Approximately 85 000 deaths globally in 2019 were due to drug-resistant tuberculosis (TB), which corresponds to 7% of global deaths attributable to bacterial antimicrobial resistance [1]. Yet concerns have been mounting that drug-resistant TB was being underestimated because the approaches to define susceptibility and resistance to anti-TB agents had not kept up with those used for other major bacterial pathogens [2–9]. Here, we outline the recent, evidence-based initiatives spearheaded by the World Health Organization (WHO) and others to update breakpoints (traditionally referred to as critical concentrations (CCs)) that are used for phenotypic antimicrobial susceptibility testing (AST), also called drug susceptibility testing in the TB literature.
Short abstract
Inappropriately high breakpoints have resulted in systematic false-susceptible AST results to anti-TB drugs. MIC, PK/PD and clinical outcome data should be combined when setting breakpoints to minimise the emergence and spread of antimicrobial resistance. https://bit.ly/3i43wb6
Approximately 85 000 deaths globally in 2019 were due to drug-resistant tuberculosis (TB), which corresponds to 7% of global deaths attributable to bacterial antimicrobial resistance [1]. Yet concerns have been mounting that drug-resistant TB was being underestimated because the approaches to define susceptibility and resistance to anti-TB agents had not kept up with those used for other major bacterial pathogens [2–9]. Here, we outline the recent, evidence-based initiatives spearheaded by the World Health Organization (WHO) and others to update breakpoints (traditionally referred to as critical concentrations (CCs)) that are used for phenotypic antimicrobial susceptibility testing (AST), also called drug susceptibility testing in the TB literature.
WHO commissioned five reports that considered studies in up to 16 languages from a wide diversity of global contributors to ensure that the compiled data were as comprehensive as possible. The first report consisted of a systematic review that covered publications relating to the CCs of the most important drugs for the treatment of multidrug-resistant (MDR) or rifampicin-resistant (RR) TB, including newly approved bedaquiline and delamanid [10]. The second report was an accompanying background document on the pharmacokinetics and pharmacodynamics (PK/PD) of those drugs, whereas the third presented the findings of a meta-analysis of clinical outcome data [11, 12]. The fourth was a systematic review of the CCs for the rifamycins and isoniazid [13]. Finally, WHO released its first official catalogue of resistance mutations to inform the interpretation of genotypic AST results [14, 15]. Together, these reports prompted WHO to make major changes to its recommendations for TB treatment (e.g. kanamycin is no longer recommended for the treatment of TB (figure 1b)) and AST, as discussed below [16].
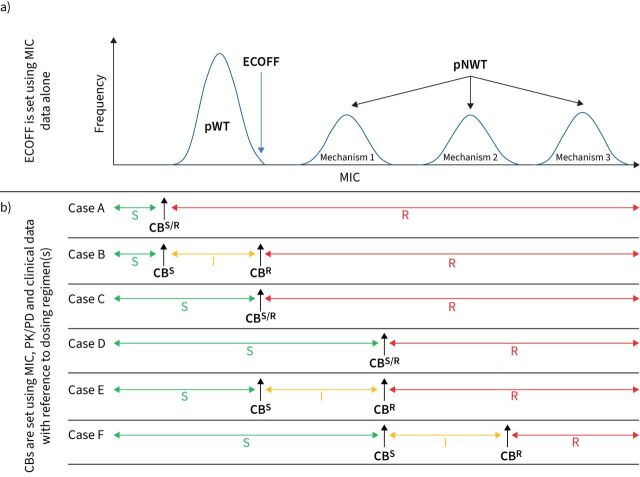
FIGURE 1.
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) approach for setting breakpoints compared with the World Health Organization (WHO). a) Four hypothetical minimum inhibitory concentration (MIC) distributions of an antibiotic for the same species. The distribution with the lowest MICs is typically the phenotypically wild-type (pWT) distribution, whereas the remaining three are phenotypically non-wild type (pNWT) with different underlying mechanisms. Notably, the upper end of the pWT distribution, which corresponds to the epidemiological cut-off value (ECOFF), does not automatically become a clinical breakpoint (CB), as shown in panel (b). Instead, pharmacokinetic/pharmacodynamic (PK/PD) and clinical data must be analysed to assess whether any of the represented populations are susceptible (S), susceptible at increased exposure (I), or resistant (R) [17, 24]. This may demonstrate that an agent offers no clinical benefits even for pWT strains at clinically attainable drug exposures, in which case the species in question would be deemed to be intrinsically resistant (case A). In 2018, WHO reached this conclusion for kanamycin and capreomycin after decades of clinical use globally, which prompted their withdrawal from clinical recommendations, although the underlying meta-analysis has attracted criticism [12, 16, 33, 34]. If a drug is clinically effective, one of five scenarios may apply. First, the pWT population may only be susceptible at increased exposure (case B). This uncommon approach is used to minimise the chance of clinicians prescribing the wrong regimen if a lower dose is commonly used for other pathogens. Second, the standard dosing regimen of the drug may be sufficient to treat only the pWT population (case C). This is the most common scenario when a drug is first approved and there is clinical outcome data to support its efficacy for the pWT population, whereas sufficient PK/PD and extensive clinical data in support of higher doses or treatment of pNWT isolates with resistance mechanisms are usually lacking. Gathering sufficient clinical outcome data for different pNWT populations is particularly challenging for TB given that multidrug regimens are always used, which may result in synergies or antagonism between one or more agents [27]. Nevertheless, the impact of individual mutations can be correlated with clinical outcomes, particularly for core drugs, provided that the studies are sufficiently powered [19, 25, 35, 36]. Third, the standard dosing regimen may also be sufficiently potent to treat strains with mechanism 1 but not strains with higher MICs because of mechanisms 2 and 3 (case D). Fourth, mechanism 1 may only be treatable at an increased exposure, as shown in case E. Finally, case F represents a hybrid between cases D and E. The current WHO definition of the critical concentration (CC) is effectively that of an ECOFF (i.e. it is set based on MIC data alone, taking genotypic information into consideration when relevant) even though the CC is actually used as a CBS/R (i.e. pWT strains are reported as susceptible and pNWT strains as resistant based on a limited review of clinical evidence and PK/PD data compared with other bacterial pathogens) [10, 12, 13, 16]. The only exception is moxifloxacin (table 1), for which the CC is used as a CBS and the CBWHO, as defined by WHO, is effectively a CBR (case E), which may cause confusion with some clinicians who rarely treat TB. Moreover, this contradicts the assertion that an “intermediate” category, which is an alternative term to describe MIC increases that can be overcome by dose increases, does not exist for TB [10, 13, 24].
For most antimicrobials with proven clinical efficacy at a specific dose, only the phenotypically wild-type (pWT) population of the bacterium in question is considered treatable (figure 1a) [17]. Thus, the main aim of the two systematic reviews commissioned by WHO was to evaluate the available minimum inhibitory concentration (MIC) data to assess whether existing CCs corresponded to epidemiological cut-off values (ECOFFs), which represent the upper end of the pWT MIC distribution (figure 1a) [10, 13]. This revealed limitations in both the quality and quantity of available MIC data in the TB field, in contrast to many other major bacterial pathogens [8, 9]. In fact, the data for most drug-medium combinations did not meet the criteria set out by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for setting ECOFFs [7, 9]. Faced with this situation and the critical, global need for AST guidance, WHO adopted a pragmatic approach and set 12 new CCs for second-line drugs based on systematic reviews of available existing data, while clearly highlighting that these decisions should be re-evaluated once additional data become available [7, 10]. Even using these less stringent criteria, three previously endorsed CCs had to be withdrawn owing to a clear lack of supporting evidence. This included the only CC for cycloserine available up to that point, which means that phenotypic AST is not possible and, consequently, patients with resistant strains are needlessly at risk of the sometimes severe side-effects of this drug [10, 16].
