Abstract
The megalencephaly-capillary malformation (MCAP) syndrome is an overgrowth disorder caused by mosaic gain-of-function variants in PIK3CA. It is characterized by megalencephaly or hemimegalencephaly, vascular malformations, somatic overgrowth, among other features. Epilepsy is commonly associated with MCAP, and a subset of individuals have cortical malformations requiring resective epilepsy surgery. Like other mosaic disorders, establishing a molecular diagnosis is largely achieved by screening lesional tissues (such as brain or skin), with a low diagnostic yield from peripheral tissues (such as blood). Therefore, in individuals with MCAP in whom lesional tissues are scarce or unavailable or those ineligible for epilepsy surgery, establishing a molecular diagnosis can be challenging. Here we report on the utility of cerebrospinal fluid (CSF)-derived cfDNA for the molecular diagnosis of an individual with MCAP syndrome harboring a mosaic PIK3CA variant (c.3139C > T, p.His1047Tyr). The proband presented with asymmetric megalencephaly without significant dysgyria. He did not have refractory epilepsy and was therefore not a candidate for epilepsy surgery. However, he developed diffuse large B-cell lymphoma (DLBCL) in late childhood, with four CSF samples obtained via lumbar puncture for cancer staging during which one sample was collected for cfDNA extraction and sequencing. PIK3CA variant allele fractions in CSF cell-free DNA (cfDNA), skin fibroblasts, and peripheral blood were 3.08%, 37.31%, and 2.04%, respectively. This report illustrates the utility of CSF-derived cfDNA in MCAP syndrome. Minimally invasive–based molecular diagnostic approaches utilizing cfDNA not only facilitate accurate genetic diagnosis but also have important therapeutic implications for individuals with refractory epilepsy as repurposed PI3K-AKT-MTOR pathway-inhibitors become more widely available.
Keywords: hemimegalencephaly, macrocephaly at birth, overgrowth
INTRODUCTION
The PIK3CA-related megalencephaly-capillary malformation (MCAP) syndrome (MIM #602501) is a multisystem overgrowth disorder caused by mosaic gain-of-function (activating) variants in PIK3CA (MIM #171834). The most common features of MCAP include diffuse or focal brain overgrowth (i.e., megalencephaly [MEG] or hemimegalencephaly [HMEG]), cortical abnormalities (predominantly polymicrogyria and focal cortical dysplasia), vascular malformations, digital anomalies (cutaneous syndactyly, polydactyly), and other skin and connective tissue abnormalities (Fig. 1; Dobyns and Mirzaa 2019). Activating PIK3CA variants cause a wide range of overgrowth phenotypes, collectively termed PIK3CA-related overgrowth spectrum (PROS). Given that these disorders are caused by mosaic variants, the yield from molecular diagnostic testing is higher when affected or lesional tissues are available for testing (Yeung et al. 2017). Previous studies have shown that affected tissues (e.g., skin fibroblast) have a higher diagnostic yield than peripheral blood or saliva (Mirzaa et al. 2016; Kuentz et al. 2017). Therefore, obtaining affected tissues in PROS is important for establishing an accurate molecular diagnosis.
Figure 1.
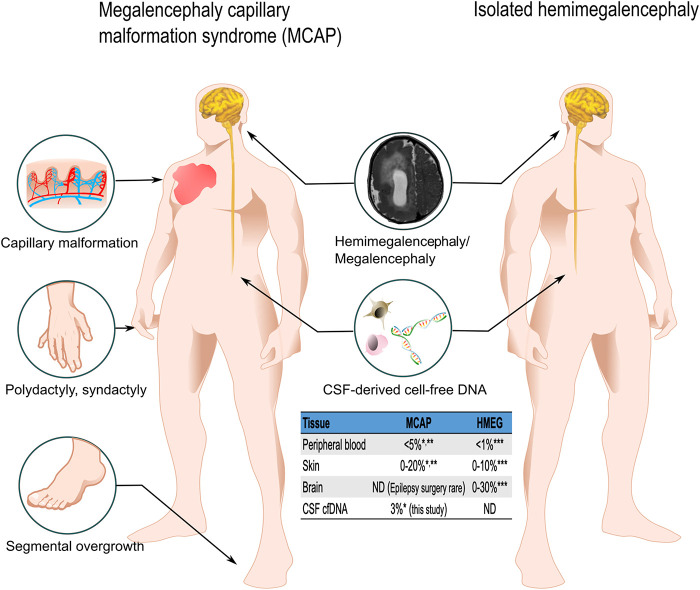
Phenotypes and variant allele fraction (VAF) in megalencephaly-capillary malformation (MCAP) and hemimegalencephaly. MCAP syndrome is characterized by megalencephaly/hemimegalencephaly and cutaneous capillary malformation with focal segmental overgrowth. This figure illustrates the phenotypical differences between MCAP syndrome and isolated hemimegalencephaly (HMEG). Although both syndromes can be caused by the same variants in PIK3CA, the distribution of the variant results in different phenotypes and molecular diagnostic yields accordingly. MCAP syndrome is caused by PIK3CA genetic variants with wider tissue distribution; thus, it can be diagnosed by sequencing affected tissues (skin) or, less reliably, peripheral blood. The table shows diagnostic yields from different tissues. However, in cases with isolated HMEG, affected brain tissue is required for molecular diagnostics because peripheral blood and skin samples have a very low yield. When cerebrospinal fluid (CSF) cell-free DNA (cfDNA) is available, hypothetically molecular diagnosis can be achieved without affected brain tissues in isolated HMEG. (ND) No data. *Mirzaa et al. 2016; **Kuentz et al. 2017; ***Pirozzi et al. 2021.
