Abstract
Sarcomatoid urothelial carcinoma (SUC) is a rare subtype of urothelial carcinoma (UC) that typically presents at an advanced stage compared to more common variants of UC. Locally advanced and metastatic UC have a poor long-term survival following progression on first-line platinum-based chemotherapy. Antibodies directed against the programmed cell death 1 protein (PD-1) or its ligand (PD-L1) are now approved to be used in these scenarios. The need for reliable biomarkers for treatment stratification is still under research. Here, we present a novel case report of the first Imaging Mass Cytometry (IMC) analysis done in SUC to investigate the immune cell repertoire and PD-L1 expression in a patient who presented with metastatic SUC and experienced a prolonged response to the anti-PD1 immune checkpoint inhibitor pembrolizumab after progression on first-line chemotherapy. This case report provides an important platform for translating these findings to a larger cohort of UC and UC variants.
Keywords: neoplasm of the genitourinary tract
INTRODUCTION
Urothelial carcinoma (UC) is a highly immunogenic malignancy (Gakis 2014). Intravesical bacillus Calmette–Guerin (BCG) therapy has been used in non-muscle-invasive bladder cancer over the last 40 years (Tripathi and Plimack 2018). Studies have demonstrated that UC has a high overall mutational burden, triggering an immune response, and tumor-associated lymphocytic infiltration (Tripathi and Plimack 2018). Programmed death-ligand-1 (PD-L1), also known as cluster of differentiation 274 (CD274), is an immunoinhibitory molecule mainly expressed on the surface of tumor cells and antigen-presenting cells in various solid malignancies that suppresses the activation of cytotoxic T cells, leading to tumor progression. Overexpression of PD-L1 in UC is associated with poor clinical outcomes, supporting the rationale for immune checkpoint blockade in advanced UC (Nakanishi et al. 2007).
Locally advanced and metastatic UC is an aggressive disease with poor long-term survival following progression on first-line platinum-based chemotherapy (De Santis et al. 2012). Recently, two antibodies directed against the programmed cell death 1 protein (PD-1) or its ligand (PD-L1) have been approved by the U.S. Food and Drug Administration for patients with advanced or metastatic UC who are ineligible to receive cisplatin-based chemotherapy and have a tumor expressing PD-L1 (pembrolizumab and atezolizumab). Five antibodies directed against the protein PD-1 or its ligand PD-L1 have been approved in patients who progress on or after a platinum-based chemotherapy. Two have recently been withdrawn from the U.S. market based on nonconfirmatory phase III studies (atezolizumab and durvalumab). However, many patients who receive checkpoint inhibitors do not achieve durable disease, and there is a need for reliable biomarkers for treatment stratification. Subgroup analyses of the KEYNOTE-045 trial showed continued clinical benefit of the anti-PD1 inhibitor pembrolizumab as second-line therapy in advanced/metastatic, platinum-refractory UC (Powles et al. 2020) over chemotherapy for efficacy and safety for treatment of locally advanced disease (Fradet et al. 2019). In KEYNOTE-045, PD-L1 was not significantly associated with improved outcomes of pembrolizumab or chemotherapy. Other recently identified potential biomarkers include tumor-infiltrating lymphocytes (TILs), microsatellite instability (MSI), tumor mutational burden (TMB), and the gut microbiome.
Sarcomatoid urothelial carcinoma (SUC) is a rare subtype of UC, with an incidence of 0.1%–0.3% across all bladder histologies (Cheng et al. 2011), whereas sarcomatoid features occur in up to 6% of UC cases. SUC typically presents at an advanced stage compared to more common variants of UC (stage T3-4, 34% vs. 28%) and is frequently associated with node-positive or metastatic disease (20% vs. 10%) (Wright et al. 2007). A retrospective analysis of 221 cases of SUC found that only 54% of patients were alive 1 year after cancer-directed surgery (Wang et al. 2010). No prospective data is available to guide management, and treatment is based on extrapolation from standard UC management (Grossman et al. 2003; Sternberg et al. 2006).
In this molecular case report, we present a patient with metastatic SUC, which demonstrated PD-L1 overexpression and gene amplification, who experienced a durable significant response to pembrolizumab. We demonstrate the combination of tissue- and plasma-based next-generation sequencing (NGS) complemented by Imaging Mass Cytometry (IMC) analysis (Giesen et al. 2014) of tissue to explore the immune cell repertoire.
RESULTS
Case Presentation
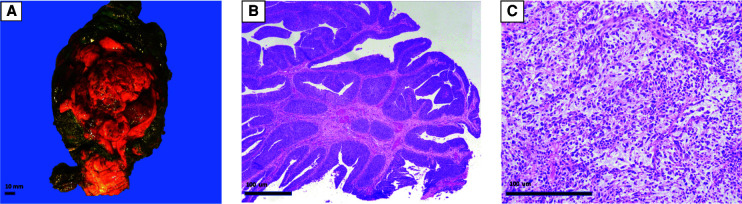
A 62-yr-old female with no significant comorbidities presented in February 2018 with hematuria. Cystoscopy revealed a noninvasive high-grade papillary urothelial carcinoma. The patient was lost to follow-up until January 2019 when she returned with difficulty voiding, pelvic pain, and gross hematuria. Cystoscopy revealed a large, organized clot in the bladder, and a computed tomography (CT) demonstrated an enhancing bladder tumor. Transurethral resection of the bladder tumor and fulguration was not successful, and the patient underwent a radical cystectomy, hysterectomy and bilateral salpingo-oophorectomy, anterior vaginectomy with pelvic lymph node dissection, and creation of a continent cutaneous urinary diversion (Fig. 1A). Pathology revealed a 6.5-cm high-grade urothelial carcinoma (Fig. 1B) with a malignant spindle cell component (Fig. 1C) consistent with sarcomatoid differentiation. The tumor invaded the superficial muscularis propria (inner half) with metastatic carcinoma identified in one of 16 pelvic lymph nodes.
