Summary:
The epidermal growth factor receptor (EGFR) is important for normal homeostasis in a variety of tissues and, when abnormally expressed or mutated, contributes to the development of many diseases. However, in vivo functional studies are hindered by the lack of adult mice lacking EGFR because of the pre- and postnatal lethality of EGFR deficient mice. We generated a conditional allele of Egfr (Egfrtm1Dwt) by flanking exon 3 with loxP sites in order to investigate tissue-specific functions of this widely expressed receptor tyrosine kinase. The activity of the Egfrtm1Dwt allele is indistinguishable from wildtype Egfr. Conversely, the EgfrΔ allele, generated by Cre-mediated deletion of exon 3 using the germline EIIa-Cre transgenic line, functions as a null allele. EgfrΔ/Δ embryos that have complete ablation of EGFR activity and die at mid-gestation with placental defects identical to those reported for mice homozygous for the Egfrtm1Mag null allele. We also inactivated the Egfrtm1Dwt allele tissue-specifically in the skin epithelium using the K14-Cre transgenic line. These mice were viable but exhibited wavy coat hair remarkably similar to mice homozygous for the Egfrwa2 hypomorphic allele or heterozygous for the EgfrWa5 antimorphic allele. These results suggest that the hairless phenotype of Egfr nullizygous mice is not solely due to absence of EGFR in the epithelium, but that EGFR activity is required also in skin stromal cells for normal hair morphogenesis. This new mouse model should have wide utility to inactivate Egfr conditionally for functional analysis of EGFR in adult tissues and disease states.
Keywords: EGFR, receptor tyrosine kinase, gene targeting, Cre/loxP, conditional allele
The epidermal growth factor receptor (EGFR) is the prototypical member of the ERBB family of tyrosine kinase receptors that also includes ERBB2, ERBB3, and ERBB4. Egfr is widely expressed and is essential for normal placental development and function of many organ systems. Many abnormal phenotypes associated with loss of EGFR are dependent on genetic background. On some backgrounds the placenta does not develop normally and has a variably reduced spongiotrophoblast layer and a disorganization of the labyrinth layer (Dackor et al., 2007; Strunk et al., 2004), while on other backgrounds EGFR deficient mice survive until term and have abnormalities of the skin and gastrointestinal tract, immature lungs, and cell death in the cortex and olfactory bulb (Miettinen et al., 1995; Sibilia and Wagner, 1995; Threadgill et al., 1995). Egfr null mice also have abnormal craniofacial development including defects in palate closure, elongated snouts, and underdeveloped lower jaws (Miettinen et al., 1999).
To study the role of EGFR in adult tissues and disease models, the Egfrwa2 hypomorphic allele is frequently used. The Egfrwa2 allele is a spontaneously arising point mutation causing a Val743Gly substitution in the ATP-binding site of the tyrosine kinase domain (Fowler et al., 1995; Luetteke et al., 1994). The receptor produced from the Egfrwa2 allele has an 80–95% reduction in EGFR activity leading to lactational defects (Fowler et al., 1995) and an increased frequency of cardiac hypertrophy and death caused by enlarged aortic valves on some genetic backgrounds (Chen et al., 2000). The only overt abnormal phenotype observed on all backgrounds is wavy coat hair, which results from loss of EGFR signaling during the anagen phase of the hair cycle (Mak and Chan, 2003).
A conditional humanized allele, EgfrKI, has also been created, containing a human EGFR cDNA that replaces the mouse Egfr allele (Sibilia et al., 2003). Mice homozygous for EgfrKI can live up to 6 months, but display a variety of organ defects making this allele unsuitable for studying the role of EGFR in specific tissues through conditional ablation. More recently, another conditional allele was reported lacking the abnormal phenotypes (Natarajan et al., 2007). This allele appears to function normally before deletion. However, exon 1 was flanked by loxP sites, which may lead to altered gene regulation on some backgrounds or under some experimental conditions.