Among the neurological phenotypes that fall under PROS, PIK3CA mutational hotspots (notably variants c.1624G > A p.Glu542Lys, c.1633G > A p.Glu545Lys, c.3140A > T p.His1047Leu, and c.3140A > G p.His1047Arg) can be associated with more severe brain phenotypes such as focal cortical dysplasia (FCD), HMEG, and dysplastic megalencephaly (DMEG) with or without severe segmental body overgrowth, whereas less-activating somatic variants cause MCAP that is most often characterized by diffuse megalencephaly and polymicrogyria (PMG) (Mirzaa et al. 2015). Among the common comorbidities associated with these PIK3CA-related brain phenotypes is epilepsy. About 30% of individuals with MCAP have seizures and refractory epilepsy is not uncommon, especially in those with cortical dysplasia (Mirzaa et al. 2012; Jansen et al. 2015).
Establishing a molecular diagnosis early in individuals with PIK3CA-related brain phenotypes is not only helpful to better understand the disorder and its prognosis, but it can also have important therapeutic implications, especially as PI3K-AKT-MTOR pathway inhibitors are beginning to show promising results in treating epilepsy and neuropsychiatric disorders in association with other disorders within this pathway such as the tuberous sclerosis complex (TSC) (French et al. 2016; Kilincaslan et al. 2017). Select MTOR, AKT, or PI3K inhibitors have been proposed as therapeutic options for treating refractory epilepsy in children with activating mutations of this pathway. However, for individuals not eligible for epilepsy surgery, establishing the molecular cause to determine whether these molecularly targeted therapies can be used poses an important diagnostic challenge (Mirzaa et al. 2015). Therefore, alternative approaches for detecting mosaic variants are warranted, especially for those with brain-restricted mosaic variants.
Free-floating cell-free DNA (cfDNA) has recently become a standard source for cancer genomic profiling and prenatal diagnostics (Wan et al. 2017; Dines et al. 2018). cfDNA from cerebrospinal fluid (CSF) has also recently emerged as an alternative source for molecular diagnostics in brain tumors (Wang et al. 2015; McEwen et al. 2020; White et al. 2021). Detection of somatic cancer variants in CSF-derived cfDNA has further encouraged researchers to investigate whether mosaic variants underlying other developmental brain disorders can be detected in CSF cfDNA. Two recent studies have shown that known somatic variants in the brain were detectable in CSF-derived cfDNA in individuals with HMEG, FCD, ganglioglioma, and subcortical band heterotopia (SBH) (Kim et al. 2021; Ye et al. 2021). These studies show that CSF-derived cfDNA can serve as a “proxy” tissue source to resected brain tissues for sequencing. Therefore, utilizing CSF-derived cfDNA has emerging potential in achieving a molecular diagnosis for individuals with mosaic brain malformations and other developmental brain disorders. Cell-free DNA obtained by minimally invasive procedures, such as lumbar puncture, can facilitate an earlier molecular diagnosis as well as consideration of medical management options (i.e., PI3K-AKT3-MTOR pathway inhibitors) prior to more invasive surgical resection. It can also offer a potential biomarker to monitor disease activity and treatment response. Here, we report the first report of an individual with MCAP syndrome secondary to a mosaic PIK3CA variant that was successfully detected in CSF-derived cfDNA, confirming his diagnosis.
RESULTS
Case Report
This boy was delivered at term following an uncomplicated pregnancy. Shortly after birth, he was identified to have macrocephaly, right-sided asymmetric overgrowth, and abnormal skin pigmentation with extensive deep purple-red vascular markings that were widely distributed over his body. He was initially clinically misdiagnosed with Klippel–Trenaunay syndrome (KTS). He also had thrombocytopenia requiring a platelet transfusion at 2 d of life. His neurological exam showed diffuse hypotonia. Cranial magnetic resonance imaging (MRI) obtained soon after birth showed asymmetric brain overgrowth and cerebellar tonsillar herniation (Fig. 2, panels). At later follow-up, he had progressive hydrocephalus requiring ventriculoperitoneal shunt placement at 6 mo of age. He later underwent laser treatment for the capillary malformation on his upper lip and right cheek. History is also notable for intestinal lymphangiectasia leading to episodes of diarrhea and nutritional deficiency during his early childhood. Developmentally, he had moderate speech delays with major delays in his gross motor skills.
Figure 2.
