Abstract
Respiratory illness caused by influenza virus is a serious public health problem worldwide. As the symptoms of influenza virus infection are similar to those of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, it is essential to distinguish these two viruses. Therefore, to properly respond to a pathogen, a detection method that is capable of rapid and accurate diagnosis in a hospital or at home is required. To satisfy this need, we applied loop-mediated isothermal amplification (LAMP) and an isothermal nucleic acid amplification technique, along with a system to analyze the results without specialized equipment, a lateral flow assay (LFA). Using the platform developed in this study, all processes, from sample preparation to detection, can be performed without special equipment. Unlike existing PCR methods, the nucleic acid amplification can be performed in the field because hot packs do not require electricity. Thus, the designed platform can provide rapid results without the need to transport the samples to a laboratory or hospital. These advantages are not limited to operations in developing countries with poor access to medical systems. In conclusion, the developed technology is a promising tool for infectious disease management that allows for rapid identification of infectious diseases and appropriate treatment of patients.
Graphical abstract

Supplementary Information
The online version contains supplementary material available at 10.1007/s00216-022-04090-8.
Keywords: Avidin-biotin complex, Hot pack, Influenza virus, Lateral flow assay, Loop-mediated isothermal amplification, Point-of-care
Introduction
Recently, the world has been threatened by the rapid increase in respiratory infectious diseases such as influenza, Middle East respiratory syndrome (MERS), Zika, and coronavirus disease 2019 (COVID-19) [1, 2]. Influenza viruses cause fatal diseases worldwide and are a source of epidemics and episodic epidemics [3–5]. The similarity between the clinical manifestations of influenza and COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which originated in Wuhan in 2019 (headache, dyspnea, fever, cough, and sore throat) can lead to confusion and difficulty in diagnosis [6–9]. The development of a detection platform that can accurately identify respiratory pathogens and can be used at home or in the field will enable appropriate countermeasures (i.e., isolation and treatment).
In general, the successful detection of pathogens in the field requires that three criteria be satisfied [2, 10]: First, a rapid and simple sample preparation process is required. Second, the nucleic acid must be amplified using simple equipment, with sensitivity and specificity that are similar to those of polymerase chain reaction (PCR). Finally, the results must be able to be immediately read with the naked eye, without the need for electrophoresis or fluorescent dyes.
In general, gold-standard methods, i.e., spin columns, cetrimonium bromide (CTAB), and magnetic beads, are used to extract DNA of high purity and at high yield [11–13]. However, these methods require both specialized equipment (a centrifuge and a magnet) and considerable time, and therefore are not suitable for point-of-care testing (POCT). In this study, we used the squeeze method for sample preparation to satisfy the criterion of a simple preparation method. Compared to the gold-standard methods, the nucleic acid obtained by the squeeze method is of relatively low purity. However, in our previous research, the concentration and elution effects of the squeeze method using a silica membrane were confirmed. No special equipment is required, and it can obtain a sample of sufficient quantity and quality for nucleic acid amplification by simply squeezing the tube [14].
Recently, reverse transcription loop-mediated isothermal amplification (RT-LAMP) has been effectively used for the rapid and accurate amplification of RNA viruses [15–18]. In RT-LAMP, RNA is amplified by adding reverse transcriptase without an additional step. This technique requires less time than PCR, but has similar specificity and sensitivity [16, 19, 20]. In addition, it can efficiently amplify nucleic acids at a constant temperature, such as in a water bath or a heat block [21–24]. In this study, a temperature suitable for amplification was achieved and maintained using a hot pack, which generates heat when iron and oxygen combine, resulting in an exothermic reaction to generate iron oxide [25–27]. It can be used as an alternative to the equipment used for PCR (a thermocycler), has excellent portability and can be applied inexpensively in places and medical environments with limited resources, and it does not require electricity.
Finally, the results were read using lateral flow assay (LFA), where our previous study was successfully performed [28]. LFA is a powerful tool for POCT requiring qualitative and quantitative analysis [29, 30]. This LFA platform is an avidin-biotin complex system and is suitable for POCT requiring qualitative and quantitative analysis [29–33].
In this study, a POCT analysis system was established by supplementing the sample preparation step, which was a limitation of previous LFA research. By detecting and distinguishing influenza A and B viruses, we demonstrated that this platform is applicable to various targets. Moreover, we confirmed its specificity for distinguishing influenza from COVID-19, which has similar symptoms. As a result, the assay is designed so that all testing processes can be performed at home or in the field, and thus it has excellent field compatibility and user-friendly advantages.