More importantly, two previous CCs for rifampicin, arguably amongst the most significant breakpoints in diagnostic microbiology, were found to be too high, leading to false-susceptible AST results for some isolates (i.e. very major diagnostic errors that increase the likelihood of treatment failure and selection of resistance to other drugs) [13, 18, 19]. In fact, the rifampicin CC for Middlebrook 7H10 medium was twice as high as the ECOFF for more than half a century [18]. The therapeutic impact of these diagnostic misclassifications was that some phenotypically non-wild type isolates were deemed treatable with the recommended 10 mg·kg−1 body weight per day dose despite a lack of PK/PD or clinical evidence (case D in figure 1b) [13, 18]. WHO, consequently, lowered the rifampicin CCs to the tentative ECOFFs based on data from the systematic review (case C in figure 1b) [13]. To minimise false-susceptible results, the WHO-endorsed CCs of three second-line drugs (amikacin, levofloxacin and moxifloxacin) were also lowered to their respective ECOFFs [10]. In the case of moxifloxacin, approximately 90% of resistant isolates could have been misclassified as susceptible using only the WHO CC of 2 mg·L−1 for BACTEC Mycobacterial Growth Indicator Tube system by Becton Dickinson that was valid between 2014 and 2018 [20]. Fortunately, a number of practical factors (table 1) meant that the clinical consequences of this incorrectly set CC were reduced considerably.
TABLE 1.
Overview of changes to moxifloxacin breakpoints
| Medium | Recommended moxifloxacin breakpoint (in mg·L −1 ); daily dose (in mg) ¶ | ||||
| CLSI | WHO | ||||
| Since 2011 [37, 43, 44] | 2008–2014 [45, 46] | 2014–2018 [47] | Since 2018/19 [10, 48] | ||
| 7H10 | CC | 0.5+; N/A§ | − |
0.5; N/Af 2; 400## |
0.5¶¶; 400 (standard dose) in longer regimen or 400–800 (high dose) in shorter regimen++ |
| CBWHO | − | − | − | 2; 400–800 (high dose) in longer regimen++ | |
| MGIT | CC | 0.25+; N/A§ | 0.25; 400 |
0.5; N/Af 2; 400## |
0.25¶¶; 400 (standard dose) in longer regimen or 400–800 (high dose) in shorter regimen++ |
| CBWHO | − | − | − | 1; 400–800 (high dose) in longer regimen++ | |
Based on World Health Organization (WHO) surveillance data#, approximately 90% of moxifloxacin-resistant isolates could have been misclassified as susceptible using the BACTEC Mycobacterial Growth Indicator Tube (MGIT) because the WHO critical concentration (CC) of 2 mg·L−1 was eight times higher than the epidemiological cut-off value (ECOFF) between 2014 and 2018. In practice, however, the rate of misclassification was far lower. First, many countries did not use moxifloxacin at all during this period and, consequently, did not use this CC. Second, even countries that prescribed moxifloxacin avoided or minimised the misclassifications because they completely or primarily relied on genotypic antimicrobial susceptibility testing, continued using the MGIT CC of 0.25 mg·L−1 in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines, used a CC for another fluoroquinolone as surrogate for moxifloxacin resistance, or relied on 0.5 mg·L−1 as the breakpoint for the standard dose of moxifloxacin in combination with 2 mg·L−1 as the breakpoint for the high dose of moxifloxacin. The theoretical rate of false-susceptible results was lower using Middlebrook 7H10 medium because the WHO CC of 2 mg·L−1 was four rather than eight times higher than the ECOFF. The clinical breakpoints introduced by WHO (CBWHO) are not recognised by CLSI. #: comparing MGIT results for 2 mg·L−1 moxifloxacin with 2 mg·L−1 ofloxacin in the WHO surveillance study, which is equivalent to testing the currently recognised levofloxacin CC of 1 mg·L−1 (i.e. 1.5 mg·L−1 tested in that study was also too high) [10, 20]. ¶: changes to WHO breakpoint/dose combinations relative to the previous guidelines are highlighted in bold. Breakpoints that correspond to ECOFFs are underlined [10]. +: can be tested as surrogate for other fluoroquinolones [37, 43–45]. §: not applicable (N/A) as CLSI does not define doses for treatment. f: 0.5 mg·L−1 moxifloxacin in 7H10 and MGIT were recommended as surrogates for resistance to ofloxacin and levofloxacin. Because the ECOFF for moxifloxacin is 0.25 mg·L−1 in MGIT, this meant that some strains resistant to ofloxacin and levofloxacin were misclassified as susceptible [10]. In effect, the surrogate breakpoints at 0.5 mg·L−1 and moxifloxacin CCs at 2 mg·L−1 were set inconsistently for both media because the 7H10 data was extrapolated to MGIT, despite the systematic differences between both media [47]. ##: the WHO-endorsed dosage for individualised multidrug-resistant/rifampicin-resistant tuberculosis (MDR/RR-TB) regimens was 400 mg [47]. However, operational research using a higher dosage of moxifloxacin (800 mg) in a standardised short-course MDR/RR-TB regimen was in progress, although not WHO-endorsed at the time [49]. ¶¶: not recommended as surrogate for other fluoroquinolones [10]. ++: levofloxacin is the preferred fluoroquinolone for the shorter all-oral bedaquiline-containing MDR/RR-TB regimen recommended by WHO in 2020, but high-dose moxifloxacin can be used instead. However, any moxifloxacin resistance, irrespective of the level, is an exclusion criterion for the shorter all-oral regimen (i.e. the CC is the relevant breakpoint) [48]. This exclusion criterion for moxifloxacin had also applied to the shorter amikacin-containing MDR/RR-TB regimen that was recommended by WHO between 2018 and 2020 [50]. High-dose moxifloxacin can only be used to treat low-level resistant strains as part of the longer MDR/RR-TB regimen, for which the CBWHO is valid [48].
When CCs are too high, they may not only result in undertreatment of the patient based on phenotypic AST, but can also adversely affect the design and interpretation of genotypic AST methods that represent the most viable option to scale up AST globally [18, 19, 21]. Between 2011 and 2014, for instance, the WHO-endorsed GenoType MTBDRplus VER 2.0 by Hain Lifescience was designed not to detect rpoB L452P because this mutation was not considered to be a rifampicin resistance mutation at that time [18, 19]. It took more than a decade for eis c-14t and rrs c1402t to be recognised as resistance mutations for amikacin [14, 15, 22]. Hence, the full potential of the GenoType MTBDRsl VER 2.0 was not exploited because these two mutations were only interpreted as markers for kanamycin and capreomycin resistance.
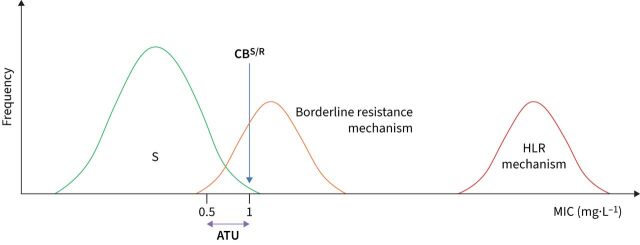
Another consequence of CCs that are too high is an unnecessarily high number of clinical isolates that are genotypically resistant (i.e. contain mutations associated with resistance) but test phenotypically susceptible. This has resulted in underestimates of the accuracy of genotypic methods when phenotypic AST has been used as a reference and, consequently, reduced the confidence in genotypic AST. This apparent discordance also obscured the fact that clinically relevant mutations for some drugs cannot be reliably confirmed by current phenotypic AST methods, even if the CC corresponds to the ECOFF, because the MIC distributions of susceptible and resistant strains overlap based on current data (figure 2) [23]. This is the case for rifampicin and, therefore, WHO has adopted a composite reference standard to ensure that borderline rpoB resistance mutations are not missed (i.e. an isolate is now considered resistant to rifampicin if it tests resistant by phenotypic AST or harbours a recognised resistance mutation, provided that the pre-test probability is considered) [13, 18, 19, 23]. However, clear and user-friendly guidance on how to resolve discordances during routine clinical care is also needed for other drugs [23].
