Abstract
Human intestinal tissue–derived enteroids (HIEs; also called organoids) are a powerful ex vivo model for gastrointestinal research. Genetic modification of these nontransformed cultures allows new insights into gene function and biological processes involved in intestinal diseases as well as gastrointestinal and donor segment-specific function. Here we provide a detailed technical pipeline and protocol for using the CRISPR–Cas9 genome editing system to knock out a gene of interest specifically in HIEs by lentiviral transduction and single-cell cloning. This protocol differs from a previously published alternative using electroporation of human colonoids to deliver piggyback transposons or CRISPR-Cas9 constructs, as this protocol uses a modified, fused LentiCRISPRv2-small-guiding RNA to express Cas9 and small-guiding RNA in a lentivirus. The protocol also includes the steps of gene delivery and subsequent single-cell cloning of the knockout cells as well as verification of clones and sequence identification of the mutation sites to establish knockout clones. An overview flowchart, step-by-step guidelines and troubleshooting suggestions are provided to aid the researcher in obtaining the genetic knockout HIE line within 2-3 months. In this protocol, we further describe how to use HIEs as an ex vivo model to assess host restriction factors for viral replication (using human norovirus replication as an example) by knocking out host attachment factors or innate immunity genes. Other applications are discussed to broaden the utility of this system, for example, to generate knockin or conditional knockout HIE lines to investigate the function of essential genes in many biological processes including other types of organoids.
Introduction
Human intestinal tissue–derived enteroids (HIEs) were first established by Sato and Clevers as an ex vivo culture model that mimics the human gastrointestinal (GI) tract epithelium1. Derived from intestinal stem cells, these nontransformed epithelial-only cultures retain the capacity for self-renewal and stem cell pluripotency. Also called organoids or miniguts, HIE cultures grow indefinitely, are multicellular and differentiated cultures, and contain enterocytes, enteroendocrine cells, goblet cells and Paneth cells found in the epithelium of the human GI tract in vivo1. Established from human biopsies or surgical tissues, HIEs recapitulate host genome variability and intestine segment specificity, and they are physiologically active, making them an exceptional ex vivo model for hostoriented translational research and precision medicine. Since 2011, HIEs have been used in GI, stem cell biology, cancer, virology, bacteriology and parasitology research for culturing human trophic microbes, including previously noncultivatable organisms2-11. In 2016, our laboratory reported HIEs as the first ex vivo intestinal cultivation model for human norovirus (HuNoV)11. HuNoV replication in HIEs recapitulates the virus tissue tropism for intestinal enterocytes observed in patient biopsies, supports reproducible replication of multiple HuNoV strains and confirms epidemiological differences in genetic susceptibility to HuNoV infection8,12,13. The HuNoV-HIE model also illustrates strain-specific requirements for replication, including the requirement of bile or components of bile for replication in these permissive cells14. In addition, the expression of genetically controlled histoblood group antigens is required for replication in HIEs15, mimicking epidemiological patterns of virus infection in people. Thus, the HIE culture system enables new studies on virus biology, GI biology, virus–host interactions, antibody-mediated neutralization and other antiviral interventions to be performed for controlling viral infections.
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 has been a well-known designer nuclease technique since 2012. Originally, the Cas9 nuclease gene was identified as a component of the CRISPR Escherichia coli bacterial innate immunity system16 to eliminate non-self DNA from bacteriophages. Cas9 encodes a novel class of sequence-specific endonuclease complexes whose target specificity is determined by an accompanying small-guiding RNA (sgRNA) sequence17-20 rather than the protein motifs as in the zinc finger nuclease and transcription activator-like effector nuclease families. Cas9 can be easily reprogrammed to target a new sequence simply by replacing the 20 bp sgRNA sequence for specificity determination. The only requirement of the target site is that it must be followed by an ‘NGG’ (N for any nucleotide) known as the protospacer adjacent motif (PAM) for Cas9 recognition17. After recognizing the sequence, the designed Cas9 can precisely modify the genome by generating a double-strand break (DSB) at the target site to activate the endogenous DNA repair system21,22. In the absence of a homologous template, the DSB can be repaired by joining the two ends together, called nonhomologous end-joining (NHEJ)23, which usually produces silent mutations, provided that the cut is clean without altering the gene function but losing the ability to be recognized by the Cas9–sgRNA complex again. In some cases, the repair is imperfect, resulting in deletion or insertion of base pairs (indels), producing frameshift mutations to truncate the target gene by generating early stop codons by chance24. If the frameshift-induced truncation occurs in both alleles, the gene will become a loss-of-function or knocked out modification in the host genome. Before Cas9 was introduced, the DSB could only be generated by chance or by engineering the protein motifs on designed zinc finger nucleases or transcription activator-like effector nucleases, which are more difficult and time-consuming to construct than ordering new primers to change the sgRNA sequence using the Cas9 system. As a result, the rapid and flexible Cas9 technique quickly dominated the fields of genome editing and genetics research.
Development of the protocol
This protocol is an updated version of the previous Cas9-mediated gene knockout (KO) method developed and used by the Estes laboratory in two publications in 202012,15. For the CRISPR system, we started from the traditional LentiCRISPRv2 system established by the Zhang laboratory at the Massachusetts Institute of Technology25, merged with a second-generation system by using pMD2.G and psPAX2 plasmids established by the Trono laboratory in École polytechnique fédérale de Lausanne for lentivirus packaging (unpublished, https://www.addgene.org/browse/article/836/). We initially tried to disperse the cells in the multicellular HIEs by pipetting with a P1000 tip and incubating with lentivirus at 37 °C overnight for transduction. However, the transduction efficiency was not ideal with our initial two trials. We then switched to another method by dispersing HIEs into single cells with trypsin–EDTA for spin inoculation, which increased the transduction efficiency and survival rate of transduced HIEs in >20 independent trials, and led to our final protocol outlined below12,15.
Applications of the method
The power of the CRISPR–Cas9 technique makes it attractive in HIE research to identify critical genes that affect stem cell differentiation, host–pathogen interactions, viral-induced innate immune responses and intestinal biology. In this protocol, we illustrate how to use CRISPR–Cas9 in lentivirus to knock out genes that affect HuNoV infection, including a host attachment factor gene FUT2 and an innate immunity gene STAT112,15. We and others have also used KO lines to investigate rotavirus infection and identified new mechanisms by which calcium waves induced by infection affect neighboring cells in KO lines26. The functional assays we used included virus replication (virus yields, viral spreading by immunofluorescent microscopy) assays, and the choice of specific assays will depend on the research field and interest of the laboratory using an enteroid line, which may be similar to the functional assays used in common cell lines. Enteroids that contain a specific gene KO can be used to identify mechanisms of pathogen entry or egress, signaling pathways, nutrient uptake and cellular metabolic processes, including lipid metabolism, energy production, phospholipid biogenesis and pathophysiological consequences. Finally, KO cultures can be used to confirm the specificity of antiviral drug treatments that target host functions by confirming host pathways that may be affected and identifying possible synergistic effects that can be detected when the drug and KO affect different cellular signaling pathways.
Potential other modifications and improvement of the protocol
As done with many plasmids used for electroporation, it would be desirable to generate lentiviruses with Cas9 that also will express a fluorescent marker such as GFP and an antibiotic marker to facilitate determining whether a final clone is a KO27. In addition, instead of using a single frameshift to generate stop codons to knock out the target gene, it might be attractive to generate a truncated protein or remove a whole exon to generate a clean KO; this would be useful when a large gene containing multiple isoforms with different start sites is targeted, or when a truncated protein is still functional compared with a clean whole exon KO for investigating a loss-of-function phenotype28. This could be achieved by generating a larger deletion on the target gene by transducing two sgRNAs together in one HIE cell to remove the whole gene fragment from the genome29. Alternatively, it may be attractive to knock in a reporter gene with stop codons in the promoter region of your target gene; this would generate a reporter KO by delivering Cas9, sgRNA and the reporter with homology arms (plasmid or dsDNA) to replace the endogenous gene30 or to profile the expression pattern of the KO gene in the whole HIE31. Moreover, it is possible to generate knockin HIEs by providing an extra template cassette with homology arms, and the DSB will induce double crossing over to replace the endogenous chromosome by the knockin cassette32.
Comparison with other genome modification methods
There have been other publications using a variety of organoid systems33, some of which we tried and found were not optimal for our nontransformed small intestinal enteroid cultures34; others lacked detailed protocols32,35,36, did not use lentiviruses37 or did not use Cas9 or a combination of lentiviruses and Cas938-40. The relative benefits of electroporation and lentiviral transduction are still debated. Electroporation has the advantage that genes can be transferred upon preparation of the plasmids, which alleviates the labor and time required to produce lentivirus34. To improve the electroporation method, GFP can be included in the plasmid as the tracing marker; for example, pL-CRISPR.EFS.GFP is easily used for sgRNA genome editing. GFP+-transfected cultures can be traced by fluorescence microscopy after single-cell seeding in 96-well plates, or by gating GFP signals in cell sorting. However, if neither screening method is available in a laboratory, it is easier to use lentiviral transduction to deliver Cas9 and sgRNA into HIEs and do antibiotic (usually puromycin) selection, since the transduction rate is relatively low to allow detection of rare GFP+ cells present following electroporation under the microscope. Since gene delivery efficiency with electroporation in HIEs is relatively low, a selection step is required to enrich for transfected or transduced HIEs. However, without genome integration of the puromycin-resistant gene, the transfected cells may lose the plasmid and get selected out during puromycin selection, which takes 2–3 weeks to eradicate nontransfected HIEs.
We use lentiviral transduction because it resulted in success to obtain Cas9-mediated KO clones compared with our trials with electroporation. In fact, we have obtained KO HIE clones for ten different genes to date using lentiviruses and only failed to generate a KO when the gene being knocked out was essential for enteroid growth. Our protocol adds value in describing detailed CRISPR-based methods in enteroids to guide other laboratories to use Cas9 to knock out their genes of interest with ease. Inclusion of practical insights will help beginners obtain KO lines that can be difficult to achieve without detailed step-by-step procedures.
Limitations of using our protocol
Our simplified CRISPR-Cas9 protocol has several limitations that need to be considered. First, we use the lentiviral transduction system, so the cassette expressing Cas9 and sgRNA can randomly insert into the host genome and disrupt host genes. Because of this, we recommend generating several independent KO clones (at least five) and comparing their phenotypes, which increases the likelihood of finding a clone in which the phenotype is caused by knocking out the gene of interest. For further validation, it also is possible to reintroduce a wild-type copy of the functional gene (e.g., complementary DNA) but with a silent mutation in the Cas9 targeting site to rescue the KO phenotype. If the phenotype can be rescued by adding a wild-type, CRISPR-resistant copy back, it indicates the phenotype is due to the loss of the target gene.
Second, the Cas9 and sgRNA will be constitutively expressed in the host genome; thus, there is a potential for causing cellular stress, DSB-induced signaling and genome instability. In fact, transient expression by transfection of the Cas9–sgRNA complex for days has been shown to be sufficient for knocking out the gene but with low efficiency41. To improve this, it would be possible to use electroporation to deliver a plasmid without integration, but antibiotic selection would need to be done before the plasmid is lost from the host cells with cell passages. It is also possible to use an inducible Cas9–sgRNA system to drive the expression of Cas9 by treating the cells with a chemical activator, such as doxycycline or tamoxifen42. Finally, it is time-consuming but recommended to introduce two loxP sites on the construct to permit removal of Cas9 and sgRNA from the genome after the KO has been confirmed43.