Experimental section
Materials
The 10 mM dNTP solution, Bst 2.0 WarmStart DNA polymerase and WarmStart RTx Reverse Transcriptase, and 10× isothermal amplification buffer (containing 2 mM MgSO4 and 0.1% Tween 20) used for RT-LAMP amplification were obtained from New England Biolabs (Ipswich, MA, USA). Phosphate-buffered saline (10 mM pH 7.4), 5 M betaine, streptavidin, Tween 20, Triton X-100, polyvinylpyrrolidone, sucrose, and agarose were purchased from Sigma-Aldrich (St. Louis, MO, USA). Biotin-11-dUTP was obtained from Jena Bioscience (Jena, Germany). AccuPower GreenStar RT-qPCR Master Mix, 100 bp DNA ladder, 1 kb DNA ladder, 6× agarose gel loading buffer, and primers were obtained from Bioneer (Seoul, Korea). Guanidine hydrochloride (7 M) and Tris-HCl (1 M, pH 8.0) were purchased from Biosesang (Seongnam, Korea). Staining STAR (20,000×) and Tris acetate-EDTA buffer (50×) were obtained from Dyne Bio (Seongnam, Korea). Borate buffer (0.1 M, pH 8.5) was supplied by Biosolution (Seoul, Korea). The QIAamp Viral RNA Mini kit and QIAamp Miniprep kit were purchased from QIAGEN (Hilden, Germany). Diethylpyrocarbonate-treated water and biotinylated bovine serum albumin (BSA) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). The hot packs were produced by GL Ltd. (Gyeonggi-do, Korea), and the silica membrane was obtained from Biocomma. Gold colloids (40 nm) were obtained from BBI Solutions (Cardiff, UK), and sample pads (grade 222), absorbent pads (grade 222), and glass fiber (grade 8964) were obtained from Boreda Biotech (Gyeonggi-do, Korea). Vivid 90 nitrocellulose was purchased from Pall Corporation (Port Washington, NY, USA), and Millipore laminated cards (6 cm × 30 cm) were obtained from Merck Millipore (Darmstadt, Germany). Influenza A and B viruses were obtained from the Korea Bank for Pathogenic Viruses. SARS-CoV-2 (NR-55245), human respiratory syncytial virus (NR-28528) and Enterovirus D68 (NR-49130) were purchased from BEI Resources. Coronavirus NL63 (NR-470) and coronavirus OC43 (NR-52705) genomic RNA were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The influenza A and B viruses received from the Korea Bank for Pathogenic Viruses were compared with a standard to determine the genomic RNA copy number. Total RNA was isolated from the influenza viruses using the QIAamp Viral RNA Mini kit.
Design of the Flu-LAMP-LFA method and mechanism
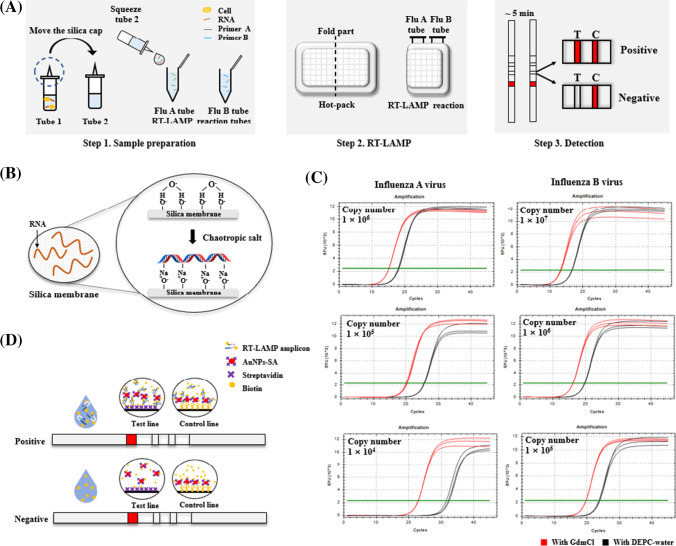
A schematic diagram of the Flu-LAMP-LFA detection platform is shown in Fig. 1a. The platform developed here consists of three steps (sample preparation, RT-LAMP, and LFA detection). First, a sample was prepared using two tubes and a screw cap with a silica membrane.
Fig. 1.
Principle and procedure of the Flu-LAMP-LFA for the detection of influenza virus. (a) The Flu-LAMP-LFA method consists of three steps: Step 1. After lysis and elution using the squeeze method, place one drop each into the Flu A and B tubes. Step 2. Amplify the nucleic acids by placing the tubes on a hot pack that has been preheated for 10 min. Step 3. Visualize the results using LFA. (b) Principle of nucleic acid binding to the silica cap: The silica membrane binds specifically to extracted nucleic acids via a chaotropic agent. (c) Comparison of nucleic acid binding capacity with silica membrane of GdmCl and DEPC-water: GdmCl was measured as low as Ct 3 at all concentrations tested. (d) Principle of LFA detection: The RT-LAMP amplicon containing biotin-dUTP is captured by immobilized avidin and visualized (test line). Immobilized avidin cannot be captured, and thus no signal is generated in the test line. Proper operation of LFA can be confirmed through visualization of immobilized biotin-BSA (control line)
Tube 1 contained 500 μL of lysis buffer (800 mM guanidine hydrochloride (GdmCl), 50 mM Tris [pH 8.0], 0.5% [v/v] Triton X-100, and 1% [v/v] Tween 20), and tube 2 contained elution buffer (10 mM Tris [pH 8.0] and 0.1% [v/v] Tween 20). The flu A and B tubes contained 15 μL of premix (10× isothermal amplification buffer [20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, and 0.1% Tween 20]), 0.8 M betaine, 1.2 mM dNTPs (0.3 mM of each dNTP), reverse transcriptase, Bst 2.0 WarmStart DNA polymerase and each primer for RT-LAMP. A primer mixture was prepared with 0.2 μM F3 and B3 primers, 1.6 μM FIP and BIP primers, and 0.4 μM LF and LB primers.
Based on the universal swab capacity, a 50 μL sample was placed into tube 1 and incubated for 3 min to lyse the sample. During lysis, a chaotropic agent breaks the hydrogen bonds of DNA and proteins and facilitates the adsorption of the nucleic acids to silica in the cap (Fig. 1b) [1, 34, 35]. The screw cap was then moved to tube 2, and the silica membrane was placed in contact with the elution buffer (Fig. 1a). Sample preparation was complete after incubation for 3 min. One drop (approximately 10 μL) was added to each RT-LAMP reaction tube to simultaneously detect influenza A and B viruses. Then, the RT-LAMP reaction tubes were wrapped in a hot pack and heated for 10 min to amplify the nucleic acid. When the RT-LAMP reaction was complete, the results were visualized using a detection strip.
