Abstract
All detectable high-molecular-mass penicillin-binding proteins (HMM PBPs) are altered in a clinical isolate of Streptococcus mitis for which the β-lactam MICs are increased from those previously reported in our region (cefotaxime MIC, 64 μg/ml). These proteins were hardly detected at concentrations that saturate all PBPs in clinical isolates and showed, after densitometric analysis, 50-fold-lower radiotracer binding. Resistance was related to mosaic structure in all HMM PBP-coding genes, where critical region replacement was complemented not only by substitutions already reported for the closely related Streptococcus pneumoniae but also by other specific replacements that are presumably close to the active-site serine. Mosaic structure was also presumed in a pbp1a-sensitive strain used for comparison, confirming that these structures do not unambiguously imply, by themselves, detectable critical changes in the kinetic properties of these proteins.
Although viridans streptococci are recognized as a common cause of endocarditis (4, 9, 25) and are an increasingly reported cause of bacteremia in neutropenic patients (2, 3, 5, 19, 25), the scientific community has lacked a clear delineation of the pathogenic potential of individual species for viridans streptococci, probably due to past and present difficulties in their taxonomy and identification (7, 25, 45, 51, 52).
However, a rising incidence of β-lactam resistance in clinical isolates of this group has often compromised patient survival (4, 5, 8, 19, 30, 49), while occurrence of pneumococcal virulence factor-encoding genes within them may imply that their true pathogenic potential should be reevaluated (51).
Interspecies gene transfer of a variety of resistance markers between different but closely related streptococcal species, all of them naturally transformable and included in the mitis group, is well documented (6, 10, 12, 13, 22). Frequently the result of this interspecific recombination is mosaic genes, composed of alternating blocks of nucleotides with different degrees of relatedness derived from a particular donor, which in turn may have been affected by the same event. The recipient may encode a protein with altered catalytic and/or immunological activity (21).
PBPs (penicillin-binding proteins), the β-lactam bacterial targets, are ubiquitous enzymes involved in late steps of peptidoglycan biosynthesis, a bacterium-specific pathway. These proteins include the monofunctional dd-carboxypeptidases, which are not essential for viability, low-molecular-mass PBPs, and multidomain, generally essential, high-molecular-mass (HMM) PBPs, in which a transmembrane spanner is linked to a non-PB module, which in turn is linked to the amino end of a PB module. It is assumed that all PBPs retain the same basic tertiary structure in their PB module, with three conserved motives forming the catalytic center: *SxxK, SxN, and KTG (x denotes a variable amino acid residue). Each organism contains a complete set of these enzymes, which are all targeted by β-lactams. PBPs interact with the antibiotic by forming a relatively stable, covalent complex via the active-site serine (15–17).
β-Lactam resistance in clinically important viridans streptococci is mostly mediated by the presence of mosaic PBP genes encoding altered PBPs with decreased affinity to antibiotics (11, 12, 30). Higher concentrations of drugs are thus both required to inhibit altered PBPs and to confer in vivo activity.
During 1994, a resistant strain (penicillin MIC, 16 μg/ml; cefotaxime MIC, 64 μg/ml) tentatively identified as Streptococcus mitis by biochemical methods, which formerly was misidentified as Streptococcus pneumoniae, was isolated in an Argentinian pediatric hospital. The misidentification was related to phenotypic characteristics (colony morphology, Gram stain, hemolytic pattern, and optochin sensitivity). Such resistance levels had never been reported before for any member of the mitis group in our area.
The aim of this work was to investigate and describe the mechanism of resistance responsible for these high MICs.
All experiments were performed simultaneously with another clinical isolate which was biochemically identical but sensitive.
In order to solve any biochemical pitfall in the identification of both isolates, a more reliable genetic identification was attempted by sequencing the gene coding for 16S rRNA and the manganese-dependent superoxide dismutase (sodA).
Their PBP pattern, relative affinity, and codifying genes, as well as their deduced amino acid sequences, were analyzed. Comparative analysis of PBP amino acid sequences coming from sensitive and resistant strains of S. mitis was done to search for particular amino acid alterations present in resistant strains of this species.
Although the participation of S. mitis in homologous recombination events leading to mosaic genes in pneumococci is well documented (11, 13, 22), reports of PBP patterns and PBP genes of sensitive and resistant strains of S. mitis are still rare. They are rare even when the appearance and spread of S. mitis clinical isolates resistant to penicillin are a serious problem, not only in areas where penicillin-resistant S. pneumoniae is common but also in those with a low incidence of resistant pneumococci (8, 19, 26, 30, 46).
(Part of this work was presented at the 100th General Meeting of the American Society for Microbiology, Los Angeles, Calif., May 2000.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains 127R and 209S were isolated in close temporal proximity (3 months) in a pediatric hospital in Buenos Aires, Argentina, during 1994 and were preliminarily identified by biochemical methods as S. mitis. Identifications were confirmed by R. Facklam (Centers for Disease Control and Prevention, Atlanta, Ga.). For most purposes both strains were grown in brain heart infusion broth (Difco) at 37°C. The microorganisms were kept frozen in 20% skim milk (Difco)–20% glycerol at −20, −70, and −196°C (the lowest temperature was for long-term storage). Escherichia coli INVαF′ and E. coli TOP 10F′ (Invitrogen, Leek, The Netherlands) were used for cloning experiments. SOC medium (1) was used during the cloning procedures.
