Abstract
Li–Fraumeni syndrome (LFS) is one of the most common cancer predisposition syndromes that affects both children and adults. Individuals with LFS are at an increased risk of developing various types of cancer over their lifetime including soft tissue sarcomas, osteosarcomas, breast cancer, leukemia, brain tumors, and adrenocortical carcinoma. Heterozygous germline pathogenic variants in the tumor suppressor gene TP53 are the known causal genetic defect for LFS. Single-nucleotide variants (SNVs) including missense substitutions that occur in the highly conserved DNA binding domain of the protein are the most common alterations, followed by nonsense and splice site variants. Gross copy-number changes in TP53 are rare and account for <1% of all variants. Using next-generation sequencing (NGS) panels, we identified a paternally inherited germline intragenic duplication of TP53 in a child with metastatic osteosarcoma who later developed acute myeloid leukemia (AML). Transcriptome sequencing (RNA-seq) demonstrated the duplication was tandem, encompassing exons 2–6 and 28 nt of the untranslated region (UTR) upstream of the start codon in exon 2. The inclusion of the 28 nt is expected to result in a frameshift with a stop codon 18 codons downstream from the exon 6, leading to a loss-of-function allele. This case highlights the significance of simultaneous identification of both significant copy-number variants as well as SNVs/indels using NGS panels.
Keywords: acute myeloid leukemia, osteosarcoma
CASE PRESENTATION
The patient was a 15-yr-old male who was first diagnosed with metastatic osteosarcoma and later developed acute myeloid leukemia (AML). He presented with leg pain and difficulty walking at the age of 14. X-ray examination showed a tumor of the right proximal tibia. Magnetic resonance imaging (MRI) displayed a large aggressive tumor mass infiltrating the metadiaphyseal region with aggressive periosteal reaction and cortical destruction. The tumor extended into surrounding muscles and soft tissues. Positron emission tomography (PET) scan exhibited skip metastasis in the mid-right tibial shaft and external iliac node. Histologic evaluation showed proliferation of large pleomorphic cells with enlarged, hyperchromatic nuclei and frequent atypical mitotic figures associated with osteoid matrix findings, consistent with a diagnosis of osteosarcoma (Fig. 1A). After initial chemotherapy, the patient was transferred to Children's Hospital of Philadelphia (CHOP) for local surgical management and concurrent chemotherapy. Seven months from the final dose of the chemotherapy, the patient was found to have pancytopenia with peripheral blasts. Bone marrow biopsy found sheets of blasts with irregular nuclei, speckled chromatin, and prominent nucleoli (Fig. 1B). Immunohistochemical stains and flow cytometry showed that the blasts were positive for CD33, MPO, lysozyme, CD117, and CD34, indicating a diagnosis of AML.
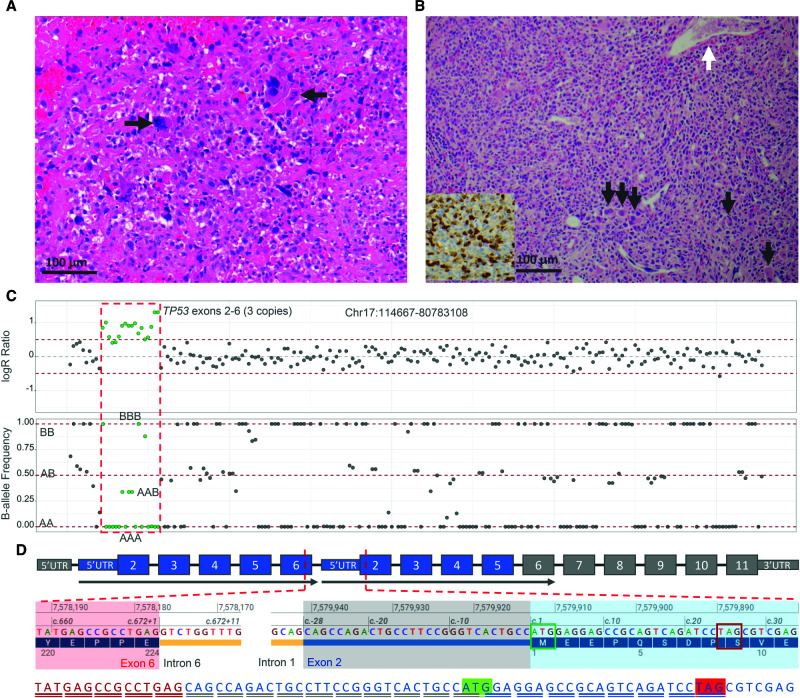
Figure 1.
