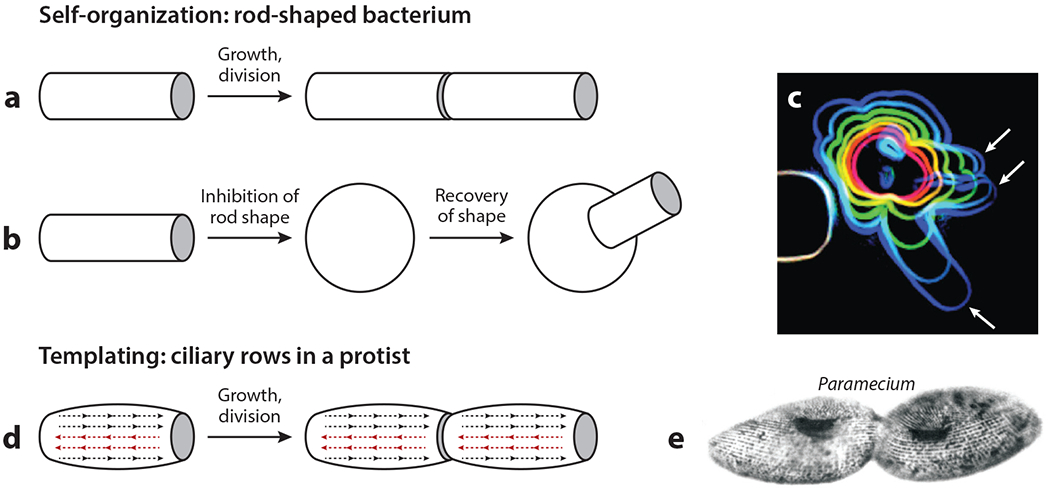
Self-organization and templating in cylindrical cell growth. (a) The growth of a rod-shaped bacterium by the elongation of a constantdiameter cylinder appears templated but is not. (b) The recovery of the cylindrical shape after experimental disruption demonstrates that this process is self-organized. Cylinders of the correct radius emerge from a disorganized starting shape. (c) An image from a shape-recovery experiment in the rod-shaped bacterium Bacillus subtilis. The red to blue outlines show the shape of a single cell at 20-min intervals after initiating recovery. White arrows indicate the emergence of multiple rods with the correct radius. Panel c image adapted with permission from Hussain et al. (2018). (d) The templated growth of ciliary rows in Paramecium, a ciliated protist. The dotted lines with arrowheads represent structurally polarized rows of basal bodies and cilia. Two of the rows (red) were experimentally inverted. The resulting abnormal architecture was inherited for hundreds of generations. Panel d adapted from Beisson & Sonneborn (1965). (e) An image of cell division in Paramecium. Note the continuity of basal body rows prior to cytokinesis. Panel e image adapted from Sonneborn (1964).
A famous example of templating happens during the growth of ciliated protists, which also occurs by extension of an approximately cylindrical shape (Figure 2d,e). Cilia grow from basal bodies that are lined up and interconnected in cortical rows that run parallel to the growth axis. Ciliary rows are structurally polarized, and their polarity controls the direction of ciliary beating. Beisson & Sonneborn (1965) followed Paramecium clones in which a few ciliary rows were mechanically reversed in the mother cell. The reversed ciliary row phenotype was inherited for hundreds of generations in a remarkable example of epigenetics by templating.
Centrioles in metazoan cells are similar to basal body duplication, suggesting that their replication might also be templated. However, we now know that centriole replication is a self-organizing process that depends on the localized activity of polo-like kinase 4 (PLK4) (Arquint & Nigg 2016, Lee et al. 2020). There is evidence for the short-term inheritance of organization in metazoan cells (Solomon 1979), but this is not considered to be a major epigenetic or organizing mechanism.
3. THE SELF-ORGANIZATION OF RADIAL MICROTUBULE ARRAYS AND CELL CENTERS
3.1. A Brief History of Microtubule-Organizing Centers
Cell centers have played a prominent role in cell biology research since its origins. Nineteenth-century cytologists noted the tendency of animal cells to adopt a radial organization around a dot in the center, or two dots in zygotes, between mitosis and cleavage. These centers were first named spheres of attraction by Van Benneden and later centrosomes [reviewed in Sluder (2014)]. The advent of electron microscopy in the 1960s revealed that most centrosomes consist of a centriole pair surrounded by pericentriolar material from which MTs emanate. In the late 1970s, the ability to reconstitute MT growth in vitro led to the discovery that centrosomes nucleate MTs with their faster-growing plus end outward (Brinkley 1985). Nucleating sites in centrosomes, and in spindle pole bodies in fungi, were found to consist of ring-shaped complexes of γ-tubulin (γTb) that template nascent MTs (Oakley et al. 1990, Liu et al. 2020). The discovery that MTs are nucleated by stable sites fostered a conceptual model in which MT-organizing centers (MTOCs) template cell organization (Figure 1c). This model was hugely influential and still dominates textbooks. The real situation is much more complicated, as we discuss in this review. Even when centrosomes are present, self-organizing mechanisms make a major contribution to defining the cell center in many cell types.
3.2. The Self-Organization of Spindle Poles
The twin poles of a mitotic spindle can be thought of as local cell centers where MT minus ends cluster. Spindle poles are defined by centrosomes in most animal cells (Figure 3a), and one of the most important functions of centrosomes is to nucleate these poles. Centrioles and centrosomes are absent in higher plant cells and during egg meiosis in many animals; spindles in these systems are termed anastral (Figure 3b). They still have distinct poles where minus ends cluster, and it has long been known that such poles must self-organize. Even when centrosomes are present, they are structurally distinct from spindle poles, as shown in experiments in which the inhibition of dynein caused centrosomes to dissociate, leaving spindle poles intact (Heald et al. 1997). Heald and colleagues proposed that centrosomes accelerate spindle pole assembly by nucleating MTs, but the spindle pole itself self-organizes in response to signals from chromatin. One of those signals is the GTP-binding nuclear protein Ran (Ran.GTP) (Karsenti & Vernos 2001). Similar conclusions were drawn from experiments in which centrosomes were removed by manipulation of embryos or laser ablation [reviewed in Sluder (2014)]. Centrioles can now be conveniently removed from proliferating mammalian cells by treatment with a PLK4 inhibitor (Wong et al. 2015). This causes a tumor protein p53–dependent cell cycle arrest in normal cells, but cancer cells continue to proliferate and can regenerate centrosomes after the drug is removed by washing the cells. In systems in which centrosomes are normally present, their removal tends to slow spindle assembly and increase segregation errors. However, most aspects of Drosophila development can adapt to the loss of centrosomes, which demonstrates the power of self-organizing mechanisms (Basto et al. 2006).
Figure 3.
