Significance
Microbes colonizing the infant gut during the first year(s) of life play an important role in immune system development. We show that after birth the (nearly) sterile gut is rapidly colonized by bacteria and their viruses (phages), which often show a strong cooccurrence. Most viruses infecting the infant do not cause clinical signs and their numbers strongly increase after day-care entrance. The infant diet is clearly reflected by identification of plant-infecting viruses, whereas fungi and parasites are not part of a stable gut microbiota. These temporal high-resolution baseline data about the gut colonization process will be valuable for further investigations of pathogenic viruses, dynamics between phages and their bacterial host, as well as studies investigating infants with a disturbed microbiota.
Keywords: infant, virota, microbiota, virome, transkingdom
Abstract
Virome and 16/18S analyses were performed on 304 longitudinal fecal samples of eight infants. The gut virota—the collection of all viruses present in the gut—was dominated by bacteriophages, which were nearly absent at birth and emerged rapidly within the first weeks after birth. Over 85% of phage reads correspond to 305 near-complete genomes, most of which (70.5%) were individual infant–specific, including two crAssphages, whereas 7.8% of phages were present in at least 50% of infants. Bacterial hosts could be predicted for 80% of phages, mainly infecting Firmicutes. Strong temporal correlations between phages and their predicted bacterial hosts were identified for >40% of our phages, and together with the observation of a decreasing fraction of phages with a temperate lifestyle further suggest that phages are induced from early-colonizing bacteria. The vast majority (>86%) of identified eukaryotic viruses, known to cause gastroenteritis, occurred without clinical signs, and an increase in the rate of infection occurred after day-care entrance. On average, 112 genomic contigs of distinct anelloviruses could be identified per infant, some of which were shed at >1 y. The identified plant viruses reflected the infant diet. Finally, the sporadic identification of fungi and parasites argues against the presence of such stable communities in the study population. Overall, this work provides a very high temporal resolution on how the different members of the infant gut microbiota, and especially the virome, develop over time in the gut of healthy infants, and might serve as valuable baseline knowledge for further studies investigating the effect of perturbations in the infant gut microbiota.
The human gut microbiota is a complex ecosystem, harboring members of several kingdoms of life, including animalia (parasites), fungi, protists, archaeabacteria, and bacteria, as well as viruses infecting all of these kingdoms in addition to the human host. All these complex interactions play an important role in health and disease (1). The bacterial component is by far the most studied and has been shown to be highly temporally stable in healthy adults (2, 3). Infants start their lives with a gut that is largely sterile (4), and prokaryotes colonize their intestinal tract in a stepwise manner (5, 6), until a stable adult-like composition is reached by the age of approximately 2 y (7). Disturbances in this primary colonization process can result in life-long health consequences and have been associated with a broad range of diseases, such as inflammatory bowel disease (IBD) (8), asthma (9), and type I diabetes (T1D) (10).
The assessment of the role of the viral component of the gut microbiota (i.e., gut virota), through the analyses of their collective genomes (i.e., the virome), in health and disease is lagging behind, but more and more associations with human diseases such as IBD (11, 12), T1D (13), and colorectal cancer (14) are being made. In terms of composition, adult and infant gut virota are dominated by bacteriophages (15–18), most of which have remained unidentified and are therefore referred to as “viral dark matter” (19, 20), complicating virome analyses. Members of different eukaryotic viral families (Adenoviridae, Anelloviridae, Astroviridae, Parvoviridae, Picornaviridae, and Reoviridae) have been described in healthy infant stool samples and, although most of them can cause human infections and disease, they are often observed in the absence of clinical signs (21, 22). Although evidence for a beneficial role of eukaryotic gut viruses in health is scarce, experiments in mice have shown a potential role of enteric viruses in the development of normal intestinal morphology and function (23). One of the most prevalent viral families of the eukaryotic virota in healthy infants is the family Anelloviridae (22, 24). This viral family has not been associated with any disease so far and a beneficial role of these viruses in human health has been suggested (25).
The role of bacteriophages in the gut virota and their link with human health remain understudied but, due to the transkingdom interactions between bacteriophages and their bacterial hosts, they are assumed to be crucial in shaping bacterial communities (26). Furthermore, their interaction dynamics remain unresolved and can only be unraveled in dense longitudinal datasets. The few studies on the virome in healthy infants and adults suggest a striking similarity to what was observed for prokaryotes: going from a (nearly) sterile composition at birth (18) toward an adult composition with a high temporal stability (15, 17, 27). Shkoporov and colleagues showed, in addition to temporal stability, also a strong individuality (17) in 10 adults sampled monthly for a period of 12 mo. Furthermore, they also conclude that phage populations are not showing classical lytic kill-the-winner dynamics in adults but rather a behavior consistent with a temperate phage lifestyle. The first longitudinal virome study in multiple healthy infants by Lim and colleagues suggested that, in contrast to increasing bacteriome diversity and richness over time, the highest richness, diversity, and abundance of phages were observed in the first months of life, followed by a significant decrease over time (22). The richness of eukaryotic viruses increased with age, suggesting that they are derived from environmental exposure (22). A recent study by Liang and colleagues investigated the infant gut virome at three time points after birth (month 0, 1, and 4) and observed a stepwise colonization of the infant gut. Samples from month 1 are dominated by phage particles induced from prophages integrated in pioneering bacteria and, in samples from month 4, eukaryotic viruses infecting human cells become more prominent (18).
For the eukaryotic component of the human gut microbiome (i.e., Eukaryota), even less is known. For particular fungi and parasites, a causal role in disease is well-appreciated (28). However, increasingly, these organisms are also considered to be commensals in the human gut and are even used in (probiotics) treatments for several diseases (29, 30). In healthy adults, the gut fungal composition [mainly composed of the phyla Ascomycota and Basiodiomycota (31)] seems very variable among individuals and over time (32, 33). Protists that have already been identified in healthy human stool samples include members of the Blastocystis, Entamoeba, and Trichomonas (34, 35). In infants, eukaryome abundance profiles have, in contrast to the stable stepwise colonization of prokaryotes, been shown to be unstable in early life (36). The lack of successional patterns and a stable presence of these eukaryotes in the infant gut led to the suggestion that in early life no stable eukaryotic community is formed (36). Some researchers even argue that such stable communities of gut eukaryotes are never formed and that they are only passengers via oral and dietary sources (37, 38).
In this study, we provide an in-depth analysis of gut microbiome dynamics, with a focus on the virome, in healthy infants during the first year of life. To study the virome, metagenomic sequencing was performed on purified virus-like particles (VLPs) of 304 samples from 8 infants (on average, 38 samples per infant), making this the most densely sampled healthy infant gut virome dataset to date. Most studies only look at a small fraction of their virome data, largely ignoring viral dark matter (19, 20). Using an elaborative bioinformatics viral characterization approach focusing not only on similarity-based methods but also including characterization at functional and specific genome structure levels (such as k-mer usage) allowed us to strongly reduce the amount of viral dark matter. To investigate transkingdom interactions, we overlaid our findings with the previously published 16S ribosomal RNA (rRNA) gene bacterial composition of these same samples (6). Furthermore, we also characterized the presence of eukaryotes using 18S rRNA gene sequencing. This study is unique because it studies gut community assembly across all kingdoms of life with a high temporal resolution.
Results and Discussion
Sample Selection.
Three hundred and four fecal samples were selected from eight healthy Belgian infants [BaBel cohort (6); SI Appendix, Fig. S1 and Dataset S1]. Shotgun sequencing was performed on purified VLPs. Quality-filtered reads were assembled into contigs, which were clustered into a set of nonredundant (NR) contigs. Of all the quality-filtered reads, 83% were found to be of viral origin (mapping to prokaryotic [71%] or eukaryotic [12%] viruses), with only a minor fraction being of bacterial or human origin (16 and 1%, respectively; Fig. 1A). The low percentage of contaminating bacterial reads in the infant data—which in adult samples is often much higher—most likely reflects the unique bacterial gut composition of the infants as reported before (6). Both 16S rRNA gene (6) and 18S rRNA gene sequencing were performed on the same fecal samples (using a different extraction protocol compared with the VLP sequencing; Methods).