Figure 1.
Histopathology of primary sarcomatoid urothelial carcinoma (UC) case. A radical cystectomy, hysterectomy and bilateral salpingo-oophorectomy, anterior vaginectomy showed a 6.5-cm bladder mass (A) with papillary high-grade urothelial carcinoma component (B) and an invasive malignant spindle cell component (C) diagnostic of sarcomatoid differentiation.
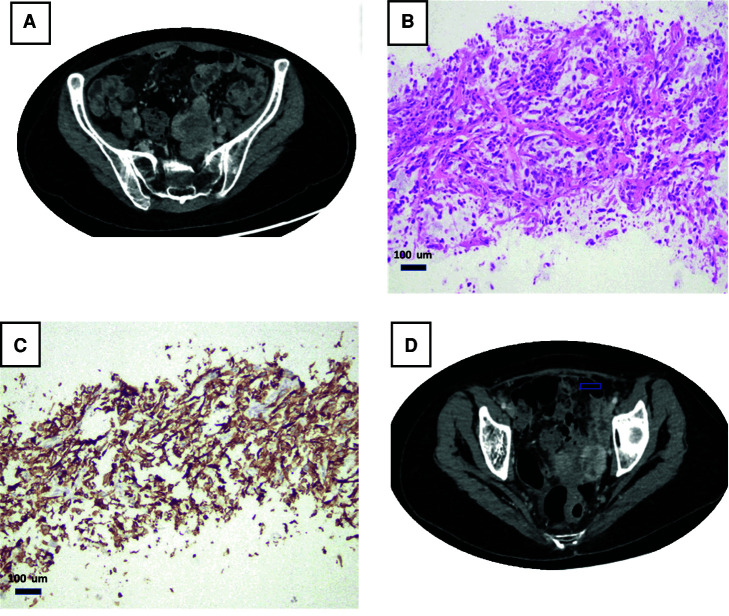
The postoperative course was complicated by hydronephrosis, pouchitis, and enteritis, and adjuvant chemotherapy was delayed. In April 2019, the patient started dose dense MVAC (methotrexate, vinblastine, doxorubicin, and Cisplatin) with granulocyte colony stimulating factor prophylaxis. She received two of four cycles and developed pseudomonas bacteremia and persistent liver function abnormalities attributed to chemotherapy. The patient was transitioned to Cisplatin and gemcitabine and within two cycles she developed new lower back pain in July 2019. CT of the abdomen and pelvis with contrast demonstrated new enhancing masses near the left pelvic sidewall and internal iliac vessels (reference mass, 3.4 × 2.8 cm; Fig. 2A). Biopsy of a pelvic mass confirmed sarcomatoid carcinoma with extensive necrosis that was histologically similar to the patient's bladder primary (Fig. 2C). Immunohistochemistry with the PDL-1 SP263 antibody revealed a combined positive score (CPS) of 100% (Fig. 2D).
Figure 2.
Clinicopathological presentation of metastatic sarcomatoid urothelial carcinoma (UC). Computed tomography (CT) with contrast showing an enhancing left pelvic mass (arrow) near the pelvic sidewall and near the left internal iliac vessels (A,B). Biopsy from that mass showed sarcomatoid carcinoma related to patient's known bladder primary (C). PDL-1 (SP263 antibody) shows a diffuse membranous staining, and the combined positive score (CPS) was 100% (D).
Because of progression on cytotoxic chemotherapy, the patient was started on pembrolizumab 200 mg intravenously every 3 wk. After three cycles, response assessment on CT scan demonstrated decrease in size and enhancement of the left lower quadrant mass (2.3 × 1.5 cm) and associated lesions. After cycle 6, the CT scan revealed further reduction of the left lower quadrant mass (1.8 × 1.3 cm), consistent with overall 65% tumor burden decrease (partial response) (Fig. 2B).
To further characterize the genetic and pathologic characteristics of the patient's metastatic tumor, we performed NGS using a commercially available platform, PGDx elio tissue complete assay (Labriola et al. 2020) (Personal Genome Diagnostics, Inc.), 100 ng DNA input with 889× distinct coverage. No actionable single-nucleotide variants (SNVs) or translocations were detected. Microsatellite analysis showed that the tumor was microsatellite stable. The TMB was 10.8 mutations per megabase (Muts/Mb), predicted to be the exome equivalent of 3.1 (Muts/Mb). This is lower than the median of 5.5 somatic Muts/Mb in 130 urothelial carcinoma samples reported by the Cancer Genome Atlas Research Network (The Cancer Genome Atlas Research Network 2013). A low-fold CD274 (PDL-1) amplification was detected in the metastatic tissue, but not in plasma cfDNA interrogated by PGDx elio plasma resolve assay (Al Zoughb et al. 2021). Blood collection occurred 2 wk after the biopsy, during which time the patient received the first cycle of pembrolizumab, which could explain the false-negative circulating free DNA (cfDNA) results.
Sequence mutation analysis in the metastatic tissue revealed variants in several cancer-related genes including oncogenes (HRAS and GLI1), tumor suppressor (FAT1), histone modifiers (KDM6A), and genes involved in various pathways (CDKN1A, cell cycle pathway; KEAP1, oxidative stress pathway). Mutations in the TERT promoter region and several other genes were detected (Tables 1 and 2). The G13P HRAS mutation was confirmed in the plasma and a F1101L GLI1 mutation with high variant allele frequency (VAF) of 51% was detected. Although no germline confirmation was performed, such a GLI1 variant was believed to represent a germline mutation given its high VAF. Based on VAF and the expected tumor purity (∼18%), the CKDN1A and KEAP1 mutations were suspected to represent a subclone of the original tumor. Plasma cfDNA sequencing confirmed G13P HRAS mutation (Tables 1 and 2).