Susceptibility tests.
β-Lactam antibiotics were tested by the agar dilution method on brain heart infusion agar (Difco) supplemented with 2.5% sheep blood. Inocula were prepared by growing the strains overnight in liquid media and diluting the cells 100-fold before plating. Incubation was performed at 37°C for at least 48 h. Antibiotics were provided by various companies: penicillin G (Eli Lilly Laboratorios, Buenos Aires, Argentina), dicloxacillin, oxacillin, and cephaloridin (Sigma Chemical Co., St. Louis, Mo.), ampicillin (Laboratorios Roemmers, Buenos Aires, Argentina), cefotaxime (Merck, Sharp and Dohme, Buenos Aires, Argentina), cefuroxime and ceftazidime (Glaxo Wellcome, Buenos Aires, Argentina), cephalotin (Roussel, Buenos Aires, Argentina), ceftriaxone and cefalexin (Laboratorios Argentina, Buenos Aires, Argentina), cefoperazone (Pfizer, Buenos Aires, Argentina), and piperacillin and aztreonam (Bristol-Myers, Buenos Aires, Argentina).
Sequencing of 16S rRNA coding and sodA genes.
Chromosomal DNA was obtained by conventional methods (43). fD1 and rD1 primers were used for amplifying 16S rRNA genes (35, 50) in a Trio-thermoblock (Biometra thermocycler). The reaction mixture (100 μl) contained 80 pmol of each primer and 2.5 μmol of each dNTP (Pharmacia Biotech, Uppsala, Sweden). PCR was performed as follows: 5 min of denaturation at 95°C and 15 min of annealing at 50°C, with 2.5 U of Taq polymerase (Pharmacia Biotech) added 6 min after the annealing temperature was reached. The reaction was followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 2 min, and extension at 72°C for 2 min, concluding with a final extension for 20 min at 72°C. The DNA fragments (1.5 kb) were purified with the Gene Clean purification kit (Bio 101, La Jolla, Calif.), digested with EcoRI and BamHI (New England Bio Labs, Beverly, Mass.), and cloned into digested pUC18.
An internal region of the sodA gene, coding for almost 85% of the manganese-dependent superoxide dismutase, was amplified by PCR using degenerate primers (40) as described above but with the annealing temperature changed to 39°C. The amplified 0.45-kb fragments were cloned into the PCR II vector (TA Cloning kit; Invitrogen) following the manufacturer's instructions.
DNA sequences were determined in at least two different clones using the method described by Sanger et al. (44) in an Alf automatic sequencer (Pharmacia Biotech).
Preparation of membranes.
Cells of exponentially growing cultures were harvested by centrifugation and washed twice in 50 mM phosphate buffer (pH 7.0). After resuspension in a minimum volume of the same buffer containing 50 μg of DNase I (Sigma Chemical Co.)/ml, cells were disrupted by five passages through a French press (SLM Instruments, Urbana, Ill.). The cell lysate was clarified by centrifugation (10 min at 5,000 rpm; SS34 rotor in a Sorvall RC5 centrifuge), and the clear supernatant was then centrifuged at 40,000 rpm for 50 min in a 50 Ti rotor (Beckman RT ultracentrifuge). The pellet obtained was resuspended in a minimum volume of 40 mM phosphate, 1 mM MgCl2, pH 7.0, and 5% glycerol buffer and was conserved at −20°C until use. All steps were performed at 4°C. Protein concentrations were determined by a modification of the method pioneered by Lowry (1).
Detection of PBPs.
Membrane samples containing approximately 200 μg of protein were labeled with the indicated concentrations of 125I-penicillin X (38) and were incubated for 15 min at 37°C. The reaction was stopped by adding an excess of nonradiolabeled antibiotic and by chilling on ice. Denaturing buffer (0.25 M Tris-HCl, 8% sodium dodecyl sulfate, 20% β-mercaptoethanol, 0.008% bromophenol blue, and 50% glycerol) was added, and the samples were immediately boiled for 5 min and centrifuged for 2 min at 14,000 rpm (Bench Eppendorf microcentrifuge).
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 20-cm-long gels. The final acrylamide concentration of the separation gel was 7% with an acrylamide/bisacrylamide ratio of 29.2:0.8. PBPs were visualized after 10 days' exposure at −20°C of an X-Omat Kodak film.
Amplification and sequencing of PBP genes.
The genes homologous to pneumococcal pbp2x and pbp2b from 127R and 209S were amplified with 2XUP and 2XDW primers (32) (corresponding to a region of the pneumococcal pbpX gene including motives 2 to 7 of class B HMM PBPs) and with 2BUP and 2BDW primers (14) (corresponding to a region coding for the transpeptidase domain). The amplified DNA fragments were purified from agarose with the Gene Clean II purification kit. Their ends were filled with T4 DNA polymerase. The fragments were then phosphorylated with T4 DNA kinase (Pharmacia Biotech), repurified, and cloned into SmaI/BAP pUC18 (Pharmacia Biotech). DNA sequences were determined in two clones, with oligonucleotides that primed at intervals along each strand of the sequence.