(A) Histologic section of the osteosarcoma prior to chemotherapy stained with hematoxylin and eosin demonstrates proliferation of large pleomorphic cells with enlarged, hyperchromatic nuclei, and frequent atypical mitotic figures (arrows) associated with osteoid matrix (arrowhead). (B) Bone marrow biopsy shows hypercellular marrow for age (>95%) with variably sized atypical mononuclear cells with irregular nuclei and ample cytoplasm. Maturing myeloid and erythroid elements are markedly decreased. Small hypolobated dysplastic megakaryocytes are seen throughout the marrow (black arrows), and underlying fibrosis is suggested by the overall streaming pattern of the cells and the dilated bone marrow sinuses (white arrow). An immunohistochemical stain for myeloperoxidase is positive in a subset of the atypical cells (inset). (C) Next-generation sequencing (NGS) copy-number analysis displays a heterozygous duplication of partial TP53 in the patient's blood sample. The green dots in the logR ratio plot represent the TP53 exon 2–6 duplication with increased logR ratio. The gray dots represent the rest of Chr 17 with normal logR ratio. In the B-allele frequency plot, green dots show the single-nucleotide polymorphism (SNP) patterns of three copies of the duplication as AAA, AAB, ABB, and BBB, whereas the rest of the SNPs in gray are two copies delineated as AA, BB, or AB alleles. (D) Schematic illustration of the tandem duplication of TP53 exons 2–6. Partial cDNA sequence identified by RNA-seq is illustrated in red with red underline (exon 6), blue with gray underline (28 nt of 5′ UTR upstream of the start codon of exon 2), and blue with blue underline (exon 2). The start codon is marked in green, and the stop codon is marked in red.
TECHNICAL ANALYSIS
Next-Generation Sequencing Panels
The CHOP Comprehensive Hematological Malignancy Panel (CHMP) evaluates 117 genes associated with pediatric hematological malignancies (Surrey et al. 2019). CHOP Comprehensive Hereditary Cancer Panel (CHCP) is a custom-designed next-generation sequencing (NGS) panel that interrogates 130 genes associated with cancer predisposition. Both panels cover all coding exons and at least 10 bp of flanking intronic sequences, certain promoter regions, and known pathogenic intronic variants. An additional 1038 common single-nucleotide polymorphisms (SNPs) were added to the CHMP and CHCP to mimic a low-density SNP array to facilitate CNV analysis. All custom-designed probes were synthesized and biotinylated to allow for target enrichment using streptavidin-conjugated beads (Agilent Technologies). An Illumina RNA Exome kit was utilized to capture the transcriptome and generate RNA-seq libraries (Illumina). Libraries were sequenced using Illumina Hiseq 2500 with 150-bp paired-end reads. Sequence data were analyzed using a homebrew NGS Analysis Pipeline, named ConcordR V1.4.2, which utilizes RSEM package for gene expression evaluation and for calculation of fold changes for gene expression levels (Li and Dewey 2011).
The CHCP test on the patient's blood sample identified a heterozygous duplication of exons 2–6 of the TP53 gene (NM_000546.5) (Fig. 1C) along with two pathogenic SNVs, each in the HFE and SBDS genes (Table 1). RNA-seq demonstrated that the TP53 duplication was tandem involving exons 2–6 and the mRNA transcripts contained 28 nt of the 5′ UTR located upstream of the start codon in exon 2 (Fig. 1D). RNA-seq also discovered reduced TP53 expression in blood cells (fold change: 69% compared to the control). Because of the later development of AML, a CHMP test was performed and disclosed a near-triploid genome with copy-number gains involving almost every chromosome. Copy-neutral loss of heterozygosity was observed for Chromosomes 7 and 17p, including the TP53 gene, which showed four copies of exons 2–6 (data not shown). A somatic frameshift variant in the NF1 gene was observed (Table 1). TP53 multiplex ligation-dependent probe amplification (MLPA) analysis on family members revealed that the TP53 duplication was found in the 46-yr-old asymptomatic father, suggesting the paternal inheritance.
Table 1.