FIGURE 2.
Strategies to minimise false-susceptible results by phenotypic antimicrobial susceptibility testing (AST) linked to borderline resistance mechanisms. Unlike the idealised scenario depicted in figure 1a, borderline resistance mechanisms exist with minimum inhibitory concentration (MIC) distributions that overlap with the susceptible distribution (e.g. the seven borderline rifampicin resistance mutations in rpoB) [10, 13, 37–39]. A clinical breakpoint (CBS/R) that corresponds to the epidemiological cut-off value (ECOFF) (case C in figure 1b) intersects the MIC distributions of such mechanisms (at 1 mg·L−1 in the hypothetical example below). Even if such an isolate is tested multiple times in the same laboratory, it will variably test susceptible and resistant because of the inherent technical variability of phenotypic AST [18]. Four measures that are not mutually exclusive can be taken to decrease such false-susceptible results. First, the optimal solution would be to eliminate or at least minimise the degree of overlap between distributions by reducing the technical variability of MIC testing as much as possible, which was one of the reasons that prompted the European Committee on Antimicrobial Susceptibility Testing (EUCAST) to develop its reference method and associated procedures to improve quality control [8, 9, 40, 41]. Second, EUCAST has introduced areas of technical uncertainty (ATUs) [24]. In this example, an MIC result of ≤0.5 mg·L−1 would be reported as susceptible, whereas MICs of >1 mg·L−1 would be resistant. By contrast, an MIC result of 1 mg·L−1 would be “uncertain” as the isolate in question could not be unequivocally classified as either susceptible or resistant based on the single MIC result because of the overlapping MIC distributions (i.e. this applies to the borderline resistance mechanism but not high-level resistance (HLR) mechanism) [18]. Although the prevalence of borderline resistance in a particular setting can give an indication of which of these possibilities is more likely, other experimental results are needed to resolve this situation conclusively. For example, if the molecular basis of the borderline resistance mechanism is known and is detected, the isolate could be reported as resistant (i.e. a composite reference standard is used, as WHO recommends for rifampicin) [18, 19, 23]. In fact, the Clinical and Laboratory Standards Institute (CLSI) has set an “inconclusive” category for ethambutol for the Sensititre MYCOTB plate by Thermo Fisher Scientific, which appears to serve as an ATU to minimise false susceptibility due to embB mutations [37, 39]. Third, adopting interpretative reading, whereby the results of two antibiotics that share at least one resistance mechanism are analysed together, may be useful (e.g. if the MICs for bedaquiline and clofazimine are equal to or just above the CBS/R, it is likely that the isolate in question has an Rv0678 mutation) [38]. Finally, a surrogate agent could be tested that provides a better resolution between the relevant distribution (e.g. CLSI and EUCAST recommend pefloxacin as a surrogate for fluoroquinolone resistance in Salmonella enterica) [42].
More fundamentally, MIC, PK/PD and clinical data should be fully integrated when setting breakpoints [11, 17]. In 2018, WHO endorsed a second breakpoint for moxifloxacin that is higher than the CC in support of high-dose moxifloxacin treatment as part of the longer individualised MDR/RR-TB regimen (table 1) [10]. A “susceptible at increased exposure” range thus was defined, though this specific terminology was not used in the report (case E in figure 1b) [24]. The primary justification for this decision relied on extrapolating clinical outcome data for high-dose gatifloxacin from a single study without data on drug exposure [10, 25]. It was not acknowledged that even high-dose gatifloxacin did not always overcome the low-level MIC increases conferred by gyrA A90V and similar mutations [10, 25]. Subsequent PK/PD modelling suggested that this second breakpoint might be clinically useful, but also reinforced the idea that low-level fluoroquinolone resistance is unlikely to be overcome by high-dose moxifloxacin in all patients because of patient-to-patient variability in the moxifloxacin exposure [26]. Nevertheless, given the potentially significant clinical value of using high-dose moxifloxacin when few other treatment options remain, this question should be prioritised for future review using additional data, including the recent studies using the hollow fibre infection model, to provide a more comprehensive and nuanced recommendation to clinicians [27–29]. Similarly, WHO has already announced that it would revisit the rifampicin breakpoint, should a higher dose of rifampicin be endorsed [13].
Considering this complex history, regulators and developers of diagnostics and drugs should fully embrace modern microbiological principles to define breakpoints and associated dosing regimens (figure 1) [17, 30, 31]. To this end, the two systematic reviews provide unprecedented detail about the underlying reasons and scientific evidence for all new recommendations by WHO, to facilitate external scrutiny and to encourage more research where the available evidence was limited [10, 13]. Where possible, these efforts should be coordinated between major regulators to minimise the burden to developers of drugs and AST devices (e.g. by recognising a single reference method against which all commercial AST methods are validated) [9]. It would also be beneficial if common AST terminology were adopted to avoid confusion. For instance, the meaning of “clinical breakpoint” differs between regulators (figure 1) and adopting the “area of technical uncertainty” for TB needs further consideration (figure 2) [10, 13]. Regulators should also review if the use of surrogate drugs can minimise false-susceptible results to provide clarity for assay developers about which agents to invest in (e.g. whether levofloxacin should be tested as the representative fluoroquinolone and whether kanamycin should be used as a surrogate for amikacin resistance (figure 2)) [22]. Although there has been a great deal of progress in the past 5 years, proactive action by the entire TB community is required to develop an updated AST framework given that the In Vitro Diagnostic Medical Device Regulation will come into effect in the European Union in May 2022. Because previous diagnostic approvals will not be automatically recognised, industry will have to invest to keep its AST devices on the market. We have an obligation to those infected and affected by TB to learn from past experiences and to make the most of this unique window of opportunity [7, 18, 32].
Shareable PDF
Acknowledgements
We thank Andrew Vernon and others who are acknowledged in the respective WHO reports covered in this viewpoint for sharing relevant data.