Only a small portion of the gene homologous to pneumococcal pbp1a could be amplified after several PCR attempts with published primers. A fragment of 0.8 kb could be obtained with primers 1AUp786 (5′-CGGCATTCGATTTGATTCGCTTCT-3′) (36) and 1A2Dw1598 (5′-GGGTCATATTGGTTTGGTGC-3′) (37). Based on the known fragment sequence, the LA PCR cloning kit (Takara Shuzo Co., Kyoto, Japan) was used following the manufacturer's instructions. Briefly, after preliminary previous hybridization experiments using the 0.8-kb fragment as the probe, genomic DNA was completely digested with PstI and was ligated with a PstI cassette (a double-stranded, synthetic oligonucleotide provided with the kit) and the corresponding PstI restriction site. A first PCR with this ligation as DNA matrix was performed by using a cassette primer (provided with the kit) and primer 1AS1 (5′-CGAACTGATTGCTGACCTTGGATCTGAAC-3′), the design of which was based upon the known external region of 0.8 kb. A second PCR was performed after dilution of the first PCR product by using a second inner cassette primer (provided) and primer 1AS2 (5′-ACTTGGTCAAGGCAATTGTGTCTATCGAAG-3′), the design of which was based on the inner 0.8 kb (an already-known region). A unique band of 2.4 kb was obtained, cloned into pUC18, and sequenced as noted above.
Nucleotide sequence accession number.
The EMBL database accession numbers for each gene are given for 127R and 209S, respectively: 16S rRNA gene, AJ295848 and AJ295853; sodA, AJ295849 and AJ295854; pen2X, AJ295850 and AJ295855; pen2b, AJ295851 and AJ295857; pen1a, AJ295852 and AJ295856.
RESULTS
Amplification and sequencing of the 16S rRNA gene.
Single bands of 1.5 kb were obtained for both strains. Their nucleotide sequence had 99% identity with homologous genes of S. mitis (GenBank accession no. D38482) (27), S. pneumoniae (GenBank accession no. AF003930) (S. Emler, N. Liassine, J. Pawlowsky, B. Hirschel, P. Rohner, and R. Auckenthaler, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. D-155, p. 235, 1997), Streptococcus oralis (GenBank accession no. AF003932) (Emler et al., Abstr. 97th Gen. Meet. Am. Soc. Microbiol.), and Streptococcus sanguinis (GenBank accession no. AF 003928) (Emler et al., Abstr. 97th Gen. Meet. Am. Soc. Microbiol.). The sequences obtained from 127R and 209S differ by only 2 nucleotides (99.94% of identity), clearly indicating that both strains belong to the same species.
Amplification and sequencing of the sodA gene.
Searching for a more discriminative target sequence to differentiate our strains from closely related bacterial species, we amplified by PCR an internal fragment of the sodA gene, obtaining a single product of 0.45 kb. Nucleotide sequences from 127R and 209S showed 97% identity with the homologous gene of S. mitis NCTC 12261 (GenBank accession no. Z95909) (40) and 94% identity with those of S. pneumoniae (GenBank accession no. 95914) (40) and S. oralis (GenBank accession no. Z99195) (40). In all cases and for both strains, only 2 of 143 amino acids changed in the analyzed fragment; showing 98% similarity to the S. mitis protein, 97% similarity to the corresponding pneumococcal protein, and 96% similarity to that of S. oralis. sodA from S. sanguinis ATCC 10556 (GenBank accession no. Z95918) (40) was 82% identical. Alignment of 127R and 209S sequences revealed a 97% identity, pointing out that the sodA heterogeneity was higher than in the 16S rRNA. Nevertheless, this identity confirms that both strains belong to the same bacterial species.
Antimicrobial susceptibility.
MICs of the various β-lactams tested, including early- and later-generation cephalosporins (Table 1) showed high-level resistance in 127R, with values far above those reported for this group of microorganisms. No correlation between chemical structure and MICs could be detected, suggesting that more than one PBP should be altered.
TABLE 1.
In vitro β-lactam antibiotic susceptibilities of strains 127R and 209Sa
| Antibiotic | MIC (μg/ml) for strain:
|
|
|---|---|---|
| 127R | 209S | |
| Penicillin G | 16 | 0.003 |
| Ampicillin | 16 | 0.003 |
| Cefotoxime | 64 | 0.003 |
| Ceftriaxone | 32 | 0.003 |
| Cefoperazone | 16 | 0.003 |
| Piperacillin | 16 | 0.006 |
| Cefuroxime | 32 | 0.003 |
| Cephalotin | 64 | 0.012 |
| Ceftazidime | 32 | 0.012 |
| Cefalexin | 32 | 0.012 |
| Cephaloridin | 16 | 0.006 |
| Dicloxacillin | 64 | 0.06 |
| Oxacillin | 64 | ND |
| Aztreonam | >64 | ND |
MICs were determined by the agar dilution method. ND, not determined.
This strain also showed, by agar diffusion tests, resistance to erythromycin, rifampin, chloramphenicol, and trimetoprim-sulfamethoxazole.
PBP pattern in sensitive and resistant strains.
