Abstract
Immunotherapies have been heavily explored in the last decade, ranging from new treatments for cancer to allergic diseases. These therapies target the immune system, a complex organ system consisting of tissues with intricate structures and cells with a multitude of functions. To better understand immune functions and develop better therapeutics, many cellular and 2-dimensional (2D) tissue models have been developed. However, research has demonstrated that the 3-dimensional (3D) tissue structure can significantly affect cellular functions, and this is not recapitulated by more traditional 2D models. Microfluidics has been used to design 3D tissue models that allow for intricate arrangements of cells and extracellular spaces, thus allowing for more physiologically relevant in vitro model systems. Here, we summarize the multitude of microfluidic devices designed to study the immune system with the ultimate goal to improve existing and design new immunotherapies. We have included models of the different immune organs, including bone marrow and lymph node (LN), models of immunity in diseases such as cancer and inflammatory bowel disease, and therapeutic models to test or engineer new immune-modulatory treatments. We particularly emphasize research on how microfluidic devices are used to better understand different physiological states and how interactions within the immune microenvironment can influence the efficacy of immunotherapies.
Keywords: Biological barriers, Devices, Disease, Model systems, In vitro
Introduction
Emerging strategies in the field of microfluidics are helping to transform the development of new immunotherapies and tailor them to address specific disease conditions. Much of the research efforts focus on advancing therapeutic solutions for cancer treatment using, e.g., immune checkpoint inhibition (ICI), monoclonal antibodies, adoptive cell transfer (ACT), and chimeric antigen receptor (CAR)-T cell therapy. These treatments take advantage of the immune system’s specificity to target diseased tissues or cellular responses. Unfortunately, inconsistent patient responses to immunotherapies make it difficult to choose the best options or predict the likelihood of treatment success. Microfluidic assays can help to assess the efficacy of immunotherapies in vitro, particularly with their potential use in individualized medicine, to predict patient-specific responses to treatment. The strategies highlighted in this review represent the latest in microfluidics used for modeling immunity and immunotherapy. We first provide an overview of the immune system and the different barriers to immunotherapy. We will then give an overview of microfluidic devices, starting with general manufacturing and finally focus on platforms designed to study and recreate the complex nature of the immune system, including immune involvement in gastrointestinal diseases and cancers, devices to study immunotherapeutic interventions, and microfluidics for single-cell analysis.
Overview of the immune system
The immune system has many layers, including physical barriers to infection and cellular and acellular immunity that comprise innate and adaptive immunity. Two key physical barriers are the skin and mucosal surfaces, which effectively trap and prevent pathogens and foreign particulates from entering the body [1]. In addition to providing a cellular barrier, mucosal surfaces are covered in a sticky mucus mesh that filters and traps pathogens and particulates, which are eventually excreted along with the mucus. The skin is coated in a layer of dead cells, which provide a similarly tight barrier to infection and prevent injury. However, some pathogens have found ways around these barriers, through a myriad of mechanisms such as bacterial flagella that effectively allow bacteria to “swim” through mucus and thus reach the underlying epithelium. Pathogens that can get past the physical barriers and infect cells are then combated via acellular and cellular immune responses.
Innate and adaptive immunity are initiated to rid the body of any harmful particles by tagging pathogens for destruction, engulfing them to be destroyed, and secreting signals to recruit other immune cells. Innate immune cells rapidly respond to a broad range of pathogen-associated molecular patterns (PAMPs), which activate innate immune cells to clear an infection [1]. Innate immune cells can engulf pathogens to degrade them, a process called phagocytosis, or release molecules that can lead to the destruction of pathogens [2]. While some of the innate cells actively fight pathogens, a subset of cells known as antigen-presenting cells will phagocytose and process pathogens into small peptide fragments to present to adaptive immune cells. Adaptive immune cells, primarily B and T cells, are educated in the LNs and spleen and are highly specific [1, 3]. The humoral, B cell-mediated response, leads to antibody production by B cells that bind to antigens and toxins on or secreted by pathogens [3]. Antigen binding can coat pathogens, which neutralizes viruses and toxins, and activates other immune cell–mediated destruction of the pathogens [3]. T cells are activated by antigen-presenting cells, provided the correct antigen fragment is presented to them, and then form a cellular or cytokine-based response that directly kills infected cells or signals for other immune cells to infiltrate the area [3]. The adaptive immune response is formed in 4–7 days, making it a slower response compared to innate immunity that can be activated within minutes. However, adaptive immunity yields memory immune cells, which are rapidly activated upon a second encounter with a pathogen and quickly eliminate the infection.
Immune organs
The repertoire of leukocytes (white blood cells) is generated in the bone marrow (BM). These cells further differentiate and either travel throughout the bloodstream to tissues (innate cells) or undergo differentiation and education via other primary and secondary immune organs including the spleen, thymus, and LNs (adaptive cells). Peripheral tissues can also contain more specialized secondary lymphoid organs, which include the mucosal associated lymphoid tissues (MALT) such as the Peyer’s patches (PPs), adenoids, and tonsils [4, 5].
Bone marrow and thymus
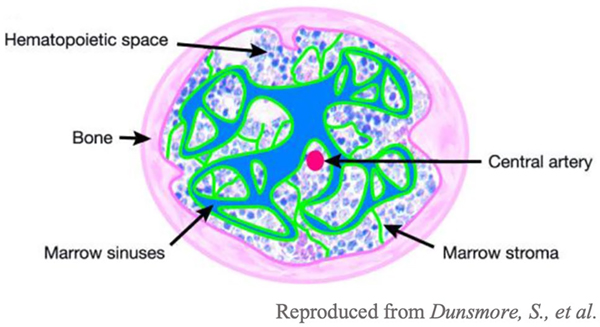
Hematopoietic stem cells (HSCs) are housed within the BM and can self-renew or differentiate into mature leukocytes, largely depending on the environmental cues they receive from the surrounding stromal cells [6, 7]. B and T cells are two key adaptive immune cells that are generated in the BM. The various components of BM within the bone can be seen in the image (Fig. 1). The primary role of B cells is the secretion of antibodies, and immunoglobulin (Ig) receptors play a key role in antibody function [6]. Ig receptors are made up of heavy and light chains that make up two regions: one that can be recognized by other immune cells and another that specifically binds to the pathogenic antigens [6].
Fig. 1.
Structure of bone [8]. All immune cells originate from the bone marrow, the spongy area within the bone
DNA segments, specifically V–D–J segments, can be recombined in numerous ways to generate the wide breadth of specificities required to maintain the antigen repertoire captured by B cells [6]. B cell maturation starts with recombination of D–J segments and is completed during the pre-B-cell stage. Here, B cells express an early form of the B cell receptor with the fully rearranged heavy chain and surrogate light chains [6]. At this point, V–J segments of the light chain will be rearranged and B cells will go through a proliferative stage, which will cause them to lose the pre-B-cell receptor and instead express a fully functional B cell receptor [6]. Any self-reactive B cells will be eliminated and any remaining B cells will travel to the spleen for further development [6]. In the spleen, B cells continue to develop and eventually will become mature B cells that express the IgD receptor on their surface [6]. At this point, B cells can recognize antigens and undergo clonal expansion, the process in which many copies of the same B cell are made to recognize antigens and produce antibodies [6].
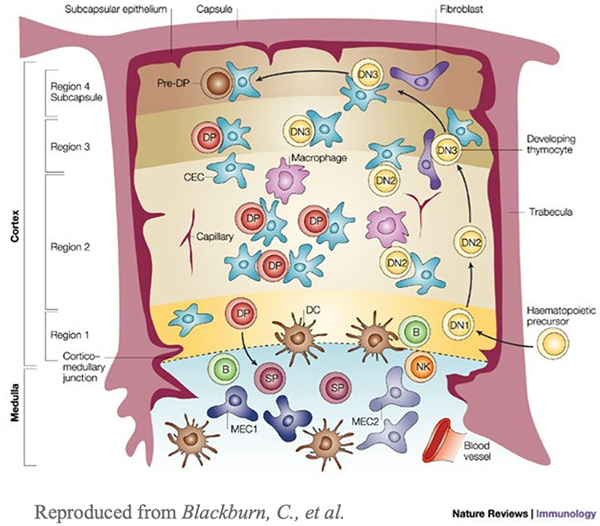
T cells are generated in the BM and travel to the thymus where they continue differentiation [6]. They travel from the outer cortex, into the thymic medulla, and exit from the corticomedullary junction [6]. These specific regions of the thymus can be seen in the cross-sectional image (Fig. 2). T cells can be categorized into CD4+ helper T cells and CD8+ cytotoxic T cells [6]. CD4+ T cells help activate B cells and CD8+ T cells directly eliminate infected cells and also secrete cytokines and chemokines at the sites of infection that help promote immune cell recruitment and activation. Early T cells lack both of these cell markers and are referred to as CD4− CD8− double-negative cells [6]. Similar to B cells, the genes of the T cell receptor (TCR) are first rearranged to get a large range of sequences that can respond to a variety of potentially antigenic peptide sequences [6]. Early T cells express TCRαβ or TCR γδ on the surface, with TCR αβ being more common [6]. Early T cells proliferate rapidly, and the β- chain genes rearrange until the T cell develops into a CD4+ CD8+ double positive cell [6]. At this point, the T cell will stop proliferation, and the α-chain is rearranged [6]. These T cells undergo both positive and negative selection before they are considered mature [6]. Positive selection occurs in the cortex of the thymus and it is the process of selecting cells that can bind various presented peptide sequences [6]. This process determines whether a T cell will be a helper or cytotoxic cell. Negative selection occurs in the medulla of the thymus and is the process of selecting against cells that have a high affinity for self-peptides, resulting in tolerance to the body’s cells [6]. T cells that have undergone both processes are considered mature and exit the thymus [6].