Footnotes
The Antimycobacterial Susceptibility Testing Group: Sophia B. Georghiou (FIND, Geneva, Switzerland), Timothy C. Rodwell (FIND, Geneva, Switzerland, and Dept of Medicine, University of California, San Diego, CA, USA), Alexei Korobitsyn (Global TB Programme, World Health Organization, Geneva, Switzerland), Said H. Abbadi (Dept of Microbiology, Faculty of Medicine, Suez University, Suez, Egypt), Kanchan Ajbani (Dept of Microbiology, P.D. Hinduja Hospital and Medical Research Centre, Mumbai, India), Jan-Willem Alffenaar (Sydney Institute for Infectious Diseases, University of Sydney, Sydney, Australia, Faculty of Medicine and Health, School of Pharmacy, The University of Sydney, Sydney, Australia, and Westmead Hospital, Sydney, Australia), David Alland (Dept of Medicine and the Public Health Research Institute, New Jersey Medical School, Rutgers University, Newark, NJ, USA), Nataly Alvarez (Unidad de Bacteriología y Micobacterias, Corporación para Investigaciones Biológicas (CIB), Medellín, Colombia), Sönke Andres (National and Supranational Reference Laboratory for Mycobacteria, Research Center Borstel, Borstel, Germany), Elisa Ardizzoni (Unit of Mycobacteriology, Dept of Biomedical Sciences, Institute of Tropical Medicine Antwerp, Antwerp, Belgium), Alexandra Aubry (Sorbonne Université, INSERM, Centre d'Immunologie et des Maladies Infectieuses, U1135, AP-HP, Hôpital Pitié-Salpêtrière, Centre National de Référence des Mycobactéries et de la Résistance des Mycobactéries aux Antituberculeux, Paris, France), Rossella Baldan (FIND, Geneva, Switzerland, and Institute for Infectious Diseases, University of Bern, Bern, Switzerland), Marie Ballif (Institute of Social and Preventive Medicine, University of Bern, Bern, Switzerland), Ivan Barilar (Molecular and Experimental Mycobacteriology, Research Center Borstel, Borstel, Germany, and German Center for Infection Research, Partner site Hamburg-Lübeck-Borstel-Riems, Borstel, Germany), Erik C. Böttger (Institut für Medizinische Mikrobiologie, Nationales Zentrum für Mykobakterien, Universität Zürich, Zürich, Switzerland), Soumitesh Chakravorty (Cepheid, Sunnyvale, CA, USA, and New Jersey Medical School, Rutgers University, Newark, NJ, USA), Pauline M. Claxton (Scottish Mycobacteria Reference Laboratory, Directorate of Laboratory Medicine, Royal Infirmary of Edinburgh, Edinburgh, UK), Daniela M. Cirillo (Emerging Bacterial Pathogens Unit, IRCCS Ospedale San Raffaele, Milan, Italy), Iñaki Comas (Instituto de Biomedicina de Valencia IBV-CSIC, Spanish National Research Council, Valencia, Spain, and CIBER in Epidemiology and Public Health, Madrid, Spain), Chris Coulter (Queensland Mycobacterium Reference Laboratory, Pathology Queensland, Herston, Australia), Claudia M. Denkinger (FIND, Geneva, Switzerland, Division of Clinical Tropical Medicine, Centre of Infectious Diseases, Heidelberg University Hospital, Heidelberg, Germany, and German Centre for Infection Research (DZIF), partner site Heidelberg University Hospital, Heidelberg, Germany), Brigitta Derendinger (DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research/South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa), Edward P. Desmond (State Laboratories Division, Department of Health, Pearl City, HI, USA), Jurriaan E.M. de Steenwinkel (Dept of Medical Microbiology and Infectious Diseases, Erasmus University Medical Centre Rotterdam, Rotterdam, The Netherlands), Keertan Dheda (Centre for Lung Infection and Immunity, Division of Pulmonology, Dept of Medicine and UCT Lung Institute, and South African MRC/UCT Centre for the Study of Antimicrobial Resistance, University of Cape Town, Cape Town, South Africa, and Faculty of Infectious and Tropical Diseases, Dept of Immunology and Infection, London School of Hygiene and Tropical Medicine, London, UK), Andreas H. Diacon (TASK, Cape Town, South Africa), David L. Dolinger (General Fluidics, Waltham, MA, USA), Kelly E. Dooley (Dept of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA), Matthias Egger (Institute of Social and Preventive Medicine, University of Bern, Bern, Switzerland, Centre for Infectious Disease Research and Epidemiology, University of Cape Town, Cape Town, South Africa, and Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK), Soudeh Ehsani (Joint Infectious Diseases Programme, Regional Office for Europe, World Health Organization, Copenhagen, Denmark), Maha R. Farhat (Dept of Biomedical Informatics, Harvard Medical School, Boston, MA, USA, and Pulmonary and Critical Care Medicine, Massachusetts General Hospital, Boston, MA, USA), Lanfranco Fattorini (Dept of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy), Iris Finci (Molecular and Experimental Mycobacteriology, Research Center Borstel, Borstel, Germany, and German Center for Infection Research, Partner site Hamburg-Lübeck-Borstel-Riems, Borstel, Germany), Laure Fournier Le Ray (Sorbonne Université, INSERM, Centre d'Immunologie et des Maladies Infectieuses, U1135, Paris, France), Victoria Furió (Instituto de Biomedicina de Valencia IBV-CSIC, Spanish National Research Council, Valencia, Spain), Ramona Groenheit (Dept of Microbiology, Public Health Agency of Sweden, Solna, Sweden), Tawanda Gumbo (Praedicare Inc, Dallas, TX, USA), Scott K. Heysell (Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, VA, USA), Doris Hillemann (National and Supranational Reference Laboratory for Mycobacteria, Research Center Borstel, Borstel, Germany), Harald Hoffmann (Institute of Microbiology and Laboratory Medicine, Dept IML Red GmbH, WHO – Supranational Tuberculosis Reference Laboratory Munich-Gauting, Gauting, Germany, and SYNLAB Gauting, SYNLAB Human Genetics, Gauting, Germany), Po-Ren Hsueh (Depts of Laboratory Medicine and Internal Medicine, China Medical University Hospital, School of Medicine, China Medical University, Taichung, Taiwan, and Depts of Laboratory Medicine and Internal Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei, Taiwan), Yi Hu (School of Public Health and Key Laboratory of Public Health Safety, Fudan University, Shanghai, China), Hairong Huang (Beijing Chest Hospital, Capital Medical University, Beijing, China), Alamdar Hussain (National TB Reference Laboratory, National TB Control Program, Pakistan), Farzana Ismail (Centre for Tuberculosis, National and Supranational TB Reference Laboratory, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa, and Dept of Medical Microbiology, University of Pretoria, Pretoria, South Africa), Kiyohiko Izumi (Dept of Epidemiology and Clinical Research, Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association, Kiyose, Japan), Tomasz Jagielski (Dept of Medical Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland), John L. Johnson (Case Western Reserve University and University Hospitals Cleveland Medical Center, Cleveland, OH, USA), Priti Kambli (Dept of Microbiology, P.D. Hinduja Hospital and Medical Research Centre, Mumbai, India), Koné Kaniga (Johnson & Johnson Global Public Health, Division of Janssen Pharmaceutica, Titusville, NJ, USA), G.H.R. Eranga Karunaratne (Dept of Microbiology, Faculty of Medicine, University of Colombo, Colombo, Sri Lanka, and Faculty of Science, Horizon Campus, Malabe, Sri Lanka), Meenu Kaushal Sharma (National Reference Centre for Mycobacteriology, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, MB, Canada), Peter M. Keller (Institute for Infectious Diseases, University of Bern, Bern, Switzerland), Ellis C. Kelly (Dept of Genetics, University of Cambridge, Cambridge, UK), Margarita Kholina (Faculty of Biology, Lomonosov Moscow State University, Moscow, Russia), Mikashmi Kohli (FIND, Geneva, Switzerland), Katharina Kranzer (Dept of Clinical Research, London School of Hygiene and Tropical Medicine, London, UK, Dept of Infectious Diseases and Tropical Medicine, Ludwig Maximilian University of Munich, Munich, Germany, and Biomedical Research and Training Institute, Harare, Zimbabwe), Ian F. Laurenson (Scottish Mycobacteria Reference Laboratory, Directorate of Laboratory Medicine, Royal Infirmary of Edinburgh, Edinburgh, UK), Jason Limberis (Dept of Medicine, Division of Experimental Medicine, University of California San Francisco, San Francisco, CA, USA), S-Y. Grace Lin (Microbial Diseases Laboratory, California Department of Public Health, Richmond, CA, USA), Yongge Liu (Otsuka Pharmaceutical Development and Commercialization, Inc., Rockville, MD, USA), Alexandre López-Gavín (Hospital Clínic, Universitat de Barcelona, Barcelona, Spain), Anna Lyander (Clinical Genomics Stockholm, Science for Life Laboratory, Solna, Sweden, and School of Engineering Sciences in Chemistry, Biotechnology and Health, KTH Royal Institute of Technology, Stockholm, Sweden), Diana Machado (Global Health and Tropical Medicine, GHTM, Instituto de Higiene e Medicina Tropical, IHMT, Universidade Nova de