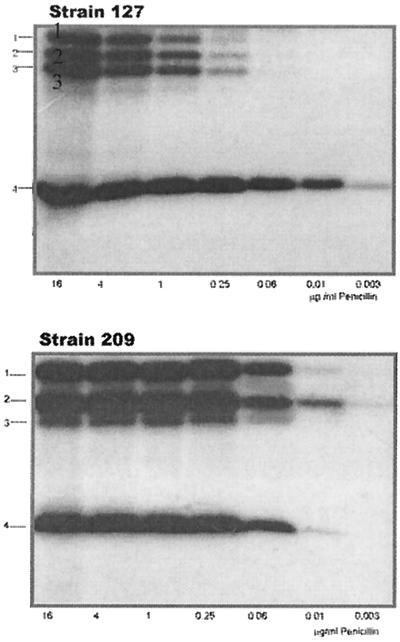
When PBPs were labeled with 125I-penicillin at concentrations that usually saturate PBPs of penicillin-resistant S. pneumoniae (23), three HMM PBPs and one low-molecular-mass PBP could be detected in both strains, but they did not show identical electrophoretic mobility (Fig. 1).
FIG. 1.
Radioactive detection of PBPs from strains 127R and 209S. Membranes containing approximately 200 μg of proteins were labeled with the indicated concentration of 125I-penicillin X and were incubated for 15 min at 37°C. Penicillin concentrations are given in micrograms per milliliter.
At a concentration that could saturate all the PBPs in the sensitive strain (125I-penicillin X, 0.25 μg/ml), HMM PBPs in the resistant strain were hardly visualized and showed, by means of densitometry, an almost 50-fold-lower binding capacity of the radiotracer (data not shown). Several PBPs are present at the same positions in S. mitis NCTC 10712 and S. pneumoniae R6 as well (30), but the relationship to these proteins is not known.
HMM PBP genes in sensitive and resistant strains.
The unusually high resistance levels measured for strain 127R and the fact that all the HMM PBPs of this strain showed a lower affinity than did those of the sensitive strain suggested that sequences of the PBP-encoding genes could give some insights about changes probably responsible for sensitivity alterations. Portions of the genes homologous to pbp2x and pbp2b of S. pneumoniae could be amplified with primers designed according to known sequences of penicillin-sensitive and -resistant S. pneumoniae strains. In the case of the pbp1a homologous genes, a different strategy was followed, as described in Materials and Methods.
pbp2x
In the resistant strain, this gene had a mosaic structure. When compared with the homologous gene of S. mitis NCTC 10712 (the type strain for the species; sensitive) (46) the C- and N-terminal coding portions (positions 1 to 400 and 1600 to 2000, respectively) showed 95% identity but diverged by 20% between positions 400 and 1000 and also between 1200 and 1600, with a short region of 90% identity between positions 1000 and 1200. Furthermore, the block between positions 540 and 780 had almost the same sequence as that of a penicillin-resistant S. pneumoniae mutant (GenBank accession no. X65132) (34), with 98% identity. At the C-terminal end encoding sequence, another 400-bp block, which started at position 1620, was identical (99% identity) to that of a highly penicillin-resistant S. pneumoniae isolate (GenBank accession no. U87092) (53). Another short sequence between positions 1260 and 1500 showed 98% identity with a penicillin-resistant strain of S. mitis (GenBank accession no. Y10535) (42).
pbp2x from 209S was 91% identical to the homologous gene of S. mitis NCTC 10712 (46) without any evidence of mosaic structure
pbp2b.
For this gene, the portion coding for the complete C-terminal transpeptidase domain was amplified. In 127R it also showed a mosaic structure. When compared to pbp2b of S. mitis NCTC 10712 (13), only a small portion of the gene, between positions 960 and 1260, showed 98% identity. The first 350-bp block diverged by 5% from that of the sensitive type strain, and blocks from positions 350 to 960 and 1260 to 1500 differed by 17 and 21%, respectively. In addition, the first block (positions 1 to 350) also showed similar percentages of identity with those of several genes of penicillin-resistant clinical isolates of S. pneumoniae (GenBank accession nos. U20076 and U20074) (46) and of a recently described (22), highly cefotaxime-resistant clinical isolate of S. mitis (GenBank accession no. AJ002289). Between positions 350 and 660 several homologous genes of penicillin-resistant S. pneumoniae strains showed 94 to 96% identity. Two blocks of 300 bases (positions 660 to 960 and 1200 to 1500) showed no divergence with those of penicillin-resistant isolates of S. pneumoniae (GenBank accession no. Z21981) (6) and S. oralis (GenBank accession no. M32228) (11).
pbp2b from 209S is 94% identical to the homologous gene of S. mitis NCTC 10712 (13), showing no evident mosaic structure.
pbp1a.
For this gene, we encountered several difficulties in the sequence analysis. First, the number of sequences available in data banks was quite low compared to those for other PBP genes, probably because PBP1A is not the primarily affected β-lactam target in clinical isolates (18). Only a few sensitive genes belonging to S. pneumoniae were available for comparison. Furthermore, to our knowledge, only a portion of a highly cefotaxime-resistant S. mitis was recently sequenced. However, analysis of both sensitive and resistant genes strongly suggests that intragenic recombination events may have occurred, since polymorphism distribution was not at random. When compared with homologous genes of S. pneumoniae, a very closely related species, blocks with clearly different rates of divergence could be detected in both resistant and sensitive genes as well.
Amino acid changes in resistant PBPs.
The deduced amino acid sequences were analyzed and aligned with homologous sequences of sensitive and resistant S. mitis strains (Table 2). Although changes already described to be involved in penicillin and cefotaxime resistance in S. pneumoniae were also found in 127R, a few extra amino acid changes highlight new interesting positions near the active site of the enzyme that are likely to lead to changes in the affinity of the PBP (Fig. 2).