Fig. 2.
Structure of the thymus. The thymus is roughly segregated into two regions: cortex and medulla. These regions can be divided into sub-regions where specific cells can be found [9]
Lymph node
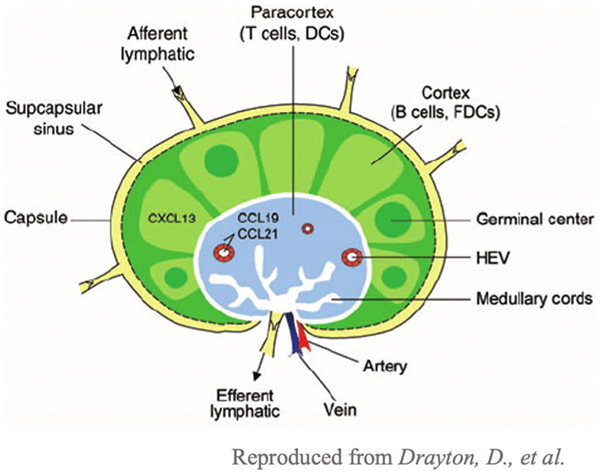
LNs scattered throughout the body are crucial to shaping adaptive immunity. The LN is a highly specialized organ with specific compartments containing immune cells. The capsule is the outer connective tissue of the LN that surrounds the inner cortex and paracortex portions of the LN [10]. Between the capsule and the cortex is the subcapsular sinus which is responsible for the transport of lymph fluid. Lymph fluid contains antigens from the periphery that travel via lymphatic vessels to the LN until they hit the subcapsular sinus [11]. The subcapsular sinus is formed by endothelial cells that form a basement membrane mesh, and size exclusion prohibits molecules larger than 70 kDa from entering [11]. Fluid and molecules then enter via the afferent lymphatic vessels and continue via the sinuses, including the trabecular and medullary sinuses [10]. Text describing the specific compartments within the LN is found in the image (Fig. 3). Antigen-presenting cells (APCs) and inflammatory mediators travel within the LN using the conduit system [11, 12]. In the LN, APCs migrate to the cortex and paracortex to educate B and T cells [11]. The compartment immediately adjacent to the subcapsular sinus is the cortex, which mainly consists of B cells assembled into follicles [10]. Immature B cells form germinal centers during inflammation, and the presence of antigens leads to B cell maturation and production of high affinity antibodies [10]. Next is the paracortex, which is rich in T cells that are here educated by APCs, primarily dendritic cells (DCs) [10, 11]. DCs present antigens in the form of peptide fragments to CD4 + or CD8 + T cells, which get activated if their T cell receptor recognizes the peptide and co-stimulatory molecules are present, yielding helper or cytotoxic T cells [6]. Helper T cells are recruited to the B cell zone to help activate B cells and secrete the antibody type appropriate to the specific immune response [6]. Activated B and T cells leave the LN and migrate to affected tissues by exiting through the efferent lymphatics.
Fig. 3.
Structure of a LN [13]. The outer capsule encloses the LN. The LN consists of two regions: the cortex (light green), the B cell zone, and the paracortex (light blue), the T cell zone
Because entrance into the LN is highly size-dependent, it is critical to study how therapeutics travel within a target environment to accurately gauge how the therapeutic will interact with the multiple cell types in the LN. Some therapeutics that are preferentially taken up into lymphatics may not be successfully delivered into the LN because of the different size restrictions these two environments have [14]. The transport and diffusion of therapeutics that are predicted and observed in vitro may be transported vastly differently in vivo, introducing the need for in vitro models that are physiologically relevant [14].
Spleen
Another key lymphoid organ is the spleen. While the spleen plays an integral role in the immune system, it functions quite differently from the LN [15]. The spleen has two specialized regions, white and red pulp. The white pulp takes in foreign antigens to be presented to B and T cells, while the red pulp removes dead, damaged, or opsonized cells from circulation while also scanning for pathogens and tissue damage [15]. Because of this dual function, models separating the two regions are integral to predict responses similar to those observed in vivo. The two compartments work together to successfully filter blood and the spleen cannot function correctly without one of the compartments. Microfluidic devices that can house these different compartments, including the numerous different cell types involved in splenic functions, could lead to new therapeutic targets and improve our ability to screen and develop such immunotherapies.
Mucosal associated lymphoid tissue
Mucosal immunity is key in surveying potentially hazardous pathogens and particles that may enter the body. The mucosal associated lymphoid tissue (MALT) is a specialized, localized secondary organ that is present throughout the mucosal surfaces of the body including the lungs and gastrointestinal (GI) tract [6]. Their function is similar to that of the LNs, in that T and B cells are educated and activated in these MALTs. Lymphoid tissues that are associated with a specific mucosal surface are called according to their region of association, e.g., the bronchial or GI associated lymphoid tissues (BALT and GALT, respectively) [6]. The GALT has both loose disorganized clusters of lymphoid cells in the lamina propria and well-organized structures like the tonsils, adenoids, appendix, and Peyer’s patches [5, 6]. The lamina propria lies just beneath the epithelial layer and contains B cells, T cells, DCs, and macrophages [6]. As part of the mucosal epithelium, the MALT is usually coated in M cells, a type of epithelial cells that transport foreign antigen from the lumen and into the MALT [6, 16]. Once foreign materials are transported across the epithelium, APCs can activate B and T cells, leading them to differentiate into effector cells [6].
Microbe entry into tissues
Microbes have established ways that help them enter the body and evade recognition from the immune system. Many microbes take advantage of the vulnerability of diseased or damaged tissue to make their way into the body. They may also have cloaking mechanisms to avoid immune responses through cell wall modifications [17]. The cell wall of microbes is their first line of defense against the body’s immune responses. By modulating properties of the cell wall they can deter immune cells from targeting and killing the microbe[17]. Escherichia coli (E. coli) and other gram-negative organisms can change a component of their cell wall changing the charge from negative to positive [17], which allows them to repel other positively charged immune components that could lead to elimination [17]. Other microbes like Neisseria meningitidis (N. meningitidsi) can create a polysaccharide capsule to cloak themselves from encountered immune cells [17]. Microbes can also produce effector proteins that directly target immune cells and inhibit their function [17]. Staphylococcus aureus (S. aureus) for example secretes staphylococcal protein A (spA) that binds to antibodies and B cell receptors, inhibiting their functions [17]. Mechanisms of evasion are one of the challenges to developing effective immunotherapies.
Challenges to effective design and delivery of immunotherapy
Structural barriers
As mentioned, many lymphoid organs are highly organized and typically house cells within compartments in the organ. The highly organized nature of lymphoid organs ensures the correct immune responses are shaped and initiated. Many models that are used to study immunotherapies do not take into account the structure of their target organs, which may lead to variable responses in vivo. A simple culture of immune cells with therapeutics only shows how cells react with a single cell when they are nearby. However, the therapeutic being studied may not even reach the area that this specific cell type is located and thus lead to less effectiveness in vivo. Additional environmental changes including the presence of mechanical forces such as increased flow during inflammation also influence how therapeutics move within a tissue, a component that is difficult to mimic in most standard 2D in vitro models [18]. Microfluidic devices can be designed to hold multiple compartments with connecting channels, providing an easy way to model both cellular interactions and barriers due to tissue structures. Such models are crucial for studying and developing more effective immunotherapies.
Cellular barriers and therapeutic targets
In lymphoid organs and other tissues, there are multiple cell types and components that are in close contact with each other. This includes immune cells and components of the extracellular matrix (ECM), cytokines, and stromal or diseased cells in the immune microenvironment. Because these different components are so close together, it makes it difficult to determine the effects of immunotherapies on specific components. Additionally, current models often that include co-culture of two cell types that are in close contact may lead to synergistic or antagonistic responses in neighboring cells that may not be physiologically relevant. Therefore, models that can support multiple cell types and acellular components within one enclosed system are critical in the development and evaluation of immunotherapies. Microfluidic devices are a promising platform as they easily incorporate supporting structures like the ECM and different cell types in appropriate tissue-like arrangements, while also allowing the introduction of mechanical forces and immunological molecules within the microenvironment such as antigens or cytokines.
Screening
To make effective immunotherapies, it is crucial to use physiologically relevant systems to study specifically induced cellular immune responses. There are numerous different immune cells (e.g., neutrophils, macrophages, monocytes, eosinophils, basophils, natural killer cells, B cells, T cells, and DCs) that can be broken down into even more subsets, each having different functionalities [19, 20]. It becomes difficult to screen for drugs or new delivery platforms in the presence of all cell types and assess the cell type that is most specifically and effectively responding to the immunotherapy. Microfluidic devices can aid the screening processes through reproducible and easy single-cell analysis. Additionally, microfluidics enables the delivery of immunotherapeutics over the course of time and cellular responses can be monitored using, for example, microscopy and timed sample analysis. Such microfluidic systems provide a more streamlined analysis of specific responses, opening the potential for individualized therapies.