Table 1.
PGDx elio tissue and plasma complete results
| Gene | Source | Alteration | Mutation | MAF (%) | Role |
|---|---|---|---|---|---|
| CARD11 | Tissue | T1022A | Missense | 56.3 | CARD-CC protein family costimulatory signal essential for T-cell receptor (TCR)-mediated T-cell activation |
| TLR4 | Tissue | N176Ffs*27 | Frameshift | 38.8 | Pathogen recognition and activation of innate immunity |
| CDKN1A | Tissue | W49S | Missense | 5.9 | Encodes cyclin-dependent kinase inhibitor |
| CDKN1A | Tissue | D52H | Missense | 5.4 | Cell cycle pathway |
| FAT1 | Tissue | Y2774* | Nonsense | 14.4 | Tumor suppressor; cell proliferation |
| GLI1 | Tissue | F1101L | Missense | 51.0 | Oncogene; Hedgehog signaling |
| HRAS | Tissue | G13P | Missense | 17.5 | Proto-oncogene |
| KDM6A | Tissue | K1018Efs*6 | Frameshift | 13.0 | Histone demethylase: chromatin-modifying or chromatin-regulatory genes |
| KEAP1 | Tissue | R272C | Missense | 7.5 | Tumor/metastasis suppressor; oxidative stress pathway |
| NCOR1 | Tissue | H1401R | Missense | 11.9 | Histones/nuclear receptors |
| PTPRD | Tissue | N1701S | Missense | 53.7 | Unclear role |
| SLX4 | Tissue | G1535E | Missense | 48.3 | DNA repair, DSBR HR |
| TERT | Tissue | n/a | Promoter | 6.7 | Telomerase complex |
| HRAS | Plasma | G13P | Missense | 1.14 | Proto-oncogene |
| HRAS | Plasma | n/a | Splice site Acceptor | 0.54 | Proto-oncogene |
(MAF) Minor allele frequency, (DSBR) double-strand break repair, (HR) homologous recombination.
Table 2.
Variant table
| Gene | Genomic Location | Reference | Mutant | Amino acid change | Transcript | Consequence | MAF (%) |
|---|---|---|---|---|---|---|---|
| CARD11 | Chr 7: 2951886–2951886 | T | C | T1022A | CCDS5336.2 | Missense | 56.3 |
| CDKN1A | Chr 6: 36652024–36652024 | G | C | W49S | CCDS4824.1 | Missense | 5.9 |
| CDKN1A | Chr 6: 36652032–36652032 | G | C | D52H | CCDS4824.1 | Missense | 5.4 |
| FAT1 | Chr 4: 187539418–187539418 | A | T | Y2774* | CCDS47177.1 | Nonsense | 14.4 |
| GLI1 | Chr 12: 57865824–57865824 | T | C | F1101L | CCDS8940.1 | Missense | 51 |
| HRAS | Chr 11: 534285–534286 | CC | GG | G13P | NM_176795 | Missense | 17.5 |
| KDM6A | Chr X:44938503–44938504 | AA | n/a | K1018Efs*6 | CCDS14265.1 | Frameshift | 13 |
| KEAP1 | Chr 19: 10602764–10602764 | G | A | R272C | CCDS12239.1 | Missense | 7.5 |
| NCOR1 | Chr 17: 15973790–15973790 | T | C | H1401R | CCDS11175.1 | Missense | 11.9 |
| PTPRD | Chr 9: 8341114–8341114 | T | C | N1701S | CCDS43786.1 | Missense | 53.7 |
| SLX4 | Chr 16: 3639035–3639035 | C | T | G1535E | CCDS10506.2 | Missense | 48.3 |
| TERT | Chr 5: 1295228–1295228 | G | A | n/a | CCDS3861.2 | Promoter | 6.7 |
| TLR4 | Chr 9: 120474928–120474946 | GACCAATCTA GAGCACTTG | n/a | N176Ffs*27 | CCDS6818.1 | Frameshift | 38.8 |
(MAF) Minor allele frequency.