Lisboa, Lisbon, Portugal), Elena Martinez (Centre for Infectious Diseases and Microbiology – Public Health, Westmead Hospital and NSW Health Pathology, Sydney, Australia), Faisal Masood (National TB Reference Laboratory, National TB Control Program, Pakistan), Satoshi Mitarai (Dept of Mycobacterium Reference and Research, Research Institute of Tuberculosis, Japan Anti-tuberculosis Association, Kiyose, Japan), Nomonde R. Mvelase (Dept of Medical Microbiology, National Health Laboratory Service, Durban, South Africa, and School of Laboratory Medicine and Medical Sciences, College of Health Sciences, University of KwaZulu-Natal, Durban, South Africa), Stefan Niemann (Molecular and Experimental Mycobacteriology, Research Center Borstel, Borstel, Germany, and German Center for Infection Research, Partner site Hamburg-Lübeck-Borstel-Riems, Borstel, Germany), Vladyslav Nikolayevskyy (Dept of Infectious Diseases, Imperial College London, London, UK), Florian P. Maurer (National and Supranational Reference Laboratory for Mycobacteria, Research Center Borstel, Borstel, Germany, German Center for Infection Research, Partner site Hamburg-Lübeck-Borstel-Riems, Borstel, Germany, and Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg-Eppendorf, Hamburg, Germany), Matthias Merker (Evolution of the Resistome, Research Center Borstel, Borstel, Germany), Paolo Miotto (Emerging Bacterial Pathogens Unit, IRCCS Ospedale San Raffaele, Milan, Italy), Shaheed V. Omar (Centre for Tuberculosis, National and Supranational TB Reference Laboratory, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa, and Dept of Molecular Medicine and Haematology, School of Pathology, Faculty of Health Sciences, University of Witwatersrand, Johannesburg, South Africa), Ralf Otto-Knapp (German Central Committee against Tuberculosis, Berlin, Germany), Moisés Palaci (Núcleo de Doenças Infecciosas, Universidade Federal do Espírito Santo, Vitória, Brazil), Juan José Palacios Gutiérrez (Unidad de Referencia Regional de Micobacterias, Hospital Universitario Central de Asturias, Instituto de Investigación Sanitaria del Principado de Asturias (ISPA), Oviedo, Spain), Sharon J. Peacock (Dept of Medicine, University of Cambridge, Cambridge, UK), Charles A. Peloquin (College of Pharmacy and Emerging Pathogens Institute, University of Florida, Gainesville, FL, USA), Jennifer Perera (Dept of Microbiology, Faculty of Medicine, University of Colombo, Colombo, Sri Lanka), Catherine Pierre-Audigier (CMIP Institut Pasteur, Paris, France, and Laboratoire de Bactériologie, Hôpital Bichat-Claude Bernard, Paris France), Suporn Pholwat (Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, VA, USA), James E. Posey (Division of Tuberculosis Elimination, National Center for HIV, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA, USA), Therdsak Prammananan (National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathum Thani, Thailand), Leen Rigouts (Unit of Mycobacteriology, Dept of Biomedical Sciences, Institute of Tropical Medicine Antwerp, Antwerp, Belgium), Jaime Robledo (Unidad de Bacteriología y Micobacterias, Corporación para Investigaciones Biológicas (CIB), Medellín, Colombia, and Escuela de Ciencias de la Salud, Universidad Pontificia Bolivariana (UPB), Medellín, Colombia), Neesha Rockwood (Dept of Microbiology, Faculty of Medicine, University of Colombo, Colombo, Sri Lanka, Dept of Infectious Diseases, Imperial College London, London, UK, and Wellcome Centre for Infectious Diseases Research in Africa, University of Cape Town, Cape Town, South Africa), Camilla Rodrigues (Dept of Microbiology, P.D. Hinduja Hospital and Medical Research Centre, Mumbai, India), Max Salfinger (University of South Florida College of Public Health and Morsani College of Medicine, Tampa, FL, USA), Marcos C. Schechter (Emory University School of Medicine, Dept of Medicine, Division of Infectious Diseases, Atlanta, GA, USA), Marva Seifert (Dept of Medicine, University of California, San Diego, CA, USA), Sarah Sengstake (Unit of Mycobacteriology, Dept of Biomedical Sciences, Institute of Tropical Medicine Antwerp, Antwerp, Belgium), Thomas Shinnick (Independent Consultant, Atlanta, GA, USA), Natalia Shubladze (National Reference Laboratory, National Center for Tuberculosis and Lung Diseases, Tbilisi, Georgia), Vitali Sintchenko (Sydney Institute for Infectious Diseases, University of Sydney, Sydney, Australia, and NSW Mycobacterium Reference Laboratory, Institute of Clinical Pathology and Medical Research, NSW Health Pathology, Sydney, Australia), Frederick Sirgel (DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research/South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa), Sulochana Somasundaram (National Institute for Research in Tuberculosis, Indian Council of Medical Research, Chennai, India), Timothy R. Sterling (Vanderbilt University Medical Center, Nashville, TN, USA), Andrea Spitaleri (Emerging Bacterial Pathogens Unit, IRCCS Ospedale San Raffaele, Milan, Italy, and Vita-Salute San Raffaele University, Milan, Italy), Elizabeth Streicher (DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research/South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa), Philip Supply (Univ. Lille, CNRS, Inserm, CHU Lille, Institut Pasteur de Lille, U1019 – UMR 9017 – CIIL – Center for Infection and Immunity of Lille, F-59000 Lille, France), Erik Svensson (International Reference Laboratory of Mycobacteriology, Statens Serum Institut, Copenhagen, Denmark), Elisa Tagliani (Emerging Bacterial Pathogens Unit, IRCCS Ospedale San Raffaele, Milan, Italy), Sabira Tahseen (National TB Reference Laboratory, National TB Control Program, Pakistan), Akiko Takaki (Dept of Mycobacterium Reference and Research, Research Institute of Tuberculosis, Japan Anti-tuberculosis Association, Kiyose, Japan), Grant Theron (DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research/South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa), Gabriela Torrea (Unit of Mycobacteriology, Dept of Biomedical Sciences, Institute of Tropical Medicine Antwerp, Antwerp, Belgium), Armand Van Deun (Independent Consultant, Leuven, Belgium), Jakko van Ingen (Radboudumc Center for Infectious Diseases, Dept of Medical Microbiology, Radboud University Medical Center, Nijmegen, The Netherlands), Annelies Van Rie (Tuberculosis Omics Research Consortium, Family Medicine and Population health, Faculty of Medicine and Health Sciences, University of Antwerp, Antwerp, Belgium), Dick van Soolingen (Tuberculosis Reference Laboratory, National Institute for Public Health and the Environment, Bilthoven, The Netherlands), Roger Vargas Jr (Dept of Biomedical Informatics, Harvard Medical School, Boston, MA, USA, and Center for Computational Biomedicine, Harvard Medical School, Boston, MA, USA), Amour Venter (TASK, Cape Town, South Africa), Nicolas Veziris (Sorbonne Université, Centre d'Immunologie et des Maladies Infectieuses (Cimi-Paris), UMR 1135, Département de Bactériologie, Hôpital Saint-Antoine, Centre National de Référence des Mycobactéries, APHP, Sorbonne Université, Paris, France), Cristina Villellas (Janssen Research and Development, Beerse, Belgium), Miguel Viveiros (Global Health and Tropical Medicine, GHTM, Instituto de Higiene e Medicina Tropical, IHMT, Universidade Nova de Lisboa, Lisbon, Portugal), Robin Warren (DSI-NRF Centre of Excellence for Biomedical Tuberculosis Research/South African Medical Research Council Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Stellenbosch, South Africa), Shu'an Wen (Beijing Chest Hospital, Capital Medical University, Beijing, China), Jim Werngren (Dept of Microbiology, Public Health Agency of Sweden, Solna, Sweden), Robert J. Wilkinson (Dept of Infectious Diseases, Imperial College London, London, UK, Wellcome Centre for Infectious Diseases Research in Africa, University of Cape Town, Cape Town, South Africa, and The Francis Crick Institute, London, UK), Caie Yang (Dept of Clinical Laboratory, The Eighth Medical Center of People's Liberation Army General Hospital, Beijing, China), F. Ferda Yılmaz (Ege University, Faculty of Pharmacy, Dept of Pharmaceutical Microbiology, Bornova, İzmir, Turkey), Tingting Zhang (Beijing Chest Hospital, Capital Medical University, Beijing, China), Danila Zimenkov (Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia), Nazir Ismail (Global TB Programme, World Health Organization, Geneva, Switzerland), Thomas Schön (Dept of Infectious Diseases, Kalmar County Hospital, Linköping University, Kalmar, Sweden, Unit of Infection and Inflammation, Dept of Biomedical and Clinical Sciences, Linköping University, Linköping, Sweden, and Dept of Infectious Diseases, Linköping University Hospital, Linköping, Sweden) and Claudio U. Köser (Dept of Genetics, University of Cambridge, Cambridge, UK). Thomas Schön and Claudio U. Köser contributed equally.