Microfluidic devices
Microfluidic devices are extremely versatile and can be designed for many purposes and applications such as studying single cells in response to various stimuli, creating organs on a chip, or screening drugs in an enclosed environment. As the name suggests, microfluidic devices are typically on the millimeter scale to hold volumes of microliters or less. Particular attention to the aspect ratio must be taken to ensure the proper function of the device, specifically fluid movement. Fluids are usually transported throughout the device using capillary action often in combination with external machines including pumps, valves, and mixers that are connected to input and output ports. Devices can also be comprised of multiple layers to control channel directions, mixing, and location of different biological or cellular compartments.
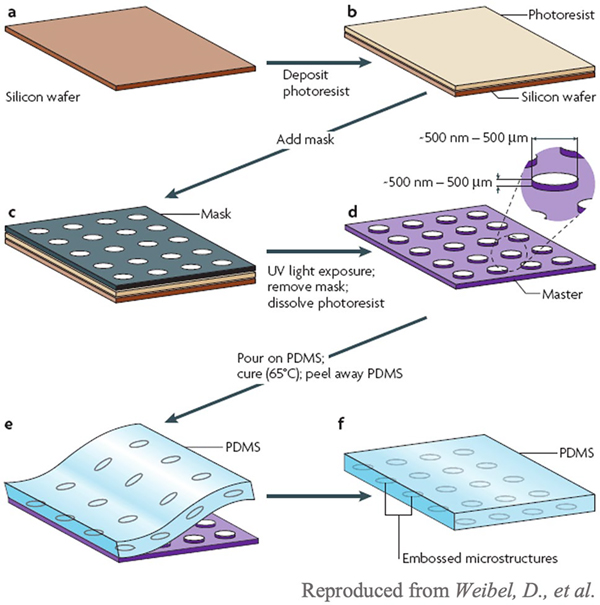
The manufacturing of microfluidic devices typically involves soft lithography (Fig. 4), where a positive mold out is created out of a silicon wafer (dark brown), a photoresist (light brown), and a mask (gray). The mask covers sections of the photoresist that will form the positive mold. Once the mold is created, a material such as poly(dimethyl sulfoxide) (PDMS) is poured onto it and cured. The final PDMS microfluidic device can then be removed from the mask. Other fabrication methods to create a positive mold include electron beam lithography, injection molding, and lamination. For electron beam lithography, a beam of electrons is focused on specific areas of the electron-sensitive film, which changes the solubility of the resist [21]. The film with the etched design is placed in a solvent within which the electron-exposed portions of the film dissolve, resulting in a positive mold [21]. For injection molding, a melted thermoplastic material is injected into a closed negative mold [22]. Lamination involves laser cutting the design on thin layers of laminate and then stacking each layer on top of each other to create a positive mold of the final device [23]. Devices can be made from a variety of materials including inorganic (silicon, glass, ceramic), polymeric (elastomers and thermoplastic polymers), and paper (cellulose-based) materials. These materials must be bio-inert to prevent interference with the molecules, cells, and fluids contained in the device.
Fig. 4.
Process of soft lithography [24]. A positive mold is first created out of a silicon wafer, a photoresist, and a mask. Once the mold is created, liquid material is poured onto the mold and cured to create the full microfluidic device
Organ-on-a-chip
Gut-on-a-chip
Mucosal surfaces play a large role in immunity, and MALTs can create a local immune response within the affected tissue. The gut is the largest of the mucosal surfaces, and an array of gut-on-a-chip (GOC) devices have been developed. Many of these models aim to recapitulate the cellular and ECM arrangement of the gut, including a mucus layer, epithelial layer, and ECM. In a study by Pocock et al., GOC technology was used to predict and analyze the transport of model therapeutic agents through the bioactive layer [25]. The chip featured three layers of PDMS and polycarbonate to separate the apical and basal chambers [25]. The authors found that the GOC model improved permeability coefficients of the model agents compared to rat tissues in an Ussing chamber [25]. The device also mimicked villi-like surfaces which are important for the movement of materials within the lumen of the gut [25]. Future iterations of this model could include immune cells, to study their behavior in a more physiologically relevant in vitro model system.
Researchers have used GOC models to better understand the progression of enterovirus infections, which commonly cause vomiting and diarrhea in patients [26]. 2D in vitro models of the gut often lack physiological fluid flow, peristalsis-like mechanical deformations, villi like structures, and the four different epithelial cell types (absorptive, mucus-secreting, enteroendocrine, and Paneth cells) [26]. To overcome some of these challenges, researchers created a microfluidic device with two chambers (apical and basal sides) separated by a thin, porous membrane. The top chamber, apical side, holds Caco2 cells, which after 3 weeks of culture in a monolayer differentiate into enterocyte-like cells. Caco2 cells were exposed to fluid flow and mechanical deformations, which encouraged villi and tight junction formation within 6 days, after which enterovirus infection could be studied [26]. Infection with Coxsackievirus B (CVB1) at the apical side of the device led to a loss of villi morphology and tight junctions and cell detachment, resulting in an overall loss of barrier function [26]. Viral load was confirmed to be higher on the apical side post infection, including when CVB1 was delivered on the basal side, suggesting that infection can occur on the basal or apical side of the cells, but virions are preferentially shed on the apical side [26]. Similarly, a higher quantity of cytokines, the chemokine (C-X-C motif) ligand (CXCL)-10 and interleukin (IL)-8, were released on the apical side regardless of where CVB1 was delivered [26]. This model could be implemented to study a variety of gut infections and diseases, particularly focusing on the effects of mechanical cues like deformation (peristalsis) or fluid flow on infection. Control over sampling and the addition of other cell types like immune cells to this system could lead to substantial advances in our understanding of gut immunity during infection.
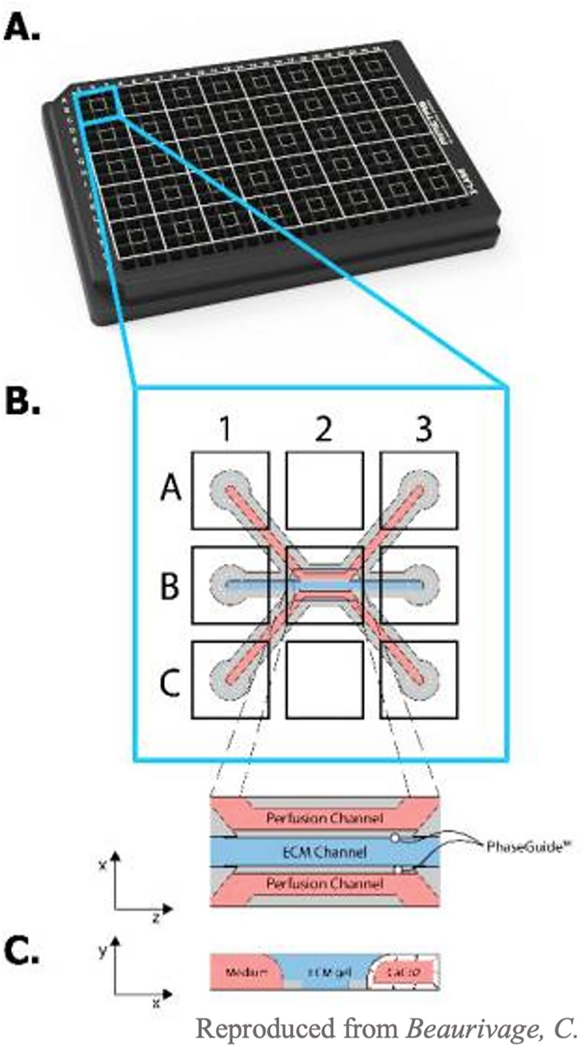
A popular method to model the epithelial barrier is the OrganoPlate [27, 28]. It has three channels, two of which are used as perfusion channels and a third is used as interstitial tissue space, containing ECM. Beaurivage et al. used the OrganoPlate to study the pathophysiology of irritable bowel disease (IBD). They seeded Caco2 cells in one of the channels, where Caco2s formed tight junctions and showed no leakiness after 4 days of culture [28]. To induce IBD, a cytokine cocktail of IL-1β, tumor necrosis factor (TNF)-α, and interferon (IFN)-γ was introduced into the basal channel. To confirm induction of IBD, cytokine secretion, barrier integrity, and adherent protein expression were quantified [28]. Exposure to the cytokine cocktail induced subsequent basal secretion of cytokines and chemokines interferon γ–induced protein (IP)-10, IL-8, and chemokine ligand (CCL)-20 was significantly higher compared to negative controls [28], which are key epithelial pro-inflammatory molecules [28]. Application of the cytokine cocktail also induced disruption of epithelial barrier integrity and reduced E-cadherin on epithelial cells [28]. The group then investigated if the effects of the cytokine cocktail can be reversed using a well-known anti-inflammatory drug, [5-(p-fluorophenyl)-2-ureido] thiophene-3-carboxamide (TPCA-1) [28, 29]. When cells were exposed to TPCA-1 for 2 h before the addition of the cytokine cocktail, secretion of CXCL-10, IL-8, and CCL-20 was reduced both on the apical and basal sides [28]. At a concentration of 1.25 mM, TPCA-1 restored barrier function and cell viability. Since IBD currently has no curative treatment, using the OrganoPlate IBD model could improve our understanding of the pathology of IBD, and, importantly, screen for therapeutic targets to reduce inflammatory responses in IBD patients.