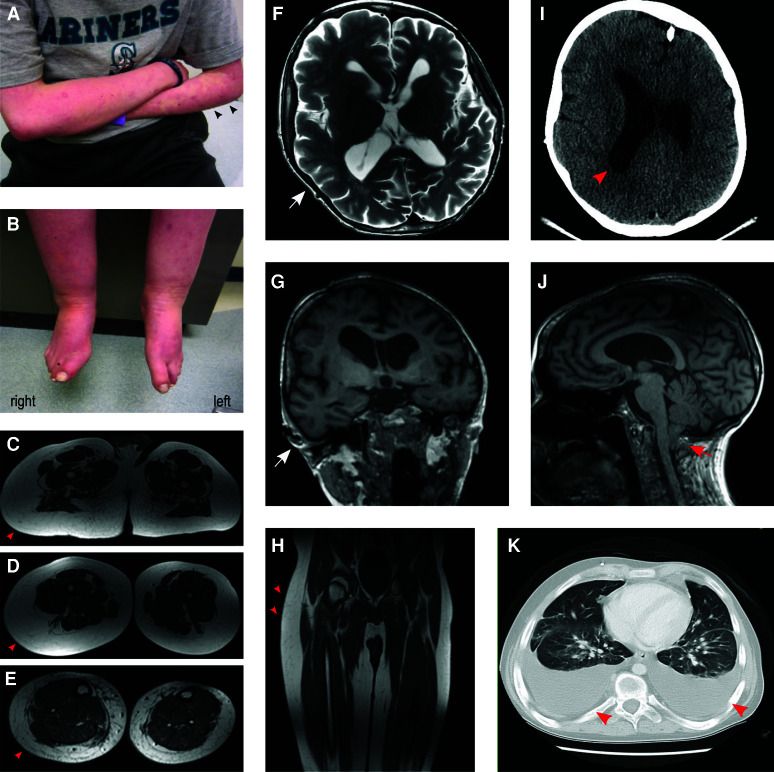
Phenotypic features observed in the proband. (A,B) Extensive capillary malformations were seen on both arms and legs, more prominent on his left arm (arrowheads). Segmental overgrowth was seen on his legs as well. (C–E,H) Lower extremity magnetic resonance imaging (MRI) showed overgrowth of adipose tissue (arrowheads) on right leg. (F,G,J) Brain MRI T2-weighted axial and T1-weighted coronal imaging showed right hemimegalencephaly and mildly enlarged and dysplastic right ventricle at age 19 yr. Arrows indicate the enlarged side. Cerebellar tonsillar ectopia was seen on T1-weighted sagittal imaging (arrow). (I) At 6 mo of age, computed tomography (CT) of head showed asymmetric brain size and hydrocephalus (arrowhead). (K) CT of chest at 19 yr showed diffuse pleural effusion (arrowheads), which later was found to have lymphoma cells in it.
Early physical examinations showed apparent macrocephaly and right-sided asymmetric overgrowth of the face and extremities. Measurements performed at 3 yr of age showed that his right ear was 6 cm (97th percentile) while the left was 4.7 cm (50th percentile). His right hand measured 9.9 cm (third percentile) from middle fingertip to wrist, whereas his left hand measured 9.3 cm (∼1st percentile) (Jones et al. 2013). His head appeared megalencephalic with a prominent venous pattern over the scalp. His occipitofrontal circumference (OFC) at 3 yr of age was 58.5 cm (+5.8 SD), and later grew to 65.5 cm (+7 SD) at 19 yr of age. He also had pinpoint elevated capillary malformations that ranged in size from 5 × 5 mm to 1 × 1 cm all over the scalp. After laser ablation, he had residual vascular staining on his right cheek, right upper lip, and more extensive irregular vascular patterns over the left arm (Fig. 2A).
In his late childhood, he developed recurrent lymphedema, protein-losing enteropathy, and pleural effusions (Fig. 2K). At age 19 yr, he was admitted to the pediatric intensive care unit (PICU) because of capillary leak syndrome with systemic inflammation. He was later found to have atypical lymphocytes in pleural and peritoneal fluid and increased fluorodeoxyglucose (FDG) uptake in bilateral cervical, mediastinal, abdominal, and pelvic lymph nodes on entire body positron emission tomography (PET) scan. Excisional biopsy of cervical lymph nodes showed sheets of atypical cells with large, vesicular nuclei with prominent nucleoli and scanty cytoplasm. Immunocytochemically, cells stained positive for CD20 and CD45. Flow cytometry immunotyping confirmed the diagnosis of diffuse large B-cell lymphoma (DLBCL). Involved sites included cervical, mediastinal, pelvic lymph nodes, and spleen with bowel wall thickening and pleural effusion (stage IIIb). He underwent lumbar puncture (LP) four times for staging and during chemotherapy with additional CSF collected for molecular diagnostics as well. There were no atypical lymphocytes found in any of the four CSF samples. He completed chemotherapy (R-CHOP [rituximab, cyclophosphamide, hydroxydaunomycin, vincristine, and prednisone]) without further remission. Cranial MRI obtained during this period showed asymmetric megalencephaly with mildly abnormal cortical gyral pattern, asymmetric dysplastic ventricles, and cerebellar tonsillar ectopia (Fig. 2F,G,J). He only had three seizures for a short period of time during chemotherapy. The semiology of his seizures included tonic seizure of left upper extremity with eye deviation to left, followed by bilateral tonic seizure, apnea, and desaturation. Seizures lasted ∼1–2 min with postictal confusion for several minutes. Seizures responded well to levetiracetam, and he therefore did not require epilepsy surgery. We previously published his clinical features as part of a large clinical-molecular series prior to his molecular diagnostic workup using cfDNA (case LR14-300) (Mirzaa et al. 2016).
Molecular Analysis
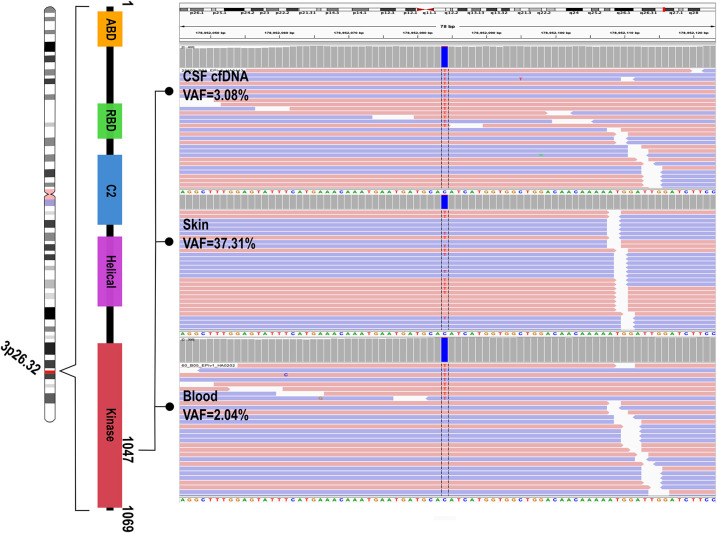
The proband underwent molecular diagnostic testing around the time when his lymphoma was diagnosed. Sequencing was performed using a clinically validated targeted multigene panel (the megaplex) performed in a College of American Pathologists (CAP)-accredited, Clinical Laboratory Improvement Amendments pf 1988 (CLIA)-certified laboratory as previously reported (Mirzaa et al. 2015). A mosaic variant in PIK3CA (NM_006218.2: c.3139C > T, p.His1047Tyr) was detected at a variant allele fraction (VAF) of 2% (variant [var]/reference [ref] reads: 8/394) in peripheral blood and 37.31% (var/ref reads: 673/1131) in cultured skin fibroblasts. The variant was identified in CSF-derived cfDNA at a VAF of 3.08% (var/ref reads: 14/440) (Fig. 3). A summary of the child's molecular findings in all samples is shown in Table 1. This variant has been previously published in association with MCAP syndrome (Mirzaa et al. 2016) and is listed as pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/39705/).