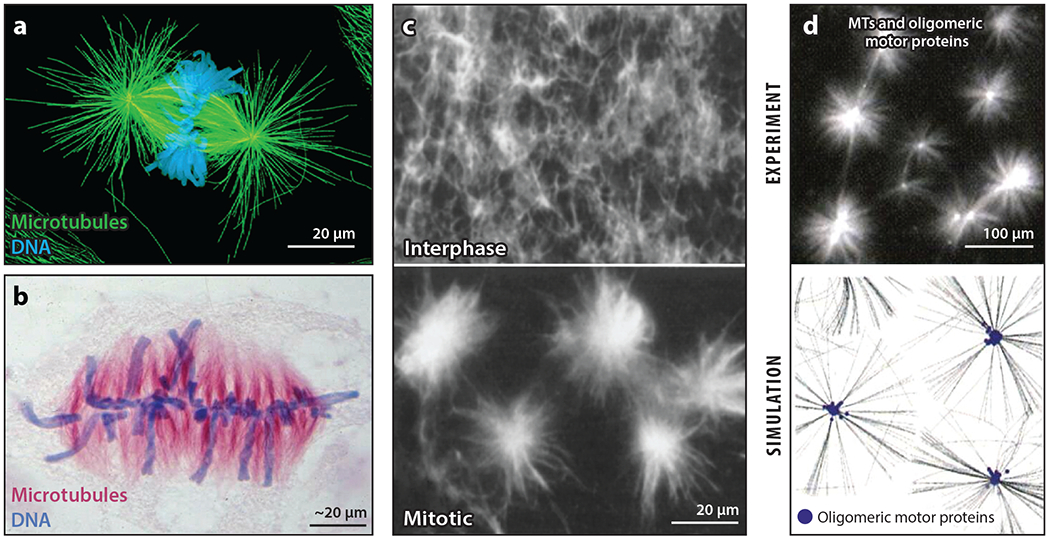
Mitotic spindle pole self-organization. (a) A metaphase spindle with centrosomes at the poles from a newt lung cell. Panel a image adapted from O’Connell & Khodjakov (2007). (b) An anastral metaphase spindle from the plant Hemanthus. Panel b image provided by De Mey et al. (1982). (c) The self-organization of asters in mitotic Xenopus egg extract after the addition of taxol. Panel c images adapted with permission from Verde et al. (1991). (d) The self-organization of asters in reconstituted reactions containing taxol-stabilized microtubules (MTs) and oligomeric motor proteins; the top image shows experimental data, while the bottom image is a simulation. Panel d images adapted with permission from Surrey et al. (2001).
3.3. The Self-Organization of Radial Microtubule Arrays by Motor Proteins
A conceptual breakthrough came from the observation that radial arrays rapidly self-organized when the MT-stabilizing drug taxol was added to mitotic Xenopus egg extract (Figure 3c). This was the first experiment to show that radial arrays could form by a process other than MTOC nucleation. At the time, whether dynein transported minus ends or MT-nucleating factors was unclear. Dynein and dynactin are now thought to transport minus ends bound to the pole-organizing protein called nuclear mitotic apparatus (NUMA) (Hueschen et al. 2017). The γTb complex accumulates at anastral spindle poles, but it is currently thought to get there by the assimilation of small asters and not by direct transport (Letort et al. 2019). Minus end–directed kinesins in the KIFC1 (HSET or NCD) family also cluster minus ends in spindles. Their role in the self-organization of spindle poles was discovered at approximately the same time as dynein’s (Hatsumi & Endow 1992). The relative importance of the two types of motors varies between systems (Borgal & Wakefield 2018). Higher plants lack dynein so their spindle poles are completely dependent on minus end–directed kinesins (Yamada & Goshima 2017). Spindle bipolarity is also a self-organizing process with a central role for tetrameric kinesins [reviewed in Kapoor (2017)].
The concept that the motor-driven transport of MT ends along MTs is sufficient to organize a radial array was proved by groundbreaking reconstitution experiments using stable MTs and artificially oligomeric motor proteins (Figure 3d) (Nédélec et al. 1997). These studies introduced computational simulation approaches that are increasingly used to interrogate self-organizing systems. Recent simulations of early steps in spindle pole self-organization have been extended to include many of the known components (Letort et al. 2019).
Minus end–directed motors can replace the minus end–focusing activity of centrosomes but not their nucleation activity. Recent work has revealed that the majority of MTs in large spindles are nucleated from the side of preexisting MTs (Alfaro-Aco et al. 2020). We review the implications of autocatalytic nucleation for self-organization in Section 5. Quantitative analysis has shown that most spindle MTs are positioned by nucleation, not transport (Decker et al. 2018). In this view, minus end–directed motors shape spindle poles more than they build them.
3.4. The Self-Organization of Organelle-Based Centers in Melanophore Fragments
MTs can self-organize into radial and more complex arrays in interphase cells by pathways that are incompletely understood. An important generalization, and a distinction from mitotic spindle poles, is that MT nucleation from membranes is strongly implicated in all cases. This line of research started with observations in fish melanophores, which are large, multi-armed cells organized by a radial array of MTs emanating from a centrally located centrosome. The MTs provide tracks for the radial transport of pigment-containing organelles called melanosomes that move inward (aggregation) or outward (dispersal) in response to signaling pathways. When McNiven et al. (1984) severed an arm of Holocentrus melanophores, MT minus ends were initially clustered at the cut site, reflecting their precut distribution and stability to cut-induced depolymerization. Over a few hours, MTs reorganized such that the minus ends were clustered at the centroid of the minicell (Figure 4a).
Figure 4.
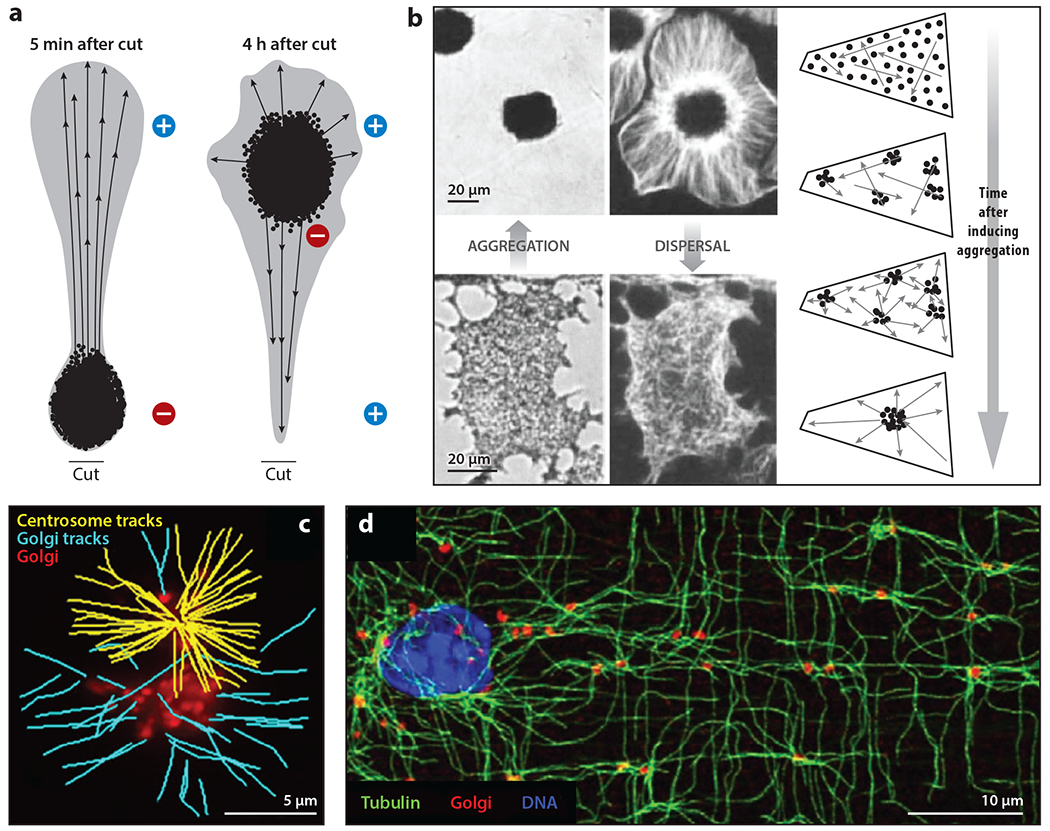
The self-organization of radial microtubules (MTs) and nucleation from organelles. (a) The reorganization of MTs in a cut arm from a Holocentrus melanophore. The black dots represent melanosomes after stimulation to aggregate, and the arrows indicate MT polarity. Panel a adapted from McNiven et al. (1984). (b) Rapid and reversible self-organization of MTs (white) and melanosomes (black) in centrosome-free fragments from Tetra melanophores. With melanosomes dispersed, MTs were randomly organized. Triggering aggregation caused MTs to organize into a radial array while melanosomes aggregated and centered over ~10 min, as illustrated in the time course. Panel b images adapted with permission from (left) Rodionov & Borisy (1997) and (right) Vorobjev et al. (2001). (c) MT nucleation from centrosomes and Golgi vesicles (red) visualized by end-binding protein 3 (EB3) comet tracking. Yellow tracks emanate from centrosomes and blue from Golgi vesicles. Panel c image provided by Efimov et al. (2007). (d) Golgi outposts in mouse skeletal muscle. Note the lattice-like arrangement of MTs centered on Golgi outposts. Panel d image adapted with permission from Oddoux et al. (2013).