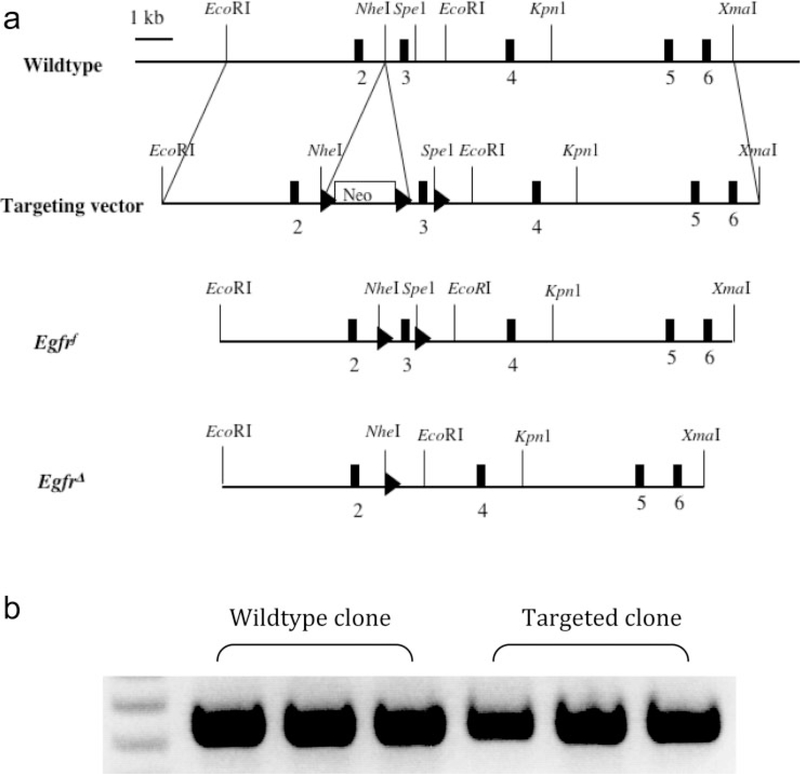
To overcome the real or potential deficiencies with existing Egfr alleles, we generated a new conditional allele of Egfr by flanking exon 3 with loxP sites (Fig. 1a). Deletion of exon 3 introduces a frameshift resulting in two stop codons in exon 4 and early termination of translation (Reiter et al., 2001). In addition, exon 3 encodes residues 57–117, which is part of the L1 subdomain of the extracellular domain that is essential for ligand-binding (Garrett et al., 2002), ensuring an inactive Egfr allele upon Cre-mediated recombination even if residual altered protein is produced. After electroporating TL1 embryonic stem (ES) cells from 129S6/SvEvTAC (129S6) mice, 13 targeted ES cell clones were identified out of 300 screened ES cell clones (4.3% targeting frequency). The targeted allele was identified as a 6.9 kbp Pfl F1 fragment by Southern blot analysis as opposed to the wildtype allele that produced a 9.4 kbp fragment. One of the clones was found to have lost the third loxP site as determined by PCR analysis, indicating that in this clone homologous recombination occurred in the region between the second and the third loxP sites. A second clone contained a correctly targeted allele but the other allele differed from the predicted wildtype allele (data not shown). The remaining 11 ES cell clones were correctly targeted.
FIG. 1.
Structure of wildtype and engineered Egfr alleles. (a) Targeting approach to flank exon 3 with loxP sites and generation of the Egfrf allele by Cre-mediated deletion of the Neo cassette. The EgfrΔ allele is generated by Cre-mediated deletion of exon 3 from the Egfrf allele. (b) RT-PCR analysis of RNA corresponding to exons 2–4 in Egfrf/+ ES cells.
On the basis of the colony morphology and karyotype analysis, two clones were chosen to remove the Neo cassette by recombination between the first and the second loxP sites using transient Cre expression. This generated the conditional Egfrtm1Dwt allele (also called Egfrf) that only has the two loxP sites franking exon 3 remaining in the Egfr locus. To verify that the remaining loxP sites do not perturb Egfr expression, RT-PCR analysis was performed to show that Egfrf transcripts are indistinguishable from those of the wildtype allele (Fig. 1b). ES cells containing Egfrf were used to produce chimeras by injection into C57BL/6J (B6) blastocysts. Ten male chimeras were obtained, five of which had high coat color chimerism (85–100%). These were mated with wildtype B6 and 129S6 females, which produced agouti pups containing the Egfrf allele as determined by PCR.