Off-target effects of CRISPR–Cas9 have been reported since the sgRNA may be able to bind to a similar sequence with around two to three mismatches to activate Cas944. It is best to predict top potential off-target sites by using the basic local alignment search tool (BLAST) and sequence them one by one to confirm there is no alteration in a KO line45,46. Comparing independent KO lines for the same phenotype also is helpful to rule out the possibility that the phenotype is caused by an off-target effect rather than by the loss of function of the gene of interest.
Overview of the procedure
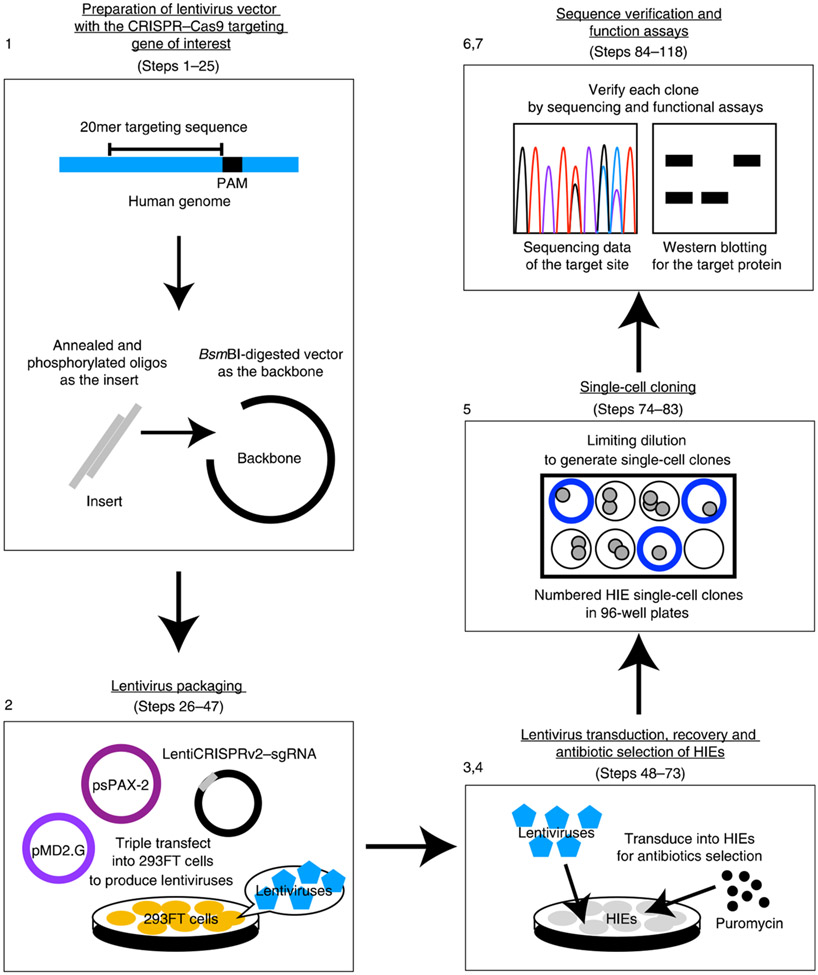
The following procedure describes how to obtain genetically modified enteroids, starting from the establishment of stably growing HIEs from human tissues. The transduced Cas9–sgRNA cassette in the host genome will execute DSB in the target sequence, which then potentially introduces frameshift mutations in both alleles to knock out the gene. Figure 1 summarizes the procedure for generating and verifying KO HIEs with seven parts: (1) preparation of lentivirus vector with the CRISPR–Cas9 targeting gene of interest (Steps 1–25); (2) lentivirus packaging (Steps 26–47); (3) lentivirus transduction of HIEs (Steps 48–64); (4) HIE recovery and antibiotic selection (Steps 65–73); (5) single-cell cloning (Steps 74–83); (6) sequencing verification of single-cell clones (Steps 84–109); and (7) using KO lines to evaluate host gene function in viral replication or other biological processes (Steps 110–118).
Fig. 1 ∣. The steps of lentivirus packaging and transduction.
The protocol has seven parts: (1) preparation of lentivirus vector with the CRISPR–Cas9 targeting gene of interest; (2) lentivirus packaging; (3) lentivirus transduction of HIEs; (4) HIE recovery and antibiotic selection; (5) single-cell cloning; (6) sequencing verification of single-cell clones; (7) using KO lines to evaluate host gene function in viral replication or other biological processes.
Experimental design
Tissue sampling and HIE culture
The small intestinal (duodenal and ileal) biopsy specimens used in this protocol were initially established from tissues obtained from adults during routine endoscopy at Baylor College of Medicine through the Texas Medical Center Digestive Diseases Center Study Design and Clinical Research Core. Jejunal tissue was obtained from patients undergoing bariatric surgery. All human tissues used to establish cultures were obtained under protocols to obtain patient consent as approved by the institutional review board. HIEs should be prepared from the tissue samples as previously described1,47, using three different types of medium to establish, maintain or differentiate HIEs as described in ‘Reagent setup’.
Investigation of the loss-of-function phenotype of the target gene in HIEs
This protocol is designed for knocking out a specific gene in the HIE genome by causing DSB-induced NHEJ frameshift mutations in both alleles of the target gene. The lentivirus can transduce the Cas9–sgRNA plasmid into both dividing and nondividing cells, which includes the intestinal stem cells. For HIEs, the goal is to transduce intestinal stem cells in healthy, proliferating cultures using high-titer lentiviruses. Only mutations occurring in the stem cells will be retained and inherited by all progeny cells. Gene functions can be evaluated by comparing the phenotypic difference between wild-type parental HIEs and KO HIEs, where the KO phenotype seen is due to a loss of function of that gene. In HuNoV research, we are knocking out host genes in HIEs that are potentially important for replication, including host glycosyltransferases, to determine if they are necessary and sufficient for HuNoV infection. Other target genes are host restriction factors including innate immunity genes, for example, the IFN pathways genes that would be expected to increase HuNoV replication and virus yields in the HIE system.
sgRNA design
Genome engineering with the CRISPR–Cas9 system is achieved via the expression of a Cas9–sgRNA fusion plasmid. As a result, an efficient sgRNA will work well to introduce NHEJ-based mutations easily in the target gene, which means the possibility of picking up a KO single-cell clone will be higher during the labor-intensive screening procedure. Based on our experience, we found the GenScript sgRNA database (https://www.genscript.com/gRNA-database.html) is a versatile, free webtool for choosing sgRNAs. Usually, you can choose one or two sgRNA sequences to order annealing primers, and then clone each into an individual LentiCRISPRv2 vector simultaneously. One out of these two sequences should work in the HIEs. Several factors should be considered when selecting a sgRNA from the database48. Generally speaking, a 20mer sgRNA sequence with an intermediate GC content from 45% to 65% usually has the highest efficiency. The sgRNA should be located within the first 10–30% of the first coding exon, rather than the last exon, to generate a shorter, loss-of-function truncated protein as early as possible. If alternative splicing occurs within the gene, the sgRNA should be designed in a common exon region among all splicing variants to knock out all splice variants at one time. The location of the sgRNA in the nontranscribed strand (the sense, positive strand) will increase the efficiency as well. Lastly, but most importantly, the sequence of the last four nucleotides of the target sequence prior to the PAM as ‘NGG’ should contain few thymine (T) nucleotides. The presence of the opposed strand poly-uridine can impair transcription activity by premature unloading of the RNA polymerase. An sgRNA that does not violate the above points will be considered efficient enough for knocking out the gene of interest in HIEs.
Important negative/mock controls for the Procedure
For the sgRNA plasmid cloning in Step 15, have a negative control by transforming 1 μL digested vector without the insert into 50 μL competent cells to check the digestion efficiency. If this transformation still leads to comparable colonies on the plate, the digestion of the backbone was incomplete and most of the bacterial colonies might be empty without an insert. During the colony PCR in Step 20, set up one PCR reaction without colony DNA to verify there was no contamination of E. coli DNA during PCR master mix preparation. A clean negative control should result in no PCR product verified by subsequent gel electrophoresis. Those two negative controls are important for generating your LentiCRISPRv2–sgRNA construct with confidence.
For the HIE transduction in Step 55, a mock control without lentivirus transduction is required to verify the selection efficiency. HIEs without lentivirus should not survive under antibiotic selection. If there is no death in the control group, the antibiotic concentration might not be optimal for killing the HIEs. In addition, it is also critical to have a backup well of transduced HIEs without antibiotic selection in Step 67, to guarantee that HIEs after transduction are able to survive and grow. If all transduced HIEs treated with antibiotics are dead in the end, you still have those backup transduced HIEs to restart a new round of selection with a lower range of antibiotic concentrations.
Using KO lines to evaluate viral replication and host restriction
The functional assays used to characterize KO lines will depend on the research field and interest of the laboratory. Using an enteroid line, the best way to verify the KO clone is to do sequencing, Western blotting or intracellular staining to see the loss of the target protein. Enteroids that contain a specific KO can be used to identify host restriction factors for viral infection. For instance, we generated STAT1-KO HIEs and found enhanced infection and spreading of a GII.3 HuNoV strain in this KO HIE line11,12. Part 7 of the Procedure outlines how to generate STAT1-KO HIEs for GII.3 infection. Moreover, a new mechanism by which calcium waves, induced by rotavirus infection, affect neighboring cells was discovered in P2Y1-KO HIEs26.
Comparisons using a one-vector versus two-vector system for lentiviruses
We have used both a one-vector and a two-vector system for modifying HIEs. For the one-vector system, as detailed in the Procedure, a single lentiCRISPRv2 vector is used to express both Cas9 and sgRNA with a single transduction and puromycin selection. For the two-vector system, which is detailed in Supplementary Methods and has been used for generating a FUT2-K015, separate Cas9-expressing and sgRNA-expressing lentiviral vectors are used. Compared with the one-vector system, the two-vector system requires one additional antibiotic (Blasticidin S) and needs more time for selection, but is more flexible by simultaneously delivering multiple sgRNAs that will have relatively smaller vector sizes for cloning and transduction. In addition, the transduced HIEs receive only one copy of the Cas9 gene after transfecting multiple vectors in the two-vector system, which therefore reduces the cellular stress in the HIEs. Besides the transduction and selection, the other steps are similar between the two systems for generating a single KO HIE line.
Generation of double- or triple-KO HIEs by re-transduction
We have tried to reintroduce a second sgRNA by retransducing lentiviruses with a different selection marker into a confirmed KO HIE clone to obtain a double-KO HIE line (e.g., STAT1 and STAT2 double KO with puromycin and hygromycin-B selection). The first selection is puromycin, the second selection can be done using hygromycin-B and the third one can be Blasticidin S or GFP sorting. However, the retransduction seems to make the HIEs less healthy with increased genome instability due to the lentiviral random insertion. The delivery of one lentiviral plasmid with two sgRNAs targeting different genes might be another way to generate a double-gene KO; however, the dual hit of both genes to generate a double KO is less likely to be successful compared with the retransduction method.
Practical insights and experimental aspects to consider before starting the protocol
The protocol outlines a step-by-step pipeline and details of how to obtain KO lines in small intestinal HIEs from jejunum and ileum. Cultures in other laboratories may be different, but our experience provides guidance to parts of the procedure that need to be optimized before starting the experiment properly and questions that may arise before starting. Many of the findings described in the below section represent our unpublished observations (S-C.L., K.H. and X-L.Z). First, it is important to produce high-titer lentiviruses for transduction owing to the low delivery rate into HIE cultures compared with common cell lines. Thus, transducing with more viruses ensures enough transduced HIEs for the subsequent single-cell cloning. Moreover, the transduction efficiency may vary in enteroid lines from different patients or intestinal segments; the health (growth rate) of the enteroid lines is a critical factor for successfully generating transduced lines. In our experience, lentiviral transduction is the best way for gene delivery. Since we transduce ~3 × 105 HIEs in one well of a 24-well plate with an excess amount of lentiviruses, several surviving enteroids are obtained in all cases after 2–3 weeks of antibiotic selection followed by single-cell cloning as described below. We use a jejunal (J2) HIE line as the example in this protocol, but the conditions may have to be optimized with different lines. Our laboratory has generated KO lines in three independent jejunal lines12,15.
