Significance
Hematopoietic stem cells (HSCs) are generated from specialized endothelial cells, called hemogenic endothelial cells (HECs). It has been debated whether HECs and non–HSC-forming conventional endothelial cells (cECs) arise from a common precursor or represent distinct lineages. Moreover, the molecular basis underlying their distinct fate determination is poorly understood. We use photoconvertible labeling, time-lapse imaging, and single-cell RNA-sequencing analysis to trace the lineage of HECs. We discovered that HECs and cECs arise from a common hemogenic angioblast precursor, and their distinct fate is determined by high or low dosage of Etv2, respectively. Our results illuminate the lineage origin and a mechanism on the fate determination of HECs, which may enhance the understanding on the ontogeny of HECs in vertebrates.
Keywords: endothelium, hemogenic endothelium, lineage origin, fate determination
Abstract
Hematopoietic stem cells (HSCs) are generated from hemogenic endothelial cells (HECs) in the floor of the dorsal aorta (DA) via endothelial-to-hematopoietic transition (EHT). Yet whether HECs and conventional endothelial cells (cECs) in the DA share a common precursor is controversial, and the molecular mechanisms governing their fate specification remain incompletely defined. Using a combination of fate mapping, time-lapse imaging, genetic manipulation, and single-cell RNA sequencing, here we show that HECs and cECs display strictly spatial separation in the DA where nearly all the endothelial cells in the floor and roof are HECs and cECs, respectively. We further show that HECs and cECs in the DA arise from a common hemogenic angioblast precursor, which differentiates into HECs and cECs during axial migration prior to the DA formation. The specification of HECs and cECs from hemogenic angioblasts is governed by the Etv2 dosage by differentially regulating Fli1a, Notch, and Sclβ. Finally, we document that pan-endothelial overexpression of transcriptional factor runx1 is sufficient to promote HEC fate in the DA roof. Our study reveals the lineage origin of HECs and cECs in the DA and uncovers the molecular network controlling their fate specification.
During vertebrate embryogenesis, hematopoietic stem cells (HSCs) are generated from a subset of specialized endothelial cells (ECs), named hemogenic endothelial cells (HECs), via endothelial-to-hematopoietic transition (EHT) in the dorsal aorta (DA) (1–4). Time-lapse imaging and in situ analysis of hematopoietic markers indicate that HECs are located exclusively in the ventral domain/floor of the DA in the aorta-gonad-mesonephros (AGM), budding toward intraaortic lumen in avians (5, 6) and mice (2, 7, 8) or budding toward subaortic space between the DA and posterior cardinal vein in zebrafish (3, 4). Although HSC-forming HECs and non–blood-forming conventional endothelial cells (cECs) are both located in the DA, whether HECs and cECs arise from a common bipotent precursor remains debatable. Several studies using human pluripotent stem cells or murine model have suggested that HECs and cECs represent distinct lineages indicated by the expression of distinct cell surface markers, such as CD73 and CD184 (9), or by their different requirements of Notch signaling strengths (10). Studies in the avian model especially suggested that different mesoderm origins, somatopleural and splanchnic mesoderm, give rise to cECs and HECs, respectively (11, 12). In contrast to the distinct lineages model, other studies suggested a common precursor model that endothelial precursors appear to adopt arterial endothelial fate before committing into HECs (13, 14). Consistent with this model, recent single-cell RNA-sequencing (scRNA-seq) studies (15–17) constructed the continuous developmental trajectory from arterial cECs to HECs, showing that this transition is accompanied with gradual loss of arterial signatures and acquirement of hematopoietic features. To clarify this controversial issue, delicate in vivo fate mapping studies are required. In parallel, although multiple signaling pathways including Hedgehog, Wnt, Vegf, Notch, and BMP are reported critical for HEC and cEC fate specification (18–21), it seems that these signaling pathways eventually converge on the ECs-autonomous requirement of Notch signaling to activate key downstream transcriptional factors, including Gata2, Runx1, and Gfi1 (22–24). Besides, these key factors are also regulated by the upstream factors, including Etv2 and Scl (25–27). Precisely how these upstream factors interplay with Notch signaling and form a genetic network to control cEC and HEC fate specification remains unexplored.
Zebrafish is a prominent model for studying many aspects of developmental biology including angiogenesis and hematopoiesis (28, 29). In zebrafish, the first wave of endothelial progenitors, named angioblasts, in the trunk region arises from the posterior lateral plate mesoderm at ∼11 h postfertilization (hpf), migrates toward the midline during 14 to 18 hpf, and finally coalesces to form the DA at 21 hpf (30, 31). Similar to mammals, HECs in zebrafish are located in the floor of the DA and express several early hematopoiesis-related genes, including gata2b, sclβ, and runx1 (13, 23, 26). During 30 to 60 hpf, HECs undergo EHT and directly bud from the floor of the DA toward the subaortic space (3, 4). Because of the unique advantages of in vivo imaging and genetic amenability, the zebrafish model system provides a powerful tool to dissect the lineage origin of HECs and cECs and the mechanism controlling their fate specification.
In this report, we combined in vivo photoconversion lineage tracing, time-lapse imaging, and scRNA-seq analysis to study the ontogeny of HECs and cECs. We showed that the ventral and dorsal endothelia of the zebrafish DA are largely HECs and cECs, respectively. We further documented that HECs and cECs arise from a common precursor, termed hemogenic angioblast, which undergoes HEC and cEC fate specification prior to the DA formation. Genetic studies further unraveled that at early stage, high and low dosages of Etv2 determine HEC and cEC fates through differentially regulating the expression and activity of fli1a, sclβ, and Notch. Finally, we showed that ectopic expression of transcription factor Runx1 in the roof of the DA can convert cECs into HECs.
Results
The Endothelia in the Floor of the DA Are HECs.
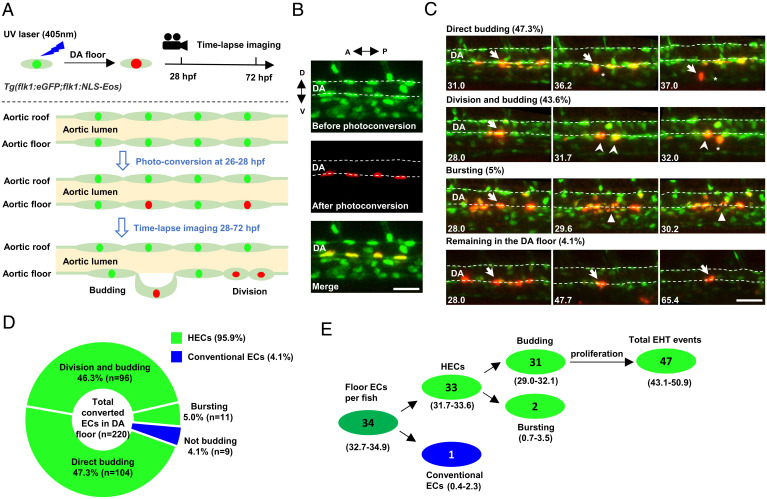
To have a better understanding of the ontogeny of HECs and cECs, we generated a double transgenic line Tg(flk1:eGFP;flk1:NLS-Eos), in which all ECs in the DA express enhanced green fluorescent protein (eGFP) in the cytoplasm and photoconvertible Eos protein in the nucleus (32). This transgenic line allows us to label ECs in the roof or the floor of the DA and follow their behavior by time-lapse imaging (Fig. 1A) during EHT from 28 to 72 hpf (3, 4). We photoconverted 5 to 10 ECs per embryo in the DA floor at 26 hpf and immediately tracked their behavior by time-lapse imaging until 72 hpf (Fig. 1B). Unexpectedly, we found that ∼96% (211 out of 220) of the photoconverted floor ECs underwent EHT (Fig. 1D), of which ∼49% (104 out of 211) of the cells underwent EHT directly, ∼45% (96 out of 211) of the cells divided at least once before initiating EHT, and ∼5% (11 out of 211) of the cells burst into pieces, a phenotype similar to abortive EHT observed in runx1 knockdown embryos (3, 26) (Fig. 1C). Whether the abortive EHT is a natural phenomenon or a consequence of experimental artifacts is unclear. Nonetheless, these data indicate that ∼96% of the ECs in DA floor undergo EHT, and most of them eventually become hematopoietic stem/progenitor cells (HSPCs). Since there are only ∼34 cells in the DA floor at 27 hpf per embryo, this result predicts that 33 of them are HECs, and only 1 cell is cEC (Fig. 1E). Considering that ECs are patterned around the DA circumference unevenly (31), we believe that the rare cECs which we observed in the DA floor in our photoconversion assay might result from the mislabeling of the lateral ECs of the DA. Based on these findings, we conclude that nearly all the ECs in the DA floor at around 26 to 28 hpf are HECs.