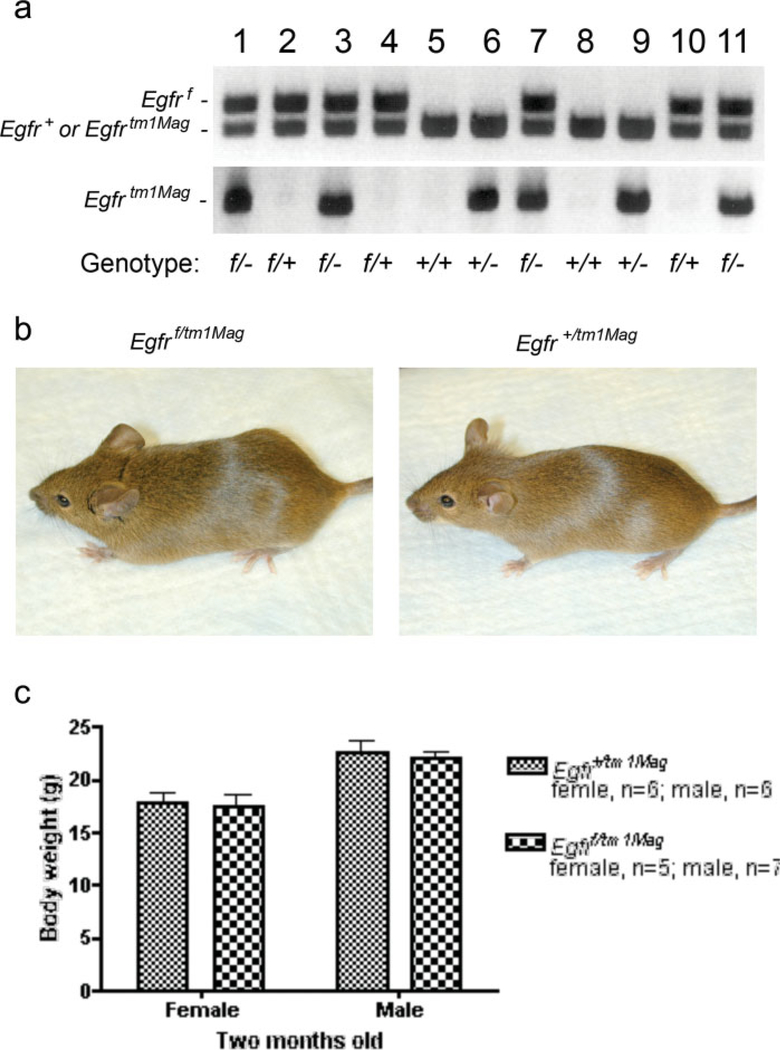
The activity of the Egfrf allele was tested by crossing Egfrf/+ mice with mice carrying the Egfrtm1Mag null allele. Progeny with Egftf/tm1Mag and Egfr+/tmIMag genotypes were born in expected Mendelian ratios (Fig. 2a). Egf//tm1Mag mice have normal coat hair (Fig. 2b), body weights (Fig. 2c), and fertility (data not shown). Statistically, there was no significant difference between genotypes. These results demonstrate that the Egfrf allele produces EGFR with wildtype activity in vivo.
FIG. 2.
Evaluation of Egfrf function. (a) Egfrf/tm1Mag mouse (left) that is indistinguishable from an Egfr+/tm1Mag mouse (right). (b) Genotypes of progeny from a cross between Egfr+/tm1Mag and Egfrf/+ mice. (c) Comparison of body weights between Egfrf/tm1Mag and Egfr+/tm1Mag mice.