Rodionov & Borisy (1997) observed more dramatic dynamics in centrosome-free fragments from Tetra melanophores. With melanosomes dispersed, MTs in the fragments were disorganized. Triggering melanosome aggregation caused the MTs to reorganize into a radial array centered on the aggregated melanosomes over ~10 min (Figure 4b). These experiments provide remarkable examples of the self-organization of a cell center and radial MT array. The full mechanism remains unclear, but two processes play central roles: dynein-mediated transport of and MT nucleation by melanosomes (Malikov et al. 2005). The reorganization of MTs into a radial array emanating from a vesicle aggregate was also observed at the tip of Aplysia neurons after axotomy in a model of recovery from neuronal injury (Kamber et al. 2009).
3.5. Microtubule Nucleation by Golgi Membranes
A breakthrough in MT biology came from observations that approximately half of the MTs in typical tissue culture cells are nucleated from Golgi membranes (Figure 4c). Golgi membranes are also dynein cargoes, so they likely contribute to the self-organization of the cell center by combining transport and nucleation. Golgi membranes tend to cluster in the G1 phase and spread out later in the cell cycle (Frye et al. 2020). These dynamics depended on MTs but not on centrosomes, suggesting that they are self-organizing. Golgi nucleation is sufficient to generate an approximately radial MT array in the absence of centrosomes in some cell types (Gavilan et al. 2018), suggesting that the melanosome mechanism may be generalizable. Golgi membranes nucleate by molecular pathways that differ from those of centrosomes and depend on A-kinase anchor protein 9 (AKAP9 or AKAP450), CLIP-associating proteins (CLASPs), and the minus end–stabilizing factor calmodulin-regulated spectrin-associated protein 2 (CAMSAP2) (Wu & Akhmanova 2017). This mechanism shares components with nucleating sites at the apical plasma membrane in epithelial cells (Wu & Akhmanova 2017), suggesting that a MT-nucleating module can be attached to diverse membranes. MT nucleation by membranes is likely to be important for the self-organization of the internal architecture in many cell types.
3.6. Golgi Outposts in Large Cells
Large, specialized cells often exhibit complex MT organization that supports their morphology and functions. In such cells, centrosomes usually lose nucleation activity and are replaced by alternative, presumably self-organizing, MTOCs (Wu & Akhmanova 2017). Neurons are the most studied example, as reviewed elsewhere (Kapitein & Hoogenraad 2015, Wilkes & Moore 2020). An emerging concept that builds on our membrane nucleation theme is that of Golgi outposts, which function as local MTOCs in muscle myotubes and oligodendrocytes (Valenzuela et al. 2020). The myotube cortex is organized by a gridlike array of dynamic MTs nucleated from the surface of nuclei and from Golgi outposts (Figure 4d) (Becker et al. 2020, Oddoux et al. 2019). The gridlike pattern may reflect the interaction of MTs with sarcomeres (Becker et al. 2020, Oddoux et al. 2019). Nucleation from Golgi outposts in muscle depends on AKAP9 and the γTb complex, consistent with mechanisms identified in tissue culture cells. Oligodendrocytes are responsible for wrapping axons in myelin using large lamellar processes whose MTs are also nucleated by Golgi outposts (Fu et al. 2019). The small, unstructured tubulin polymerization–promoting protein (TPPP) has been implicated as the MT nucleator (Fu et al. 2019), suggesting a more specialized mechanism. However, how Golgi outposts locally cluster and globally distribute is currently unclear. By analogy to melanosomes and Golgi membranes in smaller cells, we expect to find a self-organizing mechanism that depends on the combination of nucleation and dynein-mediated transport.
3.7. The Centering of Organelle-Based Microtubule-Organizing Centers
The centering of centrosomes plays a key role in organizing cells and early embryos and has been extensively reviewed (Ma et al. 2014, Pelletier et al. 2020). Broadly, centrosomes can be pushed to the middle of the cell by MT polymerization or pulled there by dynein, but in either case MTs slide as the centrosome moves. Organelle-based MTOCs appear to center by a fundamentally different mechanism. Photobleaching showed that MTs do not slide when melanosome aggregates center (Rodionov & Borisy 1997). Rather, the aggregate moves by replacing MTs as they depolymerize and are replaced with newly nucleated MTs. Microscopy and modeling supported a mechanism in which nucleating organelles slide on both MTs they nucleate and MTs that are nucleated at random. This generates a self-organizing system that exhibits a stable point with organelles clustered at the center (Cytrynbaum et al. 2006, Malikov et al. 2005). Important parameters in the model include the ratio of spontaneous to organelle-nucleated MTs and the MT turnover rate. This model provides a conceptual framework for the clustering and centering of Golgi membranes, whether these processes occur globally in small cells or locally as in myofibers and oliogodendrocytes.
4. THE SELF-ORGANIZATION OF BOUNDARIES IN SYNCYTIA
4.1. Syncytia and Energids
The self-organized units in frog egg extract that prompted this article (Figure 1a) are defined as much by their boundaries as by their centers. We proposed that these boundaries are a manifestation of a cytoplasmic-partitioning mechanism that occurs normally in frog eggs, and likely in other metazoan cells, prior to cleavage furrow ingression (Mitchison & Field 2019). This hypothesis was strengthened by recent live imaging that showed similar morphology of the boundaries formed between spontaneous asters and asters emanating from the twin poles of mitotic spindles (Afanzar et al. 2020). Here, we discuss the self-organizing biochemical modules that generate boundaries between asters in frog eggs and their possible relevance to partitioning the cytoplasm of other syncytial systems.