Fig. 1.
Time-lapse imaging indicates that the endothelium in the DA floor are HECs. (A) Schematic diagram showing the strategy of photoconversion of the floor endothelium in the DA and time-lapse imaging in Tg(flk1:eGFP;flk1:NLS-Eos) transgenic zebrafish. The green, red, and light green dots indicate the nuclei before photoconversion, the nuclei after photoconversion, and eGFP in the cytoplasm, respectively. (B) Representative images of the photoconverted floor endothelium at 26 hpf. (C) Images representing four types of behaviors of the DA floor endothelium during EHT (28 to 72 hpf) captured by time-lapse imaging. The dashed lines indicate the roof and floor boundaries of the DA, the white arrow indicates successfully converted cells, the white arrowhead indicates the descendants of labeled cells, the white triangle indicates bursting cells, and the asterisk indicates budding cells. Captured time points are labeled at the bottom left. (D) Summary of the behaviors of floor endothelium during 28 to 72 hpf captured by time-lapse imaging. (E) Estimated number of HECs and cECs in the DA floor per fish and their behaviors with 95% (CI). Data were analyzed with 220 cells by five independent experiments with 40 embryos. A, anterior; D, dorsal; P, posterior; V, ventral. (Scale bars, 30 µm.).
The Endothelium in the DA Roof Are cECs Lacking Hemogenic Potential.
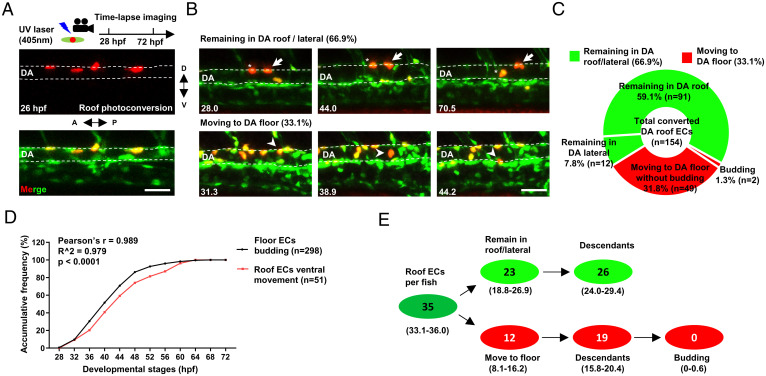
In avian and mammalian embryos, HECs are reported to undergo EHT toward the DA lumen to form intraaortic hematopoietic clusters (IAHCs) (2, 5, 33, 34). These IAHCs are not only restricted to the floor but also observed in the DA roof of murine embryos (7, 33, 35). However, whether these IAHCs in the DA roof are generated in situ or arise from the floor remains elusive. Owing to the advantages of time-lapse imaging and high resolution of ECs labeling, the Tg(flk1:eGFP;flk1:NLS-Eos) fish provides a useful tool to address this issue. Therefore, we photoconverted the ECs in the DA roof at 26 hpf and monitored their behaviors (Fig. 2A). Time-lapse imaging showed that, while the majority ( ∼67%; n = 103/154; fish = 21) of the ECs remained in the DA roof/lateral during 28 to 72 hpf, a substantial portion ( ∼33%; n = 51/154; fish = 21) of the ECs moved ventrally and repositioned to the floor of the DA, some of which underwent cell division before and during the ventral movement (Fig. 2 B and C). Remarkably, the ventral movement of the roof ECs appeared to be temporally correlated with the EHT in the floor (Fig. 2D), indicating that the loss of HECs due to EHT in the DA floor is largely replenished by the ventral movement of roof ECs, which is consistent with the previous observation (3, 36, 37). However, direct budding of ECs from the DA roof was never observed, although two EHT events ( ∼1%; n = 2/154; fish = 21) were captured after the roof ECs moved ventrally and repositioned to the floor (Fig. 2C). Given that there are ∼35 ECs in the DA roof at 27 hpf per embryo, this result means that less than 1 cell from the DA roof undergoes EHT per embryo (Fig. 2E). Taken together, these data demonstrate that the ECs in the DA roof are all cECs lacking hemogenic potential.
Fig. 2.
Time-lapse imaging reveals that the endothelium in the DA roof are cECs. (A) Schematic diagram showing the strategy of photoconversion in the DA roof endothelium and time-lapse imaging in Tg(flk1:eGFP;flk1:NLS-Eos) transgenic zebrafish. (B) Images representing two types of behaviors of the DA roof endothelium during 28 to 72 hpf captured by time-lapse imaging. The dashed line indicates the roof and floor boundary of the DA, the white arrow indicates cells remaining in the DA roof, the asterisk indicates cells remaining in the DA lateral, and the white arrowhead indicates cells moving to the DA floor. Captured time points are labeled at the bottom left. (C) Summary of the behaviors of the DA roof endothelium during 28 to 72 hpf captured by time-lapse imaging. (D) Line graph showing the accumulative frequency percentage (%) of the floor ECs budding (n = 298) and the roof ECs ventral movement (n = 51) during 28 to 72 hpf. Pearson correlation coefficient r was calculated according to the frequency distribution. (E) Estimated number of HECs and cECs in the DA roof per fish and their behaviors with 95% (CI). Data were analyzed with 154 cells by four independent experiments with 21 embryos. (Scale bars, 30 µm.).
HECs and cECs Arise from a Common Precursor and Separate prior to the DA Lumen Formation.