Figure 3.
PIK3CA c.3139C > T (p.His1047Tyr) in Integrative Genomic Viewer (IGV). IGV showed distinct variant allele fractions (VAFs) among various tissues in the proband including cerebrospinal fluid (CSF) cell-free DNA (cfDNA), skin fibroblasts, and peripheral blood.
Table 1.
Molecular findings
| HGVS DNA change (hg19/GRCh37) | HGVS protein change | Variant type | Predicted effect | dbSNP ID | Sample | DNA yield | VAF% (VAR/REF) |
|---|---|---|---|---|---|---|---|
|
NM_006218.2:c.3139C > T (Chr 3:178952084) |
p.H1047Y | Missense substitution | Gain of function | rs121913281 | CSF (cfDNA) | 29 ng (total after Ampure cleanup) | 3.08% (14/440) |
| Skin fibroblasts (cultured) | 122 ng/µL | 37.31% (673/1131) | |||||
| Peripheral blood | 172 ng/µL | 2.04% (8/384) |
Sample types, variant allele fractions of the gain of function mosaic PIK3CA variant in the proband.
(CSF) Cerebrospinal fluid, (cfDNA) cell-free DNA, (VAF) variant allele fraction, (VAR) number of variant read, (REF) number of reference reads.
DISCUSSION
Sequencing and detection of genetic variants underlying mosaic and tissue-restricted disorders typically rely on the availability of affected (or lesional) tissues. Here, we demonstrate the utility of CSF-derived cfDNA-based molecular diagnosis in PIK3CA-related MCAP syndrome. This case report has several helpful clinical implications. First, it demonstrates the utility of sequencing cfDNA from CSF to achieve a molecular diagnosis in the absence of affected or lesional brain tissues, which is particularly useful for individuals who have isolated or tissue-restricted mosaicism. Second, establishing a molecular diagnosis prior to undergoing invasive epilepsy surgery could potentially shift the paradigm of current testing and treatment strategies, especially as MTOR inhibitors are becoming more widely used (Forde et al. 2021; Garneau et al. 2021).
The PIK3CA p.His1407Tyr variant identified in this proband lies within the most commonly mutated codon within the kinase domain of the gene and has been reported multiple times as a disease-causing variant (Mirzaa et al. 2016; Kuentz et al. 2017). It has been previously identified in individuals with CLOVES (congenital lipomatous asymmetric overgrowth of the trunk, lymphatic, capillary, venous, and combined-type vascular malformations, epidermal nevi, skeletal and spinal anomalies) (MIM #612918) and MCAP syndromes (Mirzaa et al. 2015; Kuentz et al. 2017). Missense variants in this codon have been shown to cause PI3K-AKT-MTOR pathway hyperactivation (Jansen et al. 2015; Baldassari et al. 2019) and were also reported in various types of cancer tissues (Arafeh and Samuels 2019). It is uncertain whether individuals with PIK3CA-related overgrowth syndrome (PROS) are at risk for specific types of cancer. An association with Wilms’ tumor has been anecdotally suggested but not proven (Lapunzina et al. 2004; Wright et al. 2009; Gripp et al. 2016). The cancer risk in PROS in general and in MCAP in particular, however, continues to be unknown and there are no data suggesting an association between DLBCL and MCAP, with only one other individual with MCAP and leukemia diagnosed in adolescence previously reported (Moore et al. 1997). In the individual reported here, DNA extracted from lymphomatous tissue showed a 1.3% VAF (var/ref = 8/597) for the PIK3CA variant, which was similar to the level detected in the peripheral blood before the occurrence of DLBCL, and several additional somatic variants were detected in the lymphoma at a much higher VAF. Therefore, we conclude that this PIK3CA variant is unlikely the cause of his DLBCL. Moreover, earlier studies have shown activation of PI3K-AKT3-MTOR in cases with DLBCL but only a small subset harbored variants in PIK3CA (Abubaker et al. 2007; Baohua et al. 2008). Further, data from large cancer genomic databases suggest that somatic PIK3CA variants are found in hematopoietic and lymphoid cancer, including DLBCL (cBioPortal for Cancer Genomics [https://www.cbioportal.org] and COSMIC genomic mutation [https://cancer.sanger.ac.uk/cosmic]). Notably, a high burden of somatic variants was seen in the lymphomatous tissue but not in the CSF cfDNA sample. For example, copy number gain of Chromosome 1q including DNMT3A, and copy-number loss of Chromosome 9 including GNAQ and Chromosome 1p including MTOR and EPHB2 were seen in the lymphoma tissue but were absent in the CSF cfDNA sample. The lack of overlapping genomic findings provides further support of the nonlymphomatous origin of the PIK3CA variant in the CSF cfDNA sample in this individual.