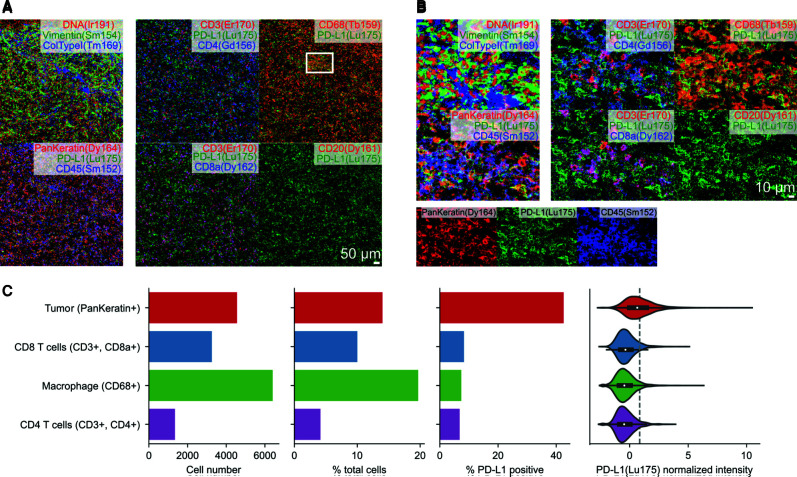
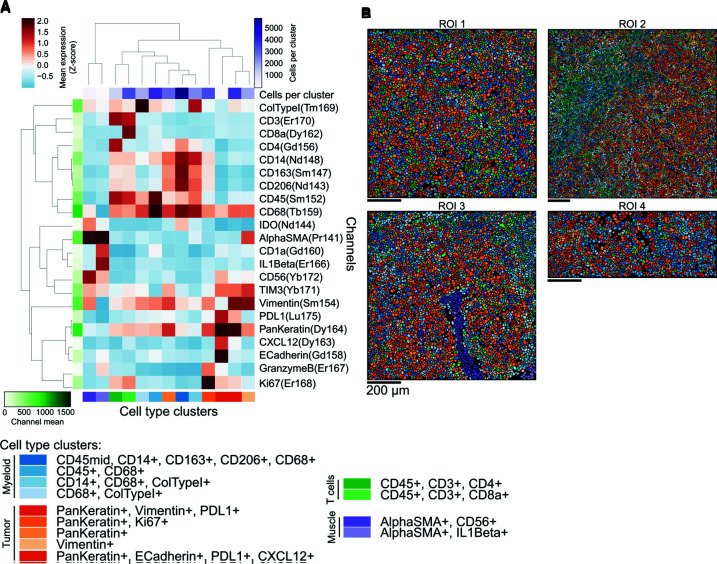
To investigate the immune cell repertoire and PD-L1 expression in the SUC tumor cells and their immune microenvironment, we next performed IMC analysis of the metastatic tissue, capturing four areas of interest in the tissue covering 4.7 mm2. We first observed the expression of tumoral (vimentin, pan-keratin) and immune markers (CD3, CD8a, CD4, CD20, CD68) in the tissue, particularly in the context of PD-L1 coexpression (Fig. 3A,B). To quantify the abundance of cell types, we detected cell boundaries in the images and quantified the intensity of these markers in each cell (Fig. 3C). We observed that macrophages were particularly abundant (20% of all cells), followed by tumor cells (14%). Intratumoral CD3 cells were also seen, with the majority being CD8a+ T cells (10% of total cells) and a smaller minority of CD4+ T cells (3% of total cells). PD-L1 expression was highest in pan-keratin-positive tumor cells (40% of PD-L1 positive cells) (Fig. 3C). PD-L1 expression was also seen at lower levels (5%–8%) in infiltrating CD68+ macrophages and CD4 and CD8 T-lymphocytes, which can help explain the immunohistochemical (IHC) findings (Fig. 3). Further characterization of cell type composition with PD-L1 expression, revealed five tumor cell groups (Fig. 4). One group of pan-keratin-positive tumor cells had high expression of PD-L1 and vimentin, but low expression of E-cadherin. A rarer group of tumor cells also had strong expression of PD-L1, along with CXCL12 and E-cadherin (Fig. 4).
Figure 3.
Tumor-immune microenvironment of metastatic sarcomatoid urothelial carcinoma (UC) revealed by Imaging Mass Cytometry. (A,B) Expression of PD-L1 in the context of coexpression of (A) tumor-specific or (B) immune-specific markers. (C) Quantification of cellular abundance in tumor and immune cells, and their expression of PD-L1.
Figure 4.
Molecular phenotype of tumor and immune cell repertoire of sarcomatoid urothelial carcinoma (UC). (A) Phenotype of discovered cellular clusters based on the profiles of marker expression as assessed by Imaging Mass Cytometry. Clusters are grouped by broad cell type ontogeny. (B) Representation of cells in the sarcomatoid carcinoma case in which each single cell is labeled with the color of the phenotypic cluster from A.
Clinical Follow-Up to Date
The patient completed 30 cycles of pembrolizumab with no issues or new complains. The CT scan shows clean chest and stable abdominal disease.
DISCUSSION
UC has a propensity for divergent differentiation and the 2016 World Health Organization (WHO) classification of UCs lists 13 different histologic variants of urothelial cancer (Humphrey et al. 2016). However, there is a paucity of UC variants included in clinical trials resulting in low-level retrospective evidence for treatment recommendations.
SUCs show evidence of both epithelial and mesenchymal differentiation either histologically or by IHC and are believed to be the final common pathway for epithelial bladder tumors (Cheng et al. 2011). Sarcomatoid subtypes can also be seen in other tumor types. Reported data from sarcomatoid renal cell carcinoma (RCC) (Debien et al. 2020) and non-small-cell lung cancer (NSCLC) (Lou et al. 2016) show frequent expression of PD-L1 and high levels of tumor-infiltrating lymphocytes with an associated increased efficacy of PDL-1/PD-1 inhibition. Li et al. (2020) evaluated the PD-L1- and CD3-immune scores among invasive UC variants and found that cases of SUC demonstrate significantly higher PD-L1 scores as compared to conventional UC and other variants with high intratumoral CD3 scores.
IMC analysis in our case revealed similar patterns with a high number of infiltrating intratumoral CD3 cells, mainly CD8a+ T cells and with high PD-L1 expression in different cell types, mainly in pan-keratin-positive tumor cells. CD8a+ T cells are a crucial component of the cellular immune system that is important for cell-mediated antitumor immune response and independently associated with prolonged survival (Teng et al. 2015). PD-L1 expression on different cellular populations (tumor cells and tumor infiltrating immune cells) might be associated with improved response to PD-1/PD-L1 inhibitors in UC patients (Wen et al. 2019) like our patient.