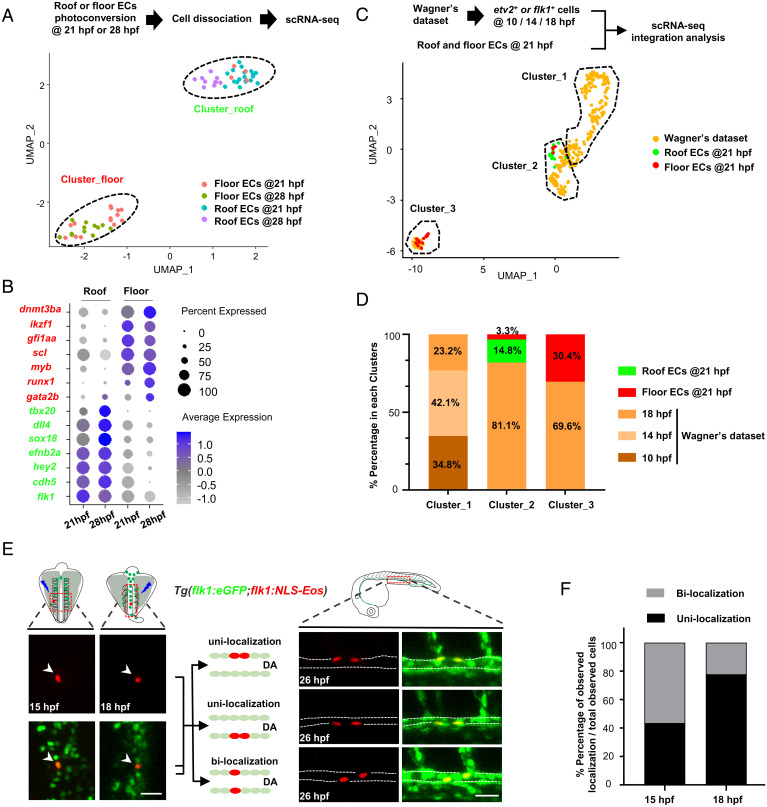
To delineate the mechanism governing the fate determination of HECs and cECs, we isolated single ECs from the DA roof and the floor respectively using the Tg(flk1:eGFP;flk1:NLS-Eos) zebrafish after photoconversion and then performed scRNA sequencing (Fig. 3A). These single ECs were isolated at two different time points, 21 and 28 hpf, when the lumen of the DA is established (31) and the EHT begins (3, 4), respectively. Uniform manifold approximation and projection (UMAP) clustering analysis identified two clusters, Cluster_roof and Cluster_floor, enriched in the ECs from the roof and floor, respectively (Fig. 3A and SI Appendix, Fig. S1A). Differentially expressed gene and gene ontology (GO) analyses showed that the feature genes, including flk1, cdh5, tbx20, sox18, efnb2a, hey2, and dll4, enriched in roof ECs (Fig. 3B) were associated with vasculogenesis, angiogenesis, and vessel development (SI Appendix, Fig. S1B), showing the vascular identity of these cells. In contrast, the enriched GO terms of floor ECs were associated with catabolic and metabolic processes (SI Appendix, Fig. S1B) and were highly enriched in HEC markers and definitive hematopoiesis-related genes, such as gfi1aa, scl, myb, dnmt3ba, ikzf1, gata2b, and runx1 (Fig. 3B), showing that they are in an active metabolic state with hematopoietic potential. These data indicate that Cluster_roof and Cluster_floor represent cECs and HECs, respectively, and that their fates have already been separated by 21 hpf. To further define the time window during which HECs and cECs are specified, we reanalyzed the Wagner’s scRNA-seq database from early developmental stages of zebrafish embryos (38) at 10-, 14-, and 18-hpf stages. By integration analysis with our single-cell RNA-seq dataset (21-hpf stage), we found that the 21-hpf cells from our dataset were predominantly (33/36; ∼92%) clustered with mesoderm cells defined in Wagner’s study (38) (SI Appendix, Fig. S1C). To gain insights into these clusters, we further extracted the information of etv2 or flk1 cells, which presumably include HECs, cECs, and their precursors, angioblasts (30, 39), from 10-, 14-, and 18-hpf stages. We then integrated the extracted etv2 and flk1 scRNA-seq dataset (referred to as Wagner’s dataset in Fig. 3C) with our scRNA-seq dataset (21-hpf stage) and reperformed clustering analysis. UMAP results identified three clusters, Cluster_1, Cluster_2, and Cluster_3 (Fig. 3C and SI Appendix, Fig. S2A). We found that the roof ECs and floor ECs from the 21-hpf embryos were predominantly clustered in Cluster_2 and Cluster_3, respectively (Fig. 3C), suggesting that Cluster_2 and Cluster_3 represent ECs which have already committed to cEC and HEC lineage, while Cluster_1 likely represents noncommitted precursors. Indeed, this notion was further supported by the findings that the Cluster_1 contained a high percentage of etv2 single positive cells with feature genes involved in early developmental process, e.g., embryonic organ morphogenesis and anterior/posterior pattern specification (SI Appendix, Fig. S2 B–D). Moreover, the cEC marker dll4 and HEC marker gata2b were highly expressed in Cluster_2 and Cluster_3, respectively, but absent in Cluster_1 (SI Appendix, Fig. S2E). Next, we examined the distribution percentage of the three clusters at different time points of zebrafish development. The results showed that Cluster_1 consists of cells from 10-, 14-, and 18-hpf stages exclusively, whereas Cluster_2 and Cluster_3 consist of cells predominantly from 18- and 21-hpf stages, with a small fraction of cells from 14 hpf ( ∼0.8%) in Cluster_2 (Fig. 3D), suggesting that cECs and HECs in the DA are likely to arise from a common angioblast precursor, and their fates are largely determined during the period of 14 to 18 hpf, which correlates with the time window when somite-derived Notch signals were shown to specify HEC formation (40, 41).
Fig. 3.
scRNA-seq and fate mapping analyses reveal that HECs and cECs arise from a common precursor and separate prior to the DA formation. (A) Isolation of single ECs from the DA roof or floor after photoconversion (21 hpf, roof/floor ECs, n = 18/18; 28 hpf, roof/floor ECs, n = 10/12) and UMAP plot for cell clustering analysis. (B) Dot plot showing representative marker genes expression enriched in the roof and floor ECs at 21 and 28 hpf. (C) UMAP plot of cell clustering analysis of integrated dataset (Wagner’s dataset at 10, 14, and 18 hpf and dataset at 21 hpf in this study). (D) Distribution percentage (%) of the cells from different time points in each cluster. (E) Single-cell photoconversion and fate mapping analysis in Tg(flk1:eGFP;flk1:NLS-Eos). (Left) Representative images showing single Eos cells photoconverted at 15 and 18 hpf. (Upper Left) Photoconverted areas (see details in Materials and Methods). (Right) Three different localization patterns at 26 hpf (roof and floor only or both in the DA) of the descendants ( 2) derived from a single converted Eos cell. (Upper Right) The observed DA region (see details in Materials and Methods). (F) Bar plot shows the percentage (%) of unilocalization and bilocalization cells in total converted cells at 15 hpf (number of cells: total n = 24; unilocalization n = 11; bilocalization n = 13) and 18 hpf (number of cells: total n = 45; unilocalization n = 35; bilocalization n = 10) stages. Data were acquired by two independent experiments (A) or analyzed by three independent photoconversion experiments (E and F). Cell numbers equal to embryo numbers for single-cell lineage tracing in F. (Scale bars, 30 µm.).
To further support our hypothesis, we performed single-cell fate mapping analysis using the Tg(flk1:eGFP;flk1:NLS-Eos) zebrafish. In this assay, we photoconverted a single flk1-Eos cell in each embryo at 15 or 18 hpf and defined the identity of the photoconverted cell by examining the distribution pattern of its progenies (if the converted single cell produced two or more progenies) in the DA roof and floor at 26 hpf (Fig. 3E). We reasoned that if cECs and HECs indeed originated from a common precursor and their fates were specified between 14 and 18 hpf, we would expect to observe two different distribution patterns, bilocalization and unilocalization. The bilocalization indicates successful labeling of a common precursor capable of giving rise to both cECs and HECs, whereas the unilocalization indicates successful labeling of a lineage committed cell capable of giving rise to either cECs or HECs (Fig. 3E). In agreement with our hypothesis, single-cell fate mapping revealed that ∼55.5 and ∼44.5% of the cells displayed bilocalization and unilocalization potential at the 15-hpf stage, respectively (Fig. 3F). As fish developed to the 18-hpf stage, the percentage of the cells with bilocalization potential dropped to ∼22.2%, accompanied by an increase of the percentage ( ∼77.8%) of the cells with unilocalization potential (Fig. 3F), indicating a gradual fate commitment process from bipotent progenitors to unipotent lineage-committed cells during this developmental time window. These results, together with the scRNA-seq data analysis, demonstrate that cECs and HECs in the DA indeed originate from a common flk1 precursor, which we refer to as hemogenic angioblast, and their fates are largely determined during the period of 14 to 18 hpf, well before the lumen formation of the DA.
Etv2 Dosage Determines HEC and cEC Fate through Differentially Regulating the Activities of Fli1a, Notch, and Sclβ.