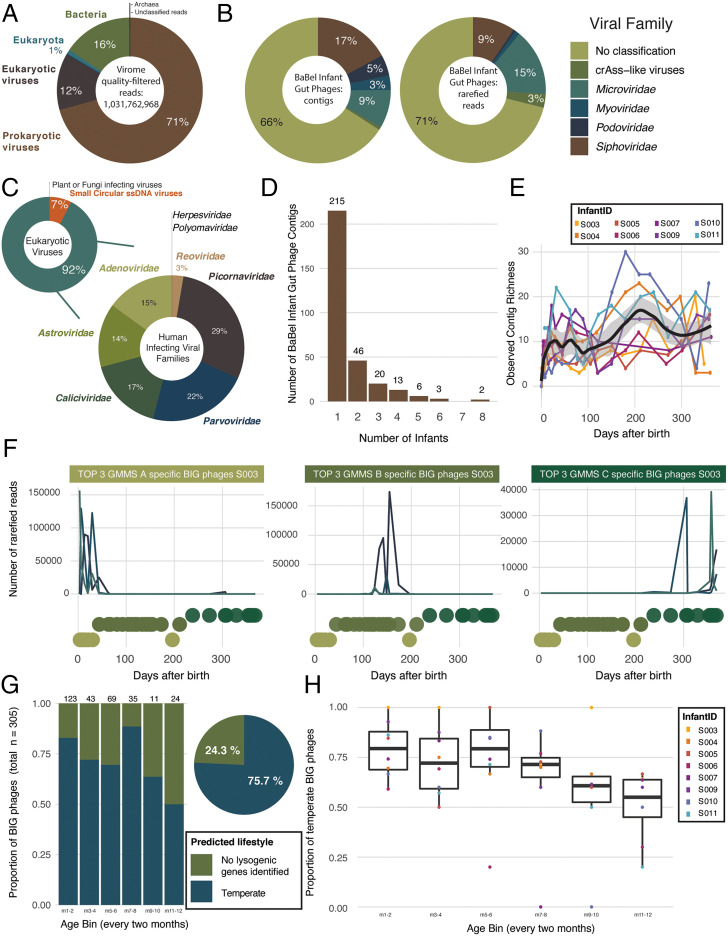
Fig. 1.
Members of the infant gut virome. (A) Overview of the distribution of the quality-filtered shotgun reads per classification category. On average, 70.7% of the quality-filtered shotgun reads represent prokaryotic viruses. (B) Taxonomic classification of the BIG phages in terms of number of reads (Left) and number of contigs (Right). (C) Overview of the taxonomic distribution of sequenced quality-filtered virome reads belonging to eukaryotic viruses. (C, Top Left) The taxonomy of all eukaryotic viruses is shown per category: human-infecting viruses, small circular ssDNA viruses, and FiVs and/or PiVs. (D) Sharing of the BIG phages by different infants. More than 70% of the BIG phages are individual infant–specific. (E) Richness of the BIG phages over time colored per infant shown here for the samples at predefined time points where the infants were not sick (n = 143; Loess smoothing with span equals 0.25). (F) Examples of BIG phages from infant S003 with a clear preference for a specific GMMS. Preferences for GMMS A are shown (Left) and for GMMS B (Middle) and GMMS C (Right). (G) Proportion of BIG phages according to the predicted lifestyle over different age bins. A BIG phage was assigned to a specific age bin based on the time point of its first detection. The numbers above the bars indicate the number of phages per age bin. (H) Proportion of temperate BIG phages over different age bins per infant separately. A BIG phage was assigned to a specific age bin based on the time point of its first detection per infant separately.
Bacteriophages Dominate the Infant Gut Virome, and the Eukaryotic Virome Is Dominated by Disease-Associated Viral Families.
Prokaryotic viruses were the most dominant member of the infant gut virome (represented by 71% of all the quality-filtered shotgun reads; Fig. 1A). Metagenomics analysis resulted in complete as well as partial genomes, which hampered reliable community-level analyses (39). Therefore, we used CheckV (40) to assess the completeness of the obtained phage genomes. Out of 3,199 phage contigs, 327 (10%) were predicted to be ≥50% complete (contig length range [2,339 to 165,703 nt], mean 35,770 nt), and only these were included in further analyses. For the analyses of the longitudinal dynamics, we only included phages present in at least two samples (which could be from the same infant or a different infant), resulting in 305 bacteriophage contigs. Despite this large reduction in phage contig number, they still represented the vast majority (>85%) of the quality-filtered rarefied bacteriophage reads. Rarefaction (i.e., a technique from numerical ecology that is often applied to operational taxonomic unit [OTU] analysis) was used to simulate even numbers of reads per sample in order to compensate for differences in sample sequencing depth. We will refer to this subset as the BaBel infant gut (BIG) phages. Assigning taxonomy to sequenced bacteriophages remains challenging (41), and only 34% of the BIG phages could be confidently assigned to a viral family (Fig. 1B). The family Siphoviridae was the most prevalent bacteriophage family but, in terms of proportional abundance, members of the Microviridae dominated (Fig. 1B).
On average, 12.3% of all quality-filtered reads represented eukaryotic viruses, mainly belonging to disease-associated mammal-infecting viruses (DaMiVs) (Fig. 1C and Table 1). All BaBel infants harbored on average eight different eukaryotic viral genera (range [5 to 10]) and six different eukaryotic viral families (range [4 to 7]) over their first year of life. Less abundant but very prevalent were the circular single-stranded DNA (ssDNA) viral families (e.g., Anelloviridae), which are regularly described in humans without any causal link to disease (25). Furthermore, a few plant- and fungus-infecting viruses (PiVs and FiVs) were identified (Table 1).
Table 1.
Overview of the eukaryotic viruses identified
| Family | Genus | Number of contigs | Number of reads | Eukaryotic viral reads, % | Number of positive samples | Positive samples, % | Number of positive infants | Positive infants, % | |
|---|---|---|---|---|---|---|---|---|---|
| DaMiVs | Adenoviridae | Mastadenovirus | 11 | 17,639,545 | 13.9 | 29 | 9.5 | 8 | 100.0 |
| Astroviridae | Mamastrovirus | 7 | 16,708,165 | 13.1 | 20 | 6.6 | 5 | 62.5 | |
| Caliciviridae | Norovirus | 8 | 10,285,164 | 8.1 | 24 | 7.9 | 7 | 87.5 | |
| Sapovirus | 3 | 9,413,164 | 7.4 | 12 | 4.0 | 5 | 62.5 | ||
| Herpesviridae | Roseolovirus | 2 | 422 | 0.0 | 1 | 0.3 | 1 | 12.5 | |
| Parvoviridae | Bocaparvovirus | 3 | 20,823,705 | 16.4 | 34 | 11.2 | 7 | 87.5 | |
| Dependoparvovirus | 1 | 5,498,014 | 4.3 | 7 | 2.3 | 2 | 25.0 | ||
| Picornaviridae | Cardiovirus | 4 | 3,912 | 0.0 | 2 | 0.7 | 1 | 12.5 | |
| Enterovirus | 35 | 16,049,833 | 12.6 | 85 | 28.0 | 8 | 100.0 | ||
| Kobuvirus | 4 | 2,311,755 | 1.8 | 3 | 1.0 | 1 | 12.5 | ||
| Parechovirus | 7 | 14,708,551 | 11.6 | 51 | 16.8 | 7 | 87.5 | ||
| Salivirus | 5 | 1,111,264 | 0.9 | 1 | 0.3 | 1 | 12.5 | ||
| Polyomaviridae | Deltapolyomavirus | 3 | 1,426 | 0.0 | 7 | 2.3 | 2 | 25.0 | |
| Reoviridae | Rotavirus | 13 | 3,144,009 | 2.5 | 42 | 13.8 | 7 | 87.5 | |
| PiVs | Endornaviridae | Alphaendornavirus | 5 | 39,728 | 0.0 | 3 | 1.0 | 3 | 37.5 |
| Luteoviridae | Polerovirus | 1 | 3,117 | 0.0 | 1 | 0.3 | 1 | 12.5 | |
| Partitiviridae | Alphapartitivirus | 1 | 245 | 0.0 | 1 | 0.3 | 1 | 12.5 | |
| Virgaviridae | Tobamovirus | 1 | 1,342 | 0.0 | 1 | 0.3 | 1 | 12.5 | |
| FiVs | Totiviridae | Unclassified | 1 | 11,218 | 0.0 | 5 | 1.6 | 3 | 37.5 |
| Small circular ssDNA | Anelloviridae | Alphatorquevirus | 6 | 74,174 | 0.1 | 30 | 9.9 | 6 | 75.0 |
| Betatorquevirus | 150 | 3,563,093 | 2.8 | 119 | 39.1 | 8 | 100.0 | ||
| Gammatorquevirus | 44 | 206,181 | 0.2 | 47 | 15.5 | 6 | 75.0 | ||
| Gyrovirus | 1 | 154 | 0.0 | 1 | 0.3 | 1 | 12.5 | ||
| Unclassified | 308 | 5,641,999 | 4.4 | 144 | 47.4 | 8 | 100.0 | ||
| Genomoviridae | Unclassified | 2 | 401 | 0.0 | 2 | 0.7 | 2 | 25.0 | |
| Unclassified CRESS (circular, rep-encoding ssDNA) viruses | Unclassified | 2 | 1,176 | 0.0 | 3 | 1.0 | 1 | 12.5 | |
| Circoviridae | Circovirus | 1 | 626 | 0.0 | 2 | 0.7 | 1 | 12.5 | |
| Unclassified | 11 | 12,596 | 0.0 | 27 | 8.9 | 5 | 62.5 | ||
| All eukaryotic viruses | 641 | 127,254,979 | 100.0 | 239/304 | 78.6 | 8 | 100.0 |
Highly Individual Phageome, but Also a Remarkable Sharing of Some Bacteriophages.