Tumors with epithelial-to-mesenchymal transition (EMT) have been shown to be associated with resistance to conventional therapy (Singh and Settleman 2010; Lou et al. 2016). EMT is a mechanism by which cancer cells can invade and metastasize by altering cellular properties such as epithelial cell–cell junctions, cytoskeletal organization, and tumor interactions with the microenvironment. Mak et al. (2016) observed a consistent and strong positive correlation between the EMT score and expression levels of immune checkpoint genes (including PD1, cytotoxic T-lymphocyte-associated protein 4, and tumor necrosis factor [TNF] receptor superfamily member 4), across all cancer types, including bladder cancer, with mesenchymal tumors expressing higher levels. Pathway analysis of those cases demonstrated enrichment of leukocyte extravasation signaling, suggesting immune activation. A study of 28 SUCs evaluating the expression of EMT markers showed that the majority of cases expressed pro-EMT transcriptional factors including FoxC2, SNAIL, and ZEB1, as well as vimentin, coupled with cadherin switching from E-cadherin to N-cadherin (Sanfrancesco et al. 2016). Although we did not evaluate N-cadherin, our case suggested that EMT may be at play, with the vast majority of tumor cells expressing vimentin and low E-cadherin expression. These same cells showed high expression of PD-L1. This is notable as the tumor demonstrated a rapid recurrence following surgical resection and resistance to cytotoxic chemotherapy while showing an excellent response to immunotherapy.
Goodman et al. (2018) studied the prevalence of PD-L1 amplification in solid tumors, and amplifications were identified in 843 (0.7%) of the samples among more than 100 types of solid tumors. Most PD-L1-amplified tumors (84.8%) had a low (<5 Muts/Mb) to intermediate TMB (6–19 Muts/Mb), as in our case of low TMB (3.1 Muts/Mb). However, in contrast to our case, PD-L1 amplification did not correlate with high-positive PD-L1 expression by immunohistochemical analysis in the same study (Goodman et al. 2018), emphasizing the need for better biomarkers. Six of nine patients (6/9; 66.7%) with PDL1-amplified solid tumors that were treated with immunotherapy had objective responses after checkpoint blockade administration, including one case of conventional UC (progression free survival of ≥17.8 mo). In another study, PD-L1 amplification was more common in squamous cell carcinoma of the bladder and included only 17 cases (1%) of urothelial cancer (Necchi et al. 2020).
Conclusion
In conclusion, we present the first IMC analysis done in SUC to investigate the immune cell repertoire and PD-L1 expression on a single-cell level. The patient presented with metastatic SUC and experienced a prolonged response to the anti-PD1 immune checkpoint inhibitor pembrolizumab. Although the analysis on the metastatic tumor showed a low TMB, it demonstrated CD274 (PD-L1) amplification by NGS and 100% PD-L1 expression by IHC. IMC showed that most of the PD-L1 expression was present in tumor cells that showed evidence of EMT, as has been previously seen in SUC. The resistance of this tumor to conventional chemotherapy and sensitivity to immune checkpoint blockade suggests the importance of identifying genetic markers such as PD-L1 and others on a larger cohort of both UC and UC variants.
METHODS
This study was approved by the Institutional Review Board at Weill Cornell Medicine (Protocol # 1305013903—Research for Precision Medicine). Under this research protocol, patient provided written consent to be part of the study.
Tissue-Based NGS for Tumor Characterization
Protocols for DNA extraction and library preparation were followed, as previously described (Beg et al. 2021). NGS was performed by using the PGDx elio tissue complete RUO assay (Personal Genome Diagnostics, Inc.), a more than 500 gene panel targeted DNA-based assay that detects SNVs, small indels, translocations in 23 genes, amplifications in 28 genes, microsatellite instability, and TMB genomic signatures. The translocations detected by the assay are ALK, AXL, BRAF, BRCA1, BRCA2, EGFR, ETV4, ETV6, EWSR1, FGFR1, FGFR2, FGFR3, MYC, NTRK1, NTRK2, NTRK3, PAX8, PDGFRA, PDGFRAB, RAF1, RET ROS1, and TMPRSS2 (Supplemental Fig. 1). Samples were processed as previously described (Labriola et al. 2020). Formalin-fixed, paraffin-embedded (FFPE) tumor tissue from this patient was evaluated by study pathologists. Sample processing from tissue, library preparation, hybrid capture, and sequencing were performed at the PGDx laboratory. Samples were run on the PGDx elio tissue complete tumor profiling NGS assay, screening for variants in the aforementioned genes. Briefly, DNA was extracted from FFPE tissue, and following shearing, genomic libraries were prepared using end repair, A tailing and adapter ligation modules. Genomic libraries were amplified and captured in-solution, targeting the predefined regions of interest across full exonic regions. Captured libraries were sequenced on an Illumina NextSeq platform (Supplemental Table S1).
Plasma-Based NGS for cfDNA Characterization
Protocols for blood fractionation, plasma cfDNA extraction, and library preparation were followed as previously described (Al Zoughbi et al. 2021). NGS was performed by using the PGDx elio plasma resolve RUO assay (Personal Genome Diagnostics, PGDx), an assay that interrogates full coding regions of 33 genes, copy-number variation of eight genes, five translocations, and microsatellite instability (MSI) (Supplemental Fig. 2).
Antibody Panel Design and Validation for IMC
Preconjugated metal labeled antibodies were bought from Fluidigm, which were tested for specificity according to the company datasheet. The rest of the antibodies were custom-conjugated following manufacturer's protocol using the Maxpar X8 Multimetal Labeling Kit (Maxpar 201300). Newly conjugated antibodies were tested for staining specificity by staining control tissues such as lymph node. The PD-L1 clone SP142 was chosen based on its staining pattern that was close enough to the IHC clone used for scoring. A pathologist verifies the staining pattern for all antibodies before proceeding with actual staining of the tissue.
Imaging Mass Cytometry
Based on the clinical and pathological characteristics and quality of the preserved tissues, suitable representative fresh-cut 4-µm-thick FFPE sections were used for IMC staining. Slides were incubated for 1 h at 60°C on a slide warmer followed by dewaxing in fresh CitriSolv (Decon Labs) twice for 10 min, rehydrated in descending series of 100%, 95%, 80%, and 75% ethanol for 5 min each. After 5 min of MilliQ water wash, the slides were treated with antigen retrieval solution (Tris-EDTA pH 9.2) for 30 min at 96°C. Slides were cooled to room temperature, washed twice in TBS, and blocked for 1.5 h in SuperBlock Solution (Thermo Fisher), followed by overnight incubation at 4°C with the prepared antibody cocktail containing all metal-labeled antibodies (Table 3). The next day, slides were washed twice in 0.2% Triton X-100 in PBS and twice in TBS. DNA staining was performed using Intercalator-Iridium in PBS solution for 30 min in a humid chamber at room temperature. Slides were washed with MilliQ water and air-dried before ablation.