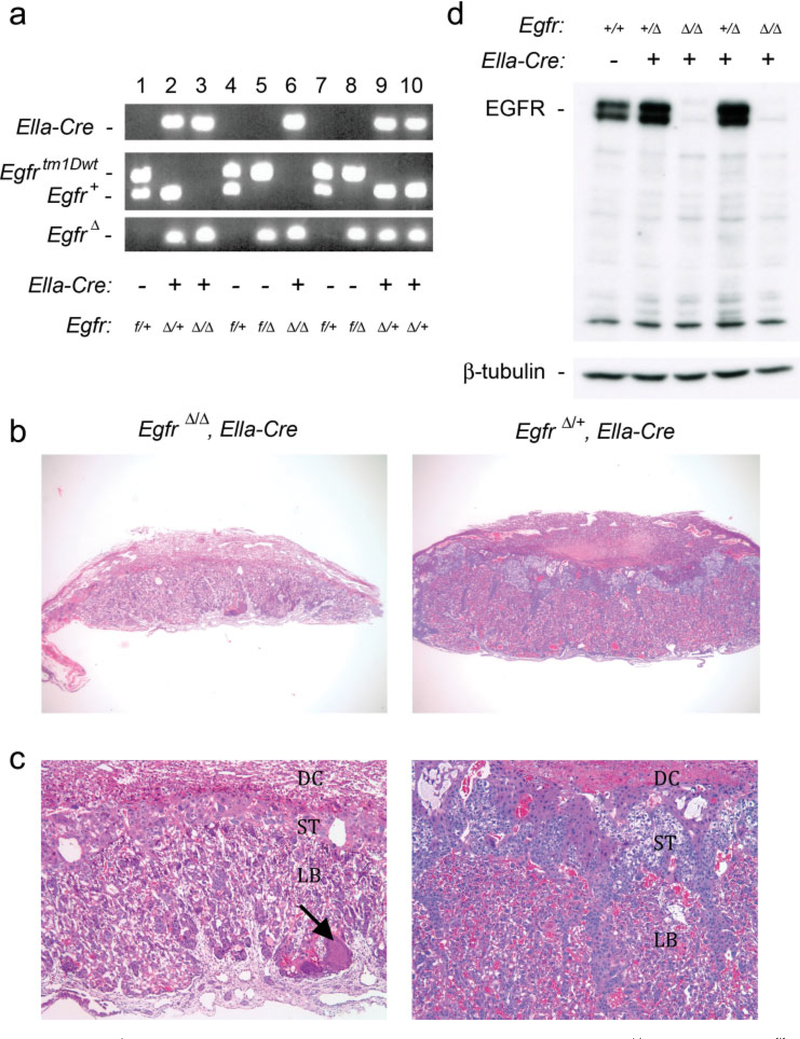
After Cre-mediated excision of exon 3, the EgfrΔ allele is produced and is predicted to act like a null allele. To test this prediction, mice carrying the EgfrΔ allele were produced by crossing Egfrf/f mice with mice carrying the EIIa-Cre transgene (Fig. 3a); the EIIa promoter expresses Cre during preimplantation development (Lakso et al., 1996). At mid-gestation, placentas from EgfrΔ/Δ, EIIa-Cre embryos were grossly smaller in size than those of their wildtype littermates (Fig. 3b,c). Histological analysis showed that the spongiotrophoblast and labyrinth layers were affected similarly to that reported for mice homozygous for the EgfrtmlMag null allele (Dackor et al., 2007; Strunk et al., 2004; Threadgill et al., 1995). In the spongiotrophoblast layer, the numbers of trophoblast and glycogen cells were moderately to severely reduced, with few spongiotrophoblast cells remaining in some EgfrΔ/Δ, EIIa-Cre placentas. The labyrinth layer in the mutant placentas had a disorganized architecture, which was also reduced in size. Western blot analysis for EGFR revealed total loss of EGFR in placentas from EgfrΔ/Δ, EIIa-Cre embryos (Fig. 3d). Taken together, these results demonstrate that the EgfrΔ allele functions identically to an Egfr null allele.
FIG. 3.