Author contributions: S.B. Georghiou, T.C. Rodwell, T. Schön and C.U. Köser wrote the initial draft of the manuscript that all other authors reviewed. The authors either contributed to at least one of the systematic reviews of the breakpoints (by reviewing the literature or providing financial support or data) and/or provided insights into the interpretation of AST results.
Conflict of interest: D. Alland receives research support and royalty payments from Cepheid, a diagnostic company that makes tests for tuberculosis. A. Aubry and N. Veziris work in a laboratory that received a grant from Janssen outside the scope of this work. R. Baldan, I. Comas, C.M. Denkinger, D.L. Dolinger, S.B. Georghiou, C.U. Köser and T.C. Rodwell are or were consultants or employees of FIND, the global alliance for diagnostics, a not-for-profit foundation that supports the evaluation of publicly prioritised tuberculosis assays and the implementation of WHO-approved (guidance and prequalification) assays using donor grants. FIND has product evaluation agreements with several private sector companies that design diagnostics for tuberculosis and other diseases. These agreements strictly define FIND's independence and neutrality with regard to these private sector companies. D.L. Dolinger works for General Fluidics and provides consulting services for Médecins Sans Frontières, Molbio Diagnostics and Partners in Health. D.L. Dolinger worked for QuantuMDx and PhAST. T. Gumbo is founder and CEO of Praedicare Inc, a pre-clinical contract research organisation and is a founder of Praedicare Africa Pvt Ltd, a clinical contract research organisation. K. Kaniga is a full-time employee of Johnson & Johnson Global Public Health. C.U. Köser is a consultant for the TB Alliance. C.U. Köser's consulting work for Becton Dickinson involves a collaboration with Janssen and Thermo Fisher Scientific. C.U. Köser is collaborating with PZA Innovation. C.U. Köser worked as a consultant for QuantuMDx, the Stop TB Partnership, the WHO Global TB Programme, and the WHO Regional Office for Europe. C.U. Köser gave a paid educational talk for Oxford Immunotec. Hain Lifescience covered C.U. Köser's travel and accommodation to present at a meeting. C.U. Köser is an unpaid advisor to BioVersys and GenoScreen. Y. Liu is an employee of Otsuka Pharmaceutical Development & Commercialization Inc, USA. V. Nikolayevskyy is employed by QIAGEN Manchester Ltd. S.V. Omar has received funding to prepare and provide training for Janssen Pharmaceutica activities. T.C. Rodwell is a cofounder, board member, and shareholder of Verus Diagnostics, a company that was founded with the intent of developing diagnostic assays. Verus Diagnostics was not involved in any way with data collection, analysis or publication of the results, and T.C. Rodwell has not received any financial support from Verus Diagnostics. University of California, San Diego (UCSD) Conflict of Interest office has reviewed and approved T.C. Rodwell's role in Verus Diagnostics. T.C. Rodwell is a coinventor of a provisional patent for a TB diagnostic assay (provisional patent 63/048.989). T.C. Rodwell is also a coinventor on a patent associated with the processing of TB sequencing data (European patent application number 14840432.0 and USSN 14/912,918), and has agreed to “donate all present and future interest in and rights to royalties from this patent” to UCSD to ensure that he does not receive any financial benefits from this patent. P. Supply is a consultant for Genoscreen. G. Theron's research group has received funding and/or in-kind donations in the last 5 years via his employer from Bruker Hain Lifesciences, Cepheid, LumiraDx, FIND, Biopromic, Newmark Diagnostics, Hemocue, Boditech and Copan. N. Veziris received travel support from Becton Dickinson for attending the Union Conference in 2018 outside the scope of this work. All other authors have nothing to disclose.
Support statement: As current or former employees or consultants for FIND, the work of R.B. Baldan, I. Comas, C.M. Denkinger, D.L. Dolinger, S.B. Georghiou, C.U. Köser and T.C. Rodwell on the systematic reviews, including this viewpoint, was supported by Unitaid (grant 2019-32-FIND MDR), BMGF (grant OPP1105925), the German Federal Ministry of Education and Research through KfW, the Dutch Ministry of Foreign Affairs, the Australian Department of Foreign Affairs and Trade, and UK aid from the British people. N. Alvarez and J. Robledo are funded by MinCiencias, Colombia (number 221389666216 CT-783-2018). A. Aubry and N. Veziris work at the Centre National de Reference des Mycobactéries, which receives an annual grant from Santé Publique France and have received research grants from Janssen for studies on bedaquiline. P. Claxton and I.F. Laurenson are funded through National Services Scotland. I. Comas was supported by PID2019-104477RB-I00 from the Spanish Science Ministry and by ERC (CoG 101001038). M. Egger is supported by the Swiss National Science Foundation (grant number 320030_153442 and 189498) and the US National Institutes of Health, National Institute of Allergy and Infectious Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Cancer Institute, the National Institute of Mental Health, the National Institute on Drug Abuse, the National Heart, Lung, and Blood Institute, the National Institute on Alcohol Abuse and Alcoholism, the National Institute of Diabetes and Digestive and Kidney Diseases, the Fogarty International Center, and the National Library of Medicine: Asia-Pacific, U01AI069907; CCASAnet, U01AI069923; Central Africa, U01AI096299; East Africa, U01AI069911; NA-ACCORD, U01AI069918; Southern Africa, U01AI069924; West Africa, U01AI069919. M.R. Farhat is supported by NIH NIAID R01AI155765. S.K. Heysell was funded by NIH NIAID grants R01 AI137080 and U01 AI150508. T. Jagielski was supported by a DAINA grant (number 2017/27/L/NZ6/03279) from the National Science Centre, Poland. J.L. Johnson was supported by contracts NO1-AI95383 and NO1-AI-70022 of the US National Institutes of Health. P.M. Keller was supported by Innosuisse 36198.1 IP-LS. C.U. Köser is a research associate at Wolfson College and visiting scientist at the Department of Genetics, University of Cambridge. The Federal Government of Germany supported C.U. Köser as part of his work for the European Laboratory Initiative, WHO Regional Office for Europe. C.U. Köser was further supported by the Royal Society of Tropical Medicine and Hygiene and the National Institute for Health Research Cambridge Biomedical Research Centre and received an observership by the European Society of Clinical Microbiology and Infectious Diseases to the EUCAST Development Laboratory for Bacteria (Växjö, Sweden), hosted by Gunnar Kahlmeter and Erika Matuschek. D. Machado and M. Viveiros are funded in part by Fundação para a Ciência e a Tecnologia, Portugal (PTDC/BIA-MIC/30692/2017, UID/Multi/04413/2020 and DL57/CEECIND/0256/2017). S. Niemann is supported by the German Center for Infection Research, Excellenz Cluster Precision Medicine in Chronic Inflammation EXC 2167, Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG). S.V. Omar has received funding to prepare and provide training for Janssen Pharmaceutica activities. L. Rigouts is supported by the Belgian Directorate General for Development. T.C. Rodwell was additionally funded in part by FIND and NIH NIAD, grants: P30 AI036214 and R21 AI135756. T. Schön is funded by the Swedish Heart and Lung Foundation and the Swedish Research Council. T.R. Sterling has received funding from the US National Institutes of Health and the Centers for Disease Control and Prevention. G. Theron and R. Warren are supported by baseline funding from the South African Medical Research Council. R.J. Wilkinson receives funding from the Wellcome Trust (203135) and from the Francis Crick Institute, which is supported by Cancer Research UK (FC0010218), UKRI (FC0010218) and the Wellcome Trust (FC0010218). The views expressed here are those of the authors and do not necessarily correspond to those of their respective employers.