Clinically significant variants identified by CHOP NGS panels
| Gene | Chr | Transcripts | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | Allele fraction | Germline/somatic | Target Coverage |
|---|---|---|---|---|---|---|---|---|---|
| TP53 | 17 | NM_000546 | c.28_672dup | p.? | CNV | LoF | Nonmosaic | Germline | 811× |
| HFE | 6 | NM_000410 | c.187c > g | p.His63Asp | SNV | Missense | 48% | Germline | 562× |
| SBDS | 7 | NM_016038 | c.258 + 2T > C | p.? | Indel | Splicing | 40% | Germline | 345× |
| NF1 | 17 | NM_001042492 | c.3884_3887dup | p.Gln1298Serfs*17 | Dup | Frameshift | 64% | Somatic | 616× |
(Chr) Chromosome, (CHOP) Children's Hospital of Philadelphia, (HGVS) Human Genome Variation Society, (NGS) next-generation sequencing, (CNV) copy-number variant, (SNV) single-nucleotide variant, (LoF) loss of function, (Dup) duplicate.
VARIANT INTERPRETATION
The tandem duplication of TP53 exons 2–6 identified in this patient led to an extra copy of exons 2–6 and the inclusion of 28 nt of the 5′ UTR in mRNA transcripts. This inclusion is expected to generate a stop codon 18 codons downstream from the exon 6, resulting in a loss-of-function allele. The reduced expression of TP53 in blood tissue is presumed to be the result of nonsense-mediated mRNA decay (Chang et al. 2007). These findings, as well as developing osteosarcoma and AML, support the diagnosis of Li–Fraumeni syndrome (LFS). The patient is also a carrier of two pathogenic variants, HFE p.His63Asp (low penetrance allele) and SBDS c.258 + 2T > C, associated with autosomal recessive hereditary hemochromatosis and Shwachman–Diamond syndrome, respectively.
SUMMARY
Here we report the first fully characterized tandem duplication of exons 2–6 of TP53 including part of the 5′ UTR in a pediatric patient with two different cancers using CHOP NGS panels, leading to the diagnosis of LFS. An exon 2–6 tandem duplication of the APC gene including 18 nucleotides of the 5′ UTR was reported in a patient with familial adenomatous polyposis (Pedace et al. 2008). The inclusion of part of the 5′ UTR introduced a premature stop codon, leading to a loss of function allele and a truncated protein. In addition, the TP53 tandem duplication in our case potentially affects multiple important p53 functional domains including the p53 tumor suppressor family domain and DNA-binding domain. The duplication was also identified in his asymptomatic father by MLPA analysis. TP53-related LFS is inherited in an autosomal dominant manner. The core LFS cancers include osteosarcomas, soft-tissue sarcomas, and central nervous system (CNS) tumors in children and breast carcinoma in adults. Missense substitutions in TP53 concentrated in the highly conserved DNA binding domain of the protein are the most reported alterations, followed by nonsense and splice-site mutations (Varley et al. 1997; Olivier et al. 2003; Varley 2003; Ruijs et al. 2010; Giacomazzi et al. 2013; Guha and Malkin 2017; Kratz et al. 2021). Additionally, identification of TP53 exons 2–4 duplication by array CGH was previously reported in an individual with LFS (Aury-Landas et al. 2013). LFS with secondary malignancy has been reported due to radiation therapy or chemotherapy (Shimatani et al. 2019; Hendrickson et al. 2020). Our patient had no prior radiation therapy, and his chemotherapy did not include alkylating agents and topoisomerase II inhibitors. His development of AML occurred ∼1 yr after the onset of chemotherapy, suggesting a second primary cancer. However, it could also be secondary leukemia resulting from the cooperation of cytotoxic osteosarcoma chemotherapy and the underlying cancer predisposition. The complexity of the genetic alterations and the biallelic loss of TP53 in the leukemia cells portend a poor prognosis. Stem cell transplantation is currently being considered as a potential cure. This case also highlights the importance of combined identification of both significant copy-number variants as well as SNVs/indels using NGS panels.
ADDITIONAL INFORMATION
Database Deposition and Access
The interpreted variant has been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV002038503, SCV002038504, and SCV002038505. The patient did not provide consent for public deposition of all raw sequencing data.
Ethics Statement
A case report does not constitute human subjects’ research. Per CHOP policy, it therefore does not require institutional review board (IRB) review. Informed consent was obtained from the legal guardian for collection of clinical information and samples. This study was conducted in accordance with the Declaration of Helsinki.