Chaotropic agents were evaluated for specific binding of nucleic acids to silica membranes. The experiment was performed using one of the chaotropic agents, GdmCl and DEPC-water. A total sample volume of 10 μL was used. Influenza A virus was used from 1 × 104 to 1 × 106 copies and influenza B virus was used from 1 × 105 to 1 × 107 copies. Based on the amplification range identified in the manuscript, we carried out tests using concentrations of 1, 10, and 100 times. We checked whether the same tendency was exhibited in the amplifiable low to high range. In order to compare only the correlation between the silica membrane and the buffer, a previously verified method, the QIAGEN spin column (QIAprep Miniprep kit), was simultaneously compared. To reproduce the same conditions as the developed platform, the silica membrane of the spin column was replaced with the silica membrane used in this study. The manufacturer's protocol was followed, and 800 μL each of GdmCl and DEPC-water were added instead of the buffers provided by QIAGEN. Ct values of the obtained eluents were compared through quantitative reverse transcription PCR (RT-qPCR). AccuPower GreenStar RT-qPCR Master Mix was used and the manufacturer's instructions were followed. The reaction was performed under the following cycling conditions: 55 °C for 15 min, 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s in a Bio-Rad CFX96 instrument (Hercules, CA, USA).
As a result, in all tested samples, the GdmCl-treated sample had a lower Ct value by 3 relative to the DEPC-water-treated sample in three out of three trials (Fig. 1c). The results are consistent with what has been reported in the literature. According to the data, it can be seen that the lower the concentration, the greater the difference in Ct values. In particular, in the influenza A virus of 1 × 104 copies, the difference in Ct values for the two buffers widens significantly. It is highly dependent on GdmCl as the concentration of the sample is lowered. Zero false positives were observed out of three trials.
The reaction on the strip was based on the avidin-biotin complex. After completion of the RT-LAMP reaction using biotin-attached dUTP (biotin-dUTP), the biotin-labeled RT-LAMP amplicon was loaded, along with 100 μL of distilled water, on the sample pad (Fig. 1d). Then, it was captured by streptavidin-conjugated gold (AuNP-SA) as it flowed along in the flow direction, captured by immobilized avidin, and the test line was visualized. A negative sample was not captured by the immobilized avidin because no biotin-dUTP was present in the reaction product. Proper operation of the LFA was confirmed by the binding of AuNP-SA to immobilized biotin-BSA (control line).
Primer design for influenza virus detection
The RT-LAMP primers were designed based on the conserved regions of the matrix gene (A/Puerto Rico/8/1934) (H1N1) for influenza A virus and NS1 (B/Lee/1940) for influenza B virus. The genomic sequences used to design the influenza A and B primers were obtained from GenBank, and the primers were designed using PrimerExplorer V5 software (http://primerexplorer.jp/lampv5e/l). The lack of primer dimer formation was confirmed, thus minimizing false positives. NCBI Primer-BLAST was used to improve target specificity. The primer sequences are shown in Table 1.
Table 1.
The sequences of the primers used to amplify influenza A and B viruses
| Target | Primer name | Sequence 5′–3′ |
|---|---|---|
| Influenza A (M1) | A-F3 | GCCAGCACTACAGCTAAGG |
| A-B3 | CACTTGAACCGTTGCATCTG | |
| A-FIP | CTTGCACCATTTGCCTAGCCTG-TGGATCGAGTGAGCAAGCA | |
| A-BIP | TGGGACTCATCCTAGCTCCAGT-CACCCCCATTCGTTTCTGA | |
| A-LF | GTCCGATCCGTTTACCACGTTC | |
| A-LB | TGGGACTCATCCTAGCTCCAGT | |
| Influenza B (NS1) | B-F3 | GAGCAACCAATGCCACTA |
| B-B3 | CCCAATTGCTTTTCTCTCTTC | |
| B-FIP | TAGTCAAGGGCTCTTTGCCA-CTTTGAAGCAGGAATTCTGGA | |
| B-BIP | CAAGACCGCCTACACAGACT-TCTCAGGCTCACTCTTGT | |
| B-LF | ACCGTTTCTCGGGAACTGAT | |
| B-LB | CAAGACCGCCTACACAGACT |
Sample preparation for POCT
Samples prepared by the squeeze method are not of high purity, because this method cannot completely remove debris or the lysis buffer components. Therefore, the amplification efficiency of RT-LAMP is comparatively decreased. To minimize the inhibitory effects on RT-LAMP and maintain nucleic acid amplification efficiency, the volume of the elution buffer (100, 200, and 400 μL) was adjusted.
To validate RNA recovery and evaluate the effect of RT-LAMP inhibitors on the Flu-LAMP-LFA platform, 50 μL RNA (4500 copies/μL) was treated to the initial sample preparation step. Then, qPCR was performed with a slight modification of the manufacturer's instructions for the AccuPower GreenStar RT-qPCR Master Mix and the amplification efficiencies were compared. Although the existing protocol requires only 5 μL of template, 10 μL of template was used to maximize the RT-LAMP inhibitor effect in a final volume of 20 μL. The F3 and B3 qPCR primers were used to amplify the influenza A and B viruses, respectively. The reaction was performed in a Bio-Rad CFX96 instrument (Hercules, CA, USA) under the following cycling conditions: 55 °C for 15 min, 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Fluorescence data were recorded every minute during amplification. All experiments were repeated three times.
Determination of the optimal RT-LAMP conditions and analysis of specificity
To optimize RT-LAMP, we evaluated the optimal reaction temperature and reaction time. The amplification efficiencies of the RT-LAMP reaction were compared at four temperatures (55 °C, 58 °C, 61 °C, and 65 °C). In addition, the lowest concentration range that can be amplified with the developed platform was analyzed. After establishing the optimal temperature conditions for both RNAs, a test was performed to determine the minimum reaction time (15, 20, and 35 min). Finally, to determine the lowest concentration range that can be amplified, the influenza A and B RNA was serially diluted (1 × 103 to 1 × 108 copies) in distilled water under the same conditions. All RT-LAMP reactions were terminated by enzyme inactivation at 80 °C. The results were analyzed using electrophoresis and LFA. All RT-LAMP assays were performed using a Bio-Rad T100 thermocycler (Hercules, CA, USA), and all experiments were repeated three times.