Contributor Information
Antimycobacterial Susceptibility Testing Group:
Sophia B. Georghiou, Timothy C. Rodwell, Alexei Korobitsyn, Said H. Abbadi, Kanchan Ajbani, Jan-Willem Alffenaar, David Alland, Nataly Alvarez, Sönke; Andres, Elisa Ardizzoni, Alexandra Aubry, Rossella Baldan, Marie Ballif, Ivan Barilar, Erik C. Böttger, Soumitesh Chakravorty, Pauline M. Claxton, Daniela M. Cirillo, Iñaki Comas, Chris Coulter, Claudia M. Denkinger, Brigitta Derendinger, Edward P. Desmond, Jurriaan E.M. de Steenwinkel, Keertan Dheda, Andreas H. Diacon, David L. Dolinger, Kelly E. Dooley, Matthias Egger, Soudeh Ehsani, Maha R. Farhat, Lanfranco Fattorini, Iris Finci, Laure Fournier Le Ray, Victoria Furió, Ramona Groenheit, Tawanda Gumbo, Scott K. Heysell, Doris Hillemann, Harald Hoffmann, Po-Ren Hsueh, Yi Hu, Hairong Huang, Alamdar Hussain, Farzana Ismail, Kiyohiko Izumi, Tomasz Jagielski, John L. Johnson, Priti Kambli, Koné Kaniga, G.H.R. Eranga Karunaratne, Meenu Kaushal Sharma, Peter M. Keller, Ellis C. Kelly, Margarita Kholina, Mikashmi Kohli, Katharina Kranzer, Ian F. Laurenson, Jason Limberis, S-Y. Grace Lin, Yongge Liu, Alexandre López-Gavín, Anna Lyander, Diana Machado, Elena Martinez, Faisal Masood, Satoshi Mitarai, Nomonde R. Mvelase, Stefan Niemann, Vladyslav Nikolayevskyy, Florian P. Maurer, Matthias Merker, Paolo Miotto, Shaheed V. Omar, Ralf Otto-Knapp, Moisés Palaci, José Juan Palacios Gutiérrez, Sharon J. Peacock, Charles A. Peloquin, Jennifer Perera, Catherine Pierre-Audigier, Suporn Pholwat, James E. Posey, Therdsak Prammananan, Leen Rigouts, Jaime Robledo, Neesha Rockwood, Camilla Rodrigues, Max Salfinger, Marcos C. Schechter, Marva Seifert, Sarah Sengstake, Thomas Shinnick, Natalia Shubladze, Vitali Sintchenko, Frederick Sirgel, Sulochana Somasundaram, Timothy R. Sterling, Andrea Spitaleri, Elizabeth Streicher, Philip Supply, Erik Svensson, Elisa Tagliani, Sabira Tahseen, Akiko Takaki, Grant Theron, Gabriela Torrea, Armand Van Deun, Jakko van Ingen, Annelies Van Rie, Dick van Soolingen, Roger Vargas Jr, Amour Venter, Nicolas Veziris, Cristina Villellas, Miguel Viveiros, Robin Warren, Shu'an Wen, Jim Werngren, Robert J. Wilkinson, Caie Yang, F. Ferda Yılmaz, Tingting Zhang, Danila Zimenkov, Nazir Ismail, Claudio U. Köser, and Thomas Schön
References
- 1.Antimicrobial Resistance Collaborators . Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchison DA. Standardisation of sensitivity tests. Int J Tuberc Lung Dis 1998; 2: 69–70. [PubMed] [Google Scholar]
- 3.Böttger EC. The ins and outs of Mycobacterium tuberculosis drug susceptibility testing. Clin Microbiol Infect 2011; 17: 1128–1134. doi: 10.1111/j.1469-0691.2011.03551.x [DOI] [PubMed] [Google Scholar]
- 4.Ängeby K, Juréen P, Kahlmeter G, et al. Challenging a dogma: antimicrobial susceptibility testing breakpoints for Mycobacterium tuberculosis. Bull World Health Organ 2012; 90: 693–698. doi: 10.2471/BLT.11.096644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heifets L. Role of phenotypic methods for drug susceptibility testing of M. tuberculosis isolates in the era of MDR and XDR epidemics. In: McHugh TD, ed. Tuberculosis: Laboratory Diagnosis and Treatment Strategies. Wallingford, CAB International, 2013. [Google Scholar]
- 6.Van Deun A, Aung KJ, Bola V, et al. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 2013; 51: 2633–2640. doi: 10.1128/JCM.00553-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köser CU, Maurer FP, Kranzer K. ‘Those who cannot remember the past are condemned to repeat it’: drug-susceptibility testing for bedaquiline and delamanid. Int J Infect Dis 2019; 80S: S32–S35. doi: 10.1016/j.ijid.2019.02.027 [DOI] [PubMed] [Google Scholar]
- 8.Schön T, Matuschek E, Mohamed S, et al. Standards for MIC testing that apply to the majority of bacterial pathogens should also be enforced for Mycobacterium tuberculosis complex. Clin Microbiol Infect 2019; 25: 403–405. doi: 10.1016/j.cmi.2019.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schön T, Köser CU, Werngren J, et al. What is the role of the EUCAST reference method for MIC testing of the Mycobacterium tuberculosis complex? Clin Microbiol Infect 2020; 26: 1453–1455. doi: 10.1016/j.cmi.2020.07.037 [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization . Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. 2018. https://apps.who.int/iris/handle/10665/260470 (Date last accessed: 13 August 2021). [Google Scholar]
- 11.World Health Organization . Technical report on the pharmacokinetics and pharmacodynamics (PK/PD) of medicines used in the treatment of drug-resistant tuberculosis. 2018. https://apps.who.int/iris/handle/10665/260440 (Date last accessed: 4 September 2021). [Google Scholar]
- 12.Collaborative Group for the Meta-Analysis of Individual Patient Data in MDR-TB treatment , Ahmad N, Ahuja SD, et al. Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 2018; 392: 821–834. doi: 10.1016/S0140-6736(18)31644-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization . Technical report on critical concentrations for drug susceptibility testing of isoniazid and the rifamycins (rifampicin, rifabutin and rifapentine). 2021. https://apps.who.int/iris/handle/10665/339275 (Date last accessed: 13 August 2021). [Google Scholar]
- 14.World Health Organization . Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. 2021. https://apps.who.int/iris/handle/10665/341981 (Date last accessed: 26 June 2021). [Google Scholar]
- 15.Walker TM, Miotto P, Köser CU, et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe 2022; 3: e265–e273. doi: 10.1016/S2666-5247(21)00301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization . WHO consolidated guidelines on tuberculosis. Module 4: treatment – drug-resistant tuberculosis treatment. 2020. https://apps.who.int/iris/handle/10665/332397 (Date last accessed: 15 June 2020). [PubMed] [Google Scholar]
- 17.Kahlmeter G. The 2014 Garrod Lecture: EUCAST – are we heading towards international agreement? J Antimicrob Chemother 2015; 70: 2427–2439. doi: 10.1093/jac/dkv145 [DOI] [PubMed] [Google Scholar]
- 18.Köser CU, Georghiou SB, Schön T, et al. On the consequences of poorly defined breakpoints for rifampin susceptibility testing of Mycobacterium tuberculosis complex. J Clin Microbiol 2021; 59: e02328-20. doi: 10.1128/JCM.02328-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Deun A, Decroo T, Aung KJM, et al. Mycobacterium tuberculosis borderline rpoB mutations: emerging from the unknown. Eur Respir J 2021; 58: 2100783. doi: 10.1183/13993003.00783-2021 [DOI] [PubMed] [Google Scholar]