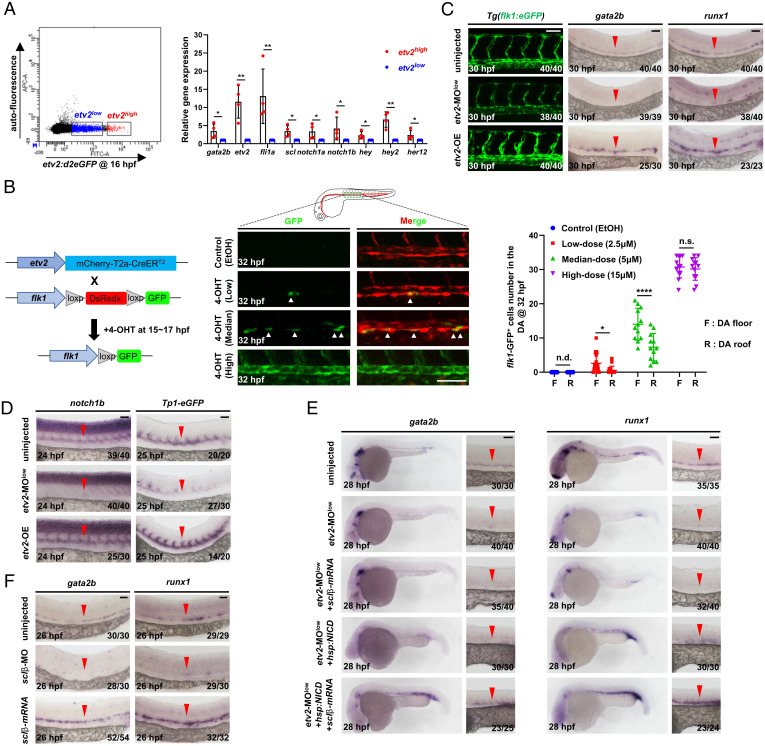
Having defined the lineage origin of HECs and cECs, we next investigated the mechanism controlling their fate specification. The Ets variant 2 (etv2) has been identified as a master regulator for HEC and cEC development (25, 42), yet the underlying mechanism remains incompletely clear. To dissect the role of Etv2 in HEC and cEC development, we generated a transgenic fish Tg(etv2:NLS-d2eGFP), in which a fast-degraded d2eGFP protein is under control of the etv2 promoter (43) (SI Appendix, Fig. S3A), and performed fluorescence-activated cell sorting (FACS) analysis at 16 hpf, when HECs and cECs are known to be specified (Fig. 3 C, D, and F). Intriguingly, two etv2 cell fractions, etv2-d2eGFPhigh and etv2-d2eGFPlow, were detected (Fig. 4A). qPCR analysis showed that the expression of the HEC marker, gata2b (23), was significantly enriched in etv2-d2eGFPhigh fraction (Fig. 4A). This observation prompted us to hypothesize that Etv2 dosage might govern the fate choice between HECs and cECs. To test this hypothesis, we generated a double transgenic line Tg(etv2:mCherry-T2a-CreER ;flk1:loxP-DsRedx-loxP-eGFP) (SI Appendix, Fig. S3A) and performed lineage tracing analysis by pulsing embryos with a range of 4-Hydroxytamoxifen (4-OHT) doses between 15 and 17 hpf when HECs and cECs are undergoing fate specification (Fig. 4B). The resulting flk1-GFP cells were examined at 32 hpf, which is the latest timing point used in the study to address the presence and localization of HECs in the aortic floor. Results showed that low (2.5 µM) and median (5 µM) dose treatment produced limited numbers of GFP cells exhibiting bias distribution pattern toward the floor of the DA (Fig. 4B). On the other hand, high-dose (15 µM) treatment led to a significant increase of GFP cells displaying comparable number in the floor and roof (Fig. 4B). In parallel, we also tested different lengths of time (1, 1.5, and 2 h) of 4-OHT treatment with median dose (5 µM). Results showed that three groups exhibited labeling bias toward floor of the DA, but the bias toward floor was decreased in the group of 2-h treatment (SI Appendix, Fig. S3B). These data suggest that the progenitors with high and low Etv2 dosages preferentially adopt the HEC and cEC fates, respectively. To further support this notion, we generated another transgenic line Tg(hsp70l:etv2-P2a-mCherry), in which Etv2-P2a-mCherry protein is under control of the hsp70l promoter (SI Appendix, Fig. S3A), and asked whether overexpression of Etv2 during 14 to 18 hpf would enhance HEC formation. Indeed, whole-mount in situ hybridization (WISH) showed a robust increase of gata2b and runx1 expression in the DA (Fig. 4C), suggesting that high Etv2 dosage promotes HEC fate. Consistent with the overexpression results, injection of low-dose etv2 MO (25, 42) into fertilized embryos at the one-cell stage (SI Appendix, Fig. S3C) dramatically reduced gata2b and runx1 expression in the DA (Fig. 4C), showing that reduced etv2 expression blocks HEC formation. Notably, the DA formation remained intact in both Etv2 overexpression and low-dose etv2 MO knockdown embryos (Fig. 4C), indicating that low Etv2 dosage is sufficient for cEC specification. Collectively, these data indicate that etv2 expression level, high and low dosage, determines HEC and cEC fate specification from hemogenic angioblasts, respectively.
Fig. 4.
Etv2 dosage determines HEC and cEC fate through differentially regulating the activities of Fli1a, Notch, and Sclβ. (A) Flow cytometric and gene expression analysis of Tg(etv2:d2eGFP) embryos. Representative results of flow cytometric analysis at 16 hpf and relative genes expression levels in the etv2-d2eGFPhigh and etv2-d2eGFPlow population. (B) Strategy to perform lineage tracing using double transgenic line Tg(etv2:mCherry-T2a-CreER ;flk1:loxp-DsRedx-loxp-eGFP). Fish were treated with EtOH or different doses of 4-OHT during 15 to 17 hpf, and GFP ECs were quantified at 32 hpf. (C) Lateral views of the DA in Tg(flk1:eGFP) embryos and WISH of gata2b and runx1 in embryos. The embryos were injected with or without a low dose of etv2 MO (etv2-MOlow) to knockdown etv2 or heat-shocked to overexpress etv2 (etv2-OE). (D) WISH of notch1b and eGFP. The embryos were injected with or without a low dose of etv2 MO to knockdown etv2 or heat-shocked to overexpress etv2. (E) WISH of gata2b and runx1 indicates rescue of HECs. The embryos were uninjected or injected with a low dose of etv2 MO, together with or without overexpressing sclβ (mRNA injection) and NICD (heat shock) alone or both. (F) WISH of gata2b and runx1 in sclβ MO knockdown and sclβ overexpression embryos. The embryos were injected with or without sclβ MO or sclβ mRNA. Data are representative of two independent experiments (four biological replicates in A; for embryos, n = 12 in B) or two different clutches of embryos (C–F). n/N reports the number of embryos with staining pattern in image/total embryos (C–F). Heat shock was performed at 14 hpf for 45 min in C–E. Student’s t tests were used in A; two-way ANOVA test was used in B. Data are represented as mean ± SD, *P 0.05, **P 0.01, ****P 0.0001; n.s., not significant (P > 0.05); n.d., not detectable. (Scale bars, 60 µm.).