TABLE 2.
Strains used for PBP sequence alignmentsa
| Accession no. | S. mitis strain | Origin | Penicillin MIC (μg/ml) | Reference(s) |
|---|---|---|---|---|
| Y10535 | Hu8 | Hungary, 1990–92 | 6–12 | 22 |
| Y10532 | B5 | Germany, 1994 | 8 | 22, 42 |
| Y10533 | 197 | Spain, 1990 | 16 | 42 |
| Y10534 | 476 | Spain, 1992 | 10 | 42 |
| AJ002288 | B6 | Germany, 1994 | 50 | 22 |
| AJ002289 | B6 | Germany, 1994 | 50 | 22 |
| Z22183 | 11189 | England, 1977 | 0.016 | 14 |
| Z22231 | NS51 | Sweden, 1965 | 0.03 | 14 |
| Z22186 | K208 | United States, 1932 | 0.03 | 14 |
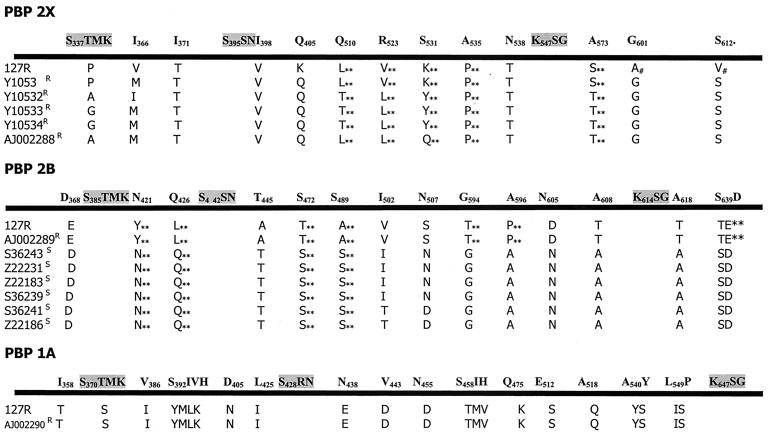
Figure 2 depicts alignments.
FIG. 2.
Protein sequence alignments from sensitive and resistant strains of S. mitis. All amino acid sequences belong to different strains of S. mitis and are named under their accession number (GenBank). 127R results are from this study. Shown in boldface are conserved motifs and selected amino acid positions where we detected variation along the complete protein sequence of sensitive strains 209S (this study) and S. mitis NCTC 10712 (there were no amino acid differences between them). R, β-lactam-resistant strain; S, β-lactam-sensitive strain; ∗∗, changes not previously reported in pneumococci; #, changes found only in strain 127R. The indicated numbers correspond to positions in PBPs from S. pneumoniae R6.
PBP2X.
For this protein sequence, alignments from resistant isolates of S. mitis showed that immediately after the active S337, T338 was changed to P, A, or G. In S. pneumoniae these changes reduced the acylation efficiency of the enzyme by cefotaxime (39) (T to A) and by penicillins (T to P or G). These changes were frequently found in clinical isolates with unusually high levels of β-lactam resistance (33, 34). Accordingly, strain 127R showed the T-to-P change, but strain 209S possessed the same T residue as the type strain. Other resistant S. mitis PBP2X analyzed also showed changes from T338 to P, A, or G, but no correlation could be established with the resistance level for any particular amino acid change. In all resistant S. mitis strains, between the conserved SxxK and SxN motives, I371 changed to T and downstream the conserved SxN motif I398 replaced V, as observed in S. pneumoniae mutants (33). In addition, we found substitutions at positions 510, 523, 531, 535, 538, and 574 in all resistant S. mitis strains, not previously reported, which could point out interesting positions influencing the affinity of the PBP.
On the other hand, as opposed to changes described for resistant pneumococci (22, 34, 46), positions 571 and 586 were not changed in resistant strains of S. mitis.
PBP2B.
In resistant S. mitis strains we found the T446-to-A and A619-to-G changes described previously in penicillin-resistant pneumococci and supposedly responsible for low-level penicillin resistance, lower lysis rate, and piperacillin resistance as well (18, 21, 24, 47). However, alignments showed additional substitutions, which were present only in resistant S. mitis strains. Upstream of the SxN motif, N421 was changed to Y and Q426 to L. Downstream of the SxN motif, the following modifications were observed: S472 to T, S489 to A, I502 to V, N507 to S, G594 to T, A596 to P, N605 to D, A608 to T, and finally, just behind the KxG triad, A618 to G. In the C-terminal portion of the protein, SD640 and NG659 were replaced by TE and KN, respectively.
PBP1A.
For this protein, the alignment includes only the sequences obtained from 127R and 209S and a partial sequence belonging to a S. mitis clinical isolate, highly resistant to cefotaxime, from Germany (22). Analyzing the PB domain of PBP1A of the resistant strains, we found that at position 358, an I changed to T in the resistant strains. Several additional substitutions were also observed in the resistant strains: V306 was changed to I, immediately after the active S370; T371 was changed to S, SIVH395 to YMLK; D405 to N and L425 to I; N438 to E; V443 to D; N455 to D; SIH460 to TMV; Q475 to K; E512 to S; A518 to Q; AY541 to YS; and finally, LP550 to IS 6 residues upstream of the KTG conserved motif. Part of the amino acid changes is present in positions already described for this and other PBPs in resistant pneumococci, but we also detected other alterations in positions not previously described.