Table 3.
Full list of antibodies and their properties
| Antibody | Metal tag | Clone | Dilution | Stock concentration (mg/mL) | Vendor | Catalog # |
|---|---|---|---|---|---|---|
| Alpha-smooth muscle actin | Pr141 | 1A4 | 1:400 | 0.5 | Fluidigm | 3141017D |
| Interferon gamma | Nd142 | IFNG/466 | 1:50 | 0.5 | Abcam | ab218890 |
| CD206 | Nd143 | E2L9N | 1:100 | 0.2 | Cell Signaling Technology | 91992BF |
| IDO | Nd144 | D5J4E | 1:50 | 0.5 | Cell Signaling Technology | 86630BF |
| LAG3 | Nd145 | D2G40 | 1:50 | 0.5 | Cell Signaling Technology | 15372BF |
| CD163 | Sm147 | EDHu-1 | 1:100 | 0.5 | Fluidigm | 3147021D |
| CD14 | Nd148 | D7A2T | 1:50 | 0.5 | Cell Signaling Technology | 56082BF |
| PD1 | Nd150 | D4W2J | 1:50 | 0.5 | Cell Signaling Technology | 86163BF |
| CD45 | Sm152 | D9M8I | 1:100 | 0.5 | Fluidigm | 3152018D |
| CD44 | Eu153 | IM7 | 1:100 | 0.5 | BioLegend | 103001 |
| Vimentin | Sm154 | D21H3 | 1:200 | 0.5 | Fluidigm | 3154014A |
| FoxP3 | Gd155 | 236A/E7 | 1:25 | 0.5 | Abcam | ab96048 |
| CD4 | Gd156 | OTI5D9 | 1:50 | 0.5 | Novus Biologicals | NBP2-70357 |
| E-cadherin | Gd158 | 2.40E + 11 | 1:100 | 0.5 | Fluidigm | 3158029D |
| CD68 | Tb159 | KP1 | 1:50 | 0.5 | Abcam | ab233172 |
| CD1a | Gd160 | O10 | 1:50 | 0.5 | Abcam | ab212980 |
| CD20 | Dy161 | L26 | 1:200 | 0.5 | Novus Biologicals | NBP2-80486 |
| CD8a | Dy162 | C8/144B | 1:100 | 0.5 | eBioscience | 14-0085-82 |
| CXCL12 | Dy163 | D7A2T | 1:50 | 0.5 | Abcam | 97958BF |
| Pan-keratin | Dy164 | AE1/AE3 | 1:100 | 0.5 | Abcam | ab80826 |
| HLADR | Ho165 | EPR3692 | 1:50 | 0.5 | Abcam | ab215985 |
| IL-1beta | Er166 | 3A6 | 1:50 | 0.5 | Cell Signaling Technology | 12242BF |
| Granzyme B | Er167 | GB11 | 1:50 | 0.5 | Fluidigm | 3167023D |
| Ki67 | Er168 | B56 | 1:50 | 0.5 | Fluidigm | 3168022D |
| ColTypeI | Tm169 | Polyclonal | 1:300 | 0.5 | Fluidigm | 3169023D |
| CD3 | Er170 | Polyclonal, carboxy-terminal | 1:100 | 0.5 | Fluidigm | 3170019D |
| TIM3 | Yb171 | D5D5R | 1:50 | 0.5 | Cell Signaling Technology | 45208BF |
| CD56 | Yb172 | 3H15L12 | 1:50 | 0.5 | Invitrogen | 701379 |
| CD45RO | Yb173 | UCHL1 | 1:100 | 0.5 | Fluidigm | 3173016D |
| HLAClassI | Yb174 | EMR8-5 | 1:50 | 0.5 | Abcam | ab70328 |
| PDL1 | Lu175 | SP142 | 1:50 | 0.5 | Abcam | ab236238 |
| CD11c | Yb176 | EP1347Y | 1:50 | 0.5 | Abcam | ab216655 |
| DNA1 | Ir191 | 1:400 | Fluidigm | 201192A | ||
| DNA2 | Ir193 | 1:400 | Fluidigm | 201192A |
The instrument was calibrated using a tuning slide to optimize the sensitivity of the detection range. Hematoxylin and eosin–stained slides were used to guide the selection of four regions of interest based on the pathological examination of the tissue. Scanning protocol was set for 200 Hz frequency and the raw MCD file was exported for downstream data analysis.
Preprocessing of IMC Data
IMD data was preprocessed as described previously (Zanotelli and Bodenmiller 2017), with some modifications. Briefly, image data was extracted from MCD files acquired with the Fluidigm Hyperion instrument. Hot pixels were removed using a fixed threshold, the image was amplified two times, and Gaussian smoothing applied. From each image a square 500-pixel crop was saved as a HDF5 file for image segmentation.
Image segmentation was performed with ilastik (Berg et al. 2019) (version 1.3.3) by manually labeling pixels as belonging to one of three classes: nuclei; cytoplasm—as the area immediately surrounding the nuclei; and background—as pixels with low signal across all channels. Outputs probabilities were used to segment the image using CellProfiler (Carpenter et al. 2006) (version 3.1.8) with the IdentifyPrimaryObjects module, and followed by the “IdentifySecondaryObjects” module in which identified nuclei are used to seed an expansion of the cell area to generate a cellular mask.