As has been described for adults (17), the infant gut showed a striking individuality, with more than 70% of the BIG phages being individual infant–specific. Strikingly, also 8% (n = 24) of the BIG contigs were present in half or more of the infants (Fig. 1D and Dataset S2), suggesting a partially conserved infant “gut core phageome,” despite the stringent cutoff criteria used to consider phages from different infants as shared [contigs were clustered at 95% nucleotide identity (41)]. Two BIG contigs (annotated as genus Skunavirus, family Siphoviridae) were shared by all eight infants (Fig. 1D and SI Appendix, Fig. S4 A and B), and were confirmed as Lactococcus phages (83.4 and 86.7% amino acid identity to GCA_003389775 and GCA_003389615, respectively), previously identified from dairy products. The abundances of both genomes were highly correlated with formula feeding, pointing to formula milk as a possible source of this phage [BaBel rarefied reads table (https://github.com/Matthijnssenslab/BabyGutVirome/tree/main/Data/ProkaryoticVirome/Rarefied); Mann–Whitney U test, n = 292, P value = 2.33e-12, R2 = 0.41; SI Appendix, Fig. S4C], which could explain the observed high infant sharing as well as their low abundances (SI Appendix, Fig. S4C). The presence of lactococcal phages has been described in the gut of Danish adults before (42). In these adults, the high prevalence of lactococcal phages was suggested to be linked to the high consumption of fermented milk products such as cheese and yogurt in Denmark. Comparing our BIG contigs with a human gut virome database (GVD) composed by Gregory et al. (43) resulted in clustering of 131 BIG phages (43%) to GVD contigs, making the majority (57%) of the BIG phages previously undescribed.
First Infant Gut Bacteriophages Are Recruited Rapidly during the First Weeks of Life.
Whether microbial colonization of the infant gut happens before, during, or after birth is still enigmatic and heavily debated (4). For the BIG phages, we observed a rapid increase in contig richness in the first weeks of life (Fig. 1E and SI Appendix, Fig. S5). Without implying sterility at birth, this pattern together with the very small number of viral reads obtained from samples from the initial days and weeks of life suggests that the initial acquisition of gut bacteriophages happened shortly after birth, in contrast to the decreasing bacteriophage richness observed by Lim et al. (22). This is in accordance with findings from Liang and colleagues showing very few or no VLPs in infant meconium or early stool samples, followed by an increasing overall VLP richness over the first months of life (18). After this rapid increase, we detected a slower increase toward the second part of the first year (peaking around day 200), after which the richness slightly decreased again to stabilize by the end of the first year (Fig. 1E and SI Appendix, Fig. S5). A possible reason for the richness peak and subsequent decrease toward a stable richness could be that in the infant gut a critical point is reached and no further colonization of bacteriophages is allowed [i.e., self-organized criticality (44)].
Most Bacteriophages Are Associated with a Single Gut Microbiota Maturation Stage.
As described previously (6), the BIG bacteriome matures through three distinct, conserved stages of ecosystem development, called gut microbiota maturation stages (GMMSs). These successional GMMSs were called GMMS A, B, and C, and reflect a strong temporal organization following a conserved pattern across infants. The bacterial genus predominance in these GMMSs was observed to shift from Escherichia over Bifidobacterium to Bacteroides, respectively. Surprisingly, 62% of our BIG phages were strictly associated with one of these stages, reflecting an association with a specific bacterial composition (Fig. 1F). Another 26% of the BIG phages were absent in the earliest (18%) or the latest GMMSs (8%), while only 12% of the BIG phages showed no preferential GMMS behavior. Note that this analysis cannot exclude that some of these phages were just nonreplicating, “passing through” the gastrointestinal tract for a short period of time (and so, by chance, associated with one of the GMMSs).
Most Early Bacteriophages Are Able to Adopt a Temperate Lifestyle.
Next, we investigated the lifestyle of the BIG phages. The vast majority (76%) of the BIG phages were predicted to have the possibility of a temperate lifestyle (Fig. 1G). There was a clear decreasing trend in the proportion of temperate phages toward the end of the first year, with a significant difference between the first and the last age bins (pairwise Fisher’s exact, month1-2:month11-12, false discovery rate [FDR]–adjusted P = 0.016; Fig. 1 G and H and Dataset S3). This observation is in line with the hypothesis that initial phages are induced from early-colonizing bacteria, as experimentally confirmed in samples from 1 mo of life (18). The fact that the fraction of temperate phages remained high during the first year, together with the correlation of the phages to specific bacterial GMMSs, suggests that over the first year, while colonization is still not complete, phages are continuously induced from the newly colonizing bacteria.
The Accumulation of DaMiVs Is Strongly Correlated with the Start of Day-Care Attendance.
In contrast to the colonization stages observed for the bacterial (6) population, DaMiVs showed short acute peaks of presence and were usually not detectable for long time periods (SI Appendix, Figs. S6 and S7). Of interest was the observation that members of the family Picornaviridae were among the first infecting DaMiVs for all infants (Dataset S4). In two of the eight BaBel infants, a wild-type Rotavirus A was detected as early as day 1 and 8 after birth, and the meconium sample of a third infant contained a herpesvirus (B6 betaherpesvirus)—as was also described previously by Liang and colleagues (18)—most likely derived nosocomially (45) or vertically (46). Interestingly, none of these early DaMiV infections was associated with clinical signs (Dataset S4). In general, a lower number of DaMiVs were observed during the first months of life, followed by an increase in subsequent months (SI Appendix, Fig. S7). The cumulative sum of individual DaMiV infections over time showed that, on average, the BaBel infants underwent 16 infections with DaMiVs in their first year of life (range [10 to 23]; Fig. 2A and SI Appendix, Fig. S8). Although viruses belonging to the DaMiV genera are known causative agents of gastroenteritis, 86% of their infections in the BaBel infants’ guts did not cause enteric signs such as vomiting or diarrhea, observed within 7 d after the start of the infection (excluding the live attenuated rotavirus vaccine strain; Fig. 2A and Dataset S5). Note that this percentage is very likely an underestimation of the percentage of asymptomatic enteric infections, since 1) symptomatic infections with multiple DaMiVs simultaneously (coinfections) were all counted separately as being associated with signs; 2) asymptomatic infections might have been missed in-between sampling points; and 3) the presence of a DaMiV in the presence of enteric signs does not prove a causal relationship. The shedding of enteric viruses with only minor and occasional symptoms has been reported previously using targeted PCRs (21, 47, 48).
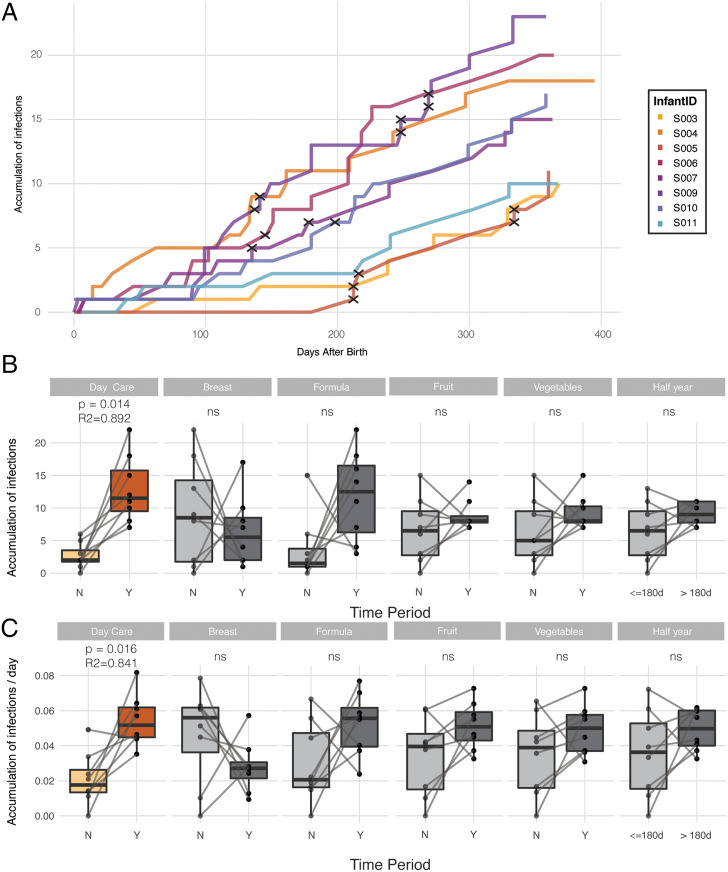
Fig. 2.