- 20.Zignol M, Dean AS, Alikhanova N, et al. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: results from a multicountry surveillance project. Lancet Infect Dis 2016; 16: 1185–1192. doi: 10.1016/S1473-3099(16)30190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed S, Köser CU, Salfinger M, et al. Targeted next-generation sequencing: a Swiss army knife for mycobacterial diagnostics? Eur Respir J 2021; 57: 2004077. doi: 10.1183/13993003.04077-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vargas R Jr, Freschi L, Spitaleri A, et al. Role of epistasis in amikacin, kanamycin, bedaquiline, and clofazimine resistance in Mycobacterium tuberculosis complex. Antimicrob Agents Chemother 2021; 65: e0116421. doi: 10.1128/AAC.01164-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köser CU, Robledo J, Shubladze N, et al. Guidance is needed to mitigate the consequences of analytic errors during antimicrobial susceptibility testing for TB. Int J Tuberc Lung Dis 2021; 25: 791–794. doi: 10.5588/ijtld.21.0428 [DOI] [PubMed] [Google Scholar]
- 24.Kahlmeter G, Giske CG, Kirn TJ, et al. Point–counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J Clin Microbiol 2019; 57: e01129-19. doi: 10.1128/JCM.01129-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigouts L, Coeck N, Gumusboga M, et al. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 2016; 71: 314–323. doi: 10.1093/jac/dkv360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifert M, Capparelli E, Catanzaro DG, et al. Using Mycobacterium tuberculosis single-nucleotide polymorphisms to predict fluoroquinolone treatment response. Antimicrob Agents Chemother 2019; 63: e00076-19. doi: 10.1128/AAC.00076-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasipanodya JG, Nuermberger E, Romero K, et al. Systematic analysis of hollow fiber model of tuberculosis experiments. Clin Infect Dis 2015; 61: Suppl. 1, S10–S17. doi: 10.1093/cid/civ425 [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs MT, Drusano GL, Brown DL, et al. Dose optimization of moxifloxacin and linezolid against tuberculosis using mathematical modeling and simulation. Int J Antimicrob Agents 2019; 53: 275–283. doi: 10.1016/j.ijantimicag.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 29.Drusano GL, Rogers S, Brown D, et al. Dose fractionation of moxifloxacin for treatment of tuberculosis: impact of dosing interval and elimination half-life on microbial kill and resistance suppression. Antimicrob Agents Chemother 2021; 65: e02533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldridge BB, Barros-Aguirre D, Barry CE 3rd, et al. The Tuberculosis Drug Accelerator at year 10: what have we learned? Nat Med 2021; 27: 1333–1337. doi: 10.1038/s41591-021-01442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization . Position statement on innovative clinical trial design for development of new TB treatments. 2021. https://apps.who.int/iris/handle/10665/342789 (Date last accessed: 19 July 2021). [Google Scholar]
- 32.Kim PS, Swaminathan S. Ending TB: the world's oldest pandemic. J Int AIDS Soc 2021; 24: e25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Deun A, Decroo T. How second-line injectable drugs work. Clin Infect Dis 2021; 72: e1167–e1168. doi: 10.1093/cid/ciaa1874 [DOI] [PubMed] [Google Scholar]
- 34.Ernest JP, Sarathy J, Wang N, et al. Lesion penetration and activity limit the utility of second-line injectable agents in pulmonary tuberculosis. Antimicrob Agents Chemother 2021; 65: e0050621. doi: 10.1128/AAC.00506-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Deun A, Decroo T, Piubello A, et al. Principles for constructing a tuberculosis treatment regimen: the role and definition of core and companion drugs. Int J Tuberc Lung Dis 2018; 22: 239–245. doi: 10.5588/ijtld.17.0660 [DOI] [PubMed] [Google Scholar]
- 36.Gausi K, Ignatius EH, Sun X, et al. A semimechanistic model of the bactericidal activity of high-dose isoniazid against multidrug-resistant tuberculosis: results from a randomized clinical trial. Am J Respir Crit Care Med 2021; 204: 1327–1335. doi: 10.1164/rccm.202103-0534OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute . Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes. 1st Edn. CLSI supplement M62. Wayne, Clinical and Laboratory Standards Institute, 2018.
- 38.Beckert P, Sanchez-Padilla E, Merker M, et al. MDR M. tuberculosis outbreak clone in Eswatini missed by Xpert has elevated bedaquiline resistance dated to the pre-treatment era. Genome Med 2020; 12: 104. doi: 10.1186/s13073-020-00793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonghanphithak D, Kaewprasert O, Chaiyachat P, et al. Whole-genome sequence analysis and comparisons between drug-resistance mutations and minimum inhibitory concentrations of Mycobacterium tuberculosis isolates causing M/XDR-TB. PLoS One 2020; 15: e0244829. doi: 10.1371/journal.pone.0244829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lounis N, Vranckx L, Gevers T, et al. In vitro culture conditions affecting minimal inhibitory concentration of bedaquiline against M. tuberculosis. Med Mal Infect 2016; 46: 220–225. doi: 10.1016/j.medmal.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 41.Mouton JW, Meletiadis J, Voss A, et al. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J Antimicrob Chemother 2018; 73: 2374–2379. doi: 10.1093/jac/dky232 [DOI] [PubMed] [Google Scholar]
- 42.Skov R, Matuschek E, Sjolund-Karlsson M, et al. Development of a pefloxacin disk diffusion method for detection of fluoroquinolone-resistant Salmonella enterica. J Clin Microbiol 2015; 53: 3411–3417. doi: 10.1128/JCM.01287-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute . Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. 2nd Edn. Approved standard. CLSI document M24-A2. Wayne, Clinical and Laboratory Standards Institute, 2011. [PubMed]
- 44.Clinical and Laboratory Standards Institute . Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes. 3rd Edn. CLSI standard M24. Wayne, Clinical and Laboratory Standards Institute, 2018. [PubMed]
- 45.World Health Organization . Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. 2008. https://apps.who.int/iris/handle/10665/70500 (Date last accessed: 10 January 2021). [PubMed] [Google Scholar]
- 46.World Health Organization . Guidelines for the programmatic management of drug-resistant tuberculosis: emergency update 2008. 2008. https://apps.who.int/iris/handle/10665/43965 (Date last accessed: 15 February 2020). [PubMed] [Google Scholar]
- 47.World Health Organization . Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. 2014. https://apps.who.int/iris/handle/10665/130918 (Date last accessed: 12 January 2020). [PubMed] [Google Scholar]
- 48.World Health Organization . WHO operational handbook on tuberculosis. Module 4: treatment – drug-resistant tuberculosis treatment. 2020. https://apps.who.int/iris/handle/10665/332398 (Date last accessed: 15 June 2020). [PubMed] [Google Scholar]
- 49.Nunn AJ, Rusen ID, Van Deun A, et al. Evaluation of a standardized treatment regimen of anti-tuberculosis drugs for patients with multi-drug-resistant tuberculosis (STREAM): study protocol for a randomized controlled trial. Trials 2014; 15: 353. doi: 10.1186/1745-6215-15-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.World Health Organization . WHO consolidated guidelines on drug-resistant tuberculosis treatment. 2019. https://apps.who.int/iris/handle/10665/311389 (Date last accessed: 21 March 2019). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00166-2022.Shareable (223.7KB, pdf)