The OrganoPlate has also been used to model neutrophil infiltration in inflammatory conditions (Fig. 5) [27]. Gjorevski et al. sought to explore how neutrophils infiltrated mucosal surfaces during inflammation to assess the potential for neutrophilic therapeutic interventions. This phenomenon is important in the inflammatory response as the migration of neutrophils to an area helps recruit other cells that propagate the inflammatory response [27]. Caco2 cells were used to model the epithelial layer, and collagen and non-activated macrophages were seeded into the ECM channel [27]. In the homeostatic condition, the epithelial barrier is connected by tight junctions and macrophages found beneath the epithelium are inactivated [27]. The presence of inactivated macrophages did not affect barrier function of the Caco2 cell model [27]. However, when macrophages were activated by bacterial components, lipopolysaccharide (LPS) and N-formylmethionine-leucyl-phenylalanine (fMLP), barrier function was compromised, modeling a leaky gut [27]. This successfully modeled an inflammatory response as evidenced by an increase in the secretion of cytokines IL-1b, Mip-1a, Mip-1B, TNF-a, and IL-6 in the basal channel [27]. The authors next looked at neutrophil infiltration in the context of mucosal inflammation. Collagen concentration was varied to assess how ECM density affects neutrophil migration. High concentrations of ECM (> 4 mg/ml) reduced neutrophil invasion, while lower concentrations (1–3 mg/ml) enhanced neutrophil infiltration. Additionally, the presence of activated macrophages in ECM with a concentration of 3 mg/ml induced neutrophil infiltration, suggesting that neutrophils could sense the inflammatory environment. It has been suggested that bioactive products of ECM degradation can also influence neutrophil migration [27]. To test this hypothesis, the authors introduced matrix metalloproteinase 9 (MMP9) and prolyl endopeptidase (PE) to induce in situ enzymatic degradation of the ECM into tripeptide Pro-GlyPro (PGP) products. The presence of PGP induced robust neutrophil infiltration compared to non-degraded control. Overall, OrganoPlate has a simple layout but can be used for many applications including disease modeling, drug screening, and studying cell interactions.
Fig. 5.
OrganoPlate setup. A Each square of the OrganoPlate is one individual gut model. B, C Each square is made up of three parallel channels: two perfusion (red) and one for ECM components
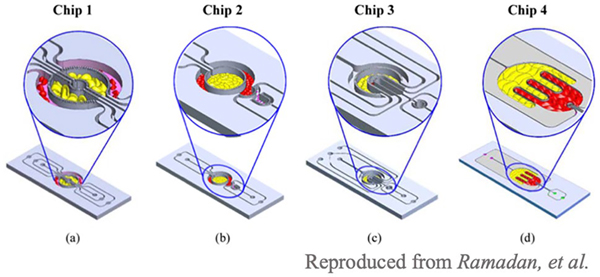
Ramadan et al. utilized a set of multiple microfluidic chips that allowed co-culturing of multiple cell types adjacent to each other to better understand the effect of obesity on insulin resistance in type 2 diabetes [30]. The device was developed from four microfluidic systems—chip 1, three circular components separated with porous barriers, chip 2, two circular concentrated compartments with a semi-circular compartment downstream, chip 3, four U-shaped compartments, and chip 4, two interdigitated compartments with a small circular compartment, as shown in Fig. 6. This paper describes four designs, but the chips can be customized to model any tissue or organ with the appropriate orientation and distribution that mimics the in vivo environment. The chips were separated by thin sidewalls with small pores that prevent cell migration and allow for continuous perfusion and diffusion in a dynamic environment between compartments, which mimics the environment in the human body. Through finite element analysis, researchers found that the shear stresses were within the range of those found in vivo. To demonstrate an example of the functionality of this microfluidic system, the role of obesity and inflammation on insulin resistance in type 2 diabetes was assessed. Research has demonstrated that inflammatory cells infiltrate fat tissues and interact with adipocytes in obese patients, ultimately leading to insulin resistance. Here, researchers co-cultured adipocytes with U937, human lymphocyte, cells in their microfluidic models. The researchers found that immune cells infiltrate into the adipocyte regions, which mimics the process that occurs in obesity. This device paves the way for immune-metabolic profiling.
Fig. 6.
3D schematic drawings of chips 1–4. Chip 1 (C1): three circular concentered compartments separated by porous barriers. Chip 2 (C2): two circular concentered compartments interface with a downstream semi-circular compartment. Chip 3 (C3): four U-shaped concentered compartments with the inner compartment connected through a channel to another circular compartment. Chip 4 (C4): two interdigitated compartments interfaced with a small circular compartment located inside the upstream compartment.
Lung-on-a-chip
Lung-on-a-chip (LOC) models have proven to be a great resource for studying immune responses to stimuli such as cytokines and therapeutics. The first LOC was described by the Ingber group which replicated a functional alveolarcapillary interface and was used to study responses to bacteria and inflammatory cytokines [31]. This model tested epithelial response to silica nanoparticles and illustrated toxic effects of ultrafine airborne particles, modeled by the nanoparticles [31]. Silica nanoparticles increased the expression levels of intercellular adhesion molecule–1 (ICAM-1) in endothelial cells, which effectively induced neutrophil capture [31]. The upregulation of ICAM-1 signifies a pulmonary immune response [31]. Since then, many iterations and applications have evolved from the original design: modeling of mechanical forces or disease and drug screening to name a few. One group designed a device that included a stretchable membrane, simulating breathing within the lung [32]. Zamprogno et al. produced a collagen-elastin membrane that can be sandwiched into a microfluidic device for further evaluation [32]. Human primary alveolar epithelial cells (hAECs) and human lung microvascular endothelial cells (VeraVec) were cultured on both sides of the membrane and were shown to withstand a 10% strain [32]. The researchers also established permeability through the cell layer, showing that smaller particles are more effectively transported. Such models allow researchers to take into account the effect that mechanical forces have on therapeutic pharmacokinetics and distribution and thus may improve the therapeutic design.
Benam et al. sought to build models of respiratory diseases including asthma and chronic obstructive pulmonary disease (COPD) using two chambers separated by a thin, porous membrane. Human airway epithelial cells were first cultured in the top chamber under flow after which the media was removed to create an air–liquid interface, which induces polarization and cilia formation in the airway epithelial cells [33]. After 5 weeks, microvascular endothelial cells were cultured on the bottom chamber to create the airway-blood interface. This model was used to study allergic airway inflammation characteristic of asthma by introducing the cytokine IL-13. IL-13 exposure in the airway epithelial cells caused an increase in goblet cells, higher production of cytokines granulocyte colony-stimulating factor (G-CSF) and granulocyte–macrophage colony-stimulating factor (GM-CSF), and a decrease in the frequency of cilia beating [33]. COPD was also successfully modeled by stimulating the cells with viral mimicking adjuvant poly(I:C) or with LPS. These molecules increase secretion of IL-8 and M-CSF, consistent with what is found in patients with COPD [33]. After successfully building models of both diseases, the group wanted to see if certain anti-inflammatory drugs could reduce cytokine secretion. Because IL-13 signals through the JAK-STAT signaling pathway in asthma, tofacitinib, an inhibitor of JAK-1, −2, and −3 was tested [33]. They found that when chips were infused with tofacitinib (1 and 10 μM), G-CSF and GM-CSF levels decreased, cilia started to beat at normal frequencies, and there was suppressed goblet cell hyperplasia [33]. To test reduce the inflammatory response in the COPD model, the group tested bromodomain-containing protein 4, an inhibitor of NFκB signaling [33]. Researchers found a reduction in the expression levels of E-selectin, vascular cell adhesion molecule (VCAM-1), and intercellular adhesion molecule (ICAM)-1, and neutrophil recruitment was reduced by a third [33]. In summary, LOC models have helped enhance our understanding of inflammatory responses and model therapeutic function in disease states.
Lymph node-on-a-chip
Several organ-on-a-chip devices try to recapitulate the 3D architecture of the LN, allowing researchers to better understand how different LN processes and interactions occur. One way to study the LN environment is to use live LN tissue slice cultures. This maintains 3D architecture and cell complexity, and when integrated into a microfluidic device, the surrounding environmental cues can be easily manipulated. Ross et al. created a device that housed the LN slice and delivered media within the top chamber [34]. The device allowed for tracking of particles and molecules in microscale areas of the slice to calculate the rate of diffusion throughout the tissue using microscopy [34]. It is important to look at diffusion within tissues because it reveals specific characteristics of the extracellular space such as the tissue-like tortuosity [34]. The researchers then used this model to study the diffusion of 10-kDa dextran within different regions of the LN. The diffusion coefficient of dextran in both the cortex and paracortex were found to be very similar, suggesting that the extracellular space is similar for these two regions [34]. The authors suggest that the model can be used for future experiments studying the diffusion of immunotherapies to assess whether or not these can reach their target cells within the LNs.