Previous studies have documented that the etv2-fli1a axis is sufficient for cEC specification (25, 44) (SI Appendix, Fig. S4A). Consistent with this notion, fli1a expression was detected, although at a lower level, in low-dose etv2 MO knockdown embryos (SI Appendix, Fig. S4B), which explained the intact vascular structure in low-dose etv2 MO knockdown embryos (Fig. 4C). Yet the mechanism controlling the HEC specification is less clear. To dissect the genetic network governing HEC fate specification behind Etv2 high dosage, we monitored the Notch signaling, which has been shown as a key regulator in HEC fate determination (20, 45). We found that the expressions of notch1 receptors (notch1a/notch1b) and Notch downstream targets (hey/hey2/her12) were significantly enriched in the etv2-d2eGFPhigh cell fraction (Fig. 4A), suggesting that high Etv2 dosage may function through regulating Notch activity. To test this possibility, we examined the expression of notch receptors and Notch activity under etv2 overexpression and low-dose etv2 MO knockdown conditions. Results showed that the expressions of arterial notch receptors (notch1a/notch1b/notch3) (46), Notch downstream targets (hey/hey2/her12), and Tp1-eGFP reporter in the DA were significantly enhanced in etv2 overexpression embryos and dramatically reduced in low-dose etv2 MO knockdown embryos (Fig. 4D and SI Appendix, Fig. S4 C and D). In contrast, etv2 expression, as indicated by the formation of the bilateral stripes of etv2 cells, was largely unaffected in Delta-Notch signaling-deficient mib mutants (47) (SI Appendix, Fig. S4E), indicating that Etv2 regulates HEC specification, at least in part, through modulating Notch activity. Notably, fli1a expression was also highly enriched in etv2-d2eGFPhigh population (Fig. 4A), which is consistent with previous findings that fli1a is a downstream target of Etv2 (44), suggesting that Fli1a may be involved in the HEC specification. Indeed, we showed that injection of fli1a messenger RNA (mRNA) into low-dose etv2 MO knockdown embryos could partially rescue the expression of Tp1-eGFP reporter (SI Appendix, Fig. S4C), suggesting that Notch signaling is regulated directly by Etv2 as well as indirectly through Fli1a. We therefore wondered whether restoration of Notch signaling is sufficient to rescue HEC formation in low-dose etv2 MO knockdown embryos, in which fli1a was expressed at low level (SI Appendix, Fig. S4B) by overexpressing Notch intracellular domain (NICD), the dominant activator of Notch pathway. Results showed that NICD overexpression only marginally rescued HECs (Fig. 4E), suggesting that additional downstream factor(s) are required for HEC specification. We then turned to sclβ, an etv2 downstream target which has been shown indispensable for runx1 expression in the DA (25, 26). We found that overexpressing Sclβ together with NICD, but not Sclβ or NICD alone, robustly rescued the HEC formation (Fig. 4E), suggesting that Sclβ and Notch are two key Etv2 downstream targets which act in parallel and collaboratively to activate gata2b-runx1 axis for HEC specification. Indeed, epistatic analysis showed that Sclβ and Notch were independent of each other as indicated by the normal expression of sclβ and Tp1-eGFP reporter in mib mutants and sclβ morphants (48), respectively (SI Appendix, Fig. S4 E and F). Consistent with this notion, gata2b and runx1 expression were diminished in sclβ MO knockdown embryos and overexpressing sclβ drastically enhanced gata2b and runx1 expression (Fig. 4F), placing sclβ upstream of the gata2b-runx1 axis. Taken together, these data demonstrate that the fate commitment of cECs and HECs from hemogenic angioblasts is determined by the Etv2 dosage, in which low dosage of Etv2 triggers low levels of Fli1a and Notch to direct cEC formation (25), whereas high dosage of Etv2 induces high levels of Fli1a, Notch, and Sclβ expression, leading to the promotion of HEC specification (SI Appendix, Fig. S5).
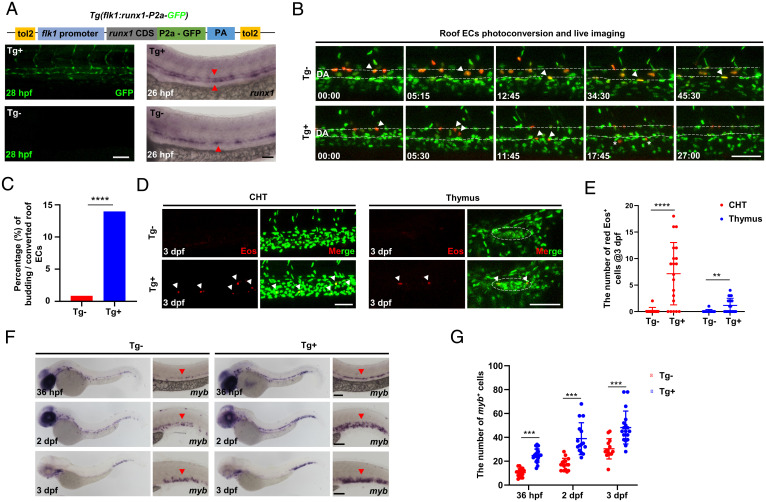
Runx1 Overexpression Promotes HEC Fate in the Roof of the DA.
Our above analysis placed the activation of the gata2b-runx1 axis as the final effector for HEC specification. Yet whether induction of gata2b-runx1 expression is sufficient to promote HEC formation in vivo remains unclear. To address this issue, we generated a transgenic line Tg(flk1:runx1-P2a-GFP), in which Runx1-P2a-GFP expression was under control of the pan-endothelial flk1 promoter (31) (Fig. 5A), and asked whether ectopically expressing runx1 in the DA roof endothelium could promote the HEC formation. To facilitate our analysis, we outcrossed the Tg(flk1:runx1-P2a-GFP) line with the Tg(flk1:NLS-Eos) fish. The resulting double transgenic fish Tg(flk1:runx1-P2a-GFP;flk1:NLS-Eos) enabled us to label the ECs in the DA roof at 26 to 28 hpf and followed their behavior by time-lapse imaging. Consistent with previous results (Fig. 2C), only 3 out of 352 roof ECs (3/352, ∼0.85%) underwent EHT in control siblings (Tg; n = 32) (Fig. 5 B and C). Strikingly, in runx1 overexpression embryos (Tg; n = 40), 57 out of 408 roof ECs analyzed underwent EHT, a 17-fold increase compared to control siblings ( ∼14% versus ∼0.85%) (Fig. 5C). We further showed that the DA roof-born Eos HECs were able to colonize the caudal hematopoietic tissue (CHT) and thymus after EHT (Fig. 5 D and E), suggesting that the roof-born HECs in runx1 overexpression fish are indeed capable of becoming hematopoietic cells. This conclusion was further supported by the findings that the Tg(flk1:runx1-P2a-GFP) fish produced significantly more myb HSPCs than control siblings (Fig. 5 F and G). Taken together, these results demonstrate that runx1 overexpression is sufficient for the conversion, at least in part, of HECs’ fate in the DA roof. Interestingly, we noticed that all 57 EHT events from the DA roof in runx1 overexpression embryos took place after they had moved ventrally and repositioned in the floor of the DA (Fig. 5D), supporting the notion that EHT occurs exclusively in the floor of the DA.
Fig. 5.
Runx1 overexpression promotes HEC fate in the DA roof. (A) Schematic graph showing the construction of Tg(flk1:runx1-P2a-GFP) with representative confocal images of stable F1 fish at 28 hpf and runx1 WISH at 26 hpf. (B) Representative behaviors of the photoconverted DA roof ECs captured by time-lapse imaging during 28 to 72 hpf in Tg (runx1 overexpression) and Tg (siblings without runx1 expression) embryos. The dashed lines denote the roof and floor walls of the DA, the triangles denote converted cells, and the asterisks denote budding cells. (C) Quantification of percentage (%) of the budding roof ECs divided by the photoconverted roof ECs (3/352 in Tg versus 57/408 in Tg). (D) Representative confocal images (after Z-projection) in CHT and thymus at 3 dpf. Fifteen DA roof ECs were photoconverted at 26 hpf in each Tg or Tg embryo, and red Eos cells were quantified at 3 dpf. The dashed circles denote the location of thymus. (E) Quantification of red Eos cells in CHT and thymus at 3 dpf. (F) WISH of myb in Tg and Tg embryos. (G) Quantification of myb cells. Data are representative of three independent experiments with 32 (Tg) and 40 (Tg) embryos in B and C or two different clutches of embryos with (36 hpf, Tg /Tg: n = 18/19; 2dpf, Tg /Tg: n = 15/15; 3dpf, Tg /Tg: n = 15/18) for myb WISH (F and G) or 11 (Tg) and 20 (Tg) embryos for photoconversion and quantification (D and E). Fisher’s exact test was used in C, unpaired Student’s t tests were used in E and G. **P 0.01, ***P 0.001, ****P 0.0001. (Scale bars, 60 µm.).
Discussion
In this study, we reported a systematic investigation on the ontogeny of cECs and HECs from early lineage fate mapping, genetic fate determination, and spatial distribution in the DA. By combining photoconversion with time-lapse imaging, we unexpectedly found that nearly the entire aortic floor endothelium are HECs, whose loss during EHT is replenished by the ventral movement and proliferation of the roof cECs in the DA. Interestingly, our above finding is similar to the previous study in the avian model that the aortic floor appears to be a transitory structure and would be completely replaced by roof and side ECs in the aorta (11), suggesting that a similar model would also be utilized in mammals.