Overview of the accumulation of DaMiV infections in the healthy infant gut. (A) The accumulation over time of the number of unique infections by DaMiVs (on a species level) detected, colored per infant. Black crosses indicate infections for which an enteric sign (diarrhea or vomiting) was detected, within 7 d after the start of that infection. Note that for the calculation of the accumulation of DaMiV infections, reads attributed to live attenuated rotavirus vaccines were disregarded. (B) The number of accumulated infections before and after the start of specific events of interest (day-care entrance, breast milk, formula milk, fruit, vegetables, the first half-year) is shown in the boxplots and statistically compared using the paired Wilcoxon test. Nonsignificant results (P > 0.05) are indicated (ns). The number of accumulated infections before and after day-care entrance was found to be significantly different (paired Wilcoxon test, n = 8 + 8, R2 = 0.892, P = 0.014). The body of the boxplots represents the first and third quartiles of the distribution and the median line. (C) The number of accumulated infections per day (i.e., infection rate) before and after the start of specific events of interest (day-care entrance, breast milk, formula milk, fruit, vegetables, the first half-year) is shown in the boxplots and statistically compared using the paired Wilcoxon test. The infection rate before and after day-care entrance was found to be significantly different (paired Wilcoxon test, n = 8 + 8, R2 = 0.841, P = 0.016). The body of the boxplots represents the first and third quartiles of the distribution and the median line.
To investigate what triggered the increase in infections observed toward the second part of the first year, we compared the number of accumulated infections before and after events such as changes in diet (stopping of breastfeeding, starting of formula feeding or solid foods) or starting in day care. Only entrance into day care was found to be significantly associated (Fig. 2B; Wilcoxon signed-rank test, n = 8 + 8, R2 = 0.89, P = 0.014; Dataset S6), with the increase of acquired DaMiVs. Also, after normalizing for the timing of every event, a significant difference in infection rate (i.e., the number of infections per day) was only observed before and after the infants went into day care (Fig. 2C; Wilcoxon signed-rank test, n = 8 + 8, R2 = 0.84, P = 0.016; Dataset S6). More specifically, before day-care entrance, the DaMiV infection rate was on average 0.021 infections per day (range [0 to 0.049]), or 1 infection every 48 d, whereas after the start of day care this rate increased to an average of 0.054 infections per day (range [0.035 to 0.082]), or 1 infection every 18.5 d (Fig. 2C). No seasonal difference was observed in terms of viral richness or accumulation of DaMiV infections. Seasonality was not further taken into account in the analyses.
Anelloviruses Are Omnipresent, but Their Role Remains Unclear.
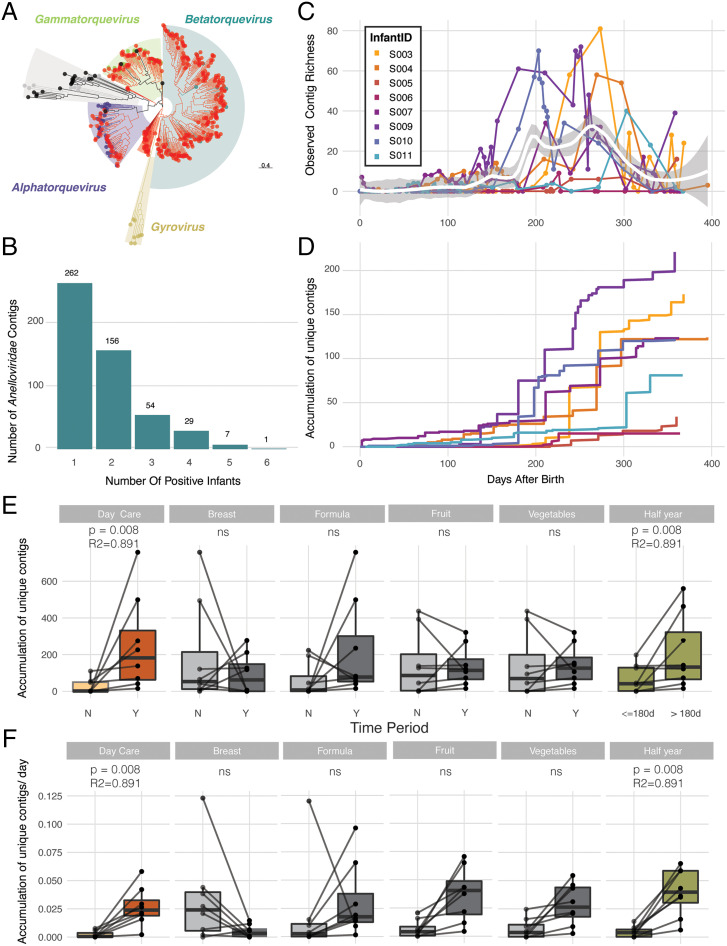
Although not very abundant (representing 7.5% of all eukaryotic viral reads), by far the most diverse eukaryotic viral family (79.5% of all eukaryotic viral contigs) was the family Anelloviridae, spanning the three known human-associated genera Alphatorquevirus, Betatorquevirus, and Gammatorquevirus (509 contigs; Fig. 3A and Table 1). This viral family is puzzling, since it has been detected in human blood, saliva, and stool; however, replication and passage of these viruses in cell culture have not been established and any causal link with human disease is lacking (25). Due to its high prevalence in healthy infants as well as immunosuppressed individuals or patients with inflammatory diseases, a role in shaping the immune system in early life is hypothesized (22, 24, 49). Anellovirus contigs were detected in all BaBel infants and in 48.7% of all BaBel samples. The majority of the members of the Anelloviridae were only present in one or two infants, whereas some contigs were present in up to six infants (Fig. 3B). The shedding of an anellovirus contig ranged from a single day to 374 d (average 25 d). The contig with the longest shedding (infant S004, from day 21 up to the last time point taken, at day 395) was present in 22 samples (out of 38), indicating a nearly constant shedding. This long-term shedding was not unique, as 25, 10, and 4 contigs were shed for periods longer than 100, 145, and 200 d, respectively. The richness of the contigs peaked between month 6 and 10 after birth, in concordance with previous reports (22) (Fig. 3C and SI Appendix, Fig. S9). Accumulation curves and rates for the anellovirus contig acquisition (Fig. 3 D–F and SI Appendix, Fig. S10) showed a much later onset compared with the accumulation of DaMiV infections (Figs. 2B and 4D). Both day-care attendance and time were found to be significantly correlated with the accumulation of unique anellovirus contigs toward the second part of the first year and their rate of accumulation (paired Wilcoxon tests, P < 0.01; Fig. 3 E and F and Dataset S7). The significance of day care for the accumulation of Anelloviridae contigs was very likely just a confounding effect for the significance of time (SI Appendix, Fig. S10). The source of the Anelloviridae contigs, their link with the developing immune system, its trigger for their increase in the second part of the first year, and the reason for the individual infant–specific patterns remain to be elucidated.
Fig. 3.