The specificity of each primer was confirmed using viruses that cause symptoms similar to those of influenza, namely SARS-CoV-2, coronavirus NL63, coronavirus OC43, Human respiratory syncytial virus and Enterovirus D68.
Manufacture and optimization of hot packs
The hot pack consists of iron powder, activated carbon, sodium chloride, vermiculite, and water. Iron, the main exothermic material, releases heat through oxidation. Activated carbon is a heat-generating and heat-promoting material, and enables high thermal stability. [26, 36, 37]. Sodium chloride and water accelerate the oxidation reaction of iron powder [38]. Vermiculite acts as heat insulator to reduce heat loss and maintain temperature [26].
In order to use the hot pack as a heat source, two pretests were carried out. Mixtures were prepared with various mass quantities of iron powder. Three different iron powder mass percentages (37.3%, 40.3%, and 50.6%) were tested for temperature screening. The second pretest was performed by modifying the composition close to the RT-LAMP reaction temperature in the first pretest. The optimized mass percentage of iron powder was fixed and the proportion of activated carbon (6.8%, 9.8%, and 12.1%) was modified. The mass percentage of the mixture used in the first and second pretests is shown in Figure S1. All experiments were repeated three times and were recorded continuously for 60 min. The temperature of the hot packs selected from one secondary pretest and the hot packs obtained from a total of three lots (nine each, a total of 27) was compared to verify the reproducibility and reliability of the hot pack.
Reproducibility in the field setting regardless of the influence of the surrounding environment is an important parameter. For the reproducibility evaluation, tests were performed assuming 10 °C, 25 °C and 37 °C environments. In order to ensure sufficient exposure to the environment, the test was performed after incubation at each temperature for 30 min. A forced-air oven (DH.WOF0715, DAIHAN Scientific) was used. All experiments were repeated three times and monitored continuously for 60 min. The average temperature was measured from 10 min after opening the hot pack.
Comparison of a hot pack and a thermocycler for RT-LAMP
After confirming the heating capacity of the hot pack, RT-LAMP was performed simultaneously using a thermocycler and a hot pack. Based on the lowest concentration successfully amplified, influenza A RNA was amplified in the range of 1 × 103 to 1 × 105 copies, and influenza B RNA was amplified in the range of 1 × 105 to 1 × 106 copies. To obtain sufficient product, the thermocycler was held at 58 °C for 35 min. Based on the minimum detection time determined in this study, the results were read at 15, 20, and 35 min for influenza A and at 20, 25, and 35 min for influenza B.
Application of Flu-LAMP-LFA for influenza virus detection
To further verify the accuracy and evaluate the overall performance of the optimized Flu-LAMP-LFA developed in this study, an evaluation was conducted using influenza A and B viruses. RT-LAMP was performed using an influenza virus sample prepared for Flu-LAMP-LFA. In this attempt, the hot pack was used as the heat source for RT-LAMP. For the evaluation, five positive samples and five negative samples were prepared. Influenza A and B viruses were prepared at 1 × 104 to 1 × 108 copies. The assay was performed according to the conditions established in this study, and the results were evaluated using LFA.
Sample preparation that omits the washing step may affect the PCR reaction. The developed sample preparation platform and the spin column method, which are gold-standard methods, were compared with nucleic acid binding and extraction efficiency. A QIAprep Miniprep kit (QIAGEN) was used and was performed according to the manufacturer's instructions. However, in order to reproduce the same conditions, the silica membrane of the spin column was replaced with the silica membrane used in this study. Experiments were performed with an optimized sample preparation method. Samples of 50 ng/μL each of influenza A and B viruses were prepared. They were used in the form of a plasmid so as not to be affected by lysis efficiency. Molecular biologists consider a method to directly evaluate nucleic acids with nanodrops as a very useful parameter [39–41]. The concentration of the obtained eluent was measured via a NanoDrop OneC spectrophotometer (Thermo Fisher Scientific). All experiments were repeated three times.
Results and discussion
Confirmation to improve POCT conformity in sample preparation
The RNA recovery rate and RT-LAMP inhibitor effect in samples prepared using the squeeze method eluted in different volumes of elution buffer (Fig. 2) were assessed by qPCR. The test results showed similar trends for the influenza A and B RNA samples. At 200 μL of elution buffer, the highest RNA recovery rate was shown. In this condition, the recovery rate was 71.1% (approximately 4000 copies). Furthermore, RNA was also diluted by the elution buffer, but sufficient RNA remained for PCR. As a result, the amplification efficiency was evaluated as the highest compared to other conditions. Although the 100 μL of elution buffer contained the most RNA, the inhibitor was not successfully removed. Therefore, a recovery rate of 44.4% (2500 copies) was observed. In contrast, 400 μL of elution buffer had the lowest amount of RNA due to the diluent effect. However, the inhibitor was sufficiently removed, and the recovery rate was 37.3% (2100 copies), which was similar to 100 μL.
Fig. 2.

RNA recovery rate according to elution buffer volume. A qPCR assay using samples with different elution buffer volumes (100, 200, and 400 μL) showed the same trend for both viruses. Approximately 2500 copies were recovered in 100 and 400 μL of elution buffer, whereas about 4000 copies were recovered in 200 μL of elution buffer, which was the best recovery rate
Our results show that by adjusting the elution buffer volume to 200 μL, samples for amplification could be obtained without additional processes such as incubation. Therefore, a simple lysis method with controlled elution volume is an optimal alternative for POCT sample preparation.