Cell-free DNA is now widely used for genomic profiling in cancer (Wang et al. 2015). Cell-free DNA refers to DNA present in body fluids after cell death (Volik et al. 2016). Plasma cfDNA in healthy individuals is mostly derived from blood cells. In individuals with cancer, the amount of plasma cfDNA increases because of high rates of apoptosis and necrosis of cancer cells (Leon et al. 1977). Therefore, plasma cfDNA has gained prominence in cancer diagnosis, treatment, and monitoring (a.k.a liquid biopsy) (Wan et al. 2017). Body fluids can also contain cfDNA from noncancerous tissues as well. For example, cfDNA from cyst fluids of lymphatic malformations has been identified as a more reliable source than plasma to diagnose PIK3CA-associated lymphatic malformations (Zenner et al. 2021). Altogether these lines of evidence suggest that cfDNA from various body fluids, such as CSF, in direct contact with pathological tissues can be utilized as a “proxy” for molecular diagnostics. Indeed, two recent studies have shown some early yet promising evidence (Kim et al. 2021; Ye et al. 2021). In one study, CSF from the epilepsy cohort (FCD, ganglioglioma, SBH, and other tumors) contained significantly more cfDNA, which demonstrated brain-specific methylation patterns, than those without epilepsy (502 copies/mL vs. 61 copies/mL). Variants in several genes (LIS1, TSC1, and BRAF) were detectable in CSF-derived cfDNA with VAFs ranging from 3.20% to 9.40% (Ye et al. 2021). The second study included individuals with HMEG, ganglioglioma, malformation of cortical development with oligodendroglial hyperplasia in epilepsy (MOGHE) and FCD, mosaic variants in PIK3CA, BRAF, and SLC35A2 were identified in CSF-derived cfDNA with VAFs ranging from 0.136% to 1.45%. Compared with VAFs in paired brain tissues (ranging from 1.00% to 24.00%), levels of detectable mosaicism CSF-derived cfDNA were lower (Kim et al. 2021). However, there seemed to be no correlation between VAFs from cfDNA to the affected brain tissues from studies on both brain tumors (Wang et al. 2015) and malformations (Kim et al. 2021).
To have a meaningful clinical impact for the diagnosis and treatment of individuals with mosaic brain disorders (i.e., pharmaceutical vs. surgical approaches), a molecular diagnosis needs to ideally precede invasive brain surgery. Mosaic variants usually cannot be reliably detected from peripheral blood given not only the low mosaicism level but also clonality of blood cells (Kuentz et al. 2017). Therefore, CSF-derived cfDNA could potentially provide a more reliable surrogate for brain-limited mosaicism. Hence, CSF-derived cfDNA-based molecular diagnostics via lumbar puncture may provide a practical and novel method for variant identification. Notably, this individual also had cutaneous capillary malformations and body overgrowth which constitute additional lesional tissues for sequencing and variant detection (Fig. 1). However, these features are highly variable among affected individuals (Mirzaa et al. 2016).
All in all, novel molecular diagnostic approaches using minimally invasive procedures can have therapeutic implications for affected individuals. Syndromes caused by genetic variants of the PI3K-AKT-MTOR pathway share many similar features associated with dysregulated overgrowth. One notable example is the tuberous sclerosis complex (TSC) characterized by cortical tubers (which shows histopathological features similar to FCD), seizures, cutaneous findings, and other systemic features (Salussolia et al. 2019). Loss of inhibition of the PI3K-AKT-MTOR pathway secondary to TSC1 or TSC2 variants results in neuronal overgrowth (Tee et al. 2016), and the FDA-approved MTOR inhibitor everolimus has shown promising results in treating TSC-related refractory epilepsy (Krueger et al. 2013; French et al. 2016; Saffari et al. 2019), which occurs in ∼30% of patients even after surgical resection (Fallah et al. 2013). Similarly, other pathway-specific drugs can be repurposed to treat PROS. For example, alpelisib, a PI3K inhibitor recently approved for the treatment of PIK3CA mutation–positive hormone receptor–positive advanced breast cancer, has been used to treat CLOVES with significant clinical improvement of body overgrowth (Venot et al. 2018; López Gutiérrez et al. 2019; Delestre et al. 2021). Whether this and other inhibitors can be used to treat epilepsy associated with PIK3CA-associated MCAP, HMEG or focal cortical dysplasia is still under preclinical investigation. Nevertheless, mouse models expressing hotspot PIK3CA variants and corresponding histopathological neuronal findings have shown dramatic antiepileptic response to other PI3K inhibitors (e.g., BKM120) (Roy et al. 2015). Although further clinical studies are needed, PI3K inhibitors or other pathway-specific drugs (such as mTOR and AKT inhibitors) might have a role in treating PI3K-AKT-MTOR pathway–related intractable or recurrent epilepsy after surgical resection.
In conclusion, CSF-derived cfDNA-based molecular diagnostics provides a new method for the detection of mosaicism in individuals with developmental brain disorders. This novel method will not only facilitate an early and minimally invasive molecular diagnosis but might also have therapeutic implications in refractory epilepsy as repurposed PI3K-AKT-MTOR pathway–specific drugs are becoming more widely used.