Our study here provides in vivo evidence that cECs and HECs in the DA arise from a bipotent common progenitor, which undergoes fate separation during the time window of 14 to 18 hpf, and finally colonize the DA roof and floor accordingly. This common progenitor, as shown in our fate mapping data, is in a previously defined flk1 angioblasts population (30, 31) and distinct from the previous conception, hemangioblasts, which refer to preprimitive wave mesodermal cells giving rise to yolk sac blood islands in mouse (49) and primitive erythrocytes and ECs in zebrafish (50). We therefore term the common precursors of cECs and HECs as hemogenic angioblasts, which represent a branch point between definitive hemogenic endothelium and conventional endothelium in the DA. Notably, our common progenitor model appears different from the previous findings in avians, which showed that distinct mesoderm regions, the somatopleural and the splanchnopleural mesoderm, give rise to aortic roof and floor endothelium, respectively (6, 11, 12, 51). The discrepancy between the avian and zebrafish models might be explained by the different methods used in the two model systems. Instead of the single-cell resolution photoconversion assay performed here, the transplantation assay employed in avian model is mainly based on anatomical recognition with lower resolution. Hence, the transplantation assay may not reflect the endogenous developmental trajectory of aortic roof and floor endothelium. Alternatively, the discrepancy is attributed to the species difference between avian and fish. Notably, several studies have suggested that, like zebrafish, the ontogeny of the DA in mice appears to arise from single mesodermal source (52, 53), implying that mammals may employ a mechanism similar to that of zebrafish.
Another interesting observation in our study is that ectopic runx1 overexpression can promote HEC formation in the roof of the DA, and yet these roof-derived runx1 overexpressing cells undergo EHT only after they have repositioned to the floor of the DA. This observation indicates that in addition to the early fate determination during the time window of 14 to 18 hpf, extrinsic signals are required for the induction of EHT. One possible mechanism is that the floor niche produces extrinsic induction signals to trigger the initiation of EHT. Alternatively, the roof environment generates extrinsic inhibitory signals to prevent EHT in the roof of the DA. It is unclear which mechanism is correct, and perhaps both mechanisms are involved. Indeed, recent studies in zebrafish have broadened the understanding of the EHT process by showing that mechanical force generated by blood flow plays a critical role in controlling the ventral emergence and EHT budding direction of the HECs (36, 37). Notably, a previous study, in which the authors utilized the whole-mount transparency method to quantify all hematopoietic clusters (c-Kit) temporally and spatially in E10.5 murine embryos, observed that some c-Kit clusters were distributed along the dorsal wall of the DA in E10.5 murine embryos, while most of the c-Kit clusters were ventrally localized (35). However, the whole-mount transparency method could not provide direct evidence supporting that the c-Kit clusters along the dorsal wall indeed arise in situ. It will be of interest to reinvestigate whether the dorsal wall of the murine DA consists of HECs. Finally, according to our data, we speculate that Runx1 expression does not seem to regulate the movements of the roof cells from the roof to the floor, since cell movements from the roof to the floor occur regardless of whether the roof cells express Runx1 or not, as indicated in runx1-overexpression or wild-type embryos, respectively. Instead, Runx1 is likely to play a role in HEC formation and EHT process (24, 54, 55).
Finally, we have proposed a model in which the Etv2 dosage plays a critical role in cEC and HEC fate specification (SI Appendix, Fig. S5). Previous studies identified that Fli1 (42, 44), Scl (25, 42), Notch1 (27), and Dll4 (56) are downstream of Etv2 in the regulation of EC development. Our study further reveals that low Etv2 dosage is sufficient to promote cEC commitment by inducing a low level of Fli1a, whereas high Etv2 dosage induces HEC lineage commitment through inducing a higher level of Fli1a and Sclβ expression and higher activity of Notch signaling. Interestingly, we also retrieved the scRNA-seq gene expression matrix of four cell populations (Conflux AE, pre-HE, HE, and IAC) from a murine scRNA-seq dataset reported previously (17) and examined the expression of Etv2, Scl, Notch, Gata2, and Runx1. We observed an increase of both Etv2 expression level and the percentage of Etv2-positive cells between pre-HE and HE populations, accompanied with the increase of Runx1 expression (SI Appendix, Fig. S6 A–D). Likewise, the expression levels and positive cell proportions are also increased for Scl, Notch2, and Notch3 but not for Gata2 (SI Appendix, Fig. S6 D and E), which seems to be constitutively highly expressed in those four populations. The discrepancy of the Gata2 expression between zebrafish and murine datasets may be due to species difference since zebrafish contain duplicated gata2a and gata2b genes, and only gata2b shows HEC-specific expression (23). Nevertheless, the overall expression of the key transcriptional regulators seems to be highly conserved between these two datasets, suggesting a similar mechanism may be applied to the murine model. It will be of great interest to further investigate this issue in the murine model. Another interesting finding is that our epistatic analysis indicates Sclβ act upstream of gata2b and is required for gata2b expression in a Notch-independent manner. However, how Sclβ regulates gata2b expression remains unknown. Given the fact that the enhancers of Gata2, Fli1, and Scl are shown to be active during murine HSC ontogeny (57) and they have also been shown to form a recursively wired gene-regulatory circuit during early mouse hematopoietic development (58), we speculate that Sclβ may act collaboratively with Fli1a to regulate gata2b expression in zebrafish. Further studies will be required to clarify these issues.
Materials and Methods
Zebrafish Strains.
Zebrafish were maintained according to standard protocols (59). Zebrafish were kept at 28.5 ∘C in a 14-h light and 10-h dark cycle. After natural spawning, embryos were collected and raised at 28.5 ∘C in 0.5 × E2 medium containing methylene blue (egg water). To avoid pigmentation, embryos were changed to 0.003% N-Phenylthiourea (P7629, Sigma) in egg water at 1 dpf. The following strains were used: Tg(flk1:eGFP) (31), Tg(flk1:NLS-Eos) (32), Tg(Tp1:eGFP) (60), Tg(hsp70l:NICD-eGFP) (61), Tg(fli1:DsRed) (62), Tg(flk1:loxP-DsRedx-loxP-eGFP) (63), mib (64), Tg(etv2:NLS-d2eGFP), Tg(hsp70l:etv2-P2a-mCherry), Tg(etv2:mCherry-T2a-CreER ), Tg(flk1:loxP-DsRedx-loxP-eGFP), and Tg(flk1:runx1-P2a-GFP). All animal experiments were conducted according to the guidelines of the Animal and Plant Care Facility and approved by the Animal Ethics Committee of the Hong Kong University of Science and Technology.
Generation of Transgenic Lines.
The destabilized d2eGFP with nuclear localization sequence (NLS-d2eGFP) and mCherry-T2a-CreER sequence were placed under the c2 etv2 promoter (43). Zebrafish full-length coding sequence of etv2 gene plus P2a-mCherry was cloned under hsp70l promoter (65). Zebrafish full-length coding sequence of runx1 gene plus P2a-GFP was cloned under ∼6.5 kb flk1 promoter (31). The above sequences were inserted into the modified pBluescript II SK(+) vector containing two arms of Tol2 sequences. The resulting etv2:NLS-d2eGFP, etv2:mCherry-T2a-CreER, hsp70l:etv2-P2a-mCherry, and flk1:runx1-P2a-GFP constructs were then injected into one-cell–stage embryos with transposase mRNA. The injected embryos were raised to adulthood for germline transmission screening.
Photoconversion Fate Mapping, Time-Lapse Imaging, Image Processing, and Unilocalization/Bilocalization Identification.
For early fate mapping analysis, a single flk1-Eos cell of Tg(flk1:eGFP;flk1:NLS-Eos) was converted by 405-nm UV laser at 15- and 18-hpf stages in each embryo as described (66). Single EC was randomly photoconverted within the 8- to 13-somite region at 15 hpf or 6- to 18-somite region at 18 hpf. With exception of three embryos, all photoconverted ECs could give rise to ECs of the DA in the AGM region (8- to 17-somite region) at 26 hpf (pretested in this study). For time-lapse imaging to capture EHT, the ECs (5 to 10 cells) in the DA of Tg(flk1:eGFP;flk1:NLS-Eos) embryos were converted by 405-nm UV laser at around 26 hpf. The embryos were subsequently mounted in 1% low–melting-point agarose in E3 embryo medium with 0.01% tricaine for anesthetization as described (66). Time-lapse imaging was carried out on SP8 confocal microscope using a 20× dry objective (Leica). A heated microscope chamber at 28.5 ∘C was used for recording time-lapse videos. Stacks were taken every 15 min with a step size of 3.0 to 5.0 µm. Confocal stacks and time-lapse videos were analyzed using ImageJ (NIH). For unilocalization/bilocalization identification of descendant cells, Z projections of images were first performed. The roof/floor (cEC/HEC) ECs were identified according to the same criterion (0 to 45∘ from the dorsal–ventral axis of the DA) relative to the roof/floor boundary indicated by the flk1-eGFP expression (37).