Optimization of RT-LAMP for the Flu-LAMP-LFA platform
Determination of the optimal temperature and time using a thermocycler
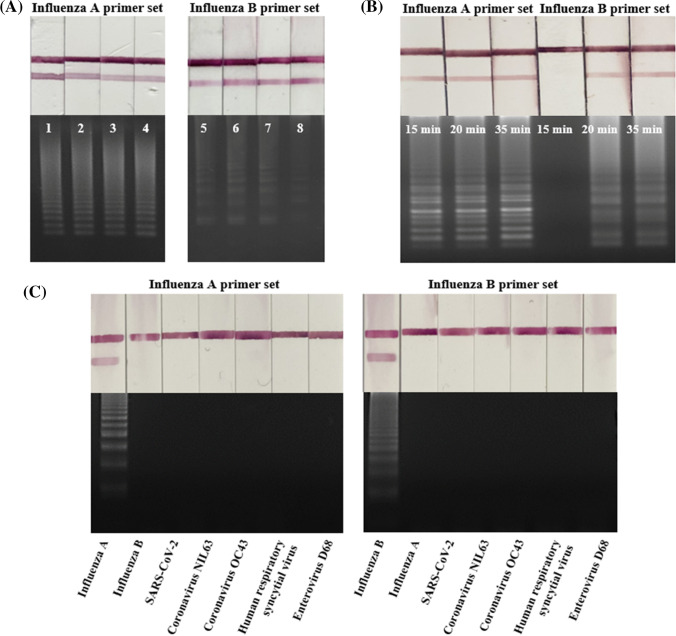
The efficiency of influenza A and B RNA (1000 copies/μL) amplification at temperatures from 55 °C to 65 °C was compared (Fig. 3a). Influenza A RNA showed excellent amplification efficiency at all tested temperatures. In contrast, influenza B RNA showed the highest amplification efficiency at 58 °C and 61 °C. Zero false positives were observed out of three trials. Since both viruses need to be detected at the same time, the temperature was selected based on influenza B, which is influenced by temperature. Therefore, an optimization experiment was performed by setting the reaction temperature to 58 °C.
Fig. 3.
Optimization of the RT-LAMP reaction temperature and time and specificity testing. (a) Influenza A was amplified efficiently at all tested temperatures (55 °C–65 °C). In contrast, although influenza B was amplified at all temperatures, the amplification was most efficient at 58 °C and 61 °C. Lanes 1–4, influenza A at 55 °C, 58 °C, 61 °C, and 65 °C; lanes 5–8, influenza B at 55 °C, 58 °C, 61 °C, and 65 °C. (b) RT-LAMP amplification for 15, 20, and 35 min. Influenza A RNA was amplified starting at 15 min, whereas influenza B RNA was amplified starting at 20 min. (c) Influenza A and B primers amplify only the specific target. Lanes 1–7: Influenza A primer set; Lane 1: Influenza A, Lane 2: Influenza B, Lane 3: SARS-CoV-2, Lane 4: Coronavirus NL63, Lane 5: Coronavirus OC43, Lane 6: Human respiratory syncytial virus, Lane 7: Enterovirus D68; Lanes 8–14: Influenza B primer set, Lane 8: Influenza B, Lane 9: Influenza A, Lane 10: SARS-CoV-2, Lane 11: Coronavirus NL63, Lane 12: Coronavirus OC43, Lane 13: Human respiratory syncytial virus, Lane 14: Enterovirus D68.
At the optimal temperature of 58 °C, RT-LAMP was performed for 15, 20, and 35 min to determine the minimum amplification time. Amplified influenza A RNA was detected at 15 min, and amplified influenza B RNA was detected at 20 min (Fig. 3b). Zero false positives were observed out of three trials. Based on these results, 25 min was used to achieve simultaneous amplification and sufficient detection. In conclusion, 25 min was a suitable duration for the RT-LAMP reaction using a thermocycler set at 58 °C.
The specificity of each primer was tested using six controls; the influenza A primer only amplified influenza A (Fig. 3c), and the influenza B primer only amplified influenza B. Neither primers amplified SARS-CoV-2, coronavirus NL63, coronavirus OC43, Human respiratory syncytial virus and Enterovirus D68. Zero false positives were observed out of three trials. Therefore, the developed Flu-LAMP-LFA likely has sufficient specificity to distinguish influenza virus from SARS-CoV-2, which causes symptoms similar to those of influenza.
The lowest concentration range that amplified for influenza A and B RNA using a thermocycler
The lowest concentration range that amplified was performed under optimized RT-LAMP conditions (i.e., at 58 °C for 25 min) using a thermocycler. The results showed that influenza A RNA was detected at 1 × 104 copies, three out of three trials (Fig. 4a) and that influenza B RNA was detected at 1 × 105 copies, three out of three trials (Fig. 4b). Zero false positives were observed out of three trials. Thus, influenza A RNA detection is 10 times more sensitive than influenza B RNA detection. At the lowest concentration range influenza A RNA (1 × 104 copies) and influenza B RNA (1 × 105 copies) were both clearly detectable by LFA.
Fig. 4.
The lowest concentration that amplified for Influenza A and B virus RNA using a thermocycler. RT-LAMP results with a thermocycler. (a) Influenza A RNA was detected starting at 1 × 104 copies, and (b) influenza B RNA was detected starting at 1 × 105 copies. Even at a low RNA copy number, LFA enables clear reading of the results
Optimization and performance test of hot packs
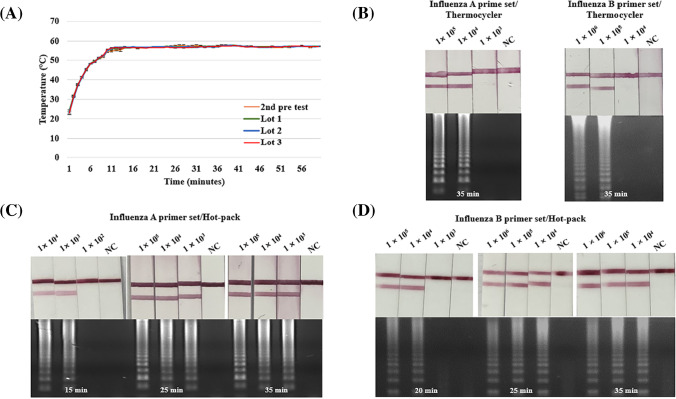
In the first pretest, the temperature according to the mass of iron powder was plotted simultaneously with the average deviation (Figure S-2): 70.8 °C for 50.6 wt% iron powder, 64.2 °C for 40.3 wt% iron powder, and 56.8 °C for 37.3 wt% iron powder. All temperature data are shown in Figure S-3 for all temperatures. It was confirmed that the temperature increased as the mass of iron powder increased. Therefore, it was selected based on C, which is the closest condition to the RT-LAMP temperature.