The idea that multiple, autonomous cellular units can share a common cytoplasm dates back to the dawn of the cell theory [reviewed in Sitte (1992)]. Sachs, one of the theory’s founders, coined the term energid to describe subcellular units containing a nucleus in syncytial giant-celled algae (Sitte 1992). The energid concept decreased in significance as it became clear that most organisms and tissues are built from cells with a single nucleus, but it remains useful for conceptualizing the organization of syncytial embryos (Mavrakis et al. 2009). Syncytial cell types in humans include muscle myofibers, osteoclasts, megakaryocytes, trophoblast cells, and polyploid giant cancer cells that may contribute to drug resistance and tumor progression (Herbein & Nehme 2020).
Syncytial cells can be conceptually divided into those in which nuclei cluster (osteoclasts, megakaryocytes, trophoblasts, and multinucleated cancer cells) versus those in which nuclei disperse (myofibers, Drosophila embryos, filamentous fungi, and giant-celled algae) (Blangy et al. 2020, Calvert et al. 2016, Mine et al. 2008, Moskovszky et al. 2010, Sullivan & Theurkauf 1995, Xiang 2018). There is no evidence for the partitioning of the cytoplasm in syncytial cells with clustered nuclei. Nuclei presumably cluster because the nuclear envelope is an important dynein cargo [reviewed in Goldberg (2017)]. In the absence of opposing motors or cytoplasmic partitioning, nuclei are drawn toward MT minus ends that cluster in the center of the cell by transport of minus ends (Figure 3) and nucleation from organelles (Figure 4). In syncytia in which nuclei disperse, the next question is if, and how, the cytoplasm is partitioned.
4.2. Partitioning of the Cytoplasm Prior to Cytokinesis
All eukaryotic cells are transiently syncytial during cell division, and their cytoskeleton is partitioned prior to cytokinesis by known mechanisms. A bipolar MT array called the midzone, or central spindle, assembles between the separating chromosomes during anaphase and serves to position the cleavage furrow and keep reforming nuclei separated (Glotzer 2009). The MT cytoskeleton is sharply partitioned at the center of the midzone (Figure 5a), which makes it a paradigm for boundary formation in syncytia.
Figure 5.
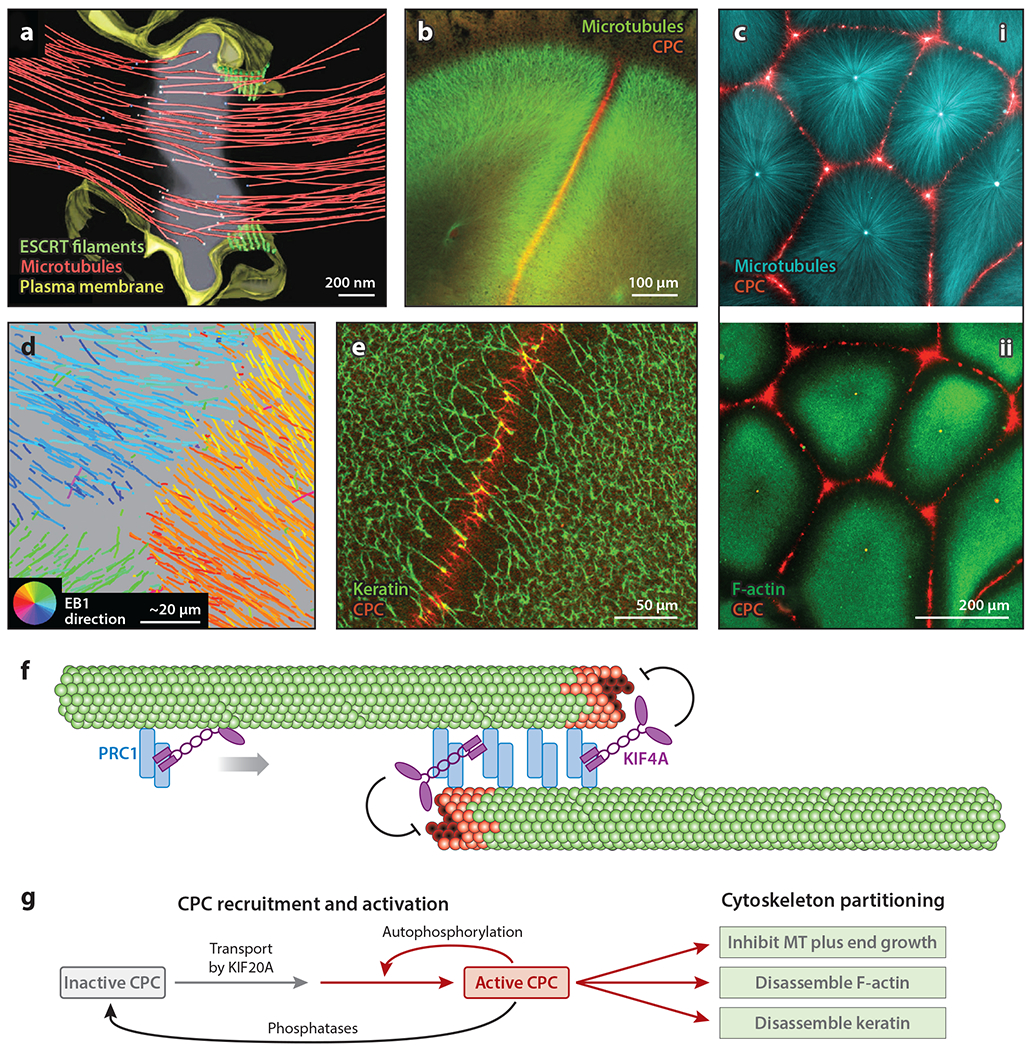
Partitioning of the cytoskeleton by cytokinesis midzone modules. (a) Cryo-electron microscopy tomogram of a HeLa cell midzone. White dots in the center of the image are MT ends, presumably plus ends. Note that MTs are sharply partitioned. This image is taken after cleavage furrow ingression and shortly before ESCRT-mediated abscission. Panel a image adapted with permission from Guizetti et al. (2011). (b) Confocal image of a frog egg fixed after first mitosis and before cleavage furrow ingression. Two large asters grow from the poles of the mitotic spindle (green). The disc-shaped boundary between these structures exhibits lower MT density and CPC-coated MT bundles (red). Panel b image provided by Nguyen et al. (2014). (c, i) Asters nucleated by artificial centrosomes in frog egg extract (cyan). CPC is recruited to aster boundaries (red). (ii) Same specimen as in subpanel i, showing that CPC recruitment to aster boundaries leads to the local disassembly of F-actin (green) and the partitioning of the cytoplasm. Panel c images provided by Field et al. (2019). (d) MT plus end growth tracks at an aster boundary in frog egg extract. The tracks are color coded by the direction of MT growth. The circular key at bottom left indicates growth direction; e.g., growth toward the top left is coded orange/yellow, while growth toward the bottom right is coded blue/cyan. Growing plus ends stop sharply at the boundary, as indicated by the color change. For a discussion of methods, see Nguyen et al. (2014). Panel d image provided by Nguyen et al. (2014). (e) Disassembly of the keratin network at a boundary between asters in frog egg extract. Panel e image provided by Field et al. (2019). (f) Schematic illustration of the PRC1-KIF4A MT-partitioning module that can self-organize from pure proteins. MT plus ends are at the center; green structures represent GDP tubulin, while red structures represent GTP tubulin. The curved inhibitory arrows indicate the slowing of plus end growth by KIF4A. Panel f adapted from Bieling et al. (2010) and Hannabuss et al. (2019). (g) Flowchart showing the process of the CPC-KIF20A module that partitions actin and keratin networks as well as MTs in frog eggs. Panel g provided by Field et al. (2019). Abbreviations: CPC, chromosome passenger complex; EB1, end-binding protein 1; ESCRT, endosomal sorting complexes required for transport; F-actin, filamentous actin; KIF, kinesin family member; MT, microtubule; PRC1, protein regulator of cytokinesis.