Analysis of IMC Data
We assessed the quality of each acquired channel as described previously (Rendeiro et al. 2021) and quantified single cells by averaging signal intensity of each channel in the area of each cell in the cell mask. We used Scanpy (Wolf et al. 2018) (version 1.5.0) to process the single-cell data. Values were log1p transformed and normalized in order for the sum of each cell to equal 1 × 104. Cell type assignment was performed by computing a neighbor graph on this standardized and centered matrix with eight neighbors per cell and by clustering the cells with the Leiden algorithm (Traag et al. 2019) in the Python implementation (leidenalg, version 0.8.0), with 0.75 as resolution parameter. To derive data-driven labels for each cluster, we used the mean intensity of each cluster in each channel, centered and scaled in both dimensions, averaged for both (Wisconsin double transformation). To those values, we fit a Gaussian Mixture model (scikit-learn, version 0.23.0) with two classes and used the decision boundary to label each cluster above that value as positive for that marker. The union of positive markers for a given cluster was used as a label for the cluster. To declare cells as positive for a certain marker, we also used a Gaussian mixture model with two classes except the ones run for each marker using the log-transformed intensity matrix.
The following additional software versions were used: Python version 3.8.2, numpy version 1.18.3, scipy version 1.4.1, and scikit-image 0.17.2.
ADDITIONAL INFORMATION
Data Deposition and Access
IMC data can be found at the following publicly accessible repository: https://doi.org/10.5281/zenodo.6251220. Source code used to analyze IMC data are available at https://github.com/ElementoLab/msuc-imc. Additional data that support the findings of this study are available on request from the corresponding author (J.M.M.). Interpreted variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV002500974–SCV002500987.
Ethics Statement
This study was approved by the Institutional Review Board (Protocol # 1305013903). Written informed consent was obtained from the patient for publication of this case report and any accompanying images.
Acknowledgments
We thank Christen Vinson and Leandra Slayton from Personal Genome Diagnostics, Inc. for their significant discussion of the manuscript.
Author Contributions
J.M.M. devised the project, the main conceptual ideas and proof outline. H.R. performed IMC experiments. A.F.R. analyzed IMC data. H.A. wrote the manuscript with input from all authors. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by the Caryl and Israel Englander Institute for Precision Medicine. A.F.R. is supported by a T32 grant from the National Cancer Institute. This work was supported in part by the Cornell Center for Immunology Core Facilities Seed Grant (PrincipaI Investigator, J.M.M.; Co-PrincipaI Investigator, K.O.; and Co-Investigators, O.E. and B.F.).
Competing Interest Statement
J.M.M. had a research collaboration agreement with Personal Genome Diagnostics Inc. during 2019–2020.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Al Zoughbi W, Fox J, Beg SB, Papp E, Hissong E, Ohara K, Keefer L, Sigouros M, Kane T, Bockelman D, et al. 2021. Validation of a ctDNA-based next-generation sequencing assay in a cohort of solid tumor patients: a proposed solution for decentralized plasma testing. Oncologist 26: e1971–e1981. 10.1002/onco.13905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg S, Bareja R, Ohara K, Eng KW, Wilkes DC, Pisapia DJ, Al Zoughbi W, Kudman S, Zhang W, Rao R, et al. 2021. Integration of whole-exome and anchored PCR-based next generation sequencing significantly increases detection of actionable alterations in precision oncology. Transl Oncol 14: 1. 10.1016/j.tranon.2020.100944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. 2019. ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16: 1226–1232. 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang S, Alexander R, MacLennan GT, Hodges KB, Harrison BT, Lopez-Beltran A, Montironi R. 2011. Sarcomatoid carcinoma of the urinary bladder: the final common pathway of urothelial carcinoma dedifferentiation. Am J Surg Pathol 35: e34–e36. 10.1097/PAS.0b013e3182159dec [DOI] [PubMed] [Google Scholar]
- Debien V, Thouvenin J, Lindner V, Barthélémy P, Lang H, Flippot R, Malouf GG. 2020. Sarcomatoid dedifferentiation in renal cell carcinoma: from novel molecular insights to new clinical opportunities. Cancers (Basel) 12: 99. 10.3390/cancers12010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis M, Bellmunt J, Mead G, Kerst JM, Leahy M, Maroto P, Gil T, Marreaud S, Daugaard G, Skoneczna I, et al. 2012. Randomized phase II/III trial assessing gemcitabine/carboplatin and methotrexate/carboplatin/vinblastine in patients with advanced urothelial cancer who are unfit for cisplatin-based chemotherapy: eORTC study 30986. J Clin Oncol 30: 191–199. 10.1200/JCO.2011.37.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet Y, Bellmunt J, Vaughn DJ, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, Necchi A, et al. 2019. Randomized phase III KEYNOTE-045 trial of pembrolizumab versus paclitaxel, docetaxel, or vinflunine in recurrent advanced urothelial cancer: results of >2 years of follow-up. Ann Oncol 30: 970–976. 10.1093/ANNONC/MDZ127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakis G. 2014. The role of inflammation in bladder cancer. Adv Exp Med Biol 816: 183–196. 10.1007/978-3-0348-0837-8_8 [DOI] [PubMed] [Google Scholar]
- Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11: 417–422. 10.1038/NMETH.2869 [DOI] [PubMed] [Google Scholar]