METHODS
CSF was collected in a centrifuge tube and processed immediately after collection. CSF was centrifuged for 10 min at 400g and 4°C and then the supernatant was transferred to 2 mL cryovial for further centrifuge (10 min at 16,000g and 4°C). The cell pellet was discarded, and the supernatant was frozen. Quality-control (QC) data showed DNA fragment sizes ranging from 147 to 167 bp, consistent with cfDNA (Volik et al. 2016). Sequencing libraries were prepared from DNA samples and hybridized to a custom set of complementary RNA (cRNA) biotinylated oligonucleotides targeting the exons of 63 genes in a panel including PTEN, PIK3CA, AKT1, AKT3, and PIK3R2, among others, and select intronic regions for targeted DNA sequencing (Megaplex, Agilent SureSelect, Agilent Technologies, Inc). The panel is a targeted, massively parallel gene sequencing assay (https://testguide.labmed.uw.edu/public/view/MEGPX). The test uses next-generation “deep” sequencing to detect mutations including single-nucleotide variants (SNVs), indels, and copy-number changes including gene amplifications. DNA was extracted from CSF cfDNA, peripheral blood, and fresh tissue samples using a purification kit (QIAsymphony Circulating DNA Kit; QIAsymphony 93756), Gentra Puregene DNA Isolation Kit (Gentra 158489), QIASymphony DSP DNA Midi QIAGEN Kit (QIAGEN 937255) (Kolarova et al. 2021). Sequencing libraries were constructed from DNA using KAPA Hyper Prep kits (Kapa Biosystems Inc.) and hybridization was performed with custom oligonucleotide probes (Agilent SureSelect, Agilent Technologies). DNA sequencing was performed on a massively parallel instrument (HiSeq2500 sequencing system, Illumina) with 2 × 101-bp, paired-end reads according to the manufacturer's instructions.
Initial read mapping against the human reference genome (hg19/GRCh37) and alignment processing was performed using BWA version 0.6.1 (http://sourceforge.net/projects/bio-bwa/files) and SAMtools version 1.3.1 (http://sourceforge.net/projects/samtools/files), respectively. Sample-level, fully local indel realignment was then performed using GATK version 2.4.9 (Broad Institute). Duplicate reads were removed using PICARD version 1.72 (http:// broadinstitute.github.io/picard). Quality score recalibration was then performed using GATK. This realigned and recalibrated alignment was used for all subsequent analyses. SNV and indel calling were performed through the GATK Universal Genotyper using default parameters and VarScan version 2.3.6 (http://dkoboldt.github.io/varscan). For indel calling through VarScan, the minimum variant frequency was set to 0.01 reads, and the minimum number of variant reads was set to 4, whereas for SNV calling, the minimum variant frequency was set to 0.03, and the minimum number of variant reads was set to 5, with default parameters for all other settings. Variants identified by VarScan alone were manually reviewed using the Integrated Genomics Viewer version 2.3 (Broad Institute) to assess the quality of base calls, the mapping quality for the reads, and the overall read depth at the site.
PINDEL version 0.2.570 was used to identify tandem duplications and indels >10 bp in length. Structural variants were identified using CREST version 1.0 and BreakDancer version 1.1.1.71 For CNV analysis, copy number states for individual probes were initially called using CONTRA version 2.0.5 (http://sourceforge.net/projects/contra-cnv/files) with reference to a CNV control comprising reads from two independent rounds of library preparation and sequencing of the HapMap individual NA12878. CNV calls were made at the resolution of individual exons using custom Perl scripts.
ADDITIONAL INFORMATION
Data Deposition
The PIK3CA variant identified in this patient (NM_006218.2: c.3139C > T, p.His1047Tyr) has been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV002104174.1 and submitted to the Leiden Open Variation Database (LOVD; https://www.lovd.nl/) under submission number 0000406049.
Ethics Statement
This patient was prospectively enrolled in the Developmental Brain Disorders Research Study under an Institutional Review Board (IRB)-approved protocol at Seattle Children's Hospital (IRB#13291). Written informed consent was obtained from parents.
Acknowledgments
We thank the family and referring providers for their contribution to this study.
Author Contributions
W.-L.C., C.L., and G.M.M. conceived the designed the study, acquired and analyzed the data, and drafted the manuscript and figures. E.P., J.O., I.G., C.P., and B.H.S. contributed to acquisition, analysis of the data, and manuscript write-up.
Funding
Research reported in this publication was supported by Jordan's Guardian Angels, the Sunderland Foundation and the Brotman Baty Institute (BBI) (to G.M.M.). W.-L.C. was supported by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) Postdoctoral Fellowship in Medical Genetics 5T32GM007454. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Abubaker J, Bavi PP, Al-Harbi S, Siraj AK, Al-Dayel F, Uddin S, Al-Kuraya K. 2007. PIK3CA mutations are mutually exclusive with PTEN loss in diffuse large B-cell lymphoma. Leukemia 21: 2368–2370. 10.1038/sj.leu.2404873 [DOI] [PubMed] [Google Scholar]
- Arafeh R, Samuels Y. 2019. PIK3CA in cancer: the past 30 years. Semin Cancer Biol 59: 36–49. 10.1016/j.semcancer.2019.02.002 [DOI] [PubMed] [Google Scholar]