Second, it may be necessary to optimize the antibiotic concentration used for selection for different enteroid lines. One should test and find the highest concentration of antibiotic that kills all nontransduced cells. We have transduced different enteroids with lentiviruses containing the same plasmid construct (LentiCRISPRv2) for generating KOs using selection with 2 μg/mL puromycin. So far, we have not observed a need to vary the antibiotic selection concentrations in our different HIE cultures since this optimal concentration can induce cell death in all nontransduced cells. However, other lines in other laboratories might have different antibiotic sensitivities.
Finally, the most technically difficult step involves obtaining and handling single-cell clones for generating a KO line. When seeding the single cells, each clone expands differently, so the initial screening size is important. We generally start with two 96-well plates of enteroids (5 × 103 cells) to obtain 20–40 single-cell clones for screening. If the first round fails, we will take out initial transduced enteroids after selection from the frozen stock to plate them into single-cell clones again, but increase the number of plates used to be four 96-well plates to obtain ~40–80 single-cell clones. However, the outcome really depends on the gene of interest and the luck to find one KO clone during the screening step. We have found obtaining >40 single-cell clones after selection is critical because the clones that are not growing or expanding cannot be maintained or passaged even if they are KO clones. Our protocol details the critical steps for generating single-cell HIE clones with pipettes and non-sticky tips. A talented researcher should be able to plate high enough numbers of pure single-cell clones at the initial step to obtain multiple independent KO HIE lines for cross-validation of the loss-of-function phenotypes.
Materials
Reagents
Plasmid LentiCRISPRv2 (RRID: Addgene_52961)
Plasmid Hygro-LentiCRISPRv2 (RRID: Addgene_98291; Cell-Based Assay Screening Service Core, BCM)
Plasmid pMD2G (RRID: Addgene_12259)
Plasmid psPAX2 (RRID: Addgene_12260)
BsmBI and NEB buffer 3.1 (NEB, cat. no. R0580S)
T4 DNA ligase and buffers (NEB, cat. no. M0202S)
T4 polynucleotide kinase (PNK; NEB, cat. no. M0201S)
Calf intestinal phosphatase (CIP; NEB, cat. no. M0290S)
Zymoclean Gel DNA Recovery Kit (Zymo Research, cat. no. D4001)
Agarose (VWR, cat. no. 0710-500G)
QIAprep Spin Miniprep Kit (QIAGEN, cat. no. 27104)
QIAGEN Plasmid Midi Kit (QIAGEN, cat. no. 12143)
NucleoSpin Tissue XS Kit (Takara, cat. no. 740952.50)
2X Platinum Green Hot Start PCR Master Mix (TF Invitrogen, cat. no. 13301-012) ▲ CRITICAL High-fidelity DNA polymerase is highly recommended. Other high-fidelity DNA polymerases, such as Q5 High-Fidelity DNA Polymerase (NEB) and KAPA HiFi DNA Polymerase (Sigma-Aldrich), can be used.
One Shot Stbl3 competent cells (Thermo, cat. no. C737303) ▲ CRITICAL Other RecA− competent cells can be used as well to minimize the recombination of long terminal repeat-containing lentiviral plasmids.
Super Optimal broth with Catabolite repression (SOC) medium (Thermo, cat. no. 15544034)
Ampicillin (Sigma, cat. no. A5354-10 mL)
Luria–Bertani (LB) broth and LB agar (Thermo, cat. no. 12780052, 22700025)
Rabbit anti-STATl polyclonal antibody (Cell Signaling Technology, cat. no. 9172, RRID: AB_2198300)
Mouse anti-villin monoclonal antibody (Santa Cruz Biotechnology, cat. no. sc-373997, RRID: AB_10917911)
Guinea pig anti-HuNoV polyclonal antibody (produced by the Estes lab49)
Goat anti-guinea pig Alexa fluor-488 secondary antibody (Thermo, cat. no. A-11073, RRID: AB_2534117)
Primers
hU6 forward primer (5′-GAG GGC CTA TTT CCC ATG ATT-3′) (Integrated DNA Technologies), used in Step 20 for colony PCR and in Step 25 for plasmid sequencing
sgRNA reverse oligo (Integrated DNA Technologies) as the reverse primer used in Step 20 for colony PCR
STAT1 sgRNA forward oligo (5′-CAC CGT CAT GAC CTC CTG TCA CAG C-3′) (Integrated DNA Technologies), used in Steps 111 and 112 to generate STAT1-KO
ST ATI sgRNA reverse oligo (5′-AAA CGC TGT GAC AGG AGG TCA TGA C-3′) (Integrated DNA Technologies), used in Steps 111 and 112 to generate STAT1-KO
WPRE forward primer (5′-GTC CTT TCC ATG GCT GCT C-3′) (Integrated DNA Technologies), used in Step 45 for quantifying lentiviruses by RT-qPCR
WPRE reverse primer (5′-CCG AAG GGA CGT AGC AGA-3′) (Integrated DNA Technologies), used in Step 45 for quantifying lentiviruses by RT-qPCR
Albumin forward primer (5′-TTT GCA GAT GTC AGT GAA AGA GA-3′) (Integrated DNA Technologies), used in Step 45 for quantifying lentiviruses by RT-qPCR
Albumin reverse primer (5′-TGG GGA GGC TAT AGA AAA TAA GG-3′) (Integrated DNA Technologies), used in Step 45 for quantifying lentiviruses by RT-qPCR
STAT1 forward sequencing primer (5′-CCA CAT AAA CTG CTG CTA AA-3′) (Integrated DNA Technologies), used in Steps 111 and 112 to generate STAT1-KO
STAT1 reverse sequencing primer (5′-ACT GAT ACC TAA GAG GTG ATA CAT-3′) (Integrated DNA Technologies), used in Steps 111 and 112 to generate STAT1-KO
Cell culture reagents for both 293FT and HIEs
Dulbecco’s modified Eagle medium (DMEM; Sigma, cat. no. D6429)
Fetal bovine serum (FBS; Sigma, cat. no. F2442)
1× phosphate-buffered saline (PBS; Gibco, cat. no. 14190-144, or any brand but without calcium and magnesium, 0.01 M, pH = 7.4)
Puromycin (InvivoGen, cat. no. ant-pr-1)
Hygromycin-B (InvivoGen, cat. no. ant-hg-1)
Polyethylenimine (PEI) transfection reagent (Sigma, cat. no. 913375)
Polyethylene glycol (PEG)-6000 (Sigma, cat. no. 81260)
Sodium chloride (NaCl; Sigma, cat. no. S5886)
Sodium butyrate (Sigma, cat. no. B5887-250 MG)
0.5 M EDTA (IBI Scientific, cat. no. IB70184)
Polybrene (stock: 10 mg/mL, EMD Millipore, cat. no. TR1003G)
Advanced DMEM–F-12 medium (Gibco, cat. no. 12634-010)
Penicillin/streptomycin (Invitrogen, cat. no. 15140-122)
1 M HEPES (Invitrogen, cat. no. 15630-080)
GlutaMAX-I (Invitrogen, cat. no. 35050-061)
0.05% (wt/vol) trypsin with 0.5 mM EDTA (Gibco, cat. no. 25300-054)
TrypLE Express enzyme (1×), no phenol red (Thermo, cat. no. 12604-013)
Mouse recombinant epidermal growth factor (Invitrogen, cat. no. PMG8043)
Nicotinamide (Sigma, cat. no. N0636)
[Leu15]-Gastrin I (Sigma, cat. no. G9145)
A-83-01 (Tocris Bioscience, cat. no. 2939)
SB202190 (Sigma, cat. no. S7067)
50× B27 supplement (Invitrogen, cat. no. 17502-044)
100× N2 supplement (Invitrogen, cat. no. 17502-048)
N-acetylcysteine (Sigma, cat. no. A9165-5G)
Y-27632 stock solution (Rock inhibitor, Sigma, cat. no. Y0503–5MG, 5 mM in 1× PBS, working concentration 10 μM)
Recovery cell freezing medium (Invitrogen, cat. no. 12648-010)
40 μm cell strainer (Falcon, cat. no. 352340)
Puromycin dihydrochloride solution (10 mg/mL in H2O, Sigma, cat. no. P9620-10ML)
Hygromycin-B solution (50 mg/mL in H2O, Sigma, cat. no. H5527-250MG)
Sodium glycochenodeoxycholate (Sigma, cat. no. G0759-25MG)
Matrigel growth factor reduced basement membrane matrix (Corning, cat. no. 356231)
Biological materials
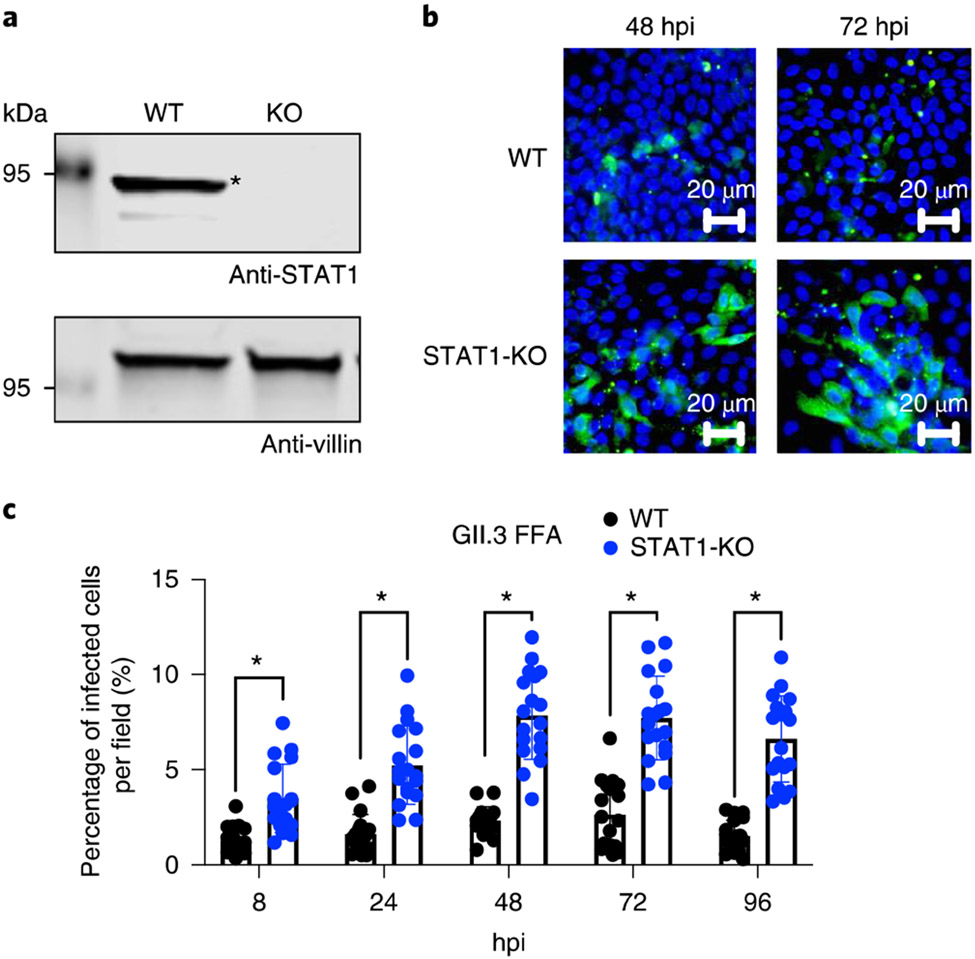
Human embryonic kidney 293FT cells (ATCC, cat. no. PTA-5077, RRID: CVCL_6911)
L-WRN cells (ATCC cat. no. CRL-3276, RRID: CVCL_DA06)
HIEs: J2 jejunal lines obtained from the Digestive Disease Core at BCM (for transduction, the HIEs were cultured in WRNE medium for >4 d for stem cell proliferation)! CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Equipment
Biosafety level 2 facility
Thermal cycler (any brand)
37 °C cell culture incubator (any brand)
Microscope (any brand to observe single-cell HIEs in 96-well plates)
Centrifuge (any brand, with a rotor for 50 mL tubes at 5,000g at 4 °C, and a swinging bucket rotor for 48-well tissue culture plates at 300g)
0.45 μm syringe filters (Corning, cat. no. 431220, surfactant-free cellulose acetate filter)
Rotator (any brand, fixed speed at 20 rpm)
Internally threaded cryogenic storage vials (Thermo, cat. no. 374080)
Six-well tissue culture plates (Corning, cat. no. 3516)
24-well tissue culture plates (Corning, cat. no. 3524)
48-well tissue culture plates (Corning, cat. no. 3548)
96-well tissue culture plates (Corning, cat. no. 3595)
10 cm cell culture dishes (Corning, cat. no. 430167)
P1000 filtered tips (Rainin Pipette Tips RT LTS 1000 μL FX 768A/8, cat. no. 30389223)
P200 filtered tips (nonsticky) (Rainin Pipette Tips RT LTS 200 μL F 960A/10, cat. no. 30389239) ▲ CRITICAL This tip is better for handling Matrigel mixtures with low retention.