- Goodman AM, Piccioni D, Kato S, Boichard A, Wang HY, Frampton G, Lippman SM, Connelly C, Fabrizio D, Miller V, et al. 2018. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 4: 1237–1244. 10.1001/jamaoncol.2018.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman HB, Natale RB, Tangen CM, Speights VO, Vogelzang NJ, Trump DL, deVere White RW, Sarosdy MF, Wood DP, Raghavan D, et al. 2003. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med 349: 859–866. 10.1056/NEJMoa022148 [DOI] [PubMed] [Google Scholar]
- Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. 2016. The 2016 WHO classification of tumours of the urinary system and male genital organs—part B: prostate and bladder tumours. Eur Urol 70: 106–119. 10.1016/j.eururo.2016.02.028 [DOI] [PubMed] [Google Scholar]
- Labriola MK, Zhu J, Gupta R, McCall S, Jackson J, Kong EF, White JR, Cerqueira G, Gerding K, Simmons JK, et al. 2020. Characterization of tumor mutation burden, PD-L1 and DNA repair genes to assess relationship to immune checkpoint inhibitors response in metastatic renal cell carcinoma. J Immunother Cancer 8: e000319. 10.1136/jitc-2019-000319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Q, Shuman L, Kaag M, Raman JD, Merrill S, DeGraff DJ, Warrick JI, Chen G. 2020. Evaluation of PD-L1 and other immune markers in bladder urothelial carcinoma stratified by histologic variants and molecular subtypes. Sci Rep 10: 1439. 10.1038/s41598-020-58351-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Diao L, Cuentas ERP, Denning WL, Chen L, Fan YH, Byers LA, Wang J, Papadimitrakopoulou VA, Behrens C, et al. 2016. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res 22: 3630–3642. 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. 2016. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res 22: 609–620. 10.1158/1078-0432.CCR-15-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi J, Wada Y, Matsumoto K, Azuma M, Kikuchi K, Ueda S. 2007. Overexpression of B7-H1 (PD-L1) significantly associates with tumor grade and postoperative prognosis in human urothelial cancers. Cancer Immunol Immunother 56: 1173–1182. 10.1007/s00262-006-0266-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necchi A, Madison R, Raggi D, Jacob JM, Bratslavsky G, Shapiro O, Elvin JA, Vergilio JA, Killian JK, Ngo N, et al. 2020. Comprehensive assessment of immuno-oncology biomarkers in adenocarcinoma, urothelial carcinoma, and squamous-cell carcinoma of the bladder. Eur Urol 77: 548–556. 10.1016/j.eururo.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Powles T, Park SH, Voog E, Caserta C, Valderrama BP, Gurney H, Kalofonos H, Radulovic S, Demey W, Ullén A, et al. 2020. Maintenance avelumab + best supportive care (BSC) versus BSC alone after platinum-based first-line (1L) chemotherapy in advanced urothelial carcinoma (UC): JAVELIN Bladder 100 phase III interim analysis. J Clin Oncol 38: LBA1. 10.1200/jco.2020.38.18_suppl.lba1 [DOI] [Google Scholar]
- Rendeiro AF, Ravichandran H, Bram Y, Chandar V, Kim J, Meydan C, Park J, Foox J, Hether T, Warren S, et al. 2021. The spatial landscape of lung pathology during COVID-19 progression. Nature 593: 564–569. 10.1038/s41586-021-03475-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfrancesco J, McKenney JK, Leivo MZ, Gupta S, Elson P, Hansel DE. 2016. Sarcomatoid urothelial carcinoma of the bladder: analysis of 28 cases with emphasis on clinicopathologic features and markers of epithelial-to-mesenchymal transition. Arch Pathol Lab Med 140: 543–551. 10.5858/arpa.2015-0085-OA [DOI] [PubMed] [Google Scholar]
- Singh A, Settleman J. 2010. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29: 4741–4751. 10.1038/onc.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg CN, De Mulder P, Schornagel JH, Theodore C, Fossa SD, Van Oosterom AT, Witjes JA, Spina M, Van Groeningen CJ, Duclos B, et al. 2006. Seven year update of an EORTC phase III trial of high-dose intensity M-VAC chemotherapy and G-CSF versus classic M-VAC in advanced urothelial tract tumours. Eur J Cancer 42: 50–54. 10.1016/j.ejca.2005.08.032 [DOI] [PubMed] [Google Scholar]
- Teng MWL, Ngiow SF, Ribas A, Smyth MJ. 2015. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res 75: 2139–2145. 10.1158/0008-5472.CAN-15-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2013. Comprehensive molecular characterization of urothelial bladder carcinoma: The Cancer Genome Atlas Research Network. Nature 507: 315–322. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9: 5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Plimack ER. 2018. Immunotherapy for urothelial carcinoma: current evidence and future directions. Curr Urol Rep 19: 109. 10.1007/s11934-018-0851-7 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang FW, Lagrange CA, Hemstreet GP Iii, Kessinger A. 2010. Clinical features of sarcomatoid carcinoma (carcinosarcoma) of the urinary bladder: analysis of 221 cases. Sarcoma 2010: 454792. 10.1155/2010/454792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Chen Y, Duan X, Zhu W, Cai C, Deng T, Zeng G. 2019. The clinicopathological and prognostic value of PD-L1 in urothelial carcinoma: a meta-analysis. Clin Exp Med 19: 407–416. 10.1007/s10238-019-00572-9 [DOI] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JL, Black PC, Brown GA, Porter MP, Kamat AM, Dinney CP, Lin DW. 2007. Differences in survival among patients with sarcomatoid carcinoma, carcinosarcoma and urothelial carcinoma of the bladder. J Urol 178: 2302–2307. 10.1016/j.juro.2007.08.038 [DOI] [PubMed] [Google Scholar]
- Zanotelli VRT, Bodenmiller B. 2017. ImcSegmentationPipeline: a pixelclassification based multiplexed image segmentation pipeline. 10.5281/ZENODO.3841961 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
IMC data can be found at the following publicly accessible repository: https://doi.org/10.5281/zenodo.6251220. Source code used to analyze IMC data are available at https://github.com/ElementoLab/msuc-imc. Additional data that support the findings of this study are available on request from the corresponding author (J.M.M.). Interpreted variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV002500974–SCV002500987.