DISCUSSION
The extensive and relatively rapid changes that have taken place in the classification of the oral streptococci and the increase in the number of species recognized in this group of bacteria have not been accompanied by the development of comprehensive species identification schemes for the ordinary laboratory (28, 29, 45, 52). However, striking species-specific variations in susceptibility, especially to β-lactams, are evident from different studies (3, 19, 41, 49), suggesting that speciation of oral streptococci must be warranted in order to evaluate results reported by different studies. Up to date, no single phenotypic or genotypic approach has demonstrated by itself its universal usefulness (28, 29, 52). Intraspecies heterogeneity and both lack of biochemical activity and genetic exchange between different species have contributed to this scenario.
The identification of our strains could be achieved only when biochemical and genetic results were taken together. Even considered as a “gold standard” for several purposes, establishment of the complete coding sequence for the 16S rRNA did not allow us to differentiate among S. mitis, S. sanguinis, S. oralis, and S. pneumoniae species, since they showed 99% identity in this gene (they differ by only 10 to 12 nucleotides). We concluded, thus, that the 16S rRNA sequence could not be applied unambiguously to the identification of this group of microorganisms.
Analysis of partial sequence of sodA has been proposed as a reliable and accurate method for members of the mitis group (28, 40). Kawamura et al. indicated that the evolutionary rate of the sodA partial gene was much higher than that of the 16S rRNA gene and that the sodA partial sequencing would be useful for differentiating closely genetically related organisms (28). In our case, sequences indicated that a higher interspecies heterogeneity than in the 16S rRNA could exist in this gene, but the identity with S. pneumoniae and S. oralis remained high (up to 95%).
The difficulties encountered for the identification of our strains could be due, in part, to the genetic exchanges that they have undergone. Further efforts should be made to improve the available methods or to find new powerful approaches in order to identify oral streptococci and particularly those of the mitis group, which, because of genetic exchange, seem to constitute a complex mosaic group more than clearly separated entities (12, 45). When studying an isolate of this group of organisms, and before defining the species, all available biochemical and genetic features must be examined and analyzed as a whole.
The β-lactam MICs determined for strain 127R are extremely high for this group of microorganisms, which are usually susceptible to penicillin concentrations as low as a few nanograms per milliliter. Such MICs, at least in pneumococci, could only be reached when more than one β-lactam target is altered (21, 22, 24). In the resistant strain all the HMM PBPs (detected with a radiotracer with a high specific activity) showed a much lower binding of the radioactive antibiotic than did those of the sensitive strain, indicating that they all had a reduced affinity for penicillin. Precompetition with other nonradioactive β-lactam antibiotics and postlabeling assays (data not shown) confirmed the observation that a reduced affinity existed in all the HMM PBPs of strain 127R. Since all of the β-lactam targets had altered affinity, higher concentrations of drugs are thus required to inhibit these enzymes.
In 127R the sequences of the HMM PBP-encoding genes display a mosaic structure, with blocks diverging up to 20% from the sensitive homologous genes.
However, this feature does not seem to be restricted to the resistant phenotype: a mosaic structure could be inferred when analyzing pbp1a from the sensitive strain 209S. Sibold et al. and Dowson et al. described this phenomenon earlier in PBP2B genes of penicillin-sensitive S. mitis (13, 46), as did Spratt et al. for penicillin-sensitive isolates of Neisseria lactamica and Neisseria polisaccharea (48), but mosaic genes, even coding for primarily affected β-lactam targets, have no selective advantage for penicillin resistance. The observation that, in pneumococci, mosaic genes are found only in penicillin-resistant isolates but not in penicillin-sensitive strains (24) seems not to be the case for S. mitis. In this species, genes displaying mosaic structures do not unequivocally code for proteins with detectable reduced affinity, and the same could be true also for pneumococci, considering that most of the analyzed PBP genes come from resistant strains. Nevertheless, PBP1A is not a primarily affected target in resistant pneumococci (18, 31), as it presumably is in the mitis group. Consequently, it can be expected that alterations in kinetic properties in this protein, if present, should not be by themselves responsible for changes in susceptibility.
With those results, we assume that precise amino acid changes should simultaneously be present along with a mosaic structure, at least in primarily affected β-lactam targets (PBP2B and PBP2X), to yield highly resistant isolates. Protein sequence alignments showed that the amino acid changes found in resistant isolates are not present in sensitive strains, so both mosaic structures and amino acid changes seem to be indispensable for β-lactam resistance in the viridans streptococci.
Furthermore, the amino acid changes that we found after protein alignment occur at positions common to all the analyzed PBPs. A few additional substitutions are seen in all resistant strains. They were not reported before in the closely related pneumococci; perhaps they could be pointing out interesting positions leading to reduced affinity of each PBP. Most of these modifications are probably responsible for increasing MICs. Those for example in the vicinity of the cavity forming the active site of the PBPs could be important for the interaction of the antibiotic with the enzyme. More information about the three-dimensional structure of these proteins is needed to validate these observations.
Further studies should be encouraged to enlarge our knowledge about particular resistance mechanisms in this species, since reports of resistant isolates are rapidly increasing. Perhaps most important is the fact that these resistant strains constitute a potential pool of genes for the development of high-level β-lactam resistance in S. pneumoniae strains and other pathogens of the mitis group, in which genetic competence has provided a powerful tool for the uptake and incorporation of resistance determinants.