In frog eggs, the interval between anaphase onset after first mitosis and the start of cleavage lasts approximately 15 min and coincides with the start of S phase, so the egg is syncytial for almost half of the cell cycle. During this syncytial interval, the egg cytoplasm is partitioned by a boundary between the asters that grow from the poles of the mitotic spindle (Figure 5b). The boundary is ~50 μm wide and has a lower MT density than does the rest of the aster (Mitchison et al. 2012). At its core is a disc of antiparallel MT bundles coated with two signaling complexes: chromosome passenger complex (CPC) and Centralspindlin (Nguyen et al. 2014). As the asters grow, this disc grows, eventually triggering furrow ingression where it touches the cortex. The aster boundary also provides spatial information that directs sister nuclei to move apart in response to dynein pulling forces (Wühr et al. 2010). In addition, boundaries form between asters from different mitotic spindles after polyspermic fertilization. These boundaries also exhibit locally low MT density and keep asters separated, but they lack CPC and Centralspindlin and cannot trigger cleavage furrows (Field et al. 2015).
Frog egg asters and the boundaries between them can be reconstituted in frog egg extract to generate model syncytia. When centrosomes are present, they dictate the number of asters (Figure 5c). Once boundaries are established, centrosomes move from random to more regular spacing in response to forces from dynein and actomyosin (Pelletier et al. 2020). Boundaries between centrosome-nucleated asters in egg extracts recruit the same signaling complexes as in eggs (Figure 5c). Aster boundaries in egg extract sharply partition all particulate components of the cytoplasm. The direction of MT growth changes sharply at aster boundaries (Figure 5d), which implies that local mechanisms block plus end growth. At the same time, the filamentous actin (F-actin) and keratin networks locally disassemble (Figure 5c, subpanel ii, e). Organelles (ER, mitochondria, and lysosomes) are transported away from aster boundaries and toward centers (Pelletier et al. 2020). The net effect is to partition the cytoplasm into discrete islands that conceptually resemble Sachs’ energids.
Partitioning of cytoplasm in somatic midzones and frog egg aster boundaries is accomplished by two self-organizing modules (Figure 5f,g). The antiparallel cross-linker PRC1 and the plus end–directed, polymerization-inhibiting kinesin KIF4A are sufficient to reconstitute minimal midzones that partition MTs into two antiparallel clusters (Figure 5f). PRC1 crosslinks MTs but leaves them free to slide, while KIF4A inhibits the growth of plus ends and compacts PRC1 toward plus ends. In frog egg extract, the PRC1-KIF4A module is required to prevent the MTs of one aster from invading their neighbors at the boundaries (Nguyen et al. 2018). This module also acts within the main body of the aster to prune out antiparallel MT interactions and thus enforce the radial polarity of the aster. In this way, the PRC1-KIF4A module allows the centrosome to dictate the radial polarity of aster MTs in the face of local nucleation, which tends to randomize it. We strongly suspect this module is involved in generating boundaries in the centrosome-free extract system (Figure 1a)
The more complicated CPC-KIF20A module partitions the cytoskeleton at midzones in somatic cells and aster boundaries in frog eggs (Figure 5g). The CPC is a complex of aurora kinase B (AURKB) and three other subunits (Ruchaud et al. 2007). This module is transported to the center of midzones and aster boundaries by KIF20A (MKLP2) (Adriaans et al. 2020, Nguyen et al. 2014). This leads to the autoactivation of CPC on MTs (Sampath et al. 2004) and the generation of a spatial gradient of AURKB phosphorylation centered on the boundary (Fuller et al. 2008). In frog eggs, this gradient partitions the cytoskeleton by inhibiting MT growth and disassembling F-actin (Figure 5c) and keratin (Figure 5e). Whether the CPC is recruited to aster boundaries in the centrosome-free system is not yet known (Figure 1a). Sharp boundaries between asters can form in frog eggs with or without the CPC present (Field et al. 2015). We do not fully understand this difference. The PRC1-KIF4A module is likely sufficient to partition MTs between asters. Addition of the CPC-KIF20A module endows the boundary with the ability to trigger cleavage furrow assembly and partition F-actin. We proposed that the CPC-KIF20A module is loaded onto the aster boundary by chromatin during anaphase and then propagates outward as the asters grow. In this way, the CPC-KIF20A module conveys the location of the mitotic chromatin to the cortex (Mitchison & Field 2017).
4.3. Partitioning of Longer-Lasting Syncytia
The next question is whether analogs of the partitioning modules shown in Figure 5f,g are relevant in cells that remain syncytial, with nuclei distributed, for multiple cell cycles. Drosophila embryos are syncytial during the first 13 cell cycles. Nuclei space out regularly and the cytoskeleton is clearly partitioned into discrete energids (Foe & Alberts 1983,Mavrakis et al. 2009, Sullivan & Theurkauf 1995). Organelles are also confined to energids (Frescas et al. 2006), but soluble proteins can diffuse, which is important for the establishment of morphogen gradients (Fradin 2017). Drosophila embryos are similar to frog eggs in their rapidly dividing nuclei and dense MT arrays that alternate between mitotic and interphase organizations. MTs and F-actin both contribute to positioning Drosophila energids (Telley et al. 2012, von Dassow & Schubiger 1994). During mitosis, MTs are too short to bridge between adjacent spindles in early cell cycles (Baker et al. 1993). After anaphase, MTs grow out from centrosomes to generate large asters that interact (Baker et al. 1993). Whether the resulting antiparallel interactions between MTs lead to recruitment of specific partitioning factors is not known. By analogy to frog eggs, we predict that a module equivalent to PRC1-KIF4A partitions Drosophila energids by blocking MT growth at boundaries. The disassembly of F-actin by AURKB may also generate asymmetries in contractility that contribute to energid partitioning and distribution.
The energid concept was originally proposed to describe organisms that grow as permanent syncytia with spaced-out nuclei (Sitte 1992). It is interesting to ask if and how the cytoplasm is partitioned in such organisms. The mechanisms that position nuclei have been extensively studied in filamentous fungi using powerful genetic approaches combined with microscopy [reviewed in Xiang (2018)]. These systems differ considerably from frog egg extract and Drosophila embryos, in part because their MT density is much lower (Gibeauxet al. 2012). To what extent the cytoplasm of fungal cells is partitioned along with nuclei is a fascinating research question. MTs are partitioned in the sense that each nucleus nucleates its own cytoplasmic MTs, but whether overlapping MTs interact and recruit active partitioning modules akin to PRC1-KIF4A is an interesting topic for future research.