- Baldassari S, Ribierre T, Marsan E, Adle-Biassette H, Ferrand-Sorbets S, Bulteau C, Dorison N, Fohlen M, Polivka M, Weckhuysen S, et al. 2019. Dissecting the genetic basis of focal cortical dysplasia: a large cohort study. Acta Neuropathol 138: 885–900. 10.1007/s00401-019-02061-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baohua Y, Xiaoyan Z, Tiecheng Z, Tao Q, Daren S. 2008. Mutations of the PIK3CA gene in diffuse large B cell lymphoma. Diagn Mol Pathol 17: 159–165. 10.1097/PDM.0b013e31815d0588 [DOI] [PubMed] [Google Scholar]
- Delestre F, Venot Q, Bayard C, Fraissenon A, Ladraa S, Hoguin C, Chapelle C, Yamaguchi J, Cassaca R, Zerbib L, et al. 2021. Alpelisib administration reduced lymphatic malformations in a mouse model and in patients. Sci Transl Med 13: eabg0809. 10.1126/scitranslmed.abg0809 [DOI] [PubMed] [Google Scholar]
- Dines JN, Eckel AM, Cheng EY, Lockwood CM. 2018. A paradigm shift: considerations in prenatal cell-free DNA screening. J Appl Lab Med 2: 784–796. 10.1373/jalm.2017.023119 [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Mirzaa GM. 2019. Megalencephaly syndromes associated with mutations of core components of the PI3K-AKT-MTOR pathway: pIK3CA, PIK3R2, AKT3, and MTOR. Am J Med Genet C Semin Med Genet 181: 582–590. 10.1002/ajmg.c.31736 [DOI] [PubMed] [Google Scholar]
- Fallah A, Guyatt GH, Snead OC, Ebrahim S, Ibrahim GM, Mansouri A, Reddy D, Walter SD, Kulkarni AV, Bhandari M, et al. 2013. Predictors of seizure outcomes in children with tuberous sclerosis complex and intractable epilepsy undergoing resective epilepsy surgery: an individual participant data meta-analysis. PLoS ONE 8: e53565. 10.1371/journal.pone.0053565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde K, Resta N, Ranieri C, Rea D, Kubassova O, Hinton M, Andrews KA, Semple R, Irvine AD, Dvorakova V. 2021. Clinical experience with the AKT1 inhibitor miransertib in two children with PIK3CA-related overgrowth syndrome. Orphanet J Rare Dis 16: 109. 10.1186/s13023-021-01745-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, Curatolo P, de Vries PJ, Dlugos DJ, Berkowitz N, et al. 2016. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet 388: 2153–2163. 10.1016/S0140-6736(16)31419-2 [DOI] [PubMed] [Google Scholar]
- Garneau AP, Haydock L, Tremblay LE, Isenring P. 2021. Somatic non-cancerous PIK3CA-related overgrowth syndrome treated with alpelisib in North America. J Mol Med (Berl) 99: 311–313. 10.1007/s00109-020-02030-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Baker L, Kandula V, Conard K, Scavina M, Napoli JA, Griffin GC, Thacker M, Knox RG, Clark GR, et al. 2016. Nephroblastomatosis or Wilms tumor in a fourth patient with a somatic PIK3CA mutation. Am J Med Genet A 170: 2559–2569. 10.1002/ajmg.a.37758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LA, Mirzaa GM, Ishak GE, O'Roak BJ, Hiatt JB, Roden WH, Gunter SA, Christian SL, Collins S, Adams C, et al. 2015. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 138: 1613–1628. 10.1093/brain/awv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Jones M, Campo MD. 2013. Smith's recognizable patterns of human malformation. Elsevier, New York. [Google Scholar]
- Kilincaslan A, Kok BE, Tekturk P, Yalcinkaya C, Ozkara C, Yapici Z. 2017. Beneficial effects of everolimus on autism and attention-deficit/hyperactivity disorder symptoms in a group of patients with tuberous sclerosis complex. J Child Adolesc Psychopharmacol 27: 383–388. 10.1089/cap.2016.0100 [DOI] [PubMed] [Google Scholar]
- Kim S, Baldassari S, Sim NS, Chipaux M, Dorfmuller G, Kim DS, Chang WS, Taly V, Lee JH, Baulac S. 2021. Detection of brain somatic mutations in cerebrospinal fluid from refractory epilepsy patients. Ann Neurol 89: 1248–1252. 10.1002/ana.26080 [DOI] [PubMed] [Google Scholar]
- Kolarova TR, Gammill HS, Nelson JL, Lockwood CM, Shree R. 2021. At preeclampsia diagnosis, total cell-free DNA concentration is elevated and correlates with disease severity. J Am Heart Assoc 10: e021477. 10.1161/JAHA.121.021477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DA, Wilfong AA, Holland-Bouley K, Anderson AE, Agricola K, Tudor C, Mays M, Lopez CM, Kim MO, Franz DN. 2013. Everolimus treatment of refractory epilepsy in tuberous sclerosis complex. Ann Neurol 74: 679–687. 10.1002/ana.23960 [DOI] [PubMed] [Google Scholar]
- Kuentz P, St-Onge J, Duffourd Y, Courcet JB, Carmignac V, Jouan T, Sorlin A, Abasq-Thomas C, Albuisson J, Amiel J, et al. 2017. Molecular diagnosis of PIK3CA-related overgrowth spectrum (PROS) in 162 patients and recommendations for genetic testing. Genet Med 19: 989–997. 10.1038/gim.2016.220 [DOI] [PubMed] [Google Scholar]
- Lapunzina P, Gairi A, Delicado A, Mori MA, Torres ML, Goma A, Navia M, Pajares IL. 2004. Macrocephaly-cutis marmorata telangiectatica congenita: report of six new patients and a review. Am J Med Genet A 130A: 45–51. 10.1002/ajmg.a.30235 [DOI] [PubMed] [Google Scholar]
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. 1977. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 37: 646–650. [PubMed] [Google Scholar]
- López Gutiérrez JC, Lizarraga R, Delgado C, Martínez Urrutia MJ, Díaz M, Miguel M, Triana P. 2019. Alpelisib treatment for genital vascular malformation in a patient with congenital lipomatous overgrowth, vascular malformations, epidermal nevi, and spinal/skeletal anomalies and/or scoliosis (CLOVES) syndrome. J Pediatr Adolesc Gynecol 32: 648–650. 10.1016/j.jpag.2019.07.003 [DOI] [PubMed] [Google Scholar]