Heat Shock, Morpholino Injection, and mRNA Injection.
Morpholinos were purchased from Gene-Tools (USA) and dissolved in distilled water. One ng/nL (high dose) or 0.1 ng/nL (low dose) of etv2 MO (42) was injected into one-cell–stage embryos. The high dose of etv2 MO completely blocked the etv2-d2eGFP expression of the Tg(etv2:NLS-d2eGFP) reporter (SI Appendix, Fig. S3C), while the low dose of etv2 MO ( 0.3 ng/nL) partially reduced etv2-d2eGFP expression (SI Appendix, Fig. S3C). sclβ MO knockdown was performed as previously described (26), and 8 ng of sclβ MO was injected into one-cell–stage embryos. The mRNA of fli1a and sclβ were synthesized by using mMessage mMachine Kit (Ambion Inc.). The primers for amplifying fli1a and sclβ full-length coding sequences and injected dosage of fli1a (60 pg) and sclβ (200 pg) mRNA were referenced to a previous report (25). For heat shock experiments, embryos at the 10-somite stage (14 hpf) were collected in a 50-mL Falcon tube containing 5 mL of E3 embryo medium and submerged in a 40 ∘C water bath for 45 min.
WISH, Double Fluorescence In Situ Hybridization, and Immunofluorescent Antibody Staining.
WISH was performed as described (66). AP-conjugated anti-DIG antibody (11093274910; Roche) and BM purple (11442074001; Roche) were used as the substrate for color reaction. Double fluorescence in situ hybridization (dFISH) assay was performed as described (67). POD-conjugated anti-fluorescein antibody (11426346910; Roche), POD-conjugated anti-digoxigenin antibody (11207733910; Roche), and TSA-Cy3 or TSA-FITC (NEL741001KT; PerkinElmer) were used for color reaction. The RNA probes were in vitro synthesized with T7 RNA polymerase (Promega) with dig-RNA-labeling mix (11277073910; Roche) or fluorescein-RNA-labeling mix (11685619910; Roche). The probes sequences are listed below: runx1 (NM_131603.3; 385–2069), gata2b (NM_001002689.1; 809–1669), notch1b (NM_131302.2; 4757–6080), etv2 (NM_001037375.1; 65–1165), fli1a (NM_131348.2; 104–1459), scl (NM_213237.1; 518–1138), and eGFP (eGFP full-length coding sequence). For antibody staining, the pretreatment of embryos was similar to WISH as described above. Anti-cleaved caspase3 (559565, Biosciences, diluted at 1:500) and Alexa 555-anti-rabbit antibody (A31572, Invitrogen, diluted at 1:400) were used as primary and secondary antibody, respectively. WISH images were captured by Stereo Discovery V20 microscope (Zeiss) with digital camera (DS-Ri2, Nikon). dFISH and antibody staining images were captured by SP8 confocal microscope (Leica) and analyzed by ImageJ (NIH).
FACS, cDNA Synthesis, and qPCR.
The embryos at desired stages were pooled together (each biological replicate with 60 to 100 embryos) and homogenized by syringe with 27G needle. The cell suspension was prepared as previously described (68) and subjected to FACS analysis (FACS Aria IIIu). The cDNA was prepared from sorted cells (100 to 500 cells per well; two or four technical replicates per biological sample) and amplified by Smart-seq2 protocol (69). Real-time qPCR was performed to quantify the transcripts of gata2b, etv2, fli1a, scl, notch1a, notch1b, notch3, hey, hey2, and her12. Primers for qPCR used in this work are listed in SI Appendix, Table S1.
Single-Cell Isolation, cDNA Preparation, and RNA-seq.
For single-cell isolation, 10 to 20 cells in the DA roof or floor of each Tg(flk1:eGFP;flk1:NLS-Eos) embryo were photoconverted at 21 or 28 hpf under a confocal microscope. Thirty embryos were anesthetized, and the trunk regions containing the converted cells were dissected with 27G needle in ice-cold phosphate-buffered saline (PBS). The dissected tissues were dissociated into single-cell suspension according to the protocol (70). After 20 min digestion, the single-cell suspension was pushed through a 40-µm Cell Strainer (352340, BD Falcon) and transferred to the 35-mm petri dish (150460, Invitrogen). A single red Eos cell was collected manually with the micromanipulator system (NT-88-V3, Nikon) under a fluorescence microscope (Nikon), then washed by transferring the cell to a drop of 500 µL RNase-free PBS with 2% bovine serum albumin, and finally transferred into RNase-free 200 µL tube (Eppendorf) containing 4.4 µL lysis buffer (69). The yield and efficiency of manual single-cell collection were relatively low. In our hand, we were able to collect 20 to 30 single RNA-qualified cells from 300 to 600 photoconverted cells within 2 h after digestion. The Smart-seq2 protocol (69) was used for whole-transcriptome amplification, and the cDNA library was prepared as previously described (68). Library quality was assessed using the Agilent Fragment Analyzer System, and the samples were sent to Novogene Company Limited for Illumina Hiseq X Ten 150-bp paired-end sequencing with an average depth of 6 × 106 raw reads per sample.
Bioinformatic Analysis of RNA-seq.
Reads were aligned to the GRCz11.94 zebrafish reference genome using Spliced Transcripts Alignment to a Reference aligner (2.7.1a). Aligned reads were converted to counts for each gene using featureCounts (Rsubread_2.6.1). Dataset from 21- and 28-hpf roof cECs and floor HECs were analyzed by Seurat package (v.4.0.2). Cell quality control was performed before analysis based on minimal detected genes (default, n = 200). First, variable genes (default, n = 2,000) were identified based on variance stabilizing transformation using a normalized matrix. Principal component analysis was then performed (Dims = 1:20) followed by scaling scRNA-seq data. Finally, a plot graph was constructed by shared nearest neighbor–based clusters of the subtypes and transformed to UMAP for dimension reduction for intuitive visualization. The top 100 maker genes were identified based on spatial information (roof versus floor). GO analysis was performed with the clusterProfiler package (v.3.16.1) using the top 100 marker genes. For RNA-seq data integration analysis, GSE112294 (38) was reanalyzed by fetching counts data at 10, 14, and 18 hpf. Because etv2 expression decreases as endothelial precursors undergo differentiation, we set criteria as etv2 expression (counts > 0) at 10 and 14 hpf, flk1 expression (counts > 0) at 18 hpf, to extract potential endothelial/precursor cells, referred to as “Wagner’s dataset.” The Wagner’s dataset was integrated with the 21-hpf dataset by the FindIntegrationAnchors function implanted in the Seurat package. Following the standard integration analysis tutorial (https://satijalab.org/seurat/articles/integration_introduction.html), we set DefaultAssay as “integrated” and performed clustering analysis. All the analysis was performed in the R (version 4.0.2) environment.
Quantification and Statistical Analysis.
Statistical parameters including the exact value of n are reported in Figs. 1, 2, 4, and 5 and the figure legends of Figs. 1–5. All statistical analyses were performed using GraphPad-Prism version 8. Paired or unpaired Student’s t tests were used to calculate the two-tailed P value for comparisons, clarified in the figure legends of Figs. 4 and 5. Data were presented as mean ± SD. Statistical significance is shown as follows: ns, P > 0.05; *P 0.05; **P 0.01; ***P 0.001; and ****P 0.0001.