As a result of controlling the proportion of activated carbon, the average temperature was measured to be 56.7 °C for 12.1 wt% activated carbon, 53.2 °C for 9.8 wt% activated carbon, and 48.6 °C for 6.8 wt% activated carbon (Figure S-4). All temperature data are shown in Figure S-5. It showed the same tendency as iron powder. Hot pack D, which satisfies the RT-LAMP reaction temperature of 58–61 °C, was established as the final condition.
The hot pack used in the second pretest and the hot pack for each lot were compared. For a total of 28 hot packs, the analogous temperature was measured regardless of the lot (Fig. 5a). All temperature data are shown in Figure S-6. As shown in Fig. 3a, influenza virus was efficiently amplified at temperatures ranging from 55 °C to 65 °C. In other words, since the RT-LAMP is robust and does not require strict temperature control, the hot pack shows the possibility of being used as a heat source for amplification.
Fig. 5.
Performance evaluation of the assay using a thermocycler and a hot pack. (a) A graph showing the temperature of the hot pack over 1 h. It reached 50 °C within 5 min and reached the average temperature of 57 °C after 10 min. After reaching 60 °C, a constant temperature was maintained without any significant change. (b) Using a thermocycler, the assay can detect as few as 1 × 104 copies of influenza A and 1 × 105 copies of influenza B. (c) Using a hot pack, the assay can detect as few as 1 × 104 copies of influenza A. (d) Using a hot pack, the assay can detect as few as 1 × 104 copies of influenza B
It was tested whether the temperature was affected by the environment (Figure S-7). At 37 °C, an average temperature of 61 °C was recorded, and the maximum temperature was reached within 2–3 min. The average temperature was 61 °C in a 37 °C environment, 56.8 °C in a 25 °C environment, and 54.4 °C in a 10 °C environment. The optimum temperature for RT-LAMP was in the range 58–61 °C. Hot packs can be used in a 37 °C environment. All temperature data are shown in Figure S-8. The average temperature was reached within 2–3 min in a 37 °C environment, 10 min in a 25 °C environment, and 20 min in a 10 °C environment. The average temperature in the 37 °C environment was 61 °C, which increased compared to the 25 °C environment. In a 10 °C environment, the average temperature was lower than the RT-LAMP reaction temperature before reaching the maximum temperature. However, after reaching the average temperature, the average temperature was measured to be 56.2 °C. Therefore, it takes more time to heat up, but after reaching the target temperature, it heats up to a temperature similar to a 25 °C environment.
Comparison of the amplification efficiency of the hot pack and thermocycler
The hot pack used for RT-LAMP maintained an average temperature of 57 °C for over 1 h (Fig. 5a). The hot pack heated up rapidly, reached 50 °C within 5 min, and reached the average temperature within 10 min. Once it reached the average temperature, it remained constant, without any significant temperature changes.
As shown in Fig. 3a, the influenza virus was efficiently amplified at temperatures ranging from 55 °C to 65 °C. In other words, since the RT-LAMP is robust and does not require strict temperature control, the hot pack shows the possibility of being used as a heat source for amplification.
When RT-LAMP was performed with a thermocycler or a hot pack under the same conditions, the thermocycler showed the same range (1 × 104 copies for influenza A RNA, three out of three trials and 1 × 105 copies for influenza B RNA, three out of three trials) was amplified in the thermocycler (Fig. 5b). RT-LAMP performed using a hot pack showed the same results as the thermocycler (Fig. 5b–d). In particular, the minimum detection time and amplified range were consistent. Amplification of influenza A RNA was first observed at 15 min (Fig. 5c), and amplification of influenza B RNA was first observed at 20 min (Fig. 5d). In Fig. 5b–d, zero false positives were observed in each of the three trials. Therefore, a hot pack is a temperature control device that can replace a complex device, such as a thermocycler, and function as an appropriate heat source for RT-LAMP.
Performance evaluation of the Flu-LAMP-LFA
In this experiment, the overall performance of the Flu-LAMP-LFA was evaluated using influenza A and B viruses (Fig. 6a). After preheating the hot pack for 10 min, based on the temperature data shown in Fig. 5a, the RT-LAMP reaction was performed. Performing all processes, from sample preparation to detection, using the developed Flu-LAMP-LFA, influenza A virus was detected at 1 × 105 to 1 × 108 copies, three out of three trials (Fig. 6b), which was 10 times higher than the calculated concentration range that amplified. However, at 1 × 105 copies there was a slight decrease in the LFA signal. Zero false positives were observed out of three trials. All the negative samples were confirmed to be negative. Influenza B virus was detected at 1 × 106 copies to 1 × 108 copies, three out of three trials (Fig. 6c). Like influenza A, the LFA signal for influenza B virus was also decreased at 1 × 106 copies. Zero false positives were observed out of three trials. All the negative samples were confirmed to be negative. These results show that the overall performance of the Flu-LAMP-LFA was somewhat inferior to the assay using a thermocycler and RNA. The factors affecting assay performance are thought to be related to the efficiency of viral lysis in the squeeze method sample preparation. It can be confirmed through Fig. 6d. Influenza A virus was measured at an average of 36.1 ng/μL on the developed platform. The standard deviation is 0.11 and the yield is 72.1%. On the other hand, QIAGEN measures an average of 45.4 ng/μL with a standard deviation of 0.27 and a yield of 90.8%. Influenza B virus was measured at an average of 36.0 ng/μL on the developed platform. The standard deviation is 0.13 and the yield is 72.0%. QIAGEN measures an average of 45.8 ng/μL. The standard deviation is 0.16 and the yield is 91.7%. Both viruses showed values similar to the recovery rate (90%) guaranteed by QIAGEN, confirming the significance of the experiment. As the data show, the yield is about 0.8 times lower than that of QIAGEN. However, there was no significant difference in the concentration range that was amplified even though no spin column or thermocycler were used. As a result, it can be confirmed that the Flu-LAMP-LFA was successfully designed in terms of sensitivity, accuracy, and POCT.