5. PRINCIPLES IN THE MICROTUBULE-DEPENDENT SELF-ORGANIZATION OF CELLULAR UNITS
5.1. Coupling Transport to the Modulation of Microtubule-Polymerization Dynamics
A principle common to many of the systems discussed is the motor-driven transport of MT ends, or proteins that modulate the dynamic properties of MT ends. Minus ends are transported by dynein or minus end–directed kinesins to organize mitotic spindle poles (Figure 3). MT-nucleating organelles, which generate and cap new minus ends, are transported by dynein to organize organelle-based cell centers (Figure 4). Plus end regulators are transported by kinesins to partition MTs at midzone boundaries (Figure 5f,g). Coupling transport to the regulation of polymerization gives rise to complex dynamic systems whose stable points are not obvious on inspection. They require biochemical reconstitution and computational simulation to properly investigate, with the challenge that it sometimes becomes difficult to explain in words the function of individual component or emergent organizational outcomes. Coarse-grained physical models, e.g., those discussed in Cytrynbaum et al. (2006) and Ishihara et al. (2016), are one possible bridge between microscopic details and conceptual understanding.
5.2. Microtubule Nucleation from Membranes and Microtubules
Classic, unitary MTOCs epitomize a templating view of cell centers (Figure 1c). MT nucleation from membranes and from the sides of existing MTs leads to distributed nucleation that is better suited to self-organization. The importance of MT nucleation from the sides of MTs, leading to autocatalysis, is increasingly appreciated. This concept builds on the autocatalytic generation of F-actin by the actin-related proteins (Arp)2/3 complex [reviewed in Pollard & Borisy (2003)]. It fits naturally into the framework of self-organization as a positive feedback mechanism that can amplify spontaneous fluctuations. Autocatalytic nucleation, by a mechanism that is currently unknown, generates large centrosome-triggered asters in frog eggs (Figure 1d) and is probably responsible for spontaneous self-organization in the absence of centrosomes (Figure 1a). One effect of autocatalytic nucleation in asters is to ensure a constant density of MTs at the aster periphery (Ishihara et al. 2016), which is important for size scaling in huge egg cells (Mitchison et al. 2015). The achievement of a steady state in self-organization requires negative as well as positive feedback mechanisms. The most general negative feedback mechanism in assembly reactions is component limitation. Inside frog egg asters, component limitation appears to limit MT density by increasing the depolymerization rate (Ishihara et al. 2021).
Autocatalytic nucleation plays a central role in building large mitotic spindles (Decker et al. 2018), where it serves to scale spindle size to cell size (Rieckhoff et al. 2020). The best characterized molecular pathway in spindles involves the activation of the γTb complex on the sides of MTs by local aggregates of the targeting protein for Xklp2 (TPX2) and augmin (Alfaro-Aco et al. 2020). The MT branching augmin/HAUS (human augmin) complex is broadly expressed in human and mouse tissues (Sánchez-Huertas & Lüders 2015) and plays a central role in the self-organization of cortical MTs in higher plants [reviewed in Lee & Liu (2019) and Tian & Kong (2019)]. Autocatalytic nucleation likely plays a central role in the self-organization of many kinds of MT assembly.
5.3. Spatial Control by Reaction–Diffusion Gradients
Reaction–diffusion gradients play central roles in the self-organization of mitotic spindles, where the gradient is Ran.GTP concentration (Karsenti & Vernos 2001, Kapoor 2017), and of cytokinesis midzones, where the gradient is AURKB substrate phosphorylation (Field et al. 2019, Fuller et al. 2008). Such gradients provide a robust mechanism for the spatial control of multiple components over micron-length scales. An important conceptual question is the extent to which an activity gradient provides detailed positional information, as proposed in the spindle GPS (global-positioning system) model (Kalab & Heald 2008). The binding of importin cargoes to MTs makes spindle length independent of the length scale of the Ran gradient (Oh et al. 2016). This favors a model in which the Ran.GTP gradient sets a permissive radius over which spindle subunits are activated but does not directly instruct spindle geometry. It will be interesting to address similar questions for the AURKB activity gradient at aster boundaries.
5.4. An Emerging Role for Liquid Condensates
Condensates are an increasingly recognized subcellular organizing principle (Banani et al. 2017). They are implicated in the self-organization of the cytoskeleton (Tiwary & Zheng 2019) and in the local control of gene expression in fungal syncytia (Langdon et al. 2018). Condensates were proposed to concentrate unpolymerized tubulin to promote γTb-independent nucleation in Caenorhabditis elegans centrosomes (Woodruff et al. 2017) and enhance nucleation by γTb in TPX2 condensates (King & Petry 2020). They were also proposed to shape anastral spindle poles (So et al. 2019) and a matrix compartment within spindles (Tiwary & Zheng 2019). The CPC has the potential to condense (Trivedi & Stukenberg 2020), which might facilitate the aggregation of MT bundles at aster boundaries (Figure 5). Condensates have interesting biophysical differences compared to alternative organizational states such as gels and monolayer adsorbates. Notably, they possess a surface tension that promotes the rounding, fusion, and beading up of surface layers (Hyman et al. 2014, Mitchison 2020). These effects may help to shape self-organizing systems at nanometer- to micrometer-length scales. The coupling of condensate assembly to dissipative biochemistry results in active liquids with interesting biophysical properties such as the centering of passive particles (Zwicker et al. 2018). Condensate biophysics will likely play a large role in self-organization inside cells.
5.5. Coda
Much remains to be learned about the mechanisms by which cellular units self-organize and partition. Understanding these mechanisms will require the biochemical reconstitution of key modules and computational modeling as well as the imaging and perturbation of intact systems. As our knowledge of molecules and processes grows, it becomes increasingly challenging to draw out simple concepts. Nevertheless, continuing the quest is important, as it goes to the heart of what constitutes a living system.