Reagent setup
Complete medium without growth factors (CMGF(−) medium)
CMGF(−) medium, also known as basal culture medium, consists of advanced DMEM–F-12 medium supplemented with 100 U/mL penicillin–streptomycin, 10 mM HEPES buffer and 1× GlutaMAX-I1,47. This can be stored at 4 °C for up to 2 months.
50 mL of complete medium with growth factors (WRNE medium)
WRNE medium, also known as enteroids proliferation medium38,50, consists of 23 mL CMGF(−) medium with 25 mL L-WRN cell conditioned medium, supplemented with 50 ng/mL epidermal growth factor, 10 mM nicotinamide, 10 nM gastrin I, 500 nM A-83-01, 10 μM SB202190, 1× B27 supplement, 1× N2 supplement and 1 mM N-acetylcysteine. This can be stored at −80 °C for 2 months.
Differentiation medium
Differentiation medium consists of the same components as those of WRNE medium without the addition of WRN conditioned medium, nicotinamide and SB202190. This can be stored at −80 °C for 2 months.
Procedure
Part 1. Preparation of Cas9 expressing lentivirus vector with sgRNA targeting gene of interest ● Timing 8 h + overnight + 4 h
Select a CRISPR–Cas9 targeting sequence of the target gene in the genome. To identify an sgRNA target sequence using the GenScript sgRNA database (https://www.genscript.com/gRNA-database.html) or other webtools, input the gene name, symbol or ID and search for human SpCas9 gRNA sequences.
-
Select at least two sgRNAs that have been pretested to design annealing primers for your insert.
▲CRITICAL STEP The GenScript sgRNA database has tested those sgRNAs in other types of cells. If there is no choice, you need to design a sgRNA by selecting a 20 bp sequence followed by a PAM site (NGG) in the first exon of your gene of interest (look at ‘sgRNA design’ under ‘Experimental design’).
▲ CRITICAL STEP It is better to select more than one sgRNA for knocking out genes for a higher success rate. One of them often works better than the other.
For each sgRNA target sequence, design two annealing oligos according to the guidelines for lentiCRISPRv2 provided by Feng Zhang’s laboratory (Massachusetts Institute of Technology) in Addgene (Addgene, cat. no. 52961).
Add one extra 5′ G to the 20mer protospacer targeting sequence if it does not start with a G.
-
Order oligos with standard desalting, no modification and 25 μmol/each from IDT DNA. A detailed protocol can be found on the Addgene website: https://media.addgene.org/data/plasmids/52/52961/52961-attachment_B3xTwla0bkYD.pdf.
▲ CRITICAL STEP The self-annealing insert should form dsDNA with 5′ CACC at one overhang and 5′ AAAC at another overhang after annealing (Steps 10–12), which are critical for ligating with the BsmBI-digested vector.
-
Digest lentiCRISPRv2 vector with the BsmBI enzyme as follows at 55 °C for 30 min.
LentiCRISPRv2 (~100 ng/μL) 20 μL (−2 μg) BsmBI (NEB, cat. no. R0580S) 1.5 μL 10× NEB buffer 3.1 4 μL ddH2O 14.5 μL Total volume 40 μL ▲ CRITICAL STEP Restriction enzyme volume should not exceed 10% (vol/vol) of the final volume, or the glycerol in the enzyme preparations will interfere with the reaction. Do not digest >30 min or the product will degrade. Smearing in the following gel electroporation is indicative of overdigested vector.
Prevention of self-ligation of digested backbone. Add 1 μL of CIP (or the new product Quick CIP with a similar result) directly into the reaction mix from Step 6 to prevent vector self-ligation, and incubate at 37 °C for 20 min.
-
Gel extraction of LentiCRISPRv2 digested backbone. Use a 1% (wt/vol) agarose gel to separate the product (loading: 25 μL/well, voltage: 100 V for 45 min), cut the single larger band at ~11 kb (do not cut the byproduct filler band at 2 kb, referring to the guidelines from the Zhang laboratory).
? TROUBLESHOOTING
-
Purify the band using the Zymoclean Gel DNA Recovery Kit, following the manufacturer’s instructions, and elute with 30 μL ddH2O to obtain the ready-to-use lentiCRISPRv2 digested backbone.
■ PAUSE POINT The digested backbone can be stored at −20 °C for months and can be used to ligate different insert gRNAs for multiple experiments.
- Perform insert self-annealing and phosphorylation (freshly prepared). Resuspend the sgRNA oligos obtained from Step 5 with ddH2O to generate 100 μM stocks (storage: −20 °C), then perform insert annealing and phosphorylation by T4 PNK as follows:
Forward sgRNA oligo (100 μM) 1 μL Reverse sgRNA oligo (100 μM) 1 μL T4 DNA ligase buffer 1 μL T4 PNK 0.5 μL ddH2O 6.5 μL Total 10 μL - Set up the PCR machine for the annealing and cool down period as follows:
Step Temperature Time Cycle number Phosphorylation 37 °C 45 min 1 Denaturation 95 °C 2 min, 30 s 1 Cool down 1a −0.3 °C/s until 50 °C - 1 Holda 50 °C 10 s 1 Cool down 2a −0.3 °C/s until 22 °C - 1 Finish On ice - 1 aIf you do not use a PCR machine for −0.3 °C/s cooling down, you can leave the product at room temperature CRT, 20-25 °C) for 30 min for cooling down, then put the tube on ice before the next step. -
Dilute 1 μL of product into 400 μL ddH2O to obtain the ready-to-use insert solution.
▲ CRITICAL STEP Highly recommended to use freshly prepared insert immediately for ligation although it can be stored at −20 °C for <3 months as well.
-
Perform ligation reaction between the vector (from Step 9) and insert (from Step 12) at 16 °C in a PCR machine, or at RT for >4 h as follows:
LentiCRISPRv2-BsmBI digested (Step 9) 1 μL Insert (1:400 diluted; Step 12) 1 μL T4 DNA ligation buffer 0.5 μL T4 DNA ligase 0.5 μL ddH2O 2 μL Total 5 μL ▲ CRITICAL STEP For the T4 ligation buffer, wait until any white precipitate (ATPs) is completely dissolved before use.
After ligation, transform the whole 5 μL product into 50 μL of One Shot Stbl3 chemically competent cells (or other RecA− competent cells to prevent recombination of the long terminal repeat-containing lentiviral vector).
Have a negative control: put 1 μL digested vector without the insert into 50 μL competent cells to check the digestion efficiency.
Incubate the mixture on ice for 15 min, heat shock at 42 °C for 45 s and put back on ice for 2 min.
Add 250 μL of prewarmed SOC medium to each vial, and shake the vial at 37 °C, 225 rpm for 1 h for recovery.
-
Spread 100 μL of bacteria on an LB plate containing 100 μg/mL of ampicillin. Incubate the LB plates at 37 °C overnight. A well-processed vector should result in no colonies without insert ligation.
? TROUBLESHOOTING
-
After overnight culture, pick up seven colonies with a sterile loop and inoculate on a new LB/Amp plate and assign a number 1–7 to distinguish each colony, then incubate the plate at 37 °C for 8 h.
▲ CRITICAL STEP Usually the cloning efficiency is high, so picking seven colonies will result in at least one positive colony for sequencing.
- For those seven colonies from Step 19, after inoculating the plate, put the remaining portion of the assigned colony on the sterile loop into numbered PCR tubes to do colony PCR as follows. The last PCR tube is the negative control: the PCR mixture without colony.
Colony 1× 10 μM hU6-Fwd primer 0.5 μL 10 μM of sgRNA-Rev oligo (dilute from the 100 μM stock ordered in Step 5) 0.5 μL 2× Platinum Green Hot Start PCR Master Mix 10 μL ddH2O 9 μL Total 20 μL - Perform PCR on the reactions in Step 20 using the following cycling conditions:
Step Temperature Time Cycle number Pre-incubation 94 °C 5 min 1 Denature 94 °C 30 s Repeat Annealing 58 °C (adjusted by the GC contents of the primers) 30 s 39 times Extension 72 °C 30 s (product ~200 bp) Final extension 72 °C 5 min 1 Hold - 1 Electrophoresis to confirm colony PCR products. Run 10 μL per well of PCR products plus DNA sample loading buffer on a 2% (wt/vol) agarose DNA gel at 80 V for 30 min. A positive colony will result in a 200 bp PCR product.
Culture two bacterial colonies with positive PCR results from the plate in Step 19 in 3 m1 LB overnight.
-
Miniprep using QIAprep Spin Miniprep Kit, and elute in 30 μL of ddH2O.
▲ CRITICAL STEP When doing minipreps, keep some E. coli culture to make glycerol stocks (500 μL of culture with 500 μL of 50% (vol/vol) glycerol in a 2 mL screw top tube or cryovial and gently mix; store at −80 °C or liquid nitrogen).
Send plasmids from at least two individual positive colonies for sequencing using the hU6-Fwd primer.
Part 2. Lentivirus packaging and concentration • Timing Overnight + 0.5 h + 4 d + 3 h
-
26
Prepare plasmids with QIAGEN Plasmid Midi Kit: pMD2G (VSV-G envelope-expressing plasmid), psPAX2 (HIV-1 gag pol-expressing plasmid) and LentiCRISPRv2-sgRNA (cloned previously in Part 1).
▲ CRITICAL STEP It is best to use the QIAGEN Plasmid Midi Kit or other comparable midiprep kits to obtain high-quality DNA for packaging plasmids, and ensure the removal of endotoxin that is required for successful transfection into 293FT cells, which are better cells than conventional 293T cells to make lentiviruses.
▲ CRITICAL STEP The 293FT cell line is a fast-growing derivative of HEK 293T cells, which is useful for lentiviral production because lentiviral titers are around four to five times higher than titers produced from 293T cells.
-
27
Take 293FT cells out from liquid nitrogen, and passage the cells more than two times and less than ten times for the best growing and transfection potential.