Fig. 6.
Flu-LAMP-LFA application and performance evaluation using actual influenza virus. (a) Pictures of Flu-LAMP-LFA supplies and method. (b) Diagnosis results from five positive and five negative samples for influenza A virus. The LFA detected as few as 1 × 105 copies, and no signal was observed for the negative samples. (c) Diagnosis results from five positive and five negative samples for influenza B virus. The LFA detected as few as 1 × 106 copies. (d) Nucleic acid concentration and yield for two sample preparation methods (Flu-LAMP-LFA and spin column) evaluated using nanodrop.
Conclusion
In this study, we designed an influenza virus detection system that applied a sample preparation step, which was a limitation of our previous research related to LFA, and the nucleic acid amplification method for POCT. The results show that the developed LFA platform can be applied to various targets. The use of the Flu-LAMP-LFA procedure for the detection of nucleic acids with viruses and RNA obtained similar results for these two samples. In particular, our platform, which does not require specialized equipment and is performed with user-friendly methods, showed significant advantages over previously reported POCT platforms.
In addition, different temperatures were achieved using a hot pack by changing the ratio of the internal components [10, 35]. Therefore, the method can be extended to multiple pathogen detection platforms by simply changing the amplification temperature and primers. However, one limitation is that it can be easily applied only in a low-throughput POCT setting. In addition, it involves complex processes and requires training and skill for operation. Nevertheless, the purpose of this study was to develop a method for the rapid and accurate identification of the influenza virus in the field and at home. Therefore, this is a very valuable study because the developed method can be used to distinguish diseases with similar symptoms and should enable increased access to medical care through self-diagnosis. We believe that it can be used to quickly respond to new viruses and pathogens, thereby improving human health and global security against future infectious diseases.
Supplementary information
(DOCX 103 kb)
Acknowledgements
This work was supported by start-up commercialization funds from MMS (10448801, ASOC) and the Research Investment for Global Health Technology Fund (RIGHT Fund, RF-TAA-2020-D02).
Author contributions
Minju Jang: Conceptualization, methodology, validation, form analysis, investigation, data curation, visualization, writing—original draft
SeJin Kim: Conceptualization, methodology, form analysis, investigation, data curation
Junkyu Song: Methodology, form analysis, investigation, data curation
Sanghyo Kim: Supervision, funding acquisition
Declarations
Conflicts of interest
There are no conflicts to declare.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl J Med. 2020;382(21):2063. doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C, Zheng T, Wang H, Chen W, Huang X, Liang J, et al. Rapid one-pot detection of SARS-CoV-2 based on a lateral flow assay in clinical samples. Anal Chem. 2021;93(7):3325–3330. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- 3.Furuse Y, Suzuki A, Kamigaki T, Oshitani H. Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol J. 2009;6(1):1–13. doi: 10.1186/1743-422X-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee M-S, Chang P-C, Shien J-H, Cheng M-C, Shieh HK. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods. 2001;97(1-2):13–22. doi: 10.1016/S0166-0934(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 5.Claas EC, Osterhaus AD, Van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351(9101):472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 6.Karimian P, Delavar MA. Comparative Study of Clinical Symptoms, Laboratory Results and Imaging Features of Coronavirus and Influenza Virus, Including Similarities and Differences of Their Pathogenesis. Pak J Med Sci. 2020;14(3):1405–1411. [Google Scholar]
- 7.Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4):2000058. doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang N, Wang L, Deng X, Liang R, Su M, He C, et al. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92(4):408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caini S, Kroneman M, Wiegers T, El Guerche-Séblain C, Paget J. Clinical characteristics and severity of influenza infections by virus type, subtype, and lineage: a systematic literature review. Influenza Other Respir Viruses. 2018;12(6):780–792. doi: 10.1111/irv.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li RJ, Mauk MG, Seok Y, Bau HH. Electricity-free chemical heater for isothermal nucleic acid amplification with applications in COVID-19 home testing. Analyst. 2021;146(13):4212–4218. doi: 10.1039/D1AN00309G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F, Wang G, Xu W, Hong N. A rapid silica spin column-based method of RNA extraction from fruit trees for RT-PCR detection of viruses. J Virol Methods. 2017;247:61–67. doi: 10.1016/j.jviromet.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Schiebelhut LM, Abboud SS, Gómez Daglio LE, Swift HF, Dawson MN. A comparison of DNA extraction methods for high-throughput DNA analyses. Mol Ecol Resour. 2017;17(4):721–729. doi: 10.1111/1755-0998.12620. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Q-B, Huang Y-M, Wu W-B, Zuo P, Hu N, Zhou Y-Z, et al. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ Int. 2019;131:104986. doi: 10.1016/j.envint.2019.104986. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Song J, Kim S. A simple and innovative sample preparation method for on-site SARS-CoV-2 molecular diagnostics. Analyst. 2021;146(22):6917–6923. doi: 10.1039/D1AN01401C. [DOI] [PubMed] [Google Scholar]