The same group adapted this device to stimulate and observe specific points within the LN using an 80-μm port above the LN slice [35]. Using this device, the researchers illustrated how glucose conjugates move throughout and stimulate cells within a specific LN region. Glucose conjugates have been used as drug delivery systems to target cancer cells and may also be used to preferentially target lymphocytes [35]. They found that fluorescent glucose-bovine serum albumin (BSA) conjugates delivered to the T cell zone were cleared after 15 min, but stayed localized when delivered to B cell zones [35]. Glucose-BSA conjugates were delivered to the intersection of the T and B cell zones, and it was discovered that after 5 min the distribution of conjugates was equal between the two zones. However, after 15 min, over 90% of the conjugates were in the B cell zones [35]. When conjugates were delivered to splenocytes, ~23% of the T cell and 57% of the B cell population took up conjugates [35]. Devices such as these enable us to more closely study communication between the myriad of different immune cells and provide live information about the spatial arrangement and movement of immune cells throughout lymphoid organs and beyond. Such new knowledge may yield new therapeutic targets and improve our design of immunotherapies.
The LN slice culture system has also been used to explore tumor–LN communication [36]. They designed a device that could hold ex vivo slices of LNs in one compartment that is connected through tubing with another compartment containing tumor slices. Their system was able to recirculate flow from tumor to LN, allowing the capture and recirculation of proteins via flow across the tumor slice in their model system. The authors confirmed the functionality by demonstrating that co-culture of tumor slices with draining LNs (tumor draining and non-tumor draining) led to immunosuppression as demonstrated by the reduced capacity to produce IFNγ in both compartments compared to IFNγ production in a naïve LN. Interestingly, co-culture of tumor-draining LNs with non-tumor draining LNs further suppressed non-tumor-draining LN IFNγ secretion [36]. This microfluidic system could be applied to study effects of peripheral tissues or tumors on LN responses and including spatially dependent changes such as cell migration.
DC migration and homing within the LN play a large role in the adaptive immune response [37]. This migration is largely directed by gradients of CCL19 and CCL21, both of which are ligands for the CCR7 receptor [37]. Haessler et al. developed a microfluidic device to study DC migration in 3D. Their device consisted of a central channel that housed DCs and two side channels that housed CCL19 and CCL21, both of which were filled with ECM filled with collagen I and Matrigel [37]. The group first sought out to see the effects of CCR7 signaling on the chemokinetics of CCL19 and CCL20. They found that there was only a small affect on DCs and found that increases in the number of migrating cells and average cell speed only occurred at CCL19 concentrations above 50 nM and CCL21 concentrations above 100 nM [37]. When the DCs were exposed to an increasing concentration gradient, the chemotactic response decreased for CCL19, but not for CCL21 [37]. This observation may be explained by the fact that CCL21 ligands are recycled, while CCL19 is not [37]. When the concentration gradient was kept constant and the average concentration was varied, the chemosensitivity decreased as average concentration decreased only for CCL21 [37]. Lastly, when percent gradient was increased, there was a positive correlation with chemotaxis found in CCL21, but a flat trend for CCL19 [37]. This device revealed key parameters that affect DC migration that has not been seen in a 3D model. Studying the transport and migration of key cells and molecules will help ensure that immunotherapies are properly translated from bench to clinic.
For more information and LN modeling including the use of organoid systems, readers are referred to the review by Kim et al. [38].
Tonsil-on-a-chip
One challenge that many researchers face is the lack of translation from a mouse model to humans. Many adaptive immune responses to vaccines and other immunotherapies in mice are not predictive of the human response. While 3D models that utilize ex vivo samples of tissues reveal a lot of information about how molecules move within the tissue, they cannot maintain the architecture past 4 days and typically are not representative of the full adaptive immune response. A key component in lymphoid organs is the germinal center where B cells mature and differentiate. Species variation in adaptive immune responses can be alleviated by using models that utilize human samples. Wagar et al. successfully created tonsil organoids from single-cell suspensions of human tonsil tissue [39]. After several days in culture, cells began to form clusters [39]. After 7 days in culture, the organoid system was stimulated with live attenuated influenza vaccine (LAIV) and was found to have an increase in B cell differentiation, plasmablast frequencies, and antibody secretion compared to unstimulated organoids [39]. The group was able to track the progression of both B and T cell differentiation within the organoid. As time progressed, more B cells were found to express CD38 and CD27, illustrating the shift to a pre-germinal center phenotype [39].
To confirm the structural integrity of these organoids, the authors found that there were distinct aggregates of B and T cells in both vaccine stimulated and unstimulated organoids [39]. Additionally, within the B cells aggregates, two distinct B cell subsets (CD83 and CXCR4) were found to form dark and light regions, which is distinctive of germinal centers within other lymphoid organs. Affinity maturation was confirmed by depleting the system of hemagglutinin (HA)+ B cells, stimulated with LAIV, and showing that more HA+ B cells after 10 days[39]. Somatic hypermutation of B cells was also observed to increase at day 7 of the vaccine-stimulated systems [39]. Human responses are extremely variable, and the use of an organoid system similar to the one presented here can help adjust for this variability and develop immunotherapies that can be more broadly delivered.
Bone marrow-on-a-chip
At the time when bone-marrow-on-a-chip (BMOC) was first created, the previously used 2D models did not recapitulate the topography, cell–cell interactions, architecture, and stiffness found within the in vivo environment [40]. The first group that successfully created a BMOC implanted a hollow cylinder in a mouse containing HSC, to mimic bone marrow, and once differentiated, placed it into a microfluidic chamber to produce blood cells [41]. They found that culturing BM cells in stroma-supported media, the current standard method for maintaining HSCs and hematopoietic progenitor cells in vitro, resulted in fewer HSCs and more hematopoietic progenitor cells after 4–7 days compared to freshly excised BM [41]. To create this culture system, bone marrow stromal cells are cultured on a dish for 3 weeks until a monolayer is formed [41]. Cells are then irradiated, and BM cells are cultured on top of the irradiated cell layer [41]. In contrast, when BM was placed into the microfluidic device, HSC numbers were maintained similar to those found in mice [41]. Even though cytokines are required for HSCs to be maintained in 2D cultures in vitro, removing cytokines from culture media did not affect the distribution of HSCs and hematopoietic progenitors in the engineered BM, thus suggesting that the device could maintain the BM niche [41]. This group then wanted to see if their BMOC can be used as an in vitro model for γ-radiation, which currently can only be done to live mice [41]. γ-Radiation has been shown to produce toxic effects on BM cells of mice [41]. When the engineered BM in the microfluidic device was exposed to radiation, the number of HSCs, hematopoietic progenitors, lymphoid cells, and myeloid cells were similar to that found in mice exposed to the same amount of radiation, but this was not the case for cells in the stroma-supported culture [41].
Another group created a device with a hematopoietic top chamber and vascular bottom chamber to simulate vascularized BM [42]. They used this device to explore the cytotoxic effects of various drugs on the BM including 5-fluorouracil (5-FU) and a small molecule inhibitor AZD2811. 5-FU is a chemotherapeutic that causes DNA damage and apoptosis in proliferating cells [42]. Researchers found that the hematotoxic effect in their model was similar to that of clinical studies when cells that had been in culture in the device for 10–12 days were exposed to 5-FU for a total of 2 days [42]. Another drug that has been explored for the treatment of hematological cancers is AZD2811, an inhibitor of Aurora B kinase, which helps chromosomes align and segregate during mitosis. While AZD2811 has been shown to cause apoptosis in some cancer cells, its effect on bone marrow cells has not fully been explored [42]. Exposure for 2 h at higher concentrations caused neutropenia and anemia, while exposure for 48 h at lower concentrations caused severe neutropenia and no anemia [42]. BMOC holds the potential of accurately assessing how treatments affect BM function. Because immune cells originate from the BM, exploring how potential immunotherapies affecting their functionality will be crucial to understand how immune cell differentiation will propagate.
A device from Aleman et al. sought to create multiple niches within a device and observed where different cells, i.e., hematopoietic stem and progenitor cell (HSPC), lymphoma cells, and leukemia cells, preferentially migrated [40]. They found that HSPCs migrated to the mesenchymal and sinusoidal areas, lymphoma cells migrated to the arterial niche, and leukemia cells migrated most frequently to the osteoblastic niche [40]. Studying cellular migration in this controlled environment could enable us to also screen for therapeutics that inhibit, e.g., leukemia and lymphoma cell migration but not HSPC migration. Thus, BMOC models could be crucial for not only biological understanding but also therapeutic design.
While these models are a great first stem in BM modeling, it does not include niches that are found in vivo. Stem cells must be located within these niches so that they can propagate, differentiate, and function properly outside of their respective niches[43]. The endosteal, central marrow, and perivascular niches are all crucial for stem cell development and are not included in microfluidic devices. Bone-forming osteoblasts and bone-resorbing osteoclasts are found in the endosteal niche[44]. Osteoblasts are of particular importance for stem cell development, as it has been suggested that they influence HSC maturation [44]. As HSC mature, they move towards the central marrow niche where the oxygen gradient is at its lowest [44]. This hypoxic environment plays a large role in regulating stem cell function [44]. The perivascular niche spans between the vascular and endosteal niche [43]. This niche is particularly important in maintaining the stem cell pool within the BM [45]. Integration of niches, particularly the three mentioned here, will be important when studying the development of cells in the BM. So many processes and responses are shaped due to the environment and their inclusion in future models will help recapitulate a better in vivo response.