-
28
Coat six wells of a six-well cell culture plate (Corning) with 0.01% (wt/vol) poly-d-lysine (EMD Millipore, cat. no. A-003-E) for 15 min, then aspirate the coating solution using a P1000 tip and let the surface dry for the best cell attachment.
-
29
Plate 293FT cells at 7 × 105 cells per well, to achieve 80–90% confluency on the day of transfection; then incubate at 37 °C in 5% CO2 and wait for 16–20 h to have cells fully attached to the wells (Fig. 2a).
▲ CRITICAL STEP The transfection efficiency is critical for obtaining high-titer lentiviruses so be sure the confluency of 293FT cells at transfection is 80–90%.
? TROUBLESHOOTING
-
30PEI-mediated transfection of the required packaging plasmids. On the day of transfection, prepare PEI-DMEM in one tube as follows:
Reagent Amount 6× for six wells PEI (1 μg/μL), diluted with ddH2O 7 μL 42 μL DMEM without serum 160 μL 960 μL -
31Vortex and spin down, then add the following plasmid DNA into the PEI–DMEM tubes.
Reagent (plasmids prepared by midiprep in Step 26) Amount 6× for six wells pMD2.G (~1 μg/μL) 0.6 μg 3.6 μL psPAX2 (~1 μg/μL) 1 μg 6 μL Lentiviral vector (~1 μg/μL) 1.6 μg 9.6 μL -
32
Vortex and spin down. Incubate at RT for 20 min.
-
33
Prior to transfection, replace cell medium in plates from Step 29 with fresh DMEM with 10% (vol/vol) FBS, 2 mL per well.
-
34
After 20 min, gently dropwise add 167 μL per well of transfection mix from Step 32 into each well. Gently rock the plate by hand to mix well, and put into a 37 °C incubator in 5% CO2.
-
35
At 16 h post-transfection, replace the cell medium with fresh DMEM containing 10% (vol/vol) FBS and 5 mM sodium butyrate to increase the yield (2 mL per well), but only leave cells in this medium until 24 h post-transfection since it is toxic to the cells.
▲ CRITICAL STEP After transfection, the cells will have a rounded-up shape and the medium still contains residual transfection reagent containing plasmid DNA that needs to be removed. The medium should be aspirated carefully every time when changing the medium, and new medium should be added very gently without detaching the fragile attached cells that will continue to produce lentiviruses. Detachment of the transduced 293FT cells will decrease the final titer of lentiviruses.
-
36
At 24 h post-transfection (Fig. 2b), collect the supernatant (2 ml × 6 wells = 12 mL) and store at 4 °C for <1 week before lentivirus concentration. Add 2 mL per well of fresh medium into each well after collection.
-
37
At 36 and 48 h post-transfection (Fig. 2c), collect the supernatants, pool them together with the supernatant collected at 24 h post transfection and store at 4 °C (total: 36 mL per virus). Add 2 mL per well fresh medium into each well after collection every time.
-
38
At 60 and 72 h post-transfection, collect the supernatants, pool them together with the supernatant from Step 37 and store at 4 °C. (total: ~60 mL per virus) Add 2 mL per well fresh medium into each well after collection until the last time.
-
39
Divide the 60 mL per virus pooled supernatants into two tubes, 30 mL each.
-
40
Centrifuge the divided supernatant at 300g, 4 °C for 5 min to remove cell debris, then filter the supernatant with a 0.45 μm syringe filter into a new 50 mL tube.
▲ CRITICAL STEP Change the filter if the medium is hard to pass through; do not press too hard or you might break the filter membrane.
-
41To each tube of filtered supernatant from Step 40 (two tubes per construct), add the following reagents:
Reagent Volume (total: 40 mL) Filtered supernatant 27 mL (adjust by adding more 1× DMEM) 50% (wt/vol) PEG-6000, dissolved in ddH2O and autoclaved 6.4 mL 4 M NaCI 2.9 mL 1× PBS 3.0 mL The PEG-6000, NaCI and PBS are all autoclaved or sterile.Mix well; put on a rotator to constantly rotate at 20 rpm at 4 °C overnight.
-
42
Centrifuge at 5,000g for 45 min at 4 °C, and discard the supernatant to get lentivirus pellets (white pellets).
-
43
Resuspend each pellet in 600 μL of CMGF(−) medium, and aliquot this into new tubes, 100–300 μL per tube (you will use 300 μL per transduction of a well of HIEs in a 24-well plate in Step 48). In summary, you will obtain 1,200 μL of lentiviruses per sgRNA construct from six wells of 293FT cells in a six-well plate.
-
44
Put the aliquots of viruses into −80 °C for storage. The viruses can be stored for 2–3 years with infectivity. We recommend not to freeze–thaw the viruses many times.
-
45
If desired, the lentiviruses can be titered in the following way51-53. The titer can be determined by re-infecting 293FT cells with prepared lentiviruses followed by RT-qPCR using two pairs of primers, for a lentiviral-specific transgene (WPRE gene) and an endogenous reference gene (albumin gene). Plasmids carrying a single copy of the WPRE gene (e.g., LentiCRISPRv2) can be used as the standard curve to determine the expression amount of the WPRE gene normalized by the albumin gene in transduced 293FT cells52.
-
46
Serially dilute the tested lentivirus stock if necessary to obtain a Ct value within the linear range of the standard curve.
-
47
Normalize the batch-to-batch variation of lentiviruses by the titers determined by RT-qPCR, allowing for the adjustment of the amount of virus to use in the standard inoculation volume (~300 μL per well) for HIE transduction.
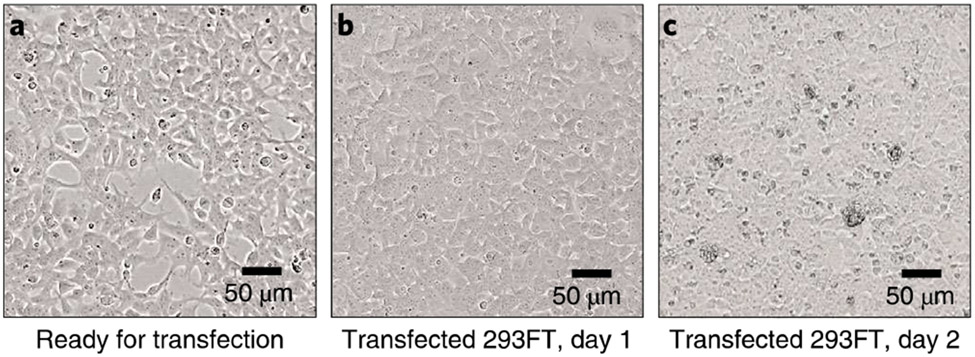
Fig. 2 ∣. The 293FT cells during lentivirus packaging.
a, Cells with 70-80% confluency before transfection. b, Cell morphology at day 1 after transfection. c, Cells start to produce lentiviruses and become less attached at day 2 after transfection. 10× objective lens.
Part 3. Lentivirus transduction of HIEs • Timing 4 d + 3.5 h
▲ CRITICAL The following steps are for transducing one well of J2 HIEs in a 24-well plate with 300 μL of lentiviruses.
-
48
Maintain and passage HIE cultures established previously from adult surgical or biopsy tissue in Matrigel matrix as multilobular 3D cultures as previously described11,14.
-
49
Culture 3D jejunal J2 HIEs in WRNE medium embedded in Matrigel as described in ref.13. Usually only 1 well of 3D HIEs in a 24-well plate is needed for transduction with each Cas9–sgRNA vector. A mock control well (HIEs without transduction but with antibiotic selection) is also required to determine the working concentration of antibiotic to kill nontransduced cells.
▲ CRITICAL STEP For optimal lentivirus transduction, HIEs should be cultured in WRNE proliferation medium for at least 4 d to ensure they are in a proliferative state. The transduction efficiency varies in different enteroid lines. The most important factor for successful transduction is the health/growth rate of the enteroid lines. Unhealthy enteroid lines are harder to cultivate and transduce.
▲ CRITICAL STEP One well of 3D HIEs in a 24-well plate can be split into two tubes in the end, so at least you need to start withN/2 wells of 3D HIEs, where N is the number of different lentiviruses you want to transduce.
-
50
Trypsin digestion of HIEs to generate single cells. For each HIE well, aspirate the WRNE medium by pipetting, and add ice-cold 1× PBS (to dissolve Matrigel) with 0.5 mM EDTA (1:1,000 dilution from the 0.5 M stock), 500 μL per well to dissolve the embedded Matrigel.
-
51
Use a P1000 tip to disrupt the Matrigel and scrape the HIEs from the wells, then pipette up and down multiple times (avoid making bubbles when pipetting).
-
52
Transfer the cells into a 15 mL tube, and centrifuge at 300g for 5 min at 4 °C.
-
53
Aspirate the supernatant, and add 500 μL/tuhe of 0.05% (wt/vol) trypsin with 0.5 mM EDTA, or use 1× TrypLE as an alternative, pipetting three to four times to dissociate the pellet, then put into a 37 °C incubator for 4 min to digest the HIEs.
-
54
Add 1 mL of CMGF(−) with 10% (vol/vol) FBS to neutralize the trypsin, then pipette 50 times to fully disperse the HIEs into single cells. Centrifuge at 300g for 5 min at 4 °C.
▲ CRITICAL STEP Pipetting 50 times is required for complete single-cell dissociation of HIEs. High numbers of HIE clusters result in decreased transduction efficiency.
? TROUBLESHOOTING
-
55
Aspirate the supernatant, and resuspend the pellets with the desired amount of CMGF(−). If you have ‘N’ lentiviruses, then you will split the digested HIEs into ‘N + 1’ microtubes to have one tube for the mock control. So, in this case, you need to resuspend with 500× (N + 1) μL of CMGF(−) medium before splitting.
-
56
Pipette and then disperse into multiple microtubes, 500 μL per tube. Pellet the digested HIEs in all tubes by centrifuging at 300g for 5 min at 4 °C, then aspirate the supernatant carefully without touching the pellet, and put all tubes on ice.
-
57
For each lentivirus transduction, prepare 300 μL of concentrated lentiviruses in a microtube from Step 43, add Y-27632 to a final concentration of 10 μM (1:500 from the 5 mM stock), 8 μL of polybrene (~1:1,200 from the 10 mM stock) and mix (still ~300 μL per tube).
-
58
Transfer this mixture to the microtube containing the digested HIE pellet from Step 56. For the control group, do not add the lentivirus, but add 300 μL of CMGF(−) with Y-27632 and polybrene into the microtube.
-
59
Pipette a few times to mix HIEs with lentiviruses, then transfer the contents of each tube to a well of a new 48-well plate.
-
60
Put the 48-well plate into a 37 °C incubator for 5 min. Seal the plate with parafilm, and centrifuge with a swinging bucket centrifuge at 300g for 60 min at RT for spin inoculation.
-
61
Aspirate and discard the supernatant by pipetting carefully without disrupting the cell layer formed at the bottom of the well in the 48-well plate.
-
62
Resuspend the cell layer in each well with 500 μL per well ice-cold CMGF(−), transfer from the plate to a new microtube and centrifuge at 400g for 5 min to pellet the transduced HIEs.
-
63
Aspirate and discard the supernatant. Resuspend the pellets in Matrigel (90 μL Matrigel for one tube, which can be seeded into 3 wells in a 24-well plate. So, for ‘N’ transduction microtubes, we can seed into ‘3N’ wells in a 24-well plate).
-
64
Incubate at 37 °C for 10 min to solidify the Matrigel, then add 500 μL of WRNE proliferation medium containing 10 mM Y-27632 to reduce cell death into each well containing the HIEs.