Analysis of EgfrΔ allele activity. (a) Genotypes of 12.5 dpc embryos from crosses between EgfrΔ/+, Ella-Cre, and Egfrf/f mice. (b) H&E staining of EgfrΔ/Δ, Ella-Cre placenta (left, ×30) with smaller size than wildtype placenta (right, ×30). (c) H&E staining of EgfrΔ/Δ, Ella-Cre placenta (left, ×100) showing reduced spongiotrophoblast layer and disorganized labyrinth layer with amorphous material (arrow) compared to wildtype placenta (right, ×100). (d) Western blot analysis showing loss of EGFR in EgfrΔ/Δ, Ella-Cre embryos at 10.5 dpc. DC, decidua; ST, spongiotrophoblast layer; LB, labyrinth layer.
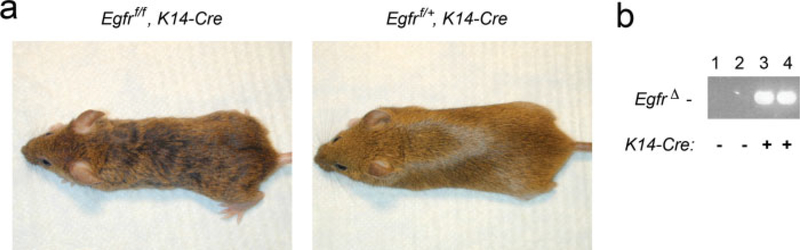
Since most uses of the Egfrf allele will be for tissue or temporal ablation of EGFR activity, Egfrf/f mice were crossed with K14-Cre transgenic mice that express Cre in the developing skin (Dassule et al., 2000). Egfrf/f, Kl4-Cre mice were obtained at Mendelian ratios and were viable and fertile (data not shown). However, unlike their wildtype littermates that had normal, straight coat hair, Egfrf/f, Kl4-Cre mice displayed wavy coat hair similar to the phenotype of Egfrwa2/wa2 mice (Fig. 4a; Keeler; 1935); as with EIIa-Cre, the EgfrΔ allele was efficiently generated in the skin of Egfrf/f, K14-Cre mice (Fig. 4b). The phenotype of Egfr nullizygous mice is absence of coat hair due to severely disrupted hair follicle morphogenesis, suggesting that deletion of Egfr in the epithelium alone is insufficient to recapitulate the null phenotype. Rather, these results suggest that disruption of hair morphogenesis seen in Egfr nullizygous mice is caused by the combined loss of EGFR in the epithelium and stroma of the skin. This result indicates that the Egfrf allele undergoes tissue-specific Cre-mediated excision.
FIG. 4.
Tissue-specific deletion of Egfr. (a) Egfrf/f, K14-Cre mouse (left) at 3 months of age displaying a wavy coat compared with the straight coat of wildtype mouse (right) at the same age. (b) PCR products using primers specific for EgfrΔ allele showing generation in skin from K14-Cre mice.
The development and validation of the conditional Egfrtm1Dwt allele reported here will be a valuable resource for investigating the role of EGFR in vivo, particularly at time points after mice constitutionally lacking EGFR die. The Egfr allele can be inactivated spatially and temporally, overcoming the limitations caused by embryonic pre- or early postnatal lethality of mice lacking EGFR. Since conditional alleles have been generated for all four Erbb genes (Crone et al., 2002; Long et al., 2003; Qu et al., 2006), it is now possible to contemplate generation of combinatorial tissue-specific knockouts to elucidate the role for specific ERBB combinations.
METHODS
Generation of the Egfrtm1Dwt Conditional Null Allele
BAC clone 158K10 containing the Egfr genomic locus was digested with Eco R1 to isolate DNA fragments of 6.0 kbp and 12.8 kbp surrounding exon 3 (Reiter et al., 2001). The 6.0 kbp Eco R1 fragment was further digested with Nhe 1 to produce a 4.3 kbp Eco R1-Nhe 1 fragment for the 5′ homology arm that was inserted upstream of a loxP-Neo-loxP cassette to generate 5′-loxP-Neo-loxP. The remaining 1.7 kbp Nhe 1-Eco R1 fragment was ligated with a 7.5 kbp Eco R1-Xma 1 fragment isolated from the 12.8 kbp Eco R1 fragment after digestion with Xma 1. After subcloning the 92 kbp Nhe 1-Xma 1 fragment, a loxP site was inserted into the Spe I site within intron 3 before cloning the fragment downstream of the 5′-loxP-Neo-loxP cassette to form the targeting vector (Fig. 1a).