Overview of the detected members of the family Anelloviridae. (A) Phylogenetic distribution (based on the nucleotide alignment of ORF1) of the Anelloviridae contigs identified in this study (red) and 108 RefSeq anelloviruses downloaded from the NCBI (September 2019). Anelloviridae genera are colored as follows: Alphatorquevirus (purple), Betatorquevirus (blue), Gammatorquevirus (green), Gyrovirus (yellow), and unclassified Anneloviridae (gray). (B) Barplot showing the number of Anelloviridae contigs, shared by different infants. (C) Alpha-diversity measure (observed Anelloviridae contig richness) of the samples over the first year of life (Loess smoothing). (D) The accumulation over time of the number of unique Anelloviridae contigs, colored per infant. (E) The number of accumulated unique Anelloviridae contigs before and after the start of specific events of interest (day-care entrance, breast milk, formula milk, fruit, vegetables, first half-year) is shown in the boxplots and statistically compared using the paired Wilcoxon test. The number of accumulated unique Anelloviridae contigs before and after day-care entrance as well as before and after the first half-year was found to be significantly different (paired Wilcoxon test, n = 8 + 8, P < 0.01). The body of the boxplots represents the first and third quartiles of the distribution and the median line. (F) The number of accumulated unique Anelloviridae contigs per day (i.e., accumulation rate) before and after the start of specific events of interest (day-care entrance, breast milk, formula milk, fruit, vegetables, the first half-year) is shown in the boxplots and statistically compared using the paired Wilcoxon test. The number of accumulated unique Anelloviridae contigs per day before and after day-care entrance as well as before and after the first half-year was found to be significantly different (paired Wilcoxon test, n = 8 + 8, P < 0.01). The body of the boxplots represents the first and third quartiles of the distribution and the median line.
Fig. 4.
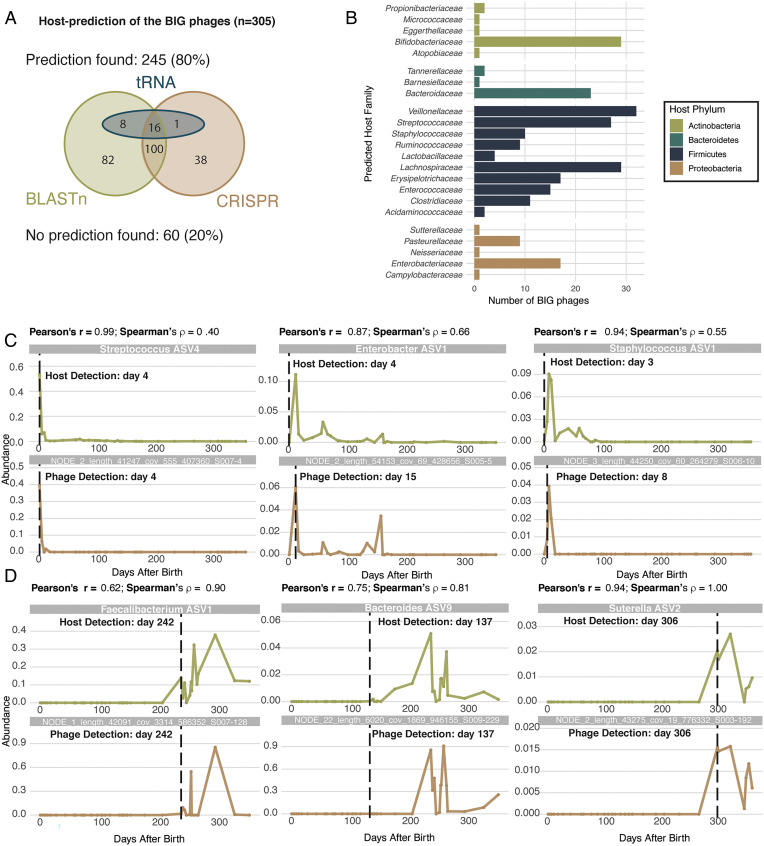
Transkingdom interactions of the BIG phages. (A) Host prediction of BIG phages according to the different in silico prediction methods used (host calling was based on CRISPR spacers, tRNAs, and BLASTn hits). (B) Distribution of the number of BIG phages for which the bacterial host could be predicted at the family level. (C) Examples of GMMS A–specific BIG phages for which the host could be successfully predicted and confirmed. The cooccurrence profiles of the bacteriophage (Bottom) and the bacterial host (Top) are shown over time. (D) Examples of GMMS C–specific BIG phages for which the host could be successfully predicted and confirmed. The cooccurrence profiles of the bacteriophage (Bottom) and the bacterial host (Top) are shown over time.
Transkingdom Interactions of the Infant Gut Bacteriophages.
Assuming that the phage–host relationships in the infant gut are mainly lysogenic, the abundances of the phages and the bacterial hosts from which they are induced should be temporally correlated. We first in silico predicted the possible bacterial hosts for the BIG phages by looking for CRISPR-spacer hits and transfer RNA (tRNA) hits complemented with information derived from BLASTn hits. For 80% of the BIG phages, a host could be predicted at family level (Fig. 4A). These predicted hosts were mainly represented by the bacterial families Bacteroidaceae, Bifidobacteriaceae, Lachnospiraceae, Streptococcaceae, and Veillonellaceae (Fig. 4B), all representing very abundant bacterial families detected in the BaBel infants’ guts by 16S sequencing (6) (Dataset S8).
To provide further support for the obtained host predictions, the phage abundances were correlated with the abundances of the amplicon-sequencing variants (ASVs) [from the 16S rRNA library (6)] of their predicted hosts (Spearman’s ρ, FDR-adjusted P < 0.05; Fig. 4B). For 51.4% of the phages with a predicted host, this prediction could be confirmed by co-occurrences (as exemplified in Fig. 4 C and D). For 94% of these confirmed host predictions, the bacteriophage was detected after or together with its predicted host (49 and 44%, respectively) and, only in 6% of cases, the bacteriophage was detected (shortly) before its predicted host. These highly correlated temporal dynamics further indicate that phage–bacteria relationships in the infant gut virome are primarily lysogenic in nature (18), whereas previous studies suggested that predator–prey dynamics were dominant (22). Note that in this study no lagged correlations were taken into account. Complex dynamics such as delayed responses or chaos could be a possible reason explaining the absence of a strong correlation between some bacteriophages and their predicted host.
CrAss-Like Viruses.
CrAssphage is the most prevalent bacteriophage associated with the human gut and is present in 87 to 100% of adult viromes (50, 51). In the BaBel cohort, reads of only two crAss-like viruses were found (96,618 and 97,133 nt in length; CheckV completeness scores of 100 and 97%, respectively) in 22 samples from two infants (14 of S006 and 8 of S010). CrAss-like viruses have been detected in infants starting from 1 mo after birth and vertical transmission from the mother has been suggested (52). Both contigs are individual infant–specific (i.e., only present in one infant) and the genera Bacteroides and Parabacteroides (phylum Bacteroidetes) were predicted as hosts (SI Appendix, Fig. S11), confirming previous studies (51, 53). Clustering both contigs to the recently composed crAss-like phage library containing 249 crAss-like phage genomes obtained from 702 human fecal samples from different studies (50) revealed that they belonged to candidate genera X and IV, including both viruses detected in samples from infants and adults.
Plant Viruses and Their Hosts.
Five eukaryotic PiVs were reported at low abundances, belonging to the plant-infecting families Endornaviridae, Luteoviridae, Partitiviridae, and Virgaviridiae (Table 1). All of these viruses were exclusively found in a single sample at a moment when the infants were weaned and, based on their annotation, they are clearly derived from the infants’ diet (infecting beans, melons, potatoes, carrots, peppers). Further validation was obtained by linking the 18S rRNA data. As could be expected, plant 18S rRNA gene reads in general (phylum Phragmoplastophyta) are very strongly correlated with weaning (Mann–Whitney U, P < 2.2e-16, R2 = 0.84; SI Appendix, Fig. S12). More specifically, plant 18S rRNA ASVs validated that four of the five above-mentioned fruits and vegetables (or at least close relatives) could be confirmed as part of the infant diet (Dataset S9). Only for the Pepper Mild Mottle Virus–positive sample was no presence of the pepper plant found in the 18S reads. This virus has been detected not only in peppers but also in products like dry spicy powders or sauces, possibly explaining the presence of the virus in the absence of the pepper (54).
One Divergent Fungus-Infecting Totivirus.
Only a single, highly divergent, fungus-infecting virus (family Totiviridae) was detected in three infants and five samples of the BaBel infants. This 5-kb-long viral contig encoded two open reading frames (ORFs), annotated as the major coat protein of Saccharomyces cerevisiae L-A Virus (InterProScan hit to pfam09220) and a viral RNA-directed RNA polymerase (57.6% amino acid similarity to Panax notoginseng Virus A [DIAMOND, YP_009225665.1]). In all five samples positive for this virus, Saccharomyces 18S rRNA gene reads are present on the same day or the day before, strongly suggesting that the host for this divergent totivirus belongs to the genus Saccharomyces. No correlation was found with the administration of the probiotics species Saccharomyces boulardii, which was administered to some infants to treat diarrhea, indicating that this fungus (and its associated totivirus) is likely derived from the diet or the environment.