Supplementary Material
Acknowledgments
We thank Dr. Fukuhara for providing Tg(flk1:NLS-Eos) fish and Dr. Feng Liu for providing Tg(hsp70l:NICD-eGFP) fish. This work was supported by grants from the National Key Research and Development Program of China (grant 2018YFA0800200), by the Research Grants Council of Hong Kong (grants C6002-17GF, N_HKUST621/17, AoE/M-09/12, T13- 605/18-W, and T13-602/21-N), by the Croucher Foundation (grant CF20SC01), and by the Innovation and Technology Commission of Hong Kong (grant ITCPD/17-9).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119051119/-/DCSupplemental.
Data and Software Availability.
The raw fastq files for each sample reported in this paper and single-cell RNA-seq data are available in the NIH Gene Expression Omnibus database (accession no. GSE197757).
References
- 1.Eilken H. M., Nishikawa S., Schroeder T., Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457, 896–900 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Boisset J. C., et al., In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kissa K., Herbomel P., Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Bertrand J. Y., et al., Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardanaud L., Dieterlen-Lièvre F., Emergence of endothelial and hemopoietic cells in the avian embryo. Anat. Embryol. (Berl.) 187, 107–114 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Jaffredo T., Gautier R., Eichmann A., Dieterlen-Lièvre F., Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125, 4575–4583 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Mizuochi C., et al., Intra-aortic clusters undergo endothelial to hematopoietic phenotypic transition during early embryogenesis. PLoS One 7, e35763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavian M., et al., Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 87, 67–72 (1996). [PubMed] [Google Scholar]
- 9.Ditadi A., et al., Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 17, 580–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gama-Norton L., et al., Notch signal strength controls cell fate in the haemogenic endothelium. Nat. Commun. 6, 8510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pouget C., Gautier R., Teillet M. A., Jaffredo T., Somite-derived cells replace ventral aortic hemangioblasts and provide aortic smooth muscle cells of the trunk. Development 133, 1013–1022 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Pardanaud L., et al., Two distinct endothelial lineages in ontogeny, one of them related to hemopoiesis. Development 122, 1363–1371 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Bonkhofer F., et al., Blood stem cell-forming haemogenic endothelium in zebrafish derives from arterial endothelium. Nat. Commun. 10, 3577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uenishi G. I., et al., NOTCH signaling specifies arterial-type definitive hemogenic endothelium from human pluripotent stem cells. Nat. Commun. 9, 1828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Y., et al., Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 29, 881–894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou S., et al., Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 30, 376–392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q., et al., Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 136, 845–856 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson R. N., et al., Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev. Cell 16, 909–916 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y., et al., Ncor2 is required for hematopoietic stem cell emergence by inhibiting Fos signaling in zebrafish. Blood 124, 1578–1585 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Kumano K., et al., Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 18, 699–711 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Clements W. K., et al., A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474, 220–224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel M. G., Rapp K., Schaniel C., Moore K. A., Induction of developmental hematopoiesis mediated by transcription factors and the hematopoietic microenvironment. Ann. N. Y. Acad. Sci. 1466, 59–72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butko E., et al., Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development 142, 1050–1061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yzaguirre A. D., Howell E. D., Li Y., Liu Z., Speck N. A., Runx1 is sufficient for blood cell formation from non-hemogenic endothelial cells in vivo only during early embryogenesis. Development 145, dev158162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren X., Gomez G. A., Zhang B., Lin S., Scl isoforms act downstream of etsrp to specify angioblasts and definitive hematopoietic stem cells. Blood 115, 5338–5346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhen F., Lan Y., Yan B., Zhang W., Wen Z., Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development 140, 3977–3985 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Liu F., et al., Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. 16, 654–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chávez M. N., Aedo G., Fierro F. A., Allende M. L., Egaña J. T., Zebrafish as an emerging model organism to study angiogenesis in development and regeneration. Front. Physiol. 7, 56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong J. L. O., Zon L. I., Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 39, 481–501 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Kohli V., Schumacher J. A., Desai S. P., Rehn K., Sumanas S., Arterial and venous progenitors of the major axial vessels originate at distinct locations. Dev. Cell 25, 196–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S. W., Beis D., Mitchell T., Chen J. N., Stainier D. Y., Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Fukuhara S., et al., Visualizing the cell-cycle progression of endothelial cells in zebrafish. Dev. Biol. 393, 10–23 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Taoudi S., Medvinsky A., Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc. Natl. Acad. Sci. U.S.A. 104, 9399–9403 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bruijn M. F. T. R., Speck N. A., Peeters M. C. E., Dzierzak E., Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19, 2465–2474 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokomizo T., Dzierzak E., Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development 137, 3651–3661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campinho P., Lamperti P., Boselli F., Vilfan A., Vermot J., Blood flow limits endothelial cell extrusion in the zebrafish dorsal aorta. Cell Rep. 31, 107505 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Lancino M., et al., Anisotropic organization of circumferential actomyosin characterizes hematopoietic stem cells emergence in the zebrafish. eLife 7, e37355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner D. E., et al., Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chestnut B., Sumanas S., Zebrafish etv2 knock-in line labels vascular endothelial and blood progenitor cells. Dev. Dyn. 249, 245–261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi I., et al., Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512, 319–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rho S. S., et al., Rap1b promotes notch-signal-mediated hematopoietic stem cell development by enhancing integrin-mediated cell adhesion. Dev. Cell 49, 681–696.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Sumanas S., Lin S., Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 4, e10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schupp M. O., Waas M., Chun C. Z., Ramchandran R., Transcriptional inhibition of etv2 expression is essential for embryonic cardiac development. Dev. Biol. 393, 71–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abedin M. J., et al., Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ. Res. 114, 1690–1699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krebs L. T., et al., Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 (2000). [PMC free article] [PubMed] [Google Scholar]
- 46.Kim A. D., et al., Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 33, 2363–2373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh M., et al., Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Qian F., et al., Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 5, e132 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huber T. L., Kouskoff V., Fehling H. J., Palis J., Keller G., Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–630 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Vogeli K. M., Jin S. W., Martin G. R., Stainier D. Y., A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature 443, 337–339 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Jaffredo T., et al., Aortic remodelling during hemogenesis: Is the chicken paradigm unique? Int. J. Dev. Biol. 54, 1045–1054 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Tanaka Y., et al., Circulation-independent differentiation pathway from extraembryonic mesoderm toward hematopoietic stem cells via hemogenic angioblasts. Cell Rep. 8, 31–39 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Tanaka Y., et al., Early ontogenic origin of the hematopoietic stem cell lineage. Proc. Natl. Acad. Sci. U.S.A. 109, 4515–4520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lancrin C., et al., GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120, 314–322 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lie-A-Ling M., et al., Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development 145, dev149419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wythe J. D., et al., ETS factors regulate Vegf-dependent arterial specification. Dev. Cell 26, 45–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao P., et al., Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev. 34, 950–964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pimanda J. E., et al., Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. U.S.A. 104, 17692–17697 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westerfield M., The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio) (University of Oregon Press, 2000). [Google Scholar]
- 60.Parsons M. J., et al., Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang C., et al., m6 A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Villefranc J. A., Amigo J., Lawson N. D., Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077–3087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He S., et al., In vivo single-cell lineage tracing in zebrafish using high-resolution infrared laser-mediated gene induction microscopy. eLife 9, e52024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang Y. J., et al., Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development 123, 205–216 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Halloran M. C., et al., Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953–1960 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Tian Y., et al., The first wave of T lymphopoiesis in zebrafish arises from aorta endothelium independent of hematopoietic stem cells. J. Exp. Med. 214, 3347–3360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lauter G., Söll I., Hauptmann G., Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev. Biol. 11, 43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu S., et al., Two phenotypically and functionally distinct microglial populations in adult zebrafish. Sci. Adv. 6, eabd1160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picelli S., et al., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Samsa L. A., Fleming N., Magness S., Qian L., Liu J., Isolation and characterization of single cells from zebrafish embryos. J. Vis. Exp. 109), 53877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw fastq files for each sample reported in this paper and single-cell RNA-seq data are available in the NIH Gene Expression Omnibus database (accession no. GSE197757).