Author Contributions
M.M.L., Y.Z., and F.X. designed the study. F.X., E.A.-E., J.S., M.M.L., and Y.Z. wrote the manuscript. F.X., J.W., J.S., M.P., K.C., A.L., Z.F., E.H.D., E.A.F., D.M.W., M.L., and L.K.C. collected and analyzed the data. G.W., T.B., J.P., A.S.D., K.Z., S.B., N.B., and S.M. provided clinical data. All authors reviewed the manuscript.
Funding
The study is partially supported by the Department of Pathology and Laboratory Medicine and Center for Childhood Cancer Research, Children's Hospital of Philadelphia.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Raymond Kim
Mahesh M Mansukhani
REFERENCES
- Aury-Landas J, Bougeard G, Castel H, Hernandez-Vargas H, Drouet A, Latouche J-B, Schouft M-T, Férec C, Leroux D, Lasset C. 2013. Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li–Fraumeni syndrome with brain tumours. Eur J Hum Genet 21: 1369–1376. 10.1038/ejhg.2013.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-F, Imam JS, Wilkinson MF. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74. 10.1146/annurev.biochem.76.050106.093909 [DOI] [PubMed] [Google Scholar]
- Giacomazzi J, Selistre S, Duarte J, Ribeiro JP, Vieira PJ, de Souza Macedo G, Rossi C, Czepielewski M, Netto CBO, Hainaut P. 2013. TP53 p.R337H is a conditional cancer-predisposing mutation: further evidence from a homozygous patient. BMC Cancer 13: 1–8. 10.1186/1471-2407-13-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha T, Malkin D. 2017. Inherited TP53 mutations and the Li–Fraumeni syndrome. Cold Spring Harb Perspect Med 7: a026187. 10.1101/cshperspect.a026187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Luo Y, Kohlmann W, Schiffman J, Maese L, Bishop AJ, Lloyd S, Kokeny KE, Hitchcock YJ, Poppe MM. 2020. Radiation therapy and secondary malignancy in Li–Fraumeni syndrome: a hereditary cancer registry study. Cancer Med 9: 7954–7963. 10.1002/cam4.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz CP, Freycon C, Maxwell KN, Nichols KE, Schiffman JD, Evans DG, Achatz MI, Savage SA, Weitzel JN, Garber JE. 2021. Analysis of the Li–Fraumeni spectrum based on an international germline TP53 variant data set: an International Agency for Research on Cancer TP53 database analysis. JAMA Oncol 7: 1800–1805. 10.1001/jamaoncol.2021.4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 1–16. 10.1186/1471-2105-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Goldgar DE, Sodha N, Ohgaki H, Kleihues P, Hainaut P, Eeles RA. 2003. Li–Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res 63: 6643–6650. [PubMed] [Google Scholar]
- Pedace L, Majore S, Megiorni F, Binni F, De Bernardo C, Antigoni I, Preziosi N, Mazzilli MC, Grammatico P. 2008. Identification of a novel duplication in the APC gene using multiple ligation probe amplification in a patient with familial adenomatous polyposis. Cancer Genet Cytogenet 182: 130–135. 10.1016/j.cancergencyto.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Ruijs MW, Verhoef S, Rookus MA, Pruntel R, van der Hout AH, Hogervorst FB, Kluijt I, Sijmons RH, Aalfs CM, Wagner A. 2010. TP53 germline mutation testing in 180 families suspected of Li–Fraumeni syndrome: mutation detection rate and relative frequency of cancers in different familial phenotypes. J Med Genet 47: 421–428. 10.1136/jmg.2009.073429 [DOI] [PubMed] [Google Scholar]
- Shimatani A, Aono M, Hoshi M, Oebisu N, Iwai T, Takada N, Hara J, Nitani C, Nakamura H. 2019. Secondary osteosarcoma in patients previously treated for childhood cancer: three case reports. Mol Clin Oncol 10: 153–158. 10.3892/mco.2018.1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey LF, MacFarland SP, Chang F, Cao K, Rathi KS, Akgumus GT, Gallo D, Lin F, Gleason A, Raman P. 2019. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med 11: 1–14. 10.1186/s13073-019-0644-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley J. 2003. Germline TP53 mutations and Li–Fraumeni syndrome. Hum Mutat 21: 313–320. 10.1002/humu.10185 [DOI] [PubMed] [Google Scholar]
- Varley J, Evans D, Birch JM. 1997. Li–Fraumeni syndrome—a molecular and clinical review. Br J Cancer 76: 1. 10.1038/bjc.1997.328 [DOI] [Google Scholar]