Eukaryome.
Seven of the 18S ASVs were of fungal origin, belonging to the fungal phyla Ascomycota and Basidiomycota (Dataset S10). Only the fungal ASV corresponding to Saccharomyces was found to be present in all infants, but all samples with a high abundance of this ASV corresponded to treatment with probiotics or incidence of thrush. Unlike in the Human Microbiome Project (55), where no associations between adult mycobiome and metadata were found, the infant gut mycobiome of the BaBel infants read like a diary, strongly corresponding to the metadata variables and events such as probiotics administration, fungal infections, and antibiotic treatment (SI Appendix, Fig. S13). Our findings indicate a very low fungal diversity in early life, and lack of a “healthy core gut mycobiome” in early life, as was suggested before (36). Reasons for this may be threefold: 1) It does not exist in this cohort, 2) it develops later in life, or 3) it depends on the diet (bread, cheese, alcohol, etc.). 18S sequencing also identified a single protist, Cryptosporidium, from the Apicomplexa phylum, in infant S009, associated with diarrhea, vomiting, and fever, reported by the parents and known to be caused by this parasite (56) (SI Appendix, Fig. S14). This infection also caused a drastic change in bacterial gut composition and even caused a regression in GMMS (6). In other studies, using the exact same wet laboratory protocol and primers, different nonfungal microeukaryotes like Blastocystis and Entamoeba were detected, indicating that the lack of nonfungal microeukaryotes in the first year of life is not a technical artifact (35).
Conclusion
To conclude, this study closely tracks the infant gut community assembly across all its kingdoms. The dense sampling and simultaneous investigation of all kingdoms of life, together with optimized bioinformatics approaches, provided unique insight into this complex ecosystem. On different levels, complex interactions between all sorts of microbes and the environment were shown. Follow-up studies with a similar setup but an increased population size and looking at other geographical regions will be of great interest to investigate whether the observed interactions are a population- and worldwide phenomenon. For future research it would be interesting to take into account host immunity-related factors (e.g., antibody levels) since, in particular in early life, the link between the microbiome and the developing immune system is of crucial importance. The data presented in this work can serve as a “healthy” comparison set for other studies investigating infants with a suggested “disturbed” microbiome or enteric disease.
Methods
Sample Collection.
Between 2013 and 2017, stool samples of eight Belgian healthy infants, the BaBel infants, were collected starting from birth at a frequency of two or three samples per week (Dataset S1). Samples were kept in −20 °C freezers at the participants’ homes and every 3 mo transported to our laboratory on dry ice, where they were stored at −80 °C until further analysis. Every time a stool sample was collected, the parents completed a questionnaire containing information about the date of sample collection, the consistency of the stool (aqueous, soft, solid), diet (breast milk, formula milk, vegetables, fruit), clinical signs or disease (diarrhea, vomiting, listless, decreased appetite, fever, etc.), and the location of the infant when the sample was taken (at home, day care, holiday location, family stay). All infants were vaginally born, the mothers did not take antibiotics during pregnancy or delivery, and no complications during pregnancy were reported. The dataset only includes one male infant (i.e., S005), who showed the same succession in terms of GMMSs as the other infants (i.e., females) (6). Also, in terms of virome, no aberrant gender-specific observations could be made. Most likely, in adult microbiota and virota studies, gender is more important, as hormonal changes could influence the composition. In infants the effect of hormones is most likely negligible.
Ethics Approval and Consent to Participate.
The study was approved by the institutional review board at KU Leuven (ML8699, S54745, B322201215465). All legal guardians of the participants gave consent to participate in this study.
Sample Selection.
To study the longitudinal dynamics in the gut microbiome, 21 stool samples from predefined days 0, 3, 7, 10, 15, 21, 30, 45, 60, 75, 80, 105, 120, 150, 180, 210, 240, 270, 300, 330, and 360 were selected from each of the 8 infants (SI Appendix, Fig. S1). When an infant showed any clinical signs at any of these time points, we selected the closest available sample of the infant without clinical signs present, or this time point was excluded. In total, we included 143 samples at predefined time points (healthy BaBel subset). In addition, we selected 161 additional samples ad hoc from before, during, and after specific external events to study how they influence the gut microbiome. A number of specific external events were recorded in detail such as vaccination history, type of food consumed, occurrence of diseases, use of antibiotics, and use of pre- or probiotics (SI Appendix, Fig. S1).
To summarize, on average, 38 samples per infant (304 in total) were selected. Of these, 143 were selected because they fell together with one of the 21 predefined time points and the infants were not showing clinical signs at these time points. The other 161 samples fell together with different external factors (SI Appendix, Fig. S1 and Dataset S1).
Purification of Viral Particles, Preparation of Viral Libraries, and Sequencing.
All selected samples were enriched for viral particles using the NetoVIR protocol (57) modified by the fact that for homogenization the Precellys homogenizer was used (15 s at 5,000 rpm). In the random amplification step the WTA2 Kit was used. This kit was tested on a mock virome comprising viruses of different genome type (ssDNA, double-stranded DNA [dsDNA], ssRNA, dsRNA) and composition (linear, circular, segmented), and no strong preferential amplification (i.e., amplification bias) toward one of the genome types or compositions was observed (57). Therefore, no major amplification bias is expected in this study.
Four negative controls were included during the whole protocol (starting from phosphate-buffered saline) and sequenced afterward. Sequencing of the samples was performed on two NextSeq 500 runs (2 × 150 paired-end [PE] reads; Illumina).
Bioinformatics Analyses of Viral Reads.
After sequencing, the individual datasets were quality-trimmed using Trimmomatic (58). This quality trimming included the removal of WTA2 and Nextera XT adapters and cropping of the leading 19 bases and tailing 15 bases. Furthermore, reads were trimmed using a sliding window of 4 with a PHRED score cutoff of 20 with a minimum size of 50 bp. metaSPAdes (59) was used to create a set of contigs per sample using a de novo assembly of the trimmed reads (metagenomic setting with k-mer sizes of 21, 33, 55, and 77). Next, from these contig sets, one NR contig set was created by clustering all contigs longer than 500 nt at 95% nucleotide identity and 80% coverage using ClusterGenomes (60), which is based on nucmer (61). The decontam (62) R package was used to remove contaminating contigs using the prevalence mode based on the assumption that contaminating sequences are prevalent in the negative-control samples. The NR contig set was compared against the National Center for Biotechnology Information (NCBI) Nucleotide database using BLASTn and against an NR protein sequence database using DIAMOND (63) for taxonomic annotation [with the lowest-common ancestor approach assigned by KronaTools (64)]. Finally, Kraken 2 (65) was used to filter out contigs mapping the human genome (default settings, with “confidence” set at 0.05 as recommended by the authors). For the identification of bacteriophages from the metagenomics data, genes from the NR contig set were predicted using Prodigal (66) (anonymous mode). The identified proteins were assigned prokaryotic virus orthologous groups (pVOGs) using HMMsearch (E value < 1e-5; http://hmmer.org/). Additionally, orthologous groups were identified in these proteins using eggNOG-mapper (67) to the viral database (default settings) and InterProScan (68) was used for further functional characterization of the proteins (default settings).
All contigs annotated as eukaryotic virus by DIAMOND and/or BLASTn were extracted (i.e., “eukaryotic viral contigs”). To identify bacteriophage contigs, a scoring scheme was developed. At four different levels, contigs were scored and if for an individual test a hit was found, a score of 1 was given, as explained in the schematic in SI Appendix, Fig. S2. First, homology-based classification was performed at the nucleotide level and at the protein level. A point at the nucleotide level was given if BLASTn annotated the contig as bacteriophage with E < 1e-10 or if MetaPhinder2 (69) reported an average nucleotide identity (ANI) >10% (only if length >2,500 nt), and another point was given at the protein level if DIAMOND (63) (with the “sensitive” option) or CAT (70) annotated the contig as bacteriophage. Second, a score of 2 was given based on the genome structure of the contig defined at the k-mer level [DeepVirFinder (71) score > 0.95 and P < 0.01] and a gene-to-length ratio of above 1.6 per kilobase. Third, at the functional level, the presence of virus-specific hallmark genes (Dataset S11) was scored, as well as a pVOG/gene ratio above 0.6. Fourth, all contigs were scored based on their VirSorter (72) category. Finally, a contig was classified as bacteriophage if it scored 3 or more of 8 or if its VirSorter category was 1 or 2. For additional taxonomic classification of the bacteriophages, vConTACT2.0 was run with references v88 (73). Bacteriophages falling in a vConTACT cluster with one of the reference strains were given the taxonomy of this reference. For final functional annotation of the BIG phages, Cenote-Taker 2 (74) was run (https://github.com/mtisza1/Cenote-Taker2.git).