The targeting vector was electroporated into TL1 ES cells (derived from 129S6/SvEvTAC mice and provided by Dr. Brigid Hogan, Duke University) as previously described (Threadgill et al., 1995). After selection with G418, resistant ES cell clones were digested with Pfl F1 and subjected to agarose gel electrophoresis before placing the gel in denaturation solution (0.4M NaOH, 3M NaCl). The restriction fragments were then Southern blotted by capillary action to a nylon membrane (Nytran Supercharge, Schleicher & Schuell, Keene, NH). After UV cross-linking, a 1.3 kbp 32P-labeled probe corresponding to a region upstream of the 5′ homology arm was hybridized to the membrane. The membrane was washed and exposed to X-ray film to reveal bands of 6.9 kbp Pfl F1 for a correctly targeted allele and 9.4 kbp for the wildtype allele. Additional confirmation of putatively targeted clones was performed using Southern blots with Eco N1 and Nde 1 digested ES cell DNA. A correctly targeted allele produced bands of 10.9 kbp and 11.2 kbp, respectively, while the wildtype allele produced bands of 8.8 kbp and 91 kbp, respectively.
Two correctly targeted ES cell clones were used by the University of North Carolina Animal Models Core for injection into C57BL/6J blastocysts and re-implantation into surrogate dams. After weaning, male chimeras were tested for germline transmission by mating to C57BL/6J females. Tail DNA samples from agouti offspring were genotyped for the Egfrtm1Dwt allele (also called Egfrf) by PCR. Conditions were 35 cycles (30 s at 94°C, 1 min at 60°C and 1 min at 72°C) with Taq DNA polymerase (Qiagen, Valencia, CA). The primers were lox3s 5′-CTTTGGA GAACCTGCAGATC-3′ and lox3as 5′-CTGCTACTGGCT CAAGTTTC-3′. A 375 bp PCR product is generated from the Egfrf allele and a 320 bp PCR product from the wildtype allele. The Cre-reduced EgfrΔ allele was detected by PCR using 40 cycles (30 s at 94°C, 20 s at 60°C and 20 s at 72°C) with primers Delta-3 5′-CTCAGCCAGAT GATGTTGAC-3′ and Delta-4 5′-CCTCGTCTGTGGAA GAACTA-3′. A 129 bp PCR fragment is amplified from the EgfrΔ allele. The Egfr null allele, EgfrtmIMag (Threadgill et al., 1995), was detected by amplifying DNA for 40 cycles (30 s at 94°C, 30 s at 55°C and 1 min at 72°C). The primers were EGFR common 5′-GCCCTGCCTTTCCCACCATA-3′ and EGFR knockout 5′-AACGTCGTGACTGGGAAAAC-3′. A 450 bp PCR product is detected from the Egfrtm1Mag allele.
RT-PCR
Total RNA were extracted from targeted and wildtype ES cell clones using TRizol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of RNA was used as template for synthesis of cDNA using random primers (Gibco-BRL, Carlsbad, CA) and SuperScript II RT (Gibco-BRL) in a total reaction volume of 20 μl. The cDNA was used as template for PCR amplification of exons 2–4 using 35 cycles (30 s at 93°C, 1 min at 58°C and 1 min at 72°C) with primers RNA2 5′-TGCCAAGGCACAAG-TAACAG-3′ and RNA4 5′-GCTCGGATGGCTCTGTAAGT-3′. The predicted wildtype band is 465 bp.