ACKNOWLEDGMENTS
T.J.M. and C.M.F. are supported by National Institute of General Medical Sciences grant R35GM131753. We thank Amy Gladfelter (at the University of North Carolina at Chapel Hill) for constructive feedback.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Glossary
- MT
microtubule
- MTOC
microtubule-organizing center
- CPC
chromosome passenger complex
LITERATURE CITED
- Adriaans IE, Hooikaas PJ, Aher A, Vromans MJM, van Es RM, et al. 2020. MKLP2 is a motile kinesin that transports the chromosomal passenger complex during anaphase. Curr. Biol 30:2628–37.e9 [DOI] [PubMed] [Google Scholar]
- Afanzar O, Buss GK, Stearns T, Ferrell JE. 2020. The nucleus serves as the pacemaker for the cell cycle. eLife 9:e59989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Aco R, Thawani A, Petry S. 2020. Biochemical reconstitution of branching microtubule nucleation. eLife 9:e49797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. 2016. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans 44:1253–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Theurkauf WE, Schubiger G. 1993. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J. Cell Biol 122:113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18:285–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW 2006. Flies without centrioles. Cell 125:1375–86 [DOI] [PubMed] [Google Scholar]
- Becker R, Leone M, Engel FB. 2020. Microtubule organization in striated muscle cells. Cells 9:1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J, Sonneborn TM. 1965. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. PNAS 53:275–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkemoun L, Saupe SJ. 2006. Prion proteins as genetic material in fungi. Fungal Genet. Biol 43:789–803 [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell 142:420–32 [DOI] [PubMed] [Google Scholar]
- Blangy A, Bompard G, Guerit D, Marie P, Maurin J, et al. 2020. The osteoclast cytoskeleton – current understanding and therapeutic perspectives for osteoporosis. J. Cell Sci 133:jcs244798. [DOI] [PubMed] [Google Scholar]
- Borgal L, Wakefield JG. 2018. Context-dependent spindle pole focusing. Essays Biochem. 62:803–13 [DOI] [PubMed] [Google Scholar]
- Brinkley BR. 1985. Microtubule organizing centers. Annu. Rev. Cell Biol 1:145–72 [DOI] [PubMed] [Google Scholar]
- Calvert SJ, Longtine MS, Cotter S, Jones CJP, Sibley CP, et al. 2016. Studies of the dynamics of nuclear clustering in human syncytiotrophoblast. Reproduction 151:657–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ferrell JE. 2019. Spontaneous emergence of cell-like organization in Xenopus egg extracts. Science 366:631–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytrynbaum EN, Rodionov V, Mogilner A. 2006. Nonlocal mechanism of self-organization and centering of microtubule asters. Bull. Math. Biol 68:1053–72 [DOI] [PubMed] [Google Scholar]
- De Mey J, Lambert AM, Bajer AS, Moeremans M, De Brabander M. 1982. Visualization of microtubules in interphase and mitotic plant cells of Haemanthus endosperm with the immuno-gold staining method. PNAS 79:1898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker F, Oriola D, Dalton B, Brugués J. 2018. Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. eLife 7:e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kapoor M, Sun Y, Wilson S, Ryan J, et al. 2019. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol 4:1294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12:917–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Groen AC, Nguyen PA, Mitchison TJ. 2015. Spindle-to-cortex communication in cleaving, polyspermic Xenopus eggs. Mol. Biol. Cell 26:3628–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Pelletier JF, Mitchison TJ. 2019. Disassembly of actin and keratin networks by Aurora B kinase at the midplane of cleaving Xenopus laevis eggs. Curr. Biol 29:1999–2008.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci 61:31–70 [DOI] [PubMed] [Google Scholar]
- Fradin C 2017. On the importance of protein diffusion in biological systems: the example of the Bicoid morphogen gradient. Biochim. Biophys. Acta Proteins Proteom 1865:1676–86 [DOI] [PubMed] [Google Scholar]
- Frescas D, Mavrakis M, Lorenz H, Delotto R, Lippincott-Schwartz J. 2006. The secretory membrane system in the Drosophila syncytial blastoderm embryo exists as functionally compartmentalized units around individual nuclei. J. Cell Biol 173:219–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye K, Renda F, Fomicheva M, Zhu X, Gong L, et al. 2020. Cell cycle-dependent dynamics of the Golgicentrosome association in motile cells. Cells 9:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M-M, McAlear TS,Nguyen H, Oses-Prieto JA, Valenzuela A, et al. 2019. The Golgi outpost protein TPPP nucleates microtubules and is critical for myelination. Cell 179:132–46.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, et al. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 453:1132–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilan MP, Gandolfo P, Balestra FR, Arias F, Bornens M, Rios RM. 2018. The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep. 19:e45942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux R, Lang C, Politi AZ, Jaspersen SL, Philippsen P, Antony C. 2012. Electron tomography of the microtubule cytoskeleton in multinucleated hyphae of Ashbya gossypii. J. Cell Sci 125:5830–39 [DOI] [PubMed] [Google Scholar]
- Glotzer M 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol 10:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW. 2017. Nuclear pore complex tethers to the cytoskeleton. Semin. Cell Dev. Biol 68:52–58 [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, et al. 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331:1616–20 [DOI] [PubMed] [Google Scholar]
- Hannabuss J, Lera-Ramirez M, Cade NI, Fourniol FJ, Nédélec F, Surrey T. 2019. Self-organization of minimal anaphase spindle midzone bundles. Curr. Biol 29:2120–30.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsumi M, Endow SA. 1992. Mutants of the microtubule motor protein, nonclaret disjunctional, affect spindle structure and chromosome movement in meiosis and mitosis. J. Cell Sci 101(Part 3):547–59 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. 1997. Spindle assemblyin Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol 138:615–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Nehme Z. 2020. Polyploid giant cancer cells, a hallmark of oncoviruses and a new therapeutic challenge. Front. Oncol 10:567116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueschen CL, Kenny SJ, Xu K, Dumont S. 2017. NuMA recruits dynein activity to microtubule minus-ends at mitosis. eLife 6:e29328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, et al. 2018. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 7:e32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30:39–58 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Decker F, Caldas P, Pelletier JF, Loose M, et al. 2021. Spatial variation of microtubule depolymerization in large asters. Mol. Biol. Cell 10.1091/mbc.E20-11-0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Korolev KS, Mitchison TJ. 2016. Physical basis of large microtubule aster growth. eLife 5:e19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Nguyen PA, Groen AC, Field CM, Mitchison TJ. 2014. Microtubule nucleation remote from centrosomes may explain how asters span large cells. PNAS 111:17715–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. 2008. The RanGTP gradient – a GPS for the mitotic spindle. J. Cell. Sci 121:1577–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber D, Erez H, Spira ME. 2009. Local calcium-dependent mechanisms determine whether a cut axonal end assembles a retarded endbulb or competent growth cone. Exp. Neurol 219:112–25 [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Hoogenraad CC. 2015. Building the neuronal microtubule cytoskeleton. Neuron 87:492–506 [DOI] [PubMed] [Google Scholar]
- Kapoor TM. 2017. Metaphase spindle assembly. Biology 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. 2001. The mitotic spindle: a self-made machine. Science 294:543–47 [DOI] [PubMed] [Google Scholar]
- King MR, Petry S. 2020. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun 11:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landge AN, Jordan BM, Diego X, Müller P. 2020. Pattern formation mechanisms of self-organizing reaction-diffusion systems. Dev. Biol 460:2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360:922–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Park J-E, Ahn JI, Wei Z, Zhang L. 2020. A self-assembled cylindrical platform for Plk4-induced centriole biogenesis. Open. Biol 10:200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Liu B. 2019. Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells. New Phytol. 222:1705–18 [DOI] [PubMed] [Google Scholar]