Microfluidics for Tumor Models and Cancer Immunotherapy
Although science and technology have made great strides in cancer treatment, cancer remains the second leading cause of death in the USA. Traditional animal models and in vitro cell culture models have helped shape our understanding of the cellular and genetic components affecting tumorigenesis. While indispensable for basic cancer research, these models do not reliably recapitulate the physical environments of human tumors that can significantly affect cancer pathology. Microfluidic devices can be used to recreate these tumor microenvironments with relevant biological features like fluid flow and matrix stiffness to better model cellular behaviors and tumor responses to treatment. In addition to screening for pharmaceutical efficacy and helping improve therapeutic design, microfluidic models can also shed light on important cell–cell interactions that can affect successful immunotherapy. Modified or engineered lymphocytes, like in adoptive cell therapy (ACT) or chimeric antigen receptor-T (CAR-T) cell therapy, perform better against blood cancers than solid tissue tumors [46]. Recently, researchers have investigated ways to expand the effectiveness of these cell therapies in the treatment of cancers [47–50]. In microfluidics, the move away from 2D and towards 3D models has led to simulations that better elucidate how cells interact within the tumor microenvironment (TME). An added benefit of these 3D tumor models is the ability to modify EMC composition and mechanical properties to test the effects of various TME characteristics on immune cell function and cytotoxicity.
For breast cancer, several studies have involved microfluidic systems. For instance, Lugo-Cintrón et al. observed the effects of ECM density on lymphatic vessel function [51]. Lymphatic vessels were examined due to the metastatic progression that occurs in breast cancer. For this, they used a microfluidic device model to replicate a lymphatic vessel in a type I collagen matrix. Morphological changes and changes in pro-inflammatory cytokine secretion were observed in the samples with higher ECM densities. More specifically, the most upregulated cytokine observed was IL-6, which serves as a regulator for endothelial cell dysfunction and tumor progression. An increase in pro-inflammatory cytokines and morphological changes are a result of dysfunction in the endothelial barrier.
Ren et al. observed natural killer (NK)–cell interactions with 4T1 breast cancer tumor cells in a microfluidic device model featuring pump-free generation [52]. They were able to track NK cell–4T1 interaction, migration, and morphology. Migrations of NK in certain conditions that mimic microenvironments for tumors can help researchers learn more about the understanding of NK immunosurveillance of cancer [38]. Interestingly, the shape of the 4T1 cells continuously changed when interacting with NK cells and some even significantly shrunk. Under the conditions of a defined chemical gradient, it was shown that LPS-activated DCs promoted activated NK cell migrations to the 4T1 cells. A better understanding of NK cell behavior in the context of cancer may lead to new NK-cell based immunotherapies.
Detection of several analytes on a chip is also possible using microfluidic devices. Gerner et al. established histocytometry to visualize and quantify cell populations directly in tissue sections through multiplexing antibody staining, high-resolution confocal microscopy, and quantitative analysis [53]. Migliozzi et al. developed a methodology to test up to ten immuno-markers on the same tissue section [54]. This method saves time and valuable samples as a 10-plex staining procedure can be completed in under 7 h, including imaging. The order of the biomarkers was PD-1, CD8, CK, CD4, CD3, FOXP3, PD-L1, CDD56, and CD20, which was found to be optimal for the multiplexing procedure. The procedure starts with the autofluorescence recorded, then the slide is stained and then imaged, goes through the elution process, and then a final imaging step. This specialized immunofluorescence protocol involves automatized microfluidic-assisted multiplexing. Co-expression and colocalization patterns were used to confirm the presence of different immune cells. Multiplexing is a great tool that can help further develop personal diagnostics and therapeutics by increasing the speed of analyzing samples.
New designs in microfluidics aim to make immunotherapy screening a quick and accessible tool for enhanced patient-tailored treatments. Moore et al. developed a microfluidic model, named EVIDENT, that incorporates up to twelve stationary tumor biopsy tissues [55]. The tumor sections are continuously perfused with autologous tumor-infiltrating lymphocytes (TILs) under biologically appropriate fluid flow. TILs are pre-treated with immunotherapies prior to circulation via the media flow of the device, along with fluorescent antibody markers and dyes. This system allows for the evaluation of lymphocyte responses to therapies like immune checkpoint inhibition using confocal microscopy to measure lymphocyte tumor infiltration and killing of tumor cells over several days [55]. The researchers found that when staurosporine, a small molecule that induces cell death, was delivered to the TILs, there was a significant increase in cell death when compared to vehicle controls [55]. When tumors were exposed to TILs, it was found that TILs previously exposed to anti-PD-1 therapy caused increased cell death in the tumor cells [55]. The use of multiple tumor sections for assessment in this device allowed researchers to better capture the heterogeneity of a patient’s tumor and makes for a scalable, high-throughput screening tool.
Recently, Beckwith et al. have improved on the design of the EVIDENT model in a microfluidic tumor analysis platform (TAP) [56]. The TAP is fabricated in a single piece of resin and can be 3D printed in 1 h without the need for further assembly. Like the EVIDENT model, TAP integrates live tumor biopsy tissue perfused with adjustable media components containing fluorescent antibody markers and dyes. Tumor fragments were viable within the device for 72 h, and viability could be monitored during the entire culture time [56]. Immunotherapy or drug molecules are introduced to the device fluid flow to evaluate the cytotoxic activity of resident TIL in the tumor tissue. The elimination of circulating, pre-treated TIL allows for faster evaluation of patient responses to therapy. Using confocal microscopy, the researchers were able to observe significant tumor tissue necrosis within 48 h of screening in response to anti-PD-1 therapy [56]. They compared the viability and T cell localization between anti-PD-1-therapy treated tumors and isotype antibody treated controls, and found that treated tumors had more dead cells [56]. They also found that T cells were localized in proximity to the dead cells, suggesting that there is an increase in T cell-mediated killing due to the presence of anti-PD-1-antibodies [56]. A drawback of this device is potential variability in the cellular composition of the biopsy sample since the device investigates anticancer properties of resident lymphocytes within tumor tissue. The addition of a standard method to confirm biopsy composition could add nuance to treatment predictions made using this device as a personalized tumor tissue model. Screening devices like this that can use freshly excised (within 24 h) tumors will help screen future immunotherapies for successful cytotoxic effects and more personalized therapies.
Another ex vivo approach to patient-specific screening for immunotherapy efficacy combines the benefits of 3D cell culture with the precise environmental controls of microfluidics. 3D tumor spheroids have been gaining recognition as useful models for research and therapeutic screening. They present an improvement over 2D cancer cell cultures by allowing for more physiologically relevant cell–cell interactions and tissue organization that more closely mimic human tumors and their resistance to therapy [57]. In their model, Jenkins et al. incorporate slightly dissociated tumor biopsy tissue into 3D tumor spheroids for short-term culture in a microfluidic device to assess individual responses and resistances to ICI therapy. This method of spheroid formation preserves basic elements of the tumor tissue and microenvironment, while allowing it to be viable in-vitro. They used cytokine secretions from tumor spheroids as a measure of immune activation following ICI treatment delivered into device media flow. They consistently observed more cytokine production, including CCL19 and CXC13 in tumor spheroids with phenotypically immunosuppressed infiltrating lymphocytes [58]. The various methods of data analysis available from this device highlight its versatility in providing detailed evaluations of patient biopsies. Other microfluidic-based tumor spheroid systems and their applications are described in a recent review [59]. Additionally, there are a number of non-microfluidic tumor spheroid systems detailed in another review that explores methods of characterizing their utility as cancer therapy models [60].
Park et al. used a microfluidic device to encapsulated HeLa cells in a collagen matrix and introduced NK-92 cells at the interface of the collagen matrix [61]. The more the NK-92 cells migrated towards the HeLa cells, the more cell death was observed [61]. The model incorporates HeLa cells in a collagen ECM barrier to recapitulate a fibrotic TME. The researchers tested different concentrations of collagen gel and found that with a higher concentration of collagen, a smaller amount of NK-92 cells was found deep in the collagen matrix, but at the same time, they found an increase in cytotoxic activity against the HeLa cells [61]. The ECM helped reduce migration and access to the cancer cells, which lowered overall cytotoxicity [61]. When comparing these results to a 2D model, they saw that more NK-92-mediated killing of HeLa cells occurred in the 2D model compared to their device [61]. This shows that the TME is highly influenced by the surrounding ECM and that accessibility into deeper portions of tissue is one of the limiting factors in the killing of tumor cells [61]. With some modifications, this model could be used to screen and enrich for activated lymphocytes with greater cytotoxicity against solid tumors for ACT [62].
Pavesi et al. developed a microfluidic model for the assessment of engineered T cells that express tumor-specific T cell receptors (TCR-T cells) against the physical and metabolic barriers of solid tumor microenvironments [63]. The model includes cancer cells suspended in a collagen ECM with T cells circulating through media channels on either side. Lymphocyte cytotoxicity in this model is dependent in part on the chemotaxis and motility of T cells, and thus simulates the mechanical barriers to T cell infiltration into tumors in vivo. Using this model, the cytotoxicity of engineered T cells was measured under different clinically relevant conditions such as hypoxia, systemic mTOR immunosuppression, and inflammation. Researchers found that their 3D device was able to evaluate efficacy between differentially engineered T cells activated against the same antigen. The engineered T cells, in the presence or absence of IL-2, produced different cytokine profiles in the 3D device, a distinction that was not observable in the 2D model. The response from the TCR-T cells could be heavily influenced by its preparation in a 3D system. With further validation, this 3D model could be used as a powerful and customizable tool for developing CAR-T cells better optimized for the treatment of solid tumors [63].