ACKNOWLEDGMENTS
We are grateful to Horacio Lopardo (Hosp. Nac. de Pediatría) and to P. Garrahan and Marta Altschuler (Hosp. Sor María Ludovica, Buenos Aires, Argentina) for providing clinical isolates; to Carlos Vay for helpful discussion related to bacterial identification; and to Richard Facklam, Centers for Disease Control and Prevention, Atlanta, Ga., for biochemical identification of strains. We thank Iris Thamm for technical support with automated DNA sequencing.
This work was supported in part by grants from the University of Buenos Aires, (Buenos Aires, Argentina) (TB 039) and from the CONICET (PID 4413) to G.G. and in part by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian government, Prime Minister's Office, Services fédéraux des affaires scientifiques, techniques et culturelles (PAI P4/03). A.A. was a recipient of a fellowship from the European Community, Program ALFA, Project 5.0111.9. G.G. is a member of the “Carrera de Investigador Científico,” CONICET, Buenos Aires, Argentina.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl J, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1990. [Google Scholar]
- 2.Avada A, van der Auwera P, Meunier F, Daneau D, Klastersky J. Streptococcal and enterococcal bacteremia in patients with cancer. Clin Infect Dis. 1992;15:33–48. doi: 10.1093/clinids/15.1.33. [DOI] [PubMed] [Google Scholar]
- 3.Bochud P Y, Engiman P, Calandra T, van Melle G, Saghafi L, Francioli P. Bacteremia due to viridans streptococci in neutropenic patients with cancer: clinical spectrum and risk factors. Clin Infect Dis. 1994;18:25–31. doi: 10.1093/clinids/18.1.25. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet A, Durand A, Devine C, Etienne J, Leport C. In vitro susceptibility to antibiotics of 200 strains of streptococci and enterococci isolated during infective endocarditis. In: Totolian A, editor. Pathogenic streptococci: present and future. St. Petersburg, Russia: Lancer Publications; 1994. pp. 72–73. [Google Scholar]
- 5.Carratala J, Alcaide F, Fernandez-Sevilla A, Corbella X, Liñares J, Gudiol F. Bacteremia due to viridans streptococci that are highly resistant to penicillin: increase among neutropenic patients with cancer. Clin Infect Dis. 1995;20:1169–1173. doi: 10.1093/clinids/20.5.1169. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Dowson C G, Daniels M, Spratt B G. Horizontal spread of an altered penicillin-binding protein 2B gene between Streptococcus pneumoniae and Streptococcus oralis. FEMS Microbiol Lett. 1993;110:335–339. doi: 10.1111/j.1574-6968.1993.tb06345.x. [DOI] [PubMed] [Google Scholar]
- 7.Coykendall A. Classification and identification of the viridans streptococci. Clin Microbiol Rev. 1989;2:315–328. doi: 10.1128/cmr.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doern G, Ferraro M, Brueggemann A, Ruoff K. Emergence of high rates of antimicrobial resistance among viridans group streptococci in the United States. Antimicrob Agents Chemother. 1996;40:891–894. doi: 10.1128/aac.40.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 10.Dowson C, Hutchison A, Brannigan J, George R, Hansman D, Liñares J, Tomasz A, Smith J, Spratt B. Horizonal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowson C, Hutchison A, Woodford N, Johnson A, George R, Spratt B. Penicillin resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowson C, Barcus V, King S, Pickerill P, Whatmore A, Yeo M. Horizontal gene transfer and the evolution of resistance and virulence determinants in Streptococcus. J Appl Microbiol Symp Ser. 1997;83:42S–51S. doi: 10.1046/j.1365-2672.83.s1.5.x. [DOI] [PubMed] [Google Scholar]
- 13.Dowson C, Coffey T, Kell C, Whiley R. Evolution of penicillin resistance in Streptococcus pneumoniae: the role of Streptococcus mitis in the formation of a low affinity PBP 2B in Streptococcus pneumoniae. Mol Microbiol. 1993;9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 14.Dowson C, Hutchison A, Spratt B. Extensive remodelling of the transpeptidase domain of penicillin binding protein 2B of a penicillin resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 15.Ghuysen J M. Penicillin binding proteins, wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int J Antimicrob Agents. 1997;8:45–60. doi: 10.1016/s0924-8579(96)00358-5. [DOI] [PubMed] [Google Scholar]
- 16.Ghuysen J M, Charlier P, Coyette J, Duez C, Fonzé E, Fraipont C, Goffin C, Joris B, Nguyen-Distèche M. Penicillin and beyond: protein fold, multimodular polypeptides and multiprotein complexes. Microb Drug Resist. 1996;2:163–175. doi: 10.1089/mdr.1996.2.163. [DOI] [PubMed] [Google Scholar]
- 17.Goffin C, Ghuysen J M. The multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40:829–834. doi: 10.1128/aac.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guiot H F L, Corel L J K, Vossen J M J J. Prevalence of penicillin-resistant viridans streptococci in healthy children and in patients with malignant haematological disorders. Eur J Clin Microbiol Infect Dis. 1994;13:645–650. doi: 10.1007/BF01973990. [DOI] [PubMed] [Google Scholar]
- 20.Hakenbeck R. Target-mediated resistance to β-lactam antibiotics. Biochem Pharmacol. 1995;50:1121–1127. doi: 10.1016/0006-2952(95)00158-v. [DOI] [PubMed] [Google Scholar]
- 21.Hakenbeck R. Mosaic genes and their role in penicillin-resistant Streptococcus pneumoniae. Electrophoresis. 1998;19:597–601. doi: 10.1002/elps.1150190423. [DOI] [PubMed] [Google Scholar]