Part 4. Recovery and selection for HIEs • Timing 2 weeks
-
65
Change the medium every other day for 4–5 d after the transduction to allow the HIEs to recover, and then start the antibiotic selection.
-
66
For each lentivirus construct, evenly divide the transduced, recovered HIEs into 6 different wells in a 24-well plate for evaluating antibiotic selection.
-
67
Keep one well without antibiotic as a control, and add puromycin at 0.5, 1, 2, 4 and 8 μg/mL to the remaining five wells, or hygromycin-B at 50, 100, 150, 200 and 500 μg/mL to the remaining five wells, respectively depending on the selection marker.
▲ CRITICAL STEP In our J2 jejunal HIEs, we found the following optimal concentrations: puromycin: 2 μg/mL for LentiCRISPRv2 constructs; hygromycin-B: 150 μg/mL for Hygro-LentiCRISPRv2 constructs.
-
68
Change the medium containing antibiotics every other day. HIEs without transduction will die within 2–3 weeks.
-
69
Routinely passage the surviving HIEs at least once per 10 d and before too many dead cells accumulate as observed visually13.
-
70
For the wells without antibiotic, you can freeze a part of the HIEs after passaging to make stocks with recovery cell freezing medium11,14 To do so, centrifuge HIEs at 300g for 5 min, remove supernatant then resuspend the pellet with recovery cell freezing medium, 500 μL per tube and transfer into internally threaded cryogenic storage vials (500 μL per vial). Put all vials into a cryogenic storage at −80 °C overnight, then transfer to a liquid nitrogen tank for long-term storage as for normal cell lines.
-
71
Check the surviving HIEs under the microscope every other day. Determine the optimal antibiotic concentration based on the highest concentration with surviving cells in the well after 1 week.
-
72
Select cells for a minimum of 2 weeks but optimally 3 weeks to eliminate nontransduced cells. After 3 weeks of selection, change to using medium without antibiotic to passage and expand the transduced HIEs into multiple wells.
▲ CRITICAL STEP A shorter selection time will result in too many negative (WT) clones during the subsequent single-cell cloning steps.
-
73
Freeze stocks of selected HIEs with recovery cell freezing medium11,14 as described in Step 70, and use the remaining cells for single-cell cloning as described below.
? TROUBLESHOOTING
Part 5. Single-cell cloning • Timing 3 h + 3 weeks
-
74
Passage the HIEs with trypsin digestion as when doing transduction in Step 50, but use a 40 μm cell strainer12,15 to only retain single cells (Fig. 3a).
-
75
At day 2 after passaging, HIE clones can be seen (similar to Fig. 3b) and can now be single-cell-cloned into 96-well plates. To do so, take one well of HIEs from Step 74, aspirate and discard the medium from the well and add 500 μL of ice-cold 1× PBS.
-
76
Centrifuge at 300g, 4 °C for 5 min, and then aspirate and discard the supernatant.
-
77
Resuspend the HIEs in a tube with 30 μL of Matrigel as expanded HIE clones. Make three additional 1:50 serial dilutions into Matrigel pipetting well between each dilution.
-
78
Inoculate 3 μL per well into a 96-well plate of Matrigel. This inoculation volume would ideally contain the correct amount of cells to target approximately one colony (expanded round-shaped single HIE, Fig. 3b) into each well (use nonsticky filter P200 tip).
▲ CRITICAL STEP To determine if the Matrigel cell suspensions are sufficiently diluted, start by plating 3 μL to seed one well, and check by bright-field microscopy whether the dilution is optional to obtain zero to one single cells per well as single-cell clones.
-
79
Inoculate enough wells (two 96-well plates for the first round of screening), and store at 37 °C to solidify the Matrigel for 10 min.
▲ CRITICAL STEP Depending on the efficiency of transduction and selection for an individual KO, you may need to have at least two plates of 96-well plates (192 wells) for the first round of single-cell selection. The number can be larger and depends on the difficulty of getting a KO. We generally start with two 96-well plates of enteroids (5 × 103 cells) to obtain 20–40 single-cell clones to screen. If the first round fails, we will take out the transduced enteroids from liquid nitrogen that were frozen after puromycin selection (in Step 73) to plate them into single-cell clones. We increase the number of 96-well plates to four to obtain ~40–80 single-cell clones. However, the outcome really depends on the gene of interest and the ability to find one KO clone during the screening step.
▲ CRITICAL STEP Since single cells do not survive well by themselves, it is useful to use a two-step process to obtain single-cell clones. First, perform serial dilutions that result in multiple single cells in one well; once each single cell grows into an enteroid in that well, collect and dilute those enteroids into more new wells as a single enteroid clone per well. Each well with a single large enteroid is a clone.
▲ CRITICAL STEP The growing efficiency of enteroids needs to be ideal to obtain single-cell clones from transduced cells. In addition, if the gene is an essential gene, it will be harder or impossible to get KO clones directly. You might need to generate a conditional KO line, which is more complicated.
? TROUBLESHOOTING
-
80
After solidifying the Matrigel, add 50 μL of WRNE medium with 10 μM Y-27632 (1:500 dilution from the stock solution) but without antibiotics (to reduce the growth pressure on each single HIE) and culture those single clones at 37 °C.
-
81
Culture for 5 d to obtain and visualize enlarged single clones (Fig. 4a-c). The dying single-cell clones (Fig. 4d-f) and wells with multiple clones (Fig. 4g,h) should be discarded.
-
82
Label wells with single-cell clones, and change medium every 3 d (WRNE medium with 10 μM Y-27632).
-
83
Visualize the size of single-cell clones daily by an inverted microscope. After 2–3 weeks when the single-cell-derived HIEs are big enough (>100 μm; Fig. 4c), they are ready to be passaged and expanded for downstream validation by genomic DNA extraction.
▲ CRITICAL STEP if you see multiple cells at the beginning but only one single clone grows up, you can still define it as a single-cell clone because the other dead cells will be degraded later.
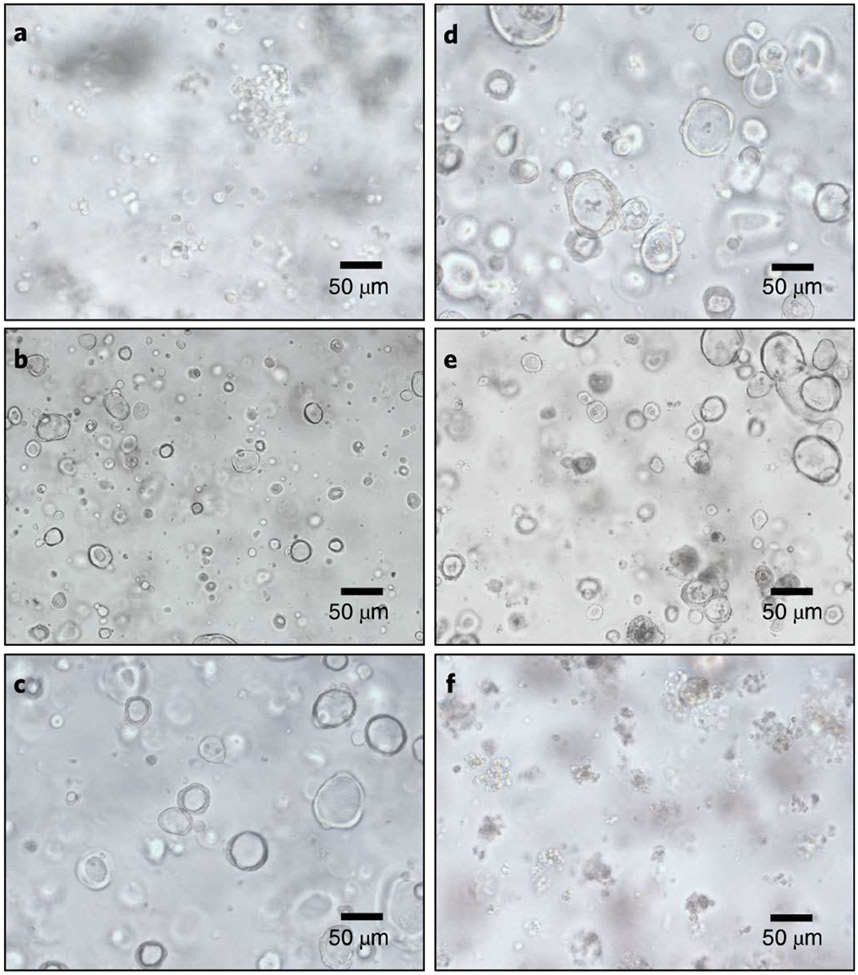
Fig. 3 ∣. HIE morphologies during transduction and puromycin selection.
a, Single-cell HIEs for lentiviral transduction at day 0. b, Expanded single-cell clones after 2 d of transduction. c,d, Expanded HIE clones 4 d after transduction, which are ready to be selected with puromycin. e, Transduced HIEs treated with puromycin for 4 d. f, Nontransduced HIEs treated with puromycin for 4 d with dying and dead cells. 20× objective lens.
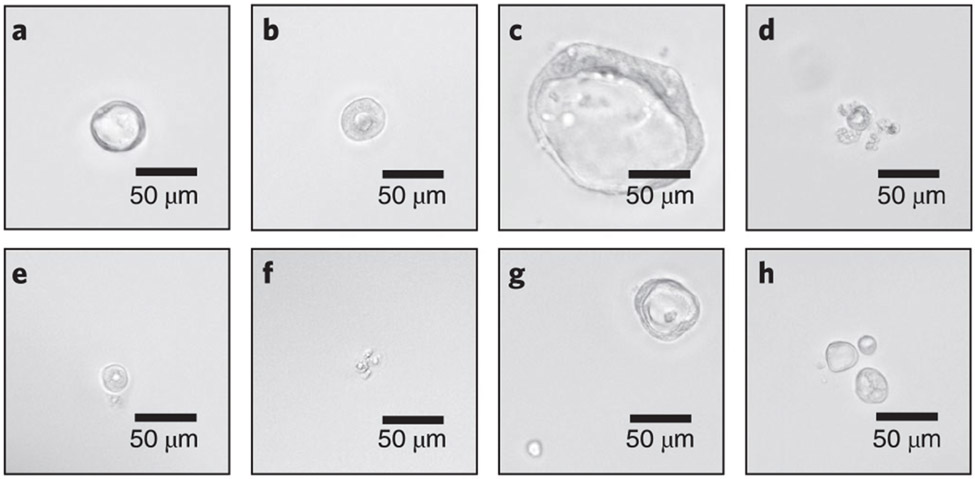
Fig. 4 ∣. The selection criteria for single-cell HIE clones.
Different HIE clones can be observed in each well of 96-well plates under a bright-field microscope. a,b, Surviving single-cell clones growing from individual transduced HIEs. c, A surviving and expanding single-cell clone that is ready for passaging. d-f, Dying or dead single-cell clones that need to be excluded. g,h, Multiple surviving clones in one well that need to be excluded as well since they are mixed populations. 20× objective lens.
Part 6. Single HIE clone passaging and sequence verification • Timing 1 week + 8 h
-
84
Aspirate and discard as much medium from the well as possible (or the FBS inside the medium will interfere with the trypsin digestion).
-
85
Wash the HIE with ice-cold 1× PBS, 50 μL per well once. Aspirate and discard as much PBS as possible, but do not disrupt the Matrigel.
-
86
Add 30 μL of 0.05% (wt/vol) trypsin with 0.5 mM EDTA, and scratch down the Matrigel with a P200 tip, then transfer the whole solution into a new 1.5 mL microtube, pipetting four to five times. Keep the used P200 tip in one hand.