Microfluidic models are also helping define the specific cell–cell interactions influencing immune responses to immunotherapy. Researchers have targeted DCs second only to T cells in their efforts to create cell-based immunotherapies [64]. Interferon-alpha–conditioned DCs (IFNα-DCs) are known for their enhanced antigen presentation and superior induction of T cell responses [65]. Parlato et al. designed a model to study the migration of IFNα-DCs, towards apoptotic, drug-treated cancer cells. The model houses the IFNα-DCs separated from cancer cells and a thin channel connects the two chambers. They found increased IFNα-DC migration into the tumor chamber was driven by secreted chemokines, like CXCL12, from the dying cancer cells. IFNα-DC phagocytosis of drug-treated cancer cells was also enhanced, which suggests that antigen presentation to T cells may also have increased. This model highlights the benefits of studying immune cell behavior in response to combination therapy. The model could also be used to study the migration of other immune cells towards tumor tissues, infiltration into TMEs, and interactions of cancer cells with immune cells the 3D tumor ECM [66].
Devices for therapeutic screening
Organ-on-a-chip devices can be used to observe and measure the antigenicity of specific proteins when in culture with immune cells. Rosa et al. explored the dynamic relationship of T cells and DCs using a device with a single channel and two inlet and outlet ports [67]. They studied CD4+/CD8+ T cell interactions with either antigenloaded DCs adhered to the bottom of the device or antigenloaded DCs suspended in solution under various levels of shear stress. CD4+ T cell–DC interactions started to break at shear stresses between 0.01 and 12.0 Dyn cm−2 and a decrease of interaction numbers was observed at higher shear stresses. In contrast, CD8+ T cell-DC interactions only broke at higher shear stresses between the range of 12.0–120.0 Dyn cm−2 [67]. Using this device, the authors applied a stream of CD4 + and CD8 + T cells over a common DC monolayer. By doing this, they found that the number of CD4 + T cell interactions stayed at a constant 6.4 × 103 CD4+ T cells to ~6.0 × 104 APCs for 80 min, compared to no attachment with the CD8 + T cells [67]. Devices such as this could pave the way for future screening cellular interactions, and how these might be affected by physiological conditions like shear stress.
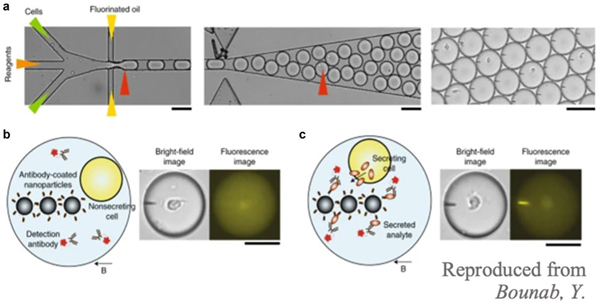
Another promising screening device is the DropMap (Fig. 7). This cell-secretion assay analyzes individual B cells using nanoparticles coated in capture antibodies [68]. When a specific cell secretes the molecule of interest, it is captured by antibodies on the nanoparticles and detected using fluorescently labeled detection antibodies [68]. Using this technology, both secretion rates and affinity can be calculated [68]. The DropMap was used to study the innate immune response in septic shock patients. A monocyte population in the DropMap was maintained by exposing cells to LPS, and it was found that there were lower rates of TNF-a secretion in the environment by septic shock patients compared to healthy controls [68]. Additionally, in septic shock patients, cells exhibit higher endocytic activity [68]. Cells activated by LPS in the device also more readily took up nanobeads than healthy cells, suggesting that they have increased phagocytic capabilities [68]. The authors also used the DropMap to quantify secretion rates of TNF-α, IFN-γ, IL-2, and IL-4 from human T cells [68]. Ex vivo peripheral blood mononuclear cells (PBMCs) were stimulated with phorbol 12-myristate 13-acetate (PMA)/ionomycin and cytokine secretion was compared to unstimulated PBMCs. They found that stimulation did not affect TNF-α, IL-2, and IL-4 secretion, but enhanced IFN-γ secretion. Screening of cell responses is critical in diagnosis, as it could allow for analysis of how well certain immunotherapy works and the type of immune response currently mounted in a patient’s body, and microfluidics could be a useful tool to achieve rapid and cheap screening.
Fig. 7.
DropMap schematic. a Cells enter the device one by one where their produced cytokines and antibodies are met with capturing antibodies on nanoparticles. b Non-secreting cells do not interact with capture and detection antibodies. c When cells are secreting target particles, they bind to capture antibodies and these bind to detection antibodies. The resulting fluorescent image can then be quantified
Microfluidics for single-cell analysis in immunotherapy
Recently, single-cell assays have been gaining traction in the field of microfluidics. These offer a novel way to study cells, individual responses to therapy, and isolated cell–cell interactions [69]. Combining single-cell screening with the high throughput screening potential available via microfluidics could revolutionize the process of discovering new therapies and enhancing existing ones. For example, one of the drawbacks of engineered cell therapies like CAR-T cells is the amount of effort and time required to produce modified T cells with high affinity and specificity for the varying tumor antigens. Segaliny et al. developed a microfluidic assay that captures individual CAR-T cells in a droplet with a target cancer cell and incubates them together in a trapping well. The assay identified activated CAR-T cells that recognized tumor cells by GFP expression. Then the activated cells were isolated from the device for analysis of T cell receptor (TCR) genetic sequences and downstream enrichment for immunotherapy [70]. The assay is enriched for engineered lymphocytes with enhanced cytotoxicity and tumor specificity, which enhances the therapeutic potential of these T cells. This platform could also be used to screen and enrich for B cells of particular specificity, crucial for the development of potent monoclonal antibodies that could be used as treatments.
Researchers have also developed devices to screen for the production of specific proteins or cytotoxic effectors. Briones et al. used a microfluidic device to screen for the production of granzyme B (GrB), which effectively induces cell apoptosis [71]. They analyzed the expression of GrB in three different cell types: GrB-transduced Jurkat T lymphocyte cell, NK-92 natural killer cell (positive control), and THP1 monocyte cell (negative control). They found that NK-92 cells produced the most GrB, consistent with findings in patients treated with immune checkpoint blockade who also had elevated GrB levels [71]. Therefore, a device that can screen protein production may be useful in enhancing our understanding of disease progression or treatment response in patients.
Conclusion and future directions
The field of microfluidic devices creates a new way of analyzing biological systems that researchers could not have in the past. They can be utilized to recreate intricate parts of the body such as the gut and lymphoid organs. These models create ways to study reactions to immunotherapies without having to use costly in vivo models and while allowing for use of patient samples (individualized medicine). Microfluidic devices can also be used to model disease and disease progression, which can lead to a new understanding of disease pathology that is crucial to effectively design therapeutics. Additionally, creating more complex devices that house multiple body systems has the potential to lead to more streamlined testing of therapeutics such as immunotherapies. Furthermore, devices that can sustain an environment for longer times can pave the way to better study disease progression. Faster and more accurate single-cell analysis via microfluidics can improve our understanding of cellular responses. Therefore, microfluidics has significant potential implications in the development and testing of immune modulatory therapeutics.
Acknowledgments
Funding This study was funded by the American Lung Association Dalsemer Award (KM), LAM Foundation Career Development Award (KM), NIH T32 (5T32GM080201, MA), and MPower Maryland (KM).
Footnotes
Declarations
Conflict of interest The authors declare no competing interests.
Data availability Data sharing not applicable to this article as no datasets were generated or analyzed during the current study. Consent for publication. All the authors whose names appear on the submission (1) made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; or the creation of new software used in the work; (2) drafted the work or revised it critically for important intellectual content; (3) approved the version to be published; and (4) agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.The innate and adaptive immune systems. [Website] 2016. August 4, 2016; Available from: https://www.ncbi.nlm.nih.gov/books/NBK279396/.