- 22.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakenbeck R, Martin C, Dowson C, Grebe T. Penicillin-binding protein 2b of Streptococcus pneumoniae in piperacillin-resistant laboratory mutants. J Bacteriol. 1994;176:5574–5577. doi: 10.1128/jb.176.17.5574-5577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakenbeck R, Grebe T, Zähner D, Stock J. β-Lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol Microbiol. 1999;33:673–678. doi: 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 25.Hardie J, Whiley R. Recent developments in streptococcal taxonomy: their relation to infections. Rev Med Microbiol. 1994;3:151–162. [Google Scholar]
- 26.Jacobs J, Schouten H, Stobberingh E, Soeters P. Viridans streptococci isolated from the bloodstream. Diagn Microbiol Infect Dis. 1995;22:267–273. doi: 10.1016/0732-8893(95)00137-y. [DOI] [PubMed] [Google Scholar]
- 27.Kawamura Y, Hou X G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 28.Kawamura Y, Whiley R, Shu S, Ezaki T, Hardie J. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi K, Ezaki T, Totsuka K-I, Shimizu K. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J Clin Microbiol. 1995;33:1215–1222. doi: 10.1128/jcm.33.5.1215-1222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.König A, Reinert R, Hakenbeck R. Streptococcus mitis with unusually high-level resistance to β-lactam antibiotics. Microb Drug Resist. 1998;4:45–49. doi: 10.1089/mdr.1998.4.45. [DOI] [PubMed] [Google Scholar]
- 31.Kraub J, van der Linden M, Grebe T, Hakenbeck R. Penicillin-binding proteins 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb Drug Resist. 1996;2:183–186. doi: 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 32.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J M. Nucleotide sequence of the pbpX gene encoding the penicillin-binding protein 2X from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 33.Laible G, Hackenbeck R. Five independent combinations of mutations can result in low-affinity penicillin-binding protein 2x of Streptococcus pneumoniae. J Bacteriol. 1991;173:6986–6990. doi: 10.1128/jb.173.21.6986-6990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 35.Lane D, Pace B, Olsen G, Stahl D, Sogin M, Pace N. Rapid determination of 16S ribosomal RNA segments for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin C, Briese T R, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1A and 2B. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin C, Sibold C, Hakenbeck R. Relatedness of penicillin-binding protein 1A genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 1992;11:3831–3836. doi: 10.1002/j.1460-2075.1992.tb05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masson J M, Labia R. Synthesis of a 125I-radiolabeled penicillin for penicillin-binding protein studies. Anal Biochem. 1983;128:164–168. doi: 10.1016/0003-2697(83)90357-3. [DOI] [PubMed] [Google Scholar]
- 39.Mouz N, Gordon E, Di Guilmi A M, Petit I, Petillot Y, Dupont Y, Hakenbeck R, Vernet T, Dideberg O. Identification of a structural determinant for resistance to β-lactam antibiotics in Gram-positive bacteria. Proc Natl Acad Sci USA. 1998;95:13403–13406. doi: 10.1073/pnas.95.23.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyart C, Quesne G, Caelon S, Berche P, Trieu-Cuot P. Identification of streptococci to species levels by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinn J P, Di Vicenzo C, Lucks D, Luskin R, Shatzer K, Lerner S. Serious infections due to penicillin-resistant strains of viridans streptococci with altered penicillin-binding proteins. J Infect Dis. 1988;157:764–769. doi: 10.1093/infdis/157.4.764. [DOI] [PubMed] [Google Scholar]
- 42.Reichmann P, König A, Liñares J, Alcaide F, Tenover F, McDugal L, Swidsinski S, Hakenbeck R. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J Infect Dis. 1997;176:1001–1012. doi: 10.1086/516532. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanger I, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlegel L, Bouvet A. Streptocoques et genres apparentés: abiotrophes et enterocoques. Bull Soc Fr Microbiol. 1998;13:7–17. [Google Scholar]
- 46.Sibold C, Henrichsen J, König A, Martin C, Chalkley L, Hakenbeck R. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol. 1994;12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 47.Smith A M, Klugman K P. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1995;39:859–867. doi: 10.1128/aac.39.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spratt B G, Bowler L D, Zhang Q-Y, Zhou J, Smith J M. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34:115–125. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 49.Tuohy M, Washington J. Antimicrobial susceptibility of viridans group streptococci. Diagn Microbiol Infect Dis. 1997;29:277–280. doi: 10.1016/s0732-8893(97)00140-5. [DOI] [PubMed] [Google Scholar]
- 50.Weisburg W, Barns S, Pelletier D, Lane D. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whatmore A M, Efstratiou A, Pickerill A P, Broughton K, Woodward G, Sturgeon D, George R, Dowson C G. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhao G, Yeh W-K, Carnahan R H, Flokowitsch J, Meier T I, Alborn W E, Jr, Becker G W, Jaskunas S R. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J Bacteriol. 1997;179:4901–4908. doi: 10.1128/jb.179.15.4901-4908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]