- 15.Sharma V, Chaudhry D, Kaushik S. Evaluation of clinical applicability of reverse transcription-loop-mediated isothermal amplification assay for detection and subtyping of Influenza A viruses. J Virol Methods. 2018;253:18–25. doi: 10.1016/j.jviromet.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nawattanapaiboon K, Pasomsub E, Prombun P, Wongbunmak A, Jenjitwanich A, Mahasupachai P, et al. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst. 2021;146(2):471–477. doi: 10.1039/D0AN01775B. [DOI] [PubMed] [Google Scholar]
- 17.Lamb LE, Bartolone SN, Tree MO, Conway MJ, Rossignol J, Smith CP, et al. Rapid detection of Zika virus in urine samples and infected mosquitos by reverse transcription-loop-mediated isothermal amplification. Sci Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-22102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirato K, Yano T, Senba S, Akachi S, Kobayashi T, Nishinaka T, et al. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol J 2014;11(1):1-11. [DOI] [PMC free article] [PubMed]
- 19.Mautner L, Baillie C-K, Herold HM, Volkwein W, Guertler P, Eberle U, et al. Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol J. 2020;17(1):1–14. doi: 10.1186/s12985-020-01435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn SJ, Baek YH, Lloren KKS, Choi W-S, Jeong JH, Antigua KJC, et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect Dis. 2019;19(1):1–12. doi: 10.1186/s12879-019-4277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63-e. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16(3):223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 23.Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015;53(1):1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 24.Njiru ZK. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl Trop Dis. 2012;6(6):e1572. doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buser JR, Zhang X, Byrnes S, Ladd P, Heiniger E, Wheeler M, et al. A disposable chemical heater and dry enzyme preparation for lysis and extraction of DNA and RNA from microorganisms. Anal Methods. 2016;8(14):2880–2886. doi: 10.1039/C6AY00107F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klarzak I, Ura-Bińczyk E, Płocińska M, Jurczyk-Kowalska M. Effect of temperature and humidity on heat effect of commercial chemical warmers based on iron powder. Therm Sci Eng Prog. 2018;6:87–94. doi: 10.1016/j.tsep.2018.03.006. [DOI] [Google Scholar]
- 27.Shah KG, Guelig D, Diesburg S, Buser J, Burton R, LaBarre P, et al. Design of a new type of compact chemical heater for isothermal nucleic acid amplification. PLoS One. 2015;10(10):e0139449. doi: 10.1371/journal.pone.0139449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang M, Kim S, Song J, Kim S. Highly sensitive and rapid detection of porcine circovirus 2 by avidin–biotin complex based lateral flow assay coupled to isothermal amplification. Anal Methods. 2021;13(38):4429–4436. doi: 10.1039/D1AY01189H. [DOI] [PubMed] [Google Scholar]
- 29.Sajid M, Kawde A-N, Daud M. Designs, formats and applications of lateral flow assay: A literature review. J Saudi Chem Soc. 2015;19(6):689–705. doi: 10.1016/j.jscs.2014.09.001. [DOI] [Google Scholar]
- 30.Posthuma-Trumpie GA, Korf J, van Amerongen A. Lateral flow (immuno) assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem. 2009;393(2):569–582. doi: 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- 31.Wilchek M, Bayer EA. The avidin-biotin complex in bioanalytical applications. Anal Biochem. 1988;171(1):1–32. doi: 10.1016/0003-2697(88)90120-0. [DOI] [PubMed] [Google Scholar]
- 32.Choi DH, Lee SK, Oh YK, Bae BW, Lee SD, Kim S, et al. A dual gold nanoparticle conjugate-based lateral flow assay (LFA) method for the analysis of troponin I. Biosens Bioelectron. 2010;25(8):1999–2002. doi: 10.1016/j.bios.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Cho I-H, Bhunia A, Irudayaraj J. Rapid pathogen detection by lateral-flow immunochromatographic assay with gold nanoparticle-assisted enzyme signal amplification. Int J Food Microbiol. 2015;206:60–66. doi: 10.1016/j.ijfoodmicro.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 34.Bienvenue JM, Duncalf N, Marchiarullo D, Ferrance JP, Landers JP. Microchip-based cell lysis and DNA extraction from sperm cells for application to forensic analysis. J Forensic Sci. 2006;51(2):266–273. doi: 10.1111/j.1556-4029.2006.00054.x. [DOI] [PubMed] [Google Scholar]
- 35.Ghadia B, Singh AK, Khatnani T, Hirpara M, Patel S, Joshi P. An improved method of DNA purification from secondary metabolites rich medicinal plants using certain chaotropic agents. Acta Physiol Plant. 2016;38(8):1–11. doi: 10.1007/s11738-016-2223-6. [DOI] [Google Scholar]
- 36.Podella CW. Chemical heating pad with differing air-admitting perforation sets for different heat-generation levels. Google Patents. 1988.
- 37.Ueki A. Disposable body warmer. Google Patents. 1994.
- 38.Glasser EC. Flameless heater, heating assembly and heating kit. Google Patents. 1967.
- 39.Gallagher SR, Desjardins PR. Quantitation of DNA and RNA with absorption and fluorescence spectroscopy. Curr Protoc Mol Biol. 2006;76(1):A. 3D. 1–A. 3D. 21. doi: 10.1002/0471142727.mba03ds76. [DOI] [PubMed] [Google Scholar]
- 40.Celis JE. Cell biology, a laboratory handbook, Vol 1. Academic Press. 2006.
- 41.Sambrook J. Molecular cloning: a laboratory manual. Synthetic oligonucleotides. 1989.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 103 kb)