- 2.Alberts BJA, Lewis J, et al. Innate immunity, in Molecular biology of the cell. Garland Science: New York. 2002. [Google Scholar]
- 3.Alberts BJA, Lewis J, et al. The adaptive immune system, in Molecular Biology of the Cell. Garland Science: New York. 2002. [Google Scholar]
- 4.Gwaltney-Brant S Chapter 22 - Immunotoxicity biomarkers. In: Gupta RC, editor. Biomarkers in Toxicology. Boston: Academic Press; 2014. p. 373–85. [Google Scholar]
- 5.Ruddle NH, Akirav EM. Secondary lymphoid organs: responding to genetic and environmental cues in ontogeny and the immune response. J Immunol (Baltimore, Md. : 1950), 2009;183(4):2205–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owen JA, et al. Kuby Immunology. New York: W.H. Freeman. 2013. [Google Scholar]
- 7.Janeway CA Jr, Walport TPM. et al. Generation of lymphocytes in bone marrow and thymus. Garland Sciences: New York. 2001. [Google Scholar]
- 8.Dunsmore SE, Shapiro SD. The bone marrow leaves its scar: new concepts in pulmonary fibrosis. J Clin Investig. 2004;113(2):180–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn CC, Manley NR. Developing a new paradigm for thymus organogenesis. Nat Rev Immunol. 2004;4(4):278–89. [DOI] [PubMed] [Google Scholar]
- 10.Bujoreanu I, Gupta V. Anatomy, lymph nodes. 2020; Available from: https://www.ncbi.nlm.nih.gov/books/NBK557717/. [PubMed]
- 11.Roozendaal R, Mebius RE, Kraal G. The conduit system of the lymph node. Int Immunol. 2008;20(12):1483–7. [DOI] [PubMed] [Google Scholar]
- 12.Willard-Mack CL. Normal structure, function, and histology of lymph nodes. Toxicol Pathol. 2006;34(5):409–24. [DOI] [PubMed] [Google Scholar]
- 13.Drayton DL, et al. Lymphoid organ development: from ontogeny to neogenesis. Nat Immunol. 2006;7(4):344–53. [DOI] [PubMed] [Google Scholar]
- 14.Martinez VG, et al. Fibroblastic reticular cells control conduit matrix deposition during lymph node expansion. Cell Rep. 2019;29(9):2810–2822.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis SM, Williams A, Eisenbarth SC. Structure and function of the immune system in the spleen. Sci Immunol, 201;4(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesta MF. Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol Pathol. 2006;34(5):599–608. [DOI] [PubMed] [Google Scholar]
- 17.Bizzell E Microbial ninja warriors: bacterial immune evasion. 2018.
- 18.Breslin JW. Mechanical forces and lymphatic transport. Microvasc Res. 2014;96:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Immunity in the tissues. Nat Immunol. 2013;14(10):977–977. [DOI] [PubMed] [Google Scholar]
- 20.Fang P, et al. Immune cell subset differentiation and tissue inflammation. J Hematol Oncol. 2018;11(1):97–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Chapa SO, Salazar A, Madou MJ. Chapter 13.2 – Two-photon polymerization as a component of desktop integrated manufacturing platforms, in Three-dimensional microfabrication using two-photon polymerization, Baldacchini T, Editor. 2016, William Andrew Publishing: Oxford. 2016:374–416. [Google Scholar]
- 22.Rosato DV, Rosato MG. The complete injection molding process, in Injection Molding Handbook. Springer; US, 2000;1460. [Google Scholar]
- 23.Weigl BH, et al. Design and rapid prototyping of thin-film laminate-based microfluidic devices. Biomed Microdevice. 2001;3(4):267–74. [Google Scholar]
- 24.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5(3):209–18. [DOI] [PubMed] [Google Scholar]
- 25.Pocock K, et al. Intestine-on-a-chip microfluidic model for efficient in vitro screening of oral chemotherapeutic uptake. ACS Biomater Sci Eng. 2017;3(6):951–9. [DOI] [PubMed] [Google Scholar]
- 26.Villenave R, et al. Human gut-on-a-chip supports polarized infection of Coxsackie B1 virus in vitro. PLoS One. 2017;12(2):e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gjorevski N, et al. Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab Chip. 2020;20(18):3365–74. [DOI] [PubMed] [Google Scholar]
- 28.Beaurivage C, et al. Development of a gut-on-a-chip model for high throughput disease modeling and drug discovery. Int J Mol Sci. 2019;20(22):5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatti FUR, Hasty KA, Cho H. Anti-inflammatory role of TPCA-1 encapsulated nanosomes in porcine chondrocytes against TNF-α stimulation. Inflammopharmacology. 2019;27(5):1011–9. [DOI] [PubMed] [Google Scholar]
- 30.Ramadan Q, Gourikutty SBN, Zhang QX. OOCHIP: Compartmentalized microfluidic perfusion system with porous barriers for enhanced cell-cell crosstalk in organ-on-a-chip. Micromachines (Basel). 2020;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamprogno P, et al. Second-generation lung-on-a-chip array with a stretchable biological membrane. bioRxiv, 2019:608919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benam KH, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13(2):151–7. [DOI] [PubMed] [Google Scholar]
- 34.Ross AE, Pompano RR. Diffusion of cytokines in live lymph node tissue using microfluidic integrated optical imaging. Anal Chim Acta. 2018;1000:205–13. [DOI] [PubMed] [Google Scholar]
- 35.Ross AE, et al. Spatially resolved microfluidic stimulation of lymphoid tissue ex vivo. Analyst. 2017;142(4):649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim S, et al. Two-way communication between ex vivo tissues on a microfluidic chip: application to tumor-lymph node interaction. Lab Chip. 2019;19(6):1013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haessler U, et al. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci U S A. 2011;108(14):5614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, et al. Multiscale engineering of immune cells and lymphoid organs. Nat Rev Mater. 2019;4(6):355–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagar LE, et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. 2021;27(1):125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aleman J et al. , Deconstructed microfluidic bone marrow on-a-chip to study normal and malignant hemopoietic cell-niche interactions. Small. 2019;15(43):e1902971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torisawa YS, et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods. 2014;11(6):663–9. [DOI] [PubMed] [Google Scholar]
- 42.Chou DB, et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat Biomed Eng. 2020;4(4):394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lévesque JP, Helwani FM, Winkler IG. The endosteal ‘osteoblastic’ niche and its role in hematopoietic stem cell homing and mobilization. Leukemia. 2010;24(12):1979–92. [DOI] [PubMed] [Google Scholar]
- 44.Tamma R, Ribatti D. Bone niches, hematopoietic stem cells, and vessel formation. Int J Mol Sci. 2017;18(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh M, Nör JE. The perivascular niche and self-renewal of stem cells. Front Physiol. 2015;6:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charrot S, Hallam S. CAR-T cells: future perspectives. HemaSphere. 2019;3(2):e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grosser R, et al. Combination immunotherapy with CAR T cells and checkpoint blockade for the treatment of solid tumors. Cancer Cell. 2019;36(5):471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang N, et al. TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI insight. 2020;5(4):e133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, et al. An IL-4/21 inverted cytokine receptor improving CAR-T cell potency in immunosuppressive solid-tumor microenvironment. Front Immunol. 2019;10(1691). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, et al. Modified CAR T cells targeting membrane-proximal epitope of mesothelin enhances the antitumor function against large solid tumor. Cell Death Dis. 2019;10(7):476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lugo-Cintrón KM, et al. Matrix density drives 3D organotypic lymphatic vessel activation in a microfluidic model of the breast tumor microenvironment. Lab Chip. 2020;20(9):1586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren X, et al. Chapter nineteen - applications of microfluidic devices in advancing NK-cell migration studies, in Methods in Enzymology, Galluzzi L and Rudqvist N-P, Editors. Academic Press. 2020;357–370. [DOI] [PubMed] [Google Scholar]
- 53.Gerner MY, et al. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Migliozzi D, et al. Microfluidics-assisted multiplexed biomarker detection for in situ mapping of immune cells in tumor sections. Microsyst Nanoeng. 2019;5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore N, et al. A multiplexed microfluidic system for evaluation of dynamics of immune-tumor interactions. Lab Chip. 2018;18(13):1844–58. [DOI] [PubMed] [Google Scholar]
- 56.Beckwith AL, Velásquez-García LF, Borenstein JT. Microfluidic model for evaluation of immune checkpoint inhibitors in human tumors. Adv Healthc Mater. 2019;8(11):e1900289. [DOI] [PubMed] [Google Scholar]
- 57.Nunes AS, et al. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng. 2019;116(1):206–26. [DOI] [PubMed] [Google Scholar]
- 58.Jenkins RW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8(2):196–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunti S, et al. Organoid and spheroid tumor models: techniques and applications. Cancers. 2021;13(4):874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinto B, et al. Three-dimensional spheroids as in vitro preclinical models for cancer research. Pharmaceutics. 2020;12(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park D, et al. High-throughput microfluidic 3D cytotoxicity assay for cancer immunotherapy (CACI-IMPACT platform). Front Immunol. 2019;10(1133). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park D, et al. High-throughput microfluidic 3D cytotoxicity assay for cancer immunotherapy (CACI-IMPACT platform). Front Immunol. 2019;10:1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pavesi A, et al. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI insight. 2017;2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez CR, De Palma M. Engineering dendritic cell vaccines to improve cancer immunotherapy. Nat Commun. 2019;10(1):5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santini SM, et al. IFN-alpha in the generation of dendritic cells for cancer immunotherapy. Handb Exp Pharmacol. 2009;188:295–317. [DOI] [PubMed] [Google Scholar]
- 66.Parlato S, et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci Rep. 2017;7(1):1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moura Rosa P, et al. The intercell dynamics of T cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip. 2016;16(19):3728–40. [DOI] [PubMed] [Google Scholar]
- 68.Bounab Y, et al. Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap. Nat Protoc. 2020;15(9):2920–55. [DOI] [PubMed] [Google Scholar]
- 69.Tavakoli H, et al. Recent advances in microfluidic platforms for single-cell analysis in cancer biology, diagnosis and therapy. Trends in analytical chemistry. TrAC. 2019;117:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segaliny AI, et al. Functional TCR T cell screening using single-cell droplet microfluidics. Lab Chip. 2018;18(24):3733–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Briones JC, et al. A microfluidic platform for single cell fluorometric Granzyme B profiling. Theranostics. 2020;10(1):123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]