-
87
Use a new P200 tip to add another 30 μL of 0.05% (wt/vol) trypsin with EDTA into the same well, then use the previous first tip to wash the well to get residual cells, transfer to the same microtube (total: 60 μL of trypsin with EDTA per tube).
-
88
Put the microtube containing the HIE into a 37 °C incubator for 5 min to digest the single HIE, then add 150 μL per tube of CMGF(−) with 10% (vol/vol) FBS to stop the reaction.
-
89
Pipette 30 times to completely dissociate the HIE into single cells. Centrifuge at 400g, 4 °C for 5 min to pellet the cells, then remove and discard the medium.
-
90
Resuspend the cells with 60 μL of Matrigel per tube, and then inoculate 15 μL into 4 individual wells in a new 48-well plate.
-
91
Solidify the Matrigel at 37 °C for 10 min, then add 200 μL per well of WRNE medium with Y-27632 for growing.
-
92
Incubate at 37 °C for 1–2 weeks, and visualize daily by an inverted microscope. After 1–2 weeks of incubation, each passaged well should have enough growing HIEs for sequencing.
▲ CRITICAL STEP By chance, 50% of single-cell clones will be lost after passaging, so you need to passage more wells at once to obtain more growing clones to be sequenced. Usually, we passage 20–40 single-cell clones at once to get at least 10–20 growing individual clones for further sequencing.
? TROUBLESHOOTING
-
93
Take one well of each clone (in the 48-well plate from Steps 90-92) to extract genomic DNA for sequencing. Remove the medium, resuspend with 1× cold PBS, 500 μL per well, centrifuge at 300g, 4 °C for 5 min to pellet the HIEs, remove supernatant and discard to obtain the HIE pellet.
-
94
Extract genomic DNA using the NucleoSpin Tissue XS Kit with its extraction protocol https://www.takarabio.com/x19686?view=documents) for animal tissue.
-
95
Resuspend the pellet in 80 μL of buffer Tl with 8 μL proteinase K (included in the kit, freshly prepared), vortex for 5 s for three times, and briefly spin down, then incubate at 56 °C for 1 h.
-
96
Add 80 μL of buffer B3, vortex and briefly spin down, and put at 70 °C for 5 min, then cool down to RT.
-
97
Add 80 μL of ethanol (96–100%, vol/vol) when the mixture has cooled down, vortex and briefly spin down, then transfer the whole mixture into one Nucleospin column tube and centrifuge at 16,000g for 5 min. Discard the flowthrough.
-
98
Add 50 μL of buffer B5, centrifuge at 16,000g for 1 min and discard the flowthrough.
-
99
Add 50 μL of buffer B5 again (second wash), centrifuge at 16,000g for 1 min, discard the flowthrough and re-centrifuge at 16,000g for 3 min to remove all residual buffer B5.
▲ CRITICAL STEP The buffer B5 contains EtOH, which will interrupt the following PCR reaction, so it needs to be completely removed.
-
100
Put the column in a new 1.5 mL microtube, add 20 μL of buffer BE, and let the column stand for 1 min, then centrifuge at 16,000g for 2 min to elute the genomic DNA product.
-
101
Measure the gDNA concentration by Nanodrop, but usually if we put 3 μL of eluted genomic DNA for one PCR reaction, there will be enough PCR product for sequencing.
-
102
Design forward and reverse primers that will be used for both amplification of the target site and sequencing. Select a forward primer ~300 bp upstream to the target site and a reverse primer ~300 bp downstream to the target site, to obtain an amplicon ~500–600 bp for sequencing. The annealing temperature should be adjusted based on the GC contents of the primers.
-
103Set up the PCR reaction as follows with the reagent 2× Platinum Green Hot Start PCR Master Mix.
DNA (100-200 ng genomic DNA) from Step 101 3 μL 10 μM Fwd primer (Step 102) 0.5 μL 10 μM Rev primer (Step 102) 0.5 μL 2× Platinum supermix 15 μL ddH2O 11 μL Total 30 μL -
104Perform PCR using the following cycling conditions:
Step Temperature Time Cycle number Pre-incubation 94 °C 5 min 1 Denature 94 °C 30 s Repeat 39 times Annealing 58 °C (adjusted by the GC content of the primers) 30 s Extension 72 °C 40 s (product ~600 bp) Final extension 72 °C 5 min 1 Hold 4 °C - 1 -
105
Run the PCR products on a 2% (wt/vol) agarose DNA gel with 30 μL per well (80 V for 45 min).
-
106
Cut the 600 bp band for gel extraction with Zymoclean Gel DNA Recovery Kit, elute with 30 μL of ddHzO and determine the DNA concentration by Nanodrop.
-
107
Send for sequencing with either Fwd or Rev primer from Step 102, then analyze using the CRISPR-ID website (http://crispid.gbiomed.kuleuven.be) with the reference wild-type sequence downloaded from PubMed or GeneCards.
-
108
By resolving the signals from the two sequenced alleles, the final goal is to identify clones that contain frame-shift mutations in both alleles of the target gene.
-
109
The sequencing data will also indicate if you have obtained a single-cell clone or a multicellular enteroid. If you see overlapping peaks in the targeting site, you might have a mixed population, which can be subcloned again to obtain single-cell clones or just be discarded.
? TROUBLESHOOTING
Part 7. Using KO lines to evaluate viral replication and host restriction • Timing 2 months + 1 week
▲ CRITICAL To identify host restriction factors, we knock out individual innate immunity genes previously discovered to be upregulated by viral infection. One example is knocking out STATl in HIEs.
-
110As an example, generate STATl-KO HIEs following the Procedure to this point by transducing the lentiCRISPRv2 vector expressing Cas9 and STATl sgRNA at the target site with the primers as follows:
Gene Target site (5′-3′) sgRNA Fwd/Rev (5′-3′) Sequencing primers, Fwd/Rev (5′-3′) STATl TCATGACCTCCT
GTCACAGC TGGCACC G TCATGACCTCCTG TCACAGC ACCACATAAACTGCTGCTAAA AAAC GCTGTGACAGGAGGTCAT GA C ACTGATACCTAAGAGGTGATACAT -
111
Verify the KO by genomic DNA sequencing (see Part 6 of the Procedure; Fig. 5a) and immunoblots (see ref. 12) with rabbit anti-STATl polyclonal antibody (Fig. 6a).
-
112
Compare phenotypes of virus replication in different jejunal HIE cultures (J2-WT, STATl-KO) infected with GII.4[P31] (Sydney 2012 variant: TCH12-580 strain) or GII.3[P21] (TCH04-577 strain) as previously described11,14. GII.4 is a pandemic strain accounting for 70% of all HuNoV cases in human patients; GII.3 is a nonpandemic HuNoV strain that requires bile acid for infection.
-
113
Dissociate 3D HIEs with 0.05% (wt/vol) trypsin/0.5 mM EDTA, and seed onto collagen IV-coated 96-well plates to make 2D monolayers.
-
114
Following culture in complete medium with growth factors (WRNE) for 24 h at 37 °C in 5% CO2, and plate monolayers in differentiation medium for 5 d.
-
115
For HuNoV infection (as discussed in Step 112), inoculate monolayers prepared in Steps 113–114 with 10% (wt/vol) stool filtrate containing 4.3 × 106 genome equivalents per well of HuNoV for 1 h at 37 °C in 5% CO2 for GII.3 fluorescent focus assay staining12.
-
116
Wash the monolayers twice with CMGF(−) medium to remove unbound viruses.
-
117
Harvest three inoculated wells for RNA extraction to determine the genome equivalents remaining in inoculated cultures at 1 h post-infection.
-
118
Further incubate three additional inoculated cultures in differentiation medium with 500 μM sodium glycochenodeoxycholate for 24 h or indicated timepoints for RT-qPCR to determine HuNoV replication or for staining of HuNoV-infected cells (Fig. 6b,c)11,54. The protocol for immunoblot and immunostaining can be obtained from ref. 12.
▲ CRITICAL STEP Each experiment should be performed at least three times with three technical replicates for each condition.
Fig. 5 ∣. The comparison between KO and wild-type sequencing results.
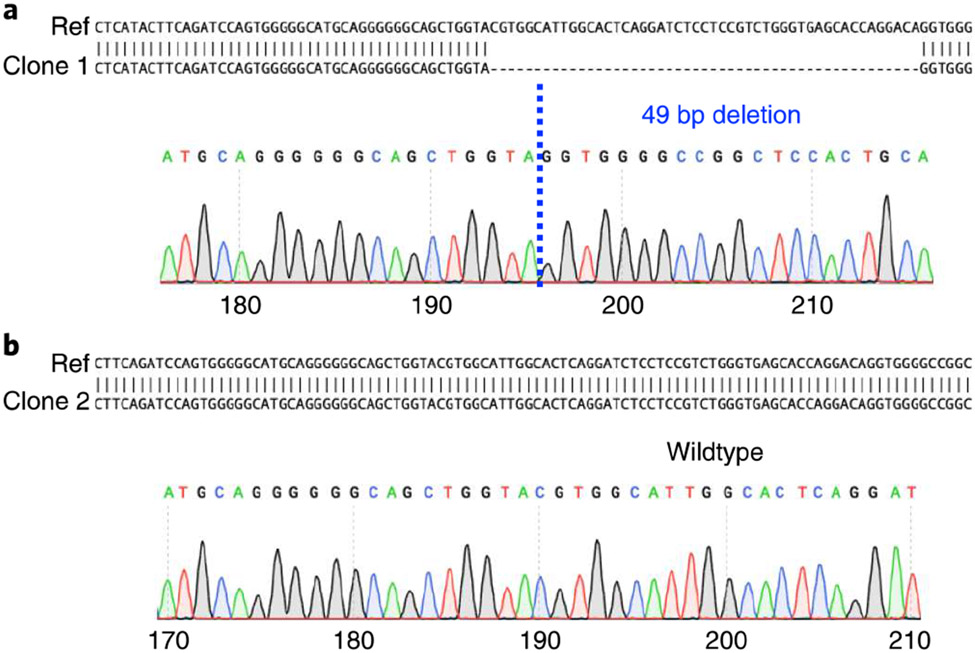
a, According to the sequencing result of the genomic DNA region around the target site compared with the reference wild-type sequence (Ref), HIE clone 1 contains a 49 bp frameshift mutation in both alleles of the target genes, indicating that the clone is a KO clone. b, HIE clone 2 contains no mutation in both alleles of the target genes, indicating that the clone is a wild-type clone.