Mouse Crosses
Mice carrying the Egfrf allele were crossed with Egfr+/tm1Mag mice to generate Egfrf/tm1Mag and Egfr+/tm1Mag F1 mice that were used as experimental and control groups, respectively. Body weights were measured and compared by gender. To evaluate whether EgfrΔ/Δ functions as a null allele, Egfrf/f mice were crossed with EIIa-Cre mice to generate EgfrΔ/+, EIIa-Cre progeny, which were then crossed with Egfrf/f mice to generate EgfrΔ/+, EIIa-Cre, and EgfrΔ/Δ, EIIa-Cre mice. To determine whether the Egfrf allele can function as a tissue-specific conditional allele, Egfrf/f mice were crossed to K14-Cre mice to obtain Egfrf/+, K14-Cre progeny, which were then crossed to Egfrf/f mice to generate Egfrf/f, K14-Cre, and Egfrf/f, K14-Cre offspring that were used as experimental and control groups, respectively. Coat phenotypes were examined by visual inspection and histology.
Egfrf/+ and Egfrf/f mice were on a 129,B6 mixed background, while Egfr+/tm1Mag were on a 129S1/SvImJ background (Threadgill et al., 1995). Ella-Cre mice, on a B6,FVB mixed background, were a gift from Dr. Mark Majesky (UNC) and K14-Cre mice, on a FVB background, were obtained from The Jackson Laboratory. Genotyping for the Ella- and K14-Cre transgenes from genomic DNA was done by PCR using 38 cycles (30 s at 94°C, 1 min at 56°C and 1 min at 72°C) with primers CRE-1 5′-GTGAT GAGGTTCGCAAGAAC-3′ and CRE-2 5′-AGCATTGCTGT CACTTGGTC-3′. A 278 bp PCR fragment is generated from the Cre transgenes.
Histological Analysis
Placentas from 12.5 days post coitus (dpc) embryos were isolated and fixed in 10% neutral buffered formalin solution (NBF) for 24 h. After fixation, the samples were dehydrated using a graded alcohol series, 70% (1 h), 85% (1 h), 95% (1 h), 100% (1 h), and 100% (1 h), followed by two washes in xylene for 1 h for each. Finally, the samples were infiltrated with paraffin at 60°C twice for 1 h each before embedding and cutting 7-μm sections. Sections were stained with hematoxylin and eosin and after mounting, examined under light microscopy as previously (Strunk et al., 2004; Threadgill et al., 1995).
Western Blot Analysis
Embryos and yolk sacs isolated at 10.5 dpc from crosses containing Egfr+/Δ, EIIa-Cre, and Egfrf/f were homogenized in 10 ml tissue lysis buffer/g of tissue. The lysis buffer contained 20 mM HEPES, pH 7.4, 150 mM MaCl, 2 mM ethylenediamine tetraaceticacid (EDTA), pH 8.0, 2 mM ethylene-bis(oxyethylenenitrilo)tetraacetic acid (EGTA), pH 8.0, 1% Triton X-100, and 10% glycerol adjusted to final pH 7.4 before adding protease inhibitors [1 mM phenyl sulfonyl fluoride (PMSF) in isopropanol, 10 μg/ml leupeptin and 10 μg/ml aprotinin] and phosphatase inhibitors [1 mM sodium orthovanadate (Na3VO4) and 1 mM NaF].
Twenty-five micrograms of total protein from each homogenized lysate were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), before transferring to a polyvinylidene fluoride (PVDF) membrane. The membrane was placed in blocking solution containing 5% milk in TBST (150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 0.1% Tween-20), and then probed with a sheep anti-EGFR polyclonal IgG primary antibody (Upstate, Billerica, MA) in blocking solution (1:500). The secondary antibody was rabbit anti-sheep polyclonal IgG conjugated with horseradish peroxidase (HRP) (1:10,000). To visualize antibody binding, the membrane was incubated with enhanced chemiluminescent (ECL) substrate (Pierce, Rockford, IL) for 1–5 min and exposed to X-ray film. The amount of β-tubulin in each sample was used as an internal control. β-tubulin was detected using a mouse primary anti-β-tubulin polyclonal IgG antibody (1:1,000), with a secondary goat anti-mouse polyclonal IgG antibody conjugated with HRP (1:15,000).
Statistical Analysis
An unpaired t-test was used for all comparisons.