To obtain relative abundances per sample, trimmed reads were mapped to a subset of the NR dataset using BWA-MEM (75). This subset consisted of cluster representatives of only those clusters containing at least one contig of this particular sample. A contig was assumed to be present if 70% of the length was covered with reads. Contigs with only 150 reads across all samples were removed from the NR contig set.
In total, for the virome analysis, 304 samples were sequenced, resulting in 1,617,700,980 raw PE reads (on average 5,321,384.8 reads per sample). After all quality control and filtering steps, we obtained an NR contig set containing 67,104 contigs to which 1,082,569,573 reads mapped in total (on average 3,561,084 reads per sample) and after decontamination 1,031,762,968 reads remained. Only few reads were mapped for all the samples collected in the first 2 d of life, ranging from 6,461 to 152,061 reads. For analysis performed at the read or abundance level (not at the presence/absence level), correction of sequencing depth was performed by rarefying all virome reads to 180,000 reads per sample (12 samples were excluded due to an insufficient number of reads; 6 were samples from the first days of life and the 6 other samples were ones with high bacterial or host contamination).
The size distribution of our 3,199 identified phage contigs ranges from 538 to 165,703 nt (mean 7,692) and the skewness toward the smaller fragments indicates the presence of a large fraction of fragmented genomes, similar to other viral metagenomic datasets (17). The quality of phage contigs was assessed using CheckV (v0.6.0), which assesses both the completeness of the contigs as well as the presence of host (bacterial) contamination (40). As expected, completeness score was correlated with both the contig length and the contig read count, indicating that for low abundant contigs there were too few reads to reach a complete assembled phage genome (CheckV completeness vs. contig length: CheckV completeness vs. 10log [rarefied contig read count], n = [2,905:2,905], Pearson correlation, r = [0.85:0.77], P = 2.2e-16; SI Appendix, Fig. S3). For community-level analyses, only the bacteriophages with a CheckV completeness score higher or equal to 50% were used (n = 327). Additionally, singleton phages (only present in one sample) were removed, resulting in a core phage set of 305 bacteriophages, BIG phages, representing 85% of all quality-filtered rarefied bacteriophage reads. For the final taxonomic assignment of the BIG phages, a combination of similarity-based searches to the NCBI Nucleotide and NR databases, CheckV database, and vConTACT2 clustering to references was used.
Comparison of the Bacteriophage Contigs with Other Datasets.
The 305 BIG phages were clustered into two different datasets, the GVD (43) and a crAss-like virus dataset (50), at 95% nucleotide identity and 80% coverage using ClusterGenomes (60).
Bacteriophage Lifestyle Prediction.
The determination of the lifestyle of the phages was based on the presence of some lysogeny-specific genes (repressor proteins, integrases, excisionases, other lysogenization-associated proteins; Dataset S12) as indicators of a lysogenic lifestyle of the phage.
Bacteriophage Host Prediction.
Hosts were predicted for the bacteriophages in silico by looking for CRISPR-spacer hits and tRNA hits, as published before (76), using an in-house-created bacterial RefSeq database (containing all available RefSeq genomes belonging to the six bacterial phyla detected in the infant gut, namely Firmicutes, Actinobacteria, Proteobacteria, Bacteroides, Fusobacteria, and Verrumicrobia, that were available in August 2019). Furthermore, BLASTn annotations were used for the annotation as well (e.g., a phage contig annotated as “Enterococcus phage [viral]” very likely has Enterococcus as a host, but also a contig annotated as “Enterococcus [bacterial]” is very likely to infect this bacterial genus).
16S rRNA Gene Library Preparation, Sequencing, and Read Analysis.
Bacterial profiling has been published before (6), and was carried out as described by Falony and colleagues (77). Briefly, nucleic acids were extracted from frozen fecal aliquots using the RNeasy PowerMicrobiome Kit (Qiagen). The manufacturer’s protocol was modified by the addition of a heating step at 90 °C for 10 min after vortexing and by the exclusion of DNA-removal steps. Microbiome characterization was performed as previously described (78); in short, the extracted DNA was further amplified in triplicate using 16S primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) targeting the V4 region, modified to contain a barcode sequence between each primer and the Illumina adaptor sequences to produce dual-barcoded libraries. Deep sequencing was performed on a MiSeq platform (2 × 250 PE reads; Illumina). All samples were randomized and negative controls were included and sequenced. Further read analysis was carried out as described in Beller et al. (6).
18S rRNA Gene Library Preparation, Sequencing, and Read Analysis.
For 18S rRNA gene characterization, the extracted DNA (obtained from frozen fecal aliquots in a similar way as for the 16S rRNA gene characterization) was amplified in duplicate using 18S primers targeting the V9 region, using the pair of primers 1389F (5′-TTGTACACACCGCCC-3′) and 1510R (5′-CCTTCYGCAGGTTCACCTAC-3′) (79), and deep sequencing was performed on a MiSeq platform (2 × 250 PE reads; Illumina). The dada2 R package was used for the analysis of the reads and the decontam R package was used to remove contaminating sequences, similar as for the 16S rRNA gene reads. Taxonomy was assigned using the Silva (silva_nr_v128_train_set) and PR2 (pr2_version_4.10.0) databases formatted for DADA2.
For the abundance analysis of the eukaryotic microbiome in the infant stool samples, we additionally removed singleton ASVs and ASVs representing less than 0.005% of the reads in all samples after decontamination. After all these filtering steps, still 99.5% of the initial reads were kept. However, 82% of the ASVs were discarded, indicating their very low abundance. Only seven of the ASVs were of fungal origin, belonging to the fungal phyla Ascomycota and Basidiomycota.
Statistical Analyses.
All statistical analyses were performed and visualized in R (http://www.R-project.org) using the ggplot2 (80), genoPlotR (81), phyloseq (82), dunn.test (83), and vegan (84) packages. To test median differences between two or more groups of continuous variables (alpha diversity measures, abundances, etc.), a Mann–Whitney U test and Kruskal–Wallis (KW) test were performed, respectively. The KW test was always followed by a post hoc Dunn’s (phD) test for all pairs of comparisons between groups. Multiple testing correction was performed where appropriate using the Benjamini–Hochberg procedure (FDR adjustment set at <0.05). Observed richness was calculated by using the phyloseq (82) package.
Longitudinal Dynamics of the BIG Phages.
For the characterization of the longitudinal profile of the BIG phages, the phage abundances were linked to the GMMS identified before (6). Since infant S011 showed a colonization distinct from the other infants, all phages only present in S011 were excluded from the longitudinal analyses (n = 25). Additionally, two phages were excluded, since they were not present in at least two samples from at least one infant. For the 278 remaining BIG phages, we identified which ones had a specific preference for one of the three GMMSs, A, B, and C; which had preference for the early GMMSs, A and B; which had preference for the late GMMSs, B and C; and which were not related to the GMMSs, by testing four different criteria (stepwise). The first criterion was an exclusive presence of the phage in only samples belonging to one of the three GMMSs; the second criterion was a significant KW with phD test indicating a significant predominance for one of the three GMMSs compared with the others (P < 0.05 and FDR < 0.05); the third criterion was exclusive absence in only samples belonging to one of the three GMMSs; and the fourth criterion was a significant KW with phD test indicating significantly lower abundances in one of the three GMMSs compared with the others (P < 0.05 and FDR < 0.05). Note that the KW with phD tests were individually tested per infant, meaning that if a phage is present in multiple infants, the KW with phD test should be significant and result in the same GMMS preference for all of the infants. The distribution of the GMMS preference for the 278 BIG phages was as follows: 71 GMMS A, 27 GMMS B, 74 GMMS C, 22 GMMSs A–B, and 51 GMMSs B–C, and 33 BIG phages were unrelated to the GMMSs.
To confirm the host predictions for the BIG phages, bacteriophage abundances were linked to the 16S ASV abundances per infant individually and Pearson’s r as well as Spearman’s ρ were calculated. A host prediction for which Spearman’s ρ was significant (P < 0.05 and FDR-adjusted P < 0.05) between the phage and an ASV from the predicted bacterial family was assumed confirmed.
Calculation of the Accumulation of DaMiV Infections.