Fig. 6 ∣. STAT1-KO HIE cells are more susceptible to GII.3 infection.
a, Western blots of cell lysates extracted from WT and STA1-KO detected the endogenous STAT1 protein by the protein-specific antibody (indicated by an asterisk) and confirmed protein loss in the isogenic KO culture. b, Gll.3 HuNoV-positive cells (green) were detected after staining with guinea pig anti-HuNoV Ab with nuclei (blue) stained by DAPI. Infected cells in clusters indicate viral spreading (40× objective lens). c, Numbers of infected cells were calculated in six independent images per condition, and each experiment was performed in triplicate (n = 18 in total). Error bars denote standard deviation of the data. Comparisons were made using the two-tailed Student's t-test. *P < 0.05. Figure reproduced with permission from ref. 12.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1 ∣.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 8 | Smearing in the digested product | Digestion time is too long | Do not digest >30 min or the product will degrade |
| 18 | No colony on LB plates (including the negative control) | The ligation or transformation efficiency is low | Check T4 DNA ligase or competent cells |
| Too many colonies in the negative control | The digestion is not complete | Redigest the vector with BsmBI | |
| 29 | The 293FT cells detach during lentiviral packaging | The cells are not healthy | Obtain a new batch of 293FT cells |
| The plate is not well coated | Double the coating time, reseed the cells and perform gently | ||
| 54 | HIEs aggregate together after digestion | The trypsin-EDTA is expired or not working well | Use freshly prepared trypsin-EDTA |
| Wash HIEs with 1× PBS before digestion to remove FBS in the well | |||
| 73 | All HIEs are dead after selection | The titer of lentiviruses is too low | Remake lentivirus with a titer determination step |
| Few HIEs die after selection in the nontransduced well | The puromycin or hygromycin-B antibiotic is expired | Prepare fresh antibiotic solution before use | |
| 79 | Hard to inoculate Matrigel in 96-well plates | The tip is not optimal | Use desired nonsticky P200 tips for inoculation |
| Too many bubbles in the Matrigel | Try not to generate bubbles in Matrigel | ||
| Put the tube on ice to remove bubbles | |||
| 92 | The single-cell clone does not grow after passaging the cells | The gene may be an essential gene | Check that the KO is viable |
| The stem cell is dying | Increase the screening size to get some surviving single-cell clones. All cells grow differently, so the initial screening size is important. We have found obtaining more growing enteroid clones is critical since the clones that are not growing or expanding cannot be maintained or passaged even if they are KO clones | ||
| 109 | No PCR product or nonspecific bands | The concentration of genomic DNA may be too low | Check the DNA concentration by Nanodrop |
| The primers are not specific enough to amplify the genomic DNA | Redesign the primer set with similar annealing temperatures | ||
| Increase primer length and annealing temperature | |||
| 109 | Obtain too many wild-type single-cell clones without indels | The gene may be an essential gene | Use knockdown or conditional KO strategy |
| The puromycin selection does not work well to kill nontransduced clones | Extend the puromycin selection time for an extra 2 weeks with freshly prepared >2 μg/mL puromycin | ||
| The sgRNA efficiency is not ideal | Switch to another sgRNA sequence, and repeat the experiment |
Timing
Part 1. Preparation of lentivirus vector with CRISPR–Cas9 targeting gene of interest
Steps 1–5, order self-annealing primers to generate the insert: 1 h
Steps 6–9, digest lentiCRISPRv2 vector with the BsmBI restriction enzyme: 3 h
Steps 10–12, perform insert self-annealing and phosphorylation: 3 h, together with Steps 6–9
Step 13, ligate digested vector, and insert together: 4 h
Steps 14–18, transform the product into E. coli: overnight
Steps 19–25, colony PCR and sequencing verification: 4 h
Part 2. Lentivirus packaging and concentration
Step 26, obtain lentivirus packaging plasmids by Maxiprep: overnight
Steps 27–29, seeding of 293FT cells for transfection: overnight, together with Step 26
Steps 30–34, transfection of required packaging plasmids with PEI: 0.5 h
Steps 35–39, lentivirus harvest from transfected 293FT cells: 3 d
Steps 40–44, concentrate lentiviruses by PEG-6000 and NaCl: overnight
Steps 45–47, lentivirus titer determination by RT-qPCR 3 h
Part 3. Lentivirus transduction of HIEs
Steps 48–49, preparation of HIE cultures in the proliferative stage using WRNE medium: 4 d
Steps 50–56, trypsin digestion of HIEs before transduction: 0.5 h
Steps 57–64, transduction of HIEs with concentrated lentiviruses and polybrene: 3 h
Part 4. Recovery and selection for HIEs
Steps 65–73, HIE recovery and antibiotic selection: 2 weeks
Part 5. Single-cell cloning
Steps 74–80, single-cell cloning: 3 h
Steps 81–83, single-cell recovery: 3 weeks
Part 6. Single HIE clone passaging and sequence verification
Steps 84–92, passage single-done-derived HIEs for splitting in 48-well plates: 1 week
Steps 93–101, take one well of each clone (in a 48-well plate) to extract genomic DNA: 3 h
Steps 102–109, PCR to amplify target sequence. Sequence the PCR product to verify KOs: 5 h
Part 7. Using KO lines to evaluate viral replication and host restriction
Steps 110–111, viruses, cells and infections: 2 months
Steps 112–118, HuNoV infection: 1 week
Anticipated results
Parts 3 and 4. Lentivirus transduction of HIEs, recovery and selection for transduced HIEs
Figure 3 illustrates typical HIE morphologies during transduction and puromycin selection. HIEs right after transduction at day 0 are shown as single cells in Fig. 3a. Antibiotic (puromycin) selection (Step 67) will remove nontransduced HIEs, and transduced HIEs will expand into larger single-cell clones at days 2–4 (Fig. 3b,c). A mixture of dead and surviving HIEs together before passage can be obtained after 4 d with puromycin selection (Fig. 3e,f).
Part 6. Single HIE clone passaging and sequence verification
According to the sequencing results of the genomic DNA region around the target site, Fig. 5a shows one HIE clone that contains a 49 bp frameshift mutation in both alleles of the target genes, indicating that the clone is a KO clone. A separate HIE clone contains no mutations in either allele of the target gene, indicating that the clone is a wild-type clone (Fig. 5b).
Part 7. Using KO lines to evaluate viral replication and host restriction
In conclusion, KO HIE clones will be obtained for further research. These KO lines are important to investigate the requirements or restriction factors for viral or other microbial infections with loss-of-function phenotypes. For instance, the STATl-KO HIEs were validated by immunoblot that slowed the loss of STATl protein with an apical plasma membrane protein (villin) still being expressed in HIEs and detected as an internal control (Fig. 6a). Later, the KO HIEs were found to be more susceptible to GII.3 HuNoV infection compared with the parental J2 line as evaluated by staining with anti-HuNoV antibody to identify infected cells at 48 and 72 hpi (Fig. 6b). The total number of antibody-labeled GII.3 infected cells in STATl-KO monolayers was significantly higher than in the parental J2 monolayers, and the virus was spreading throughout the culture as more positive cells were detected at 72 hpi compared with 48 hpi (Fig. 6c). The detailed data and protocol for immunoblot and immunostaining can be obtained from our published article12.
Supplementary Material
Acknowledgements
We thank B. Lee and the Cell-Based Assay Screening Service (C-BASS) Core in BCM for lentivirus packaging plasmids. This work was supported in part by Public Health Service grants POIAI057788, U19AI116497, Ul9AI144297, P30DK56338 and contract HHSN2722017000381 from the National Institutes of Health, by The Cancer Prevention Institute of Texas (CPRIT) RP160283—Baylor College of Medicine Comprehensive Cancer Training Program and RP17005, by NIH P30 shared resource grant CA125123, NIEHS grants P30ES030285 and P42ES0327725, and by the John S. Dunn Research Foundation.
Footnotes
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596–021-00669–0.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Competing interests
The authors declare no competing interests.
Data availability
The data are available from the authors upon request. Source data from Fig. 6 were generated using this protocol and were published in ref. 12. Source data are provided with this paper.
References
- 1.Sato T et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Heo I et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol 3, 814–823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Middendorp S et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32, 1083–1091 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Foulke-Abel J et al. Human enteroids as an ex-vivo model of host–pathogen interactions in the gastrointestinal tract. Exp. Biol Med 239, 1124–1134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajan R et al. Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio 9, e02419–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.In JG et al. Human mini-guts: new insights into intestinal physiology and host–pathogen interactions. Nat. Rev. Gastroenterol. Hepatol 13, 633–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zachos NC et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem 291, 3759–3766 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clevers H COVID-19: organoids go viral. Nat. Rev. Mol. Cell Biol 21, 355–356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geurts MH, van der Vaart J, Beumer J & Clevers H The organoid platform: promises and challenges as tools in the fight against COVID-19. Stem Cell Rep. 16, 412–418 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hares MF et al. Stem cell-derived enteroid cultures as a tool for dissecting host–parasite interactions in the small intestinal epithelium. Parasite Immunol 43, e12765 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Ettayebi K et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SC et al. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc. Natl Acad. Sci. USA 117, 23782–23793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ettayebi K et al. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 6, e01136–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami K et al. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc. Natl Acad. Sci. USA 117, 1700–1710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haga K et al. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 11, e00251–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishino Y, Shinagawa H, Makino K, Amemura M & Nakata A Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. j. Bacteriol 169, 5429–5433 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol 31, 833–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doudna JA & Charpentier E Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Paques F & Haber JE Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev 63, 349–404 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umov FD, Rebar EJ, Holmes MC, Zhang S,H, Gregory PD, Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet 11, 636–646 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Moore JK & Haber JE Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol 16, 2164–2173 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durai S et al. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 33, 5978–5990 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalem O et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang-Graham AL et al. Rotavirus induces intercellular calcium waves through ADP signaling. Science 370, eabc3621 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giuliano CJ, Lin A, Girish V & Sheltzer JM Generating single cell-derived knockout clones in mammalian cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol 128, e100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michels BE et al. Human colon organoids reveal distinct physiologic and oncogenic Wnt responses. J. Exp. Med 216, 704–720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ousterout DG et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun 6, 6244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimokawa M et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Schwank G et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Artegiani B et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol 22, 321–331 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Hendriks D, Clevers H & Artegiani B CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 27, 705–731 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Fujii M, Matano M, Nanki K & Sato T Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc 10, 1474–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Matano M et al. Modeling colorectal cancer using CRISPR–Cas9-mediated engineering of human intestinal organoids. Nat. Med 21, 256–262 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Teriyapirom I, Batista-Rocha AS & Koo BK Genetic engineering in organoids. j. Mol. Med 99,555–568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaebler AM et al. Universal and efficient electroporation protocol for genetic engineering of gastrointestinal organoids. J. Vis. Exp 10.3791/60704 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi H, Stappenbeck TS, In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc 8, 2471–2482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drost J, Artegiani B & Clevers H The generation of organoids for studying Wnt signaling. Methods Mol. Biol 1481, 141–159 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Van Lidth de Jeude JL, Vermeulen JL, Montenegro-Miranda PS, Van den Brink GR, Heijmans J, A protocol for lentiviral transduction and downstream analysis of intestinal organoids. j. Vis. Exp 10.3791/52531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merenda A et al. A protocol for multiple gene knockout in mouse small intestinal organoids using a CRISPR-concatemer. J. Vis. Exp 10.3791/55916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Chen L, Zhang J & Wang Y Drug inducible CRISPR/Cas systems. Comput. Struct. Biotechnol. J 17, 1171–1177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell 17, 233–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Hay EA et al. An analysis of possible off target effects following CAS9/CRISPR targeted deletions of neuropeptide gene enhancers from the mouse genome. Neuropeptides 64, 101–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena K et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol 90, 43–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T, Wei JJ, Sabatini DM & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang X, Wang M, Graham DY & Estes MK Expression self-assembly and antigenicity of the Norwalk virus capsid protein. J. Virol 66, 6527–6532 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanDussen KL, Sonnek NM & Stappenbeck TS L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Res. 37, 101430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sastry L, Johnson T, Hobson MJ, Smucker B & Cornetta K Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 9, 1155–1162 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Barczak W, Suchorska W, Rubiś B & Kulcenty K Universal real-time PCR-based assay for lentiviral titration. Mol. Biotechnol 57, 195–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joung J et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loisy F et al. Real-time RT-PCR for norovirus screening in shellfish. J. Virol. Methods 123, 1–7 (2005). [DOI] [PubMed] [Google Scholar]
Related links
Key references using this protocol
- lin SC et al. Proc. Natl Acad. Sci. USA 117, 23782–23793 (2020): 10.1073/pnas.2010834117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K et al. mBio 11, e00251–20 (2020): 10.1128/mBio.00251-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the authors upon request. Source data from Fig. 6 were generated using this protocol and were published in ref. 12. Source data are provided with this paper.