- Letort G, Bennabi I, Dmitrieff S, Nedelec F, Verlhac M-H, Terret M-E. 2019. A computational model of the early stages of acentriolar meiotic spindle assembly. Mol. Biol. Cell 30:863–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zupa E, Neuner A, Böhler A, Loerke J, et al. 2020. Insights into the assembly and activation of the microtubule nucleator γ-TuRC. Nature 578:467–71 [DOI] [PubMed] [Google Scholar]
- Ma R, Laan L, Dogterom M, Pavin N, Julicher F. 2014. General theory for the mechanics of confined microtubule asters. New J. Phys 16:013018 [Google Scholar]
- Malikov V, Cytrynbaum EN, Kashina A, Mogilner A, Rodionov V. 2005. Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat. Cell Biol 7:1213–18 [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Rikhy R, Lippincott-Schwartz J. 2009. Cells within a cell: insights into cellular architecture and polarization from the organization of the early fly embryo. Commun. Integr. Biol 2:313–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven MA, Wang M, Porter KR. 1984. Microtubule polarity and the direction of pigment transport reverse simultaneously in surgically severed melanophore arms. Cell 37:753–65 [DOI] [PubMed] [Google Scholar]
- Mine I, Menzel D, Okuda K. 2008. Morphogenesis in giant-celled algae. Int. Rev. Cell Mol. Biol 266:37–83 [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. 2020. Beyond Langmuir: surface-bound macromolecule condensates. Mol. Biol. Cell 31:2502–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Field CM. 2017. Spindle-to-cortex communication in cleaving frog eggs. Cold Spring Harb. Symp. Quant. Biol 82:165–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Field CM. 2019. Toward synthetic cells. Science 366:569–70 [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Ishihara K, Nguyen P, Wühr M. 2015. Size scaling of microtubule assemblies in early Xenopus embryos. Cold Spring Harb. Perspect. Biol 7:a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Wühr M, Nguyen P, Ishihara K, Groen A, Field CM. 2012. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton 69:738–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovszky L, Dezsö K, Athanasou N, Szendröi M, Kopper L, et al. 2010. Centrosome abnormalities in giant cell tumour of bone: possible association with chromosomal instability. Mod. Pathol 23:359–66 [DOI] [PubMed] [Google Scholar]
- Nédélec FJ, Surrey T, Maggs AC, Leibler S. 1997. Self-organization of microtubules and motors. Nature 389:305–8 [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Field CM, Mitchison TJ. 2018. Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol. Biol. Cell 29:304–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Groen AC, Loose M, Ishihara K, Wühr M, et al. 2014. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science 346:244–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. 1990. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 61:1289–301 [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Khodjakov AL. 2007. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci 120:1717–22 [DOI] [PubMed] [Google Scholar]
- Oddoux S, Randazzo D, Kenea A, Alonso B, Zaal KJM, Ralston E. 2019. Misplaced Golgi elements produce randomly oriented microtubules and aberrant cortical arrays of microtubules in dystrophic skeletal muscle fibers. Front. Cell Dev. Biol 7:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddoux S, Zaal KJ, Tate V, Kenea A, Nandkeolyar SA, et al. 2013. Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J. Cell Biol 203:205–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Yu C-H, Needleman DJ. 2016. Spatial organization of the Ran pathway by microtubules in mitosis. PNAS 113:8729–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JF, Field CM, Fuerthauer S, Sonnett M, Mitchison TJ. 2020. Co-movement of astral microtubules, organelles and F-actin by dynein and actomyosin forces in frog egg cytoplasm. eLife 9:e60047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–65 [DOI] [PubMed] [Google Scholar]
- Rieckhoff EM, Berndt F, Elsner M, Golfier S, Decker F, et al. 2020. Spindle scaling is governed by cell boundary regulation of microtubule nucleation. Curr. Biol 30:4973–83.e10 [DOI] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG. 1997. Self-centring activity of cytoplasm. Nature 386:170–73 [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol 8:798–812 [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118:187–202 [DOI] [PubMed] [Google Scholar]
- Sánchez-Huertas C, Lüders J. 2015. The augmin connection in the geometry of microtubule networks. Curr. Biol 25:R294—99 [DOI] [PubMed] [Google Scholar]
- Shamipour S, Caballero-Mancebo S, Heisenberg C-P. 2021. Cytoplasm’s got moves. Dev. Cell 56:213–26 [DOI] [PubMed] [Google Scholar]
- Sitte P 1992. A modern concept of the “cell theory”: a perspective on competing hypotheses of structure. Int. J. Plant Sci 153. 10.1086/297059 [DOI] [Google Scholar]
- Sluder G 2014. One to only two: a short history of the centrosome and its duplication. Philos. Trans. R. Soc. B 369. 10.1098/rstb.2013.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C, Seres KB, Steyer AM, Mönnich E, Clift D, et al. 2019. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science 364:eaat9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon F 1979. Detailed neurite morphologies of sister neuroblastoma cells are related. Cell 16:165–69 [DOI] [PubMed] [Google Scholar]
- Sonneborn TM. 1964. The determinants and evolution of life. The differentiation of cells. PNAS 51:915–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Theurkauf WE. 1995. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol 7:18–22 [DOI] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. 2001. Physical properties determining self-organization of motors and microtubules. Science 292:1167–71 [DOI] [PubMed] [Google Scholar]
- Telley IA, Gáspár I, Ephrussi A, Surrey T. 2012. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol 197:887–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Kong Z. 2019. The role of the augmin complex in establishing microtubule arrays. J. Exp. Bot 70:3035–41 [DOI] [PubMed] [Google Scholar]
- Tiwary AK, Zheng Y. 2019. Protein phase separation in mitosis. Curr. Opin. Cell Biol 60:92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, Stukenberg PT. 2020. A condensed view of the chromosome passenger complex. Trends Cell Biol. 30:676–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela A, Meservey L, Nguyen H, Fu M-M. 2020. Golgi outposts nucleate microtubules in cells with specialized shapes. Trends Cell Biol. 30:792–804 [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez JM, Antony C, Karsenti E. 1991. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. J. Cell Biol 112:1177–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. 1999. Self-polarization and directional motility of cytoplasm. Curr. Biol 9:11–20 [DOI] [PubMed] [Google Scholar]
- von Dassow G, Schubiger G. 1994. How an actin network might cause fountain streaming and nuclear migration in the syncytial Drosophila embryo. J. Cell Biol 127:1637–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I, Malikov V, Rodionov V.2001. Self-organization of a radial microtubule array by dynein-dependent nucleation of microtubules. PNAS 98:10160–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikipedia. 2021. Self-organization. Wikipedia. https://en.wikipedia.org/wiki/Self-organization [Google Scholar]
- Wilkes OR, Moore AW. 2020. Distinct microtubule organizing center mechanisms combine to generate neuron polarity and arbor complexity. Front. Cell Neurosci 14:594199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, et al. 2015. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348:1155–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. 2017. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169:1066–77.e10 [DOI] [PubMed] [Google Scholar]
- Wu J, Akhmanova A. 2017. Microtubule-organizing centers. Annu. Rev. Cell Dev. Biol 33:51–75 [DOI] [PubMed] [Google Scholar]
- Wühr M, Tan ES, Parker SK, Detrich HW 3rd, Mitchison TJ. 2010.A model for cleavage plane determination in early amphibian and fish embryos. Curr. Biol. 20:2040–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X 2018. Nuclear movement in fungi. Semin. Cell Dev. Biol 82:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. 2014. Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J. Mol. Biol 426:1004–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Goshima G. 2017. Mitotic spindle assembly in land plants: molecules and mechanisms. Biology 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker D, Baumgart J, Redemann S, Müller-Reichert T, Hyman AA, Jülicher F. 2018. Positioning of particles in active droplets. Phys. Rev. Lett 121:158102. [DOI] [PubMed] [Google Scholar]