- McEwen AE, Leary SES, Lockwood CM. 2020. Beyond the blood: cSF-derived cfDNA for diagnosis and characterization of CNS tumors. Front Cell Dev Biol 8: 45. 10.3389/fcell.2020.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Conway RL, Gripp KW, Lerman-Sagie T, Siegel DH, de Vries LS, Lev D, Kramer N, Hopkins E, Graham JM, et al. 2012. Megalencephaly-capillary malformation (MCAP) and megalencephaly-polydactyly-polymicrogyria-hydrocephalus (MPPH) syndromes: two closely related disorders of brain overgrowth and abnormal brain and body morphogenesis. Am J Med Genet A 158A: 269–291. 10.1002/ajmg.a.34402 [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Conti V, Timms AE, Smyser CD, Ahmed S, Carter M, Barnett S, Hufnagel RB, Goldstein A, Narumi-Kishimoto Y, et al. 2015. Characterisation of mutations of the phosphoinositide-3-kinase regulatory subunit, PIK3R2, in perisylvian polymicrogyria: a next-generation sequencing study. Lancet Neurol 14: 1182–1195. 10.1016/S1474-4422(15)00278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa G, Timms AE, Conti V, Boyle EA, Girisha KM, Martin B, Kircher M, Olds C, Juusola J, Collins S, et al. 2016. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight 1: e87623. 10.1172/jci.insight.87623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Toriello HV, Abuelo DN, Bull MJ, Curry CJ, Hall BD, Higgins JV, Stevens CA, Twersky S, Weksberg R, et al. 1997. Macrocephaly-cutis marmorata telangiectatica congenita: a distinct disorder with developmental delay and connective tissue abnormalities. Am J Med Genet 70: 67–73. [DOI] [PubMed] [Google Scholar]
- Pirozzi MB, Shear R, Gonzalez L, Timms AE, Sulc J, Pao E, Oyama N, Forzano F, Conti V, Guerrini R, et al. 2021. Analysis of common PI3K-AKT-MTOR mutations in pediatric surgical epilepsy by droplet digital PCR reveals novel clinical and molecular insights. medRxiv 10.1101/2021.06.09.21257462 [DOI] [Google Scholar]
- Roy A, Skibo J, Kalume F, Ni J, Rankin S, Lu Y, Dobyns WB, Mills GB, Zhao JJ, Baker SJ, et al. 2015. Mouse models of human PIK3CA-related brain overgrowth have acutely treatable epilepsy. Elife 4: e12703. 10.7554/eLife.12703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffari A, Brösse I, Wiemer-Kruel A, Wilken B, Kreuzaler P, Hahn A, Bernhard MK, van Tilburg CM, Hoffmann GF, Gorenflo M, et al. 2019. Safety and efficacy of mTOR inhibitor treatment in patients with tuberous sclerosis complex under 2 years of age—a multicenter retrospective study. Orphanet J Rare Dis 14: 96. 10.1186/s13023-019-1077-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salussolia CL, Klonowska K, Kwiatkowski DJ, Sahin M. 2019. Genetic etiologies, diagnosis, and treatment of tuberous sclerosis complex. Annu Rev Genomics Hum Genet 20: 217–240. 10.1146/annurev-genom-083118-015354 [DOI] [PubMed] [Google Scholar]
- Tee AR, Sampson JR, Pal DK, Bateman JM. 2016. The role of mTOR signalling in neurogenesis, insights from tuberous sclerosis complex. Semin Cell Dev Biol 52: 12–20. 10.1016/j.semcdb.2016.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot Q, Blanc T, Rabia SH, Berteloot L, Ladraa S, Duong JP, Blanc E, Johnson SC, Hoguin C, Boccara O, et al. 2018. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 558: 540–546. 10.1038/s41586-018-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volik S, Alcaide M, Morin RD, Collins C. 2016. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res 14: 898–908. 10.1158/1541-7786.MCR-16-0044 [DOI] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17: 223–238. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, Ptak J, Brem H, Chaichana K, Gallia GL, et al. 2015. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci 112: 9704–9709. 10.1073/pnas.1511694112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MD, Klein RH, Shaw B, Kim A, Subramanian M, Mora JL, Giobbie-Hurder A, Nagabhushan D, Jain A, Singh M, et al. 2021. Detection of leptomeningeal disease using cell-free DNA from cerebrospinal fluid. JAMA Netw Open 4: e2120040. 10.1001/jamanetworkopen.2021.20040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DR, Frieden IJ, Orlow SJ, Shin HT, Chamlin S, Schaffer JV, Paller AS. 2009. The misnomer “macrocephaly-cutis marmorata telangiectatica congenita syndrome”: report of 12 new cases and support for revising the name to macrocephaly-capillary malformations. Arch Dermatol 145: 287–293. 10.1001/archdermatol.2008.545 [DOI] [PubMed] [Google Scholar]
- Ye Z, Chatterton Z, Pflueger J, Damiano JA, McQuillan L, Harvey AS, Malone S, Do H, Maixner W, Schneider A, et al. 2021. Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain. Brain Commun 3: fcaa235. 10.1093/braincomms/fcaa235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KS, Ip JJ, Chow CP, Kuong EY, Tam PK, Chan GC, Chung BH. 2017. Somatic PIK3CA mutations in seven patients with PIK3CA-related overgrowth spectrum. Am J Med Genet A 173: 978–984. 10.1002/ajmg.a.38105 [DOI] [PubMed] [Google Scholar]
- Zenner K, Jensen DM, Cook TT, Dmyterko V, Bly RA, Ganti S, Mirzaa GM, Dobyns WB, Perkins JA, Bennett JT. 2021. Cell-free DNA as a diagnostic analyte for molecular diagnosis of vascular malformations. Genet Med 23: 123–130. 10.1038/s41436-020-00943-8 [DOI] [PMC free article] [PubMed] [Google Scholar]