ACKNOWLEDGMENTS
The authors thank Dr. Daekee Lee for discussion and suggestions during these studies.
Contract grant sponsor: NSF, Contract grant number: MCB-9729645; Contract grant sponsor: NIH, Contract grant numbers: HD39896, CA92479, CA106991
LITERATURE CITED
- Chen B, Bronson RT, Klaman LD, Hampton TG, Wang JF, Green PJ, Magnuson T, Douglas PS, Morgan JP, Neel BG. 2000. Mice mutant for Egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nat Genet 24:296–299. [DOI] [PubMed] [Google Scholar]
- Crone SA, Zhao YY, Fan L, Gu Y Minamisawa S, Liu Y, Peterson KL, Chen J, Kahn R, Condorelli G, Ross J, Chien KR, Lee KF. 2002. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med 8:459–465. [DOI] [PubMed] [Google Scholar]
- Dackor J, Strunk KE, Wehmeyer MM, Threadgill DW 2007. Altered trophoblast proliferation is insufficient to account for placental dysfunction in Egfr null embryos. Placenta 28:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127:4775–4785. [DOI] [PubMed] [Google Scholar]
- Fowler KJ, Walker F, Alexander W, Hibbs ML, Nice EC, Bohmer RM, Mann GB, Thumwood C, Maglitto R, Danks JA, Chetty R, Burgess AW, Dunn AR. 1995. A mutation in the epidermal growth factor receptor in waved-2 mice has a profound effect on receptor biochemistry that results in impaired lactation. Proc Natl Acad Sci USA 92:1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, Jorissen RN, Nice EC, Burgess AW, Ward CW. 2002. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell 110:763–773. [DOI] [PubMed] [Google Scholar]
- Keeler CE. 1935. A second rexoid coat character in the house mouse. J Hered 26:189–191. [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 93:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long W, Wagner KU, Lloyd KC, Binart N, Shillingford JM, Hennighausen L, Jones FE. 2003. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130:5257–5268. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. 1994. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev 8:399–413. [DOI] [PubMed] [Google Scholar]
- Mak KK, Chan SY 2003. Epidermal growth factor as a biologic switch in hair growth cycle. J Biol Chem 278:26120–26126. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376:337–341. [DOI] [PubMed] [Google Scholar]
- Miettinen PJ, Chin JR, Shum L, Slavkin HC, Shuler CF, Derynck R, Werb Z. 1999. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat Genet 22:69–73. [DOI] [PubMed] [Google Scholar]
- Natarajan A, Wagner B, Sibilia M. 2007. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci USA 104:17081–17086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S, Rinehart C, Wu HH, Wang SE, Carter B, Xin H, Kotlikoff M, Arteaga CL. 2006. Gene targeting of ErbB3 using a Cre-mediated unidirectional DNA inversion strategy. Genesis 44:477–486. [DOI] [PubMed] [Google Scholar]
- Reiter JL, Threadgill DW, Eley GD, Strunk KE, Danielsen AJ, Sinclair CS, Pearsall RS, Green PJ, Yee D, Lampland AL, Balasubramaniam S, Crossley TD, Magnuson TR, James CD, Maihle NJ. 2001. Comparative genomic sequence analysis and isolation of human and mouse alternative EGFR transcripts encoding truncated receptor isoforms. Genomics 71:1–20. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner B, Hoebertz A, Elliott C, Marino S, Jochum W, Wagner EF. 2003. Mice humanised for the EGF receptor display hypomorphic phenotypes in skin, bone and heart. Development 130:4515–4525. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner EE 1995. Strain-dependent epithelial defects in mice lacking the EGE receptor. Science 269:234–238. [DOI] [PubMed] [Google Scholar]
- Strunk KE, Amann V, Threadgill DW 2004. Phenotypic variation resulting from a deficiency of epidermal growth factor receptor in mice is caused by extensive genetic heterogeneity that can be genetically and molecularly partitioned. Genetics 167:1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T. 1995. Targeted disruption of mouse EGE receptor: Effect of genetic background on mutant phenotype. Science 269:230–234. [DOI] [PubMed] [Google Scholar]