The cumulative sum of DaMiV infections was measured per infant and is an indication of how many infections (or detections) with unique DaMiVs were observed in the samples of a specific infant within a specific time period. With the term “unique infection,” we mean that if the same virus (i.e., viral contig or multiple viral contigs in the case of a segmented virus) is present for subsequent samples, it is only counted as one infection, and also that if multiple different DaMiVs are detected in one sample, this is counted as multiple infections.
Phylogeny and Abundance Analysis of the Anelloviridae Contigs.
Protein alignments of ORF1 of the Anelloviridae contigs identified in this study and the 108 known RefSeq anelloviruses downloaded from the NCBI (September 2019) were built using MAFFT (85) and trimmed using trimAl (86) (gappyout setting). Model prediction and tree creation were obtained using IQ-TREE (87, 88) (bootstrap values with 1,000 replicates). Visualization was performed using the ggtree R package (89).
The duration of the presence (i.e., shedding) of an Anelloviridae contig was measured as the number of days between the first and last observations (for every contig, the infant with the longest shedding period was used).
Genotyping Rotavirus Segments.
Rotavirus segments were genotyped using the Virus Pathogen Database and Analysis Resource (ViPR) Rotavirus A genotype determination tool (https://www.viprbrc.org/brc/rvaGenotyper.spg?method=ShowCleanInputPage&decorator=reo.
Supplementary Material
Acknowledgments
We thank all participating infants and parents for their contribution. We thank Dr. Johan Nordgren for the characterization of the saliva samples. This research was supported by the Fonds Wetenschappelijk Onderzoek (FWO; Research Foundation Flanders) (L.B.: 1S61618N; R.Y.T.: 1234321N; B.V.: 1S28617N; L.V.E.: 1S25720N; D.J.: 1S78021N) and by a KU Leuven Grant (OT-14-113). The computational resources for this work were provided by the Flemish Supercomputer Center (VSC) and funded by the FWO and the Flemish Government Department of Economy, Science and Innovation (EWI).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114619119/-/DCSupplemental.
Data Availability
Virome-sequencing data and metadata used in this study have been deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA693793/) after removal of all human reads (using the Bowtie2 sensitive mode, mapping to reference genome hg38). The code to perform the analyses and create figures starting from the ASV abundance table has been made available at GitHub (https://github.com/Matthijnssenslab/BabyGutVirome/). Previously published data were used for this work (https://doi.org/10.1128/mbio.01857-21).
References
- 1.Wang B., Yao M., Lv L., Ling Z., Li L., The human microbiota in health and disease. Engineering 3, 71–82 (2017). [Google Scholar]
- 2.Poyet M., et al. , A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019). [DOI] [PubMed] [Google Scholar]
- 3.David L. A., et al. , Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Muñoz M. E., Arrieta M.-C., Ramer-Tait A. E., Walter J., A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 5, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart C. J., et al. , Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beller L., et al. , Successional stages in infant gut microbiota maturation. mBio 12, e0185721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T., et al. , Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis J. D., et al. , Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 18, 489–500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta M.-C., et al. , Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 142, 424–434.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vatanen T., et al. , The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadwell K., et al. , Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman J. M., et al. , Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao G., et al. , Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc. Natl. Acad. Sci. U.S.A. 114, E6166–E6175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannigan G. D., Duhaime M. B., Ruffin M. T. IV, Koumpouras C. C., Schloss P. D., Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9, e02248-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes A., et al. , Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minot S., et al. , The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shkoporov A. N., et al. , The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe 26, 527–541.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Liang G., et al. , The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581, 470–474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux S., Hallam S. J., Woyke T., Sullivan M. B., Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife 4, e08490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beller L., Matthijnssens J., What is (not) known about the dynamics of the human gut virome in health and disease. Curr. Opin. Virol. 37, 52–57 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Kapusinszky B., Minor P., Delwart E., Nearly constant shedding of diverse enteric viruses by two healthy infants. J. Clin. Microbiol. 50, 3427–3434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim E. S., et al. , Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kernbauer E., Ding Y., Cadwell K., An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes A., et al. , Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. U.S.A. 112, 11941–11946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaczorowska J., van der Hoek L., Human anelloviruses: Diverse, omnipresent and commensal members of the virome. FEMS Microbiol. Rev. 44, 305–313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Sordi L., Lourenço M., Debarbieux L., The battle within: Interactions of bacteriophages and bacteria in the gastrointestinal tract. Cell Host Microbe 25, 210–218 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Minot S., et al. , Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U.S.A. 110, 12450–12455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukeš J., Stensvold C. R., Jirků-Pomajbíková K., Wegener Parfrey L., Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 11, e1005039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanello G., Meurens F., Berri M., Salmon H., Saccharomyces boulardii effects on gastrointestinal diseases. Curr. Issues Mol. Biol. 11, 47–58 (2009). [PubMed] [Google Scholar]
- 30.Williamson L. L., et al. , Got worms? Perinatal exposure to helminths prevents persistent immune sensitization and cognitive dysfunction induced by early-life infection. Brain Behav. Immun. 51, 14–28 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann C., et al. , Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS One 8, e66019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallen-Adams H. E., Kachman S. D., Kim J., Legge R. M., Martínez I., Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 15, 9–17 (2015). [Google Scholar]
- 33.Raimondi S., et al. , Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping, and colonization. Front. Microbiol. 10, 1575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parfrey L. W., Walters W. A., Knight R., Microbial eukaryotes in the human microbiome: Ecology, evolution, and future directions. Front. Microbiol. 2, 153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tito R. Y., et al. , Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 68, 1180–1189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wampach L., et al. , Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front. Microbiol. 8, 738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suhr M. J., Hallen-Adams H. E., The human gut mycobiome: Pitfalls and potentials—A mycologist’s perspective. Mycologia 107, 1057–1073 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Auchtung T. A., et al. , Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3, e00092-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quince C., Walker A. W., Simpson J. T., Loman N. J., Segata N., Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 35, 833–844 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Nayfach S., Camargo A. P., Eloe-Fadrosh E., Roux S., Kyrpides N., CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2021). [DOI] [PMC free article] [PubMed]
- 41.Roux S., et al. , Minimum information about an uncultivated virus genome (MIUViG). Nat. Biotechnol. 37, 29–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waller A. S., et al. , Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 8, 1391–1402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory A. C., et al. , The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe 28, 724–740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solé R. V., Alonso D., McKane A., Self-organized instability in complex ecosystems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 667–671 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koukou D., et al. , Rotavirus gastroenteritis in a neonatal unit of a Greek tertiary hospital: Clinical characteristics and genotypes. PLoS One 10, e0133891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall C. B., et al. , Congenital infections with human herpesvirus 6 (HHV6) and human herpesvirus 7 (HHV7). J. Pediatr. 145, 472–477 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Ye S., et al. , Detection of viruses in weekly stool specimens collected during the first 2 years of life: A pilot study of five healthy Australian infants in the rotavirus vaccine era. J. Med. Virol. 89, 917–921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hebbelstrup Jensen B., et al. , Children attending day care centers are a year-round reservoir of gastrointestinal viruses. Sci. Rep. 9, 3286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCann A., et al. , Viromes of one year old infants reveal the impact of birth mode on microbiome diversity. PeerJ 6, e4694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guerin E., et al. , Biology and taxonomy of crAss-like bacteriophages, the most abundant virus in the human gut. Cell Host Microbe 24, 653–664.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Dutilh B. E., et al. , A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siranosian B. A., Tamburini F. B., Sherlock G., Bhatt A. S., Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat. Commun. 11, 280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shkoporov A. N., et al. , ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9, 4781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colson P., et al. , Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One 5, e10041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nash A. K., et al. , The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerace E., Lo Presti V. D. M., Biondo C., Cryptosporidium infection: Epidemiology, pathogenesis, and differential diagnosis. Eur. J. Microbiol. Immunol. (Bp) 9, 119–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conceição-Neto N., et al. , Modular approach to customise sample preparation procedures for viral metagenomics: A reproducible protocol for virome analysis. Sci. Rep. 5, 16532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nurk S., Meleshko D., Korobeynikov A., Pevzner P. A., metaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolduc B., Roux S., Clustering Viral Genomes in iVirus (The Ohio State University, 2017). https:// 10.17504/protocols.io.gwebxbe. Accessed 1 September 2020. [DOI]
- 61.Delcher A. L., Salzberg S. L., Phillippy A. M., Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics chap. 10, unit 10.3 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Callahan B., Davis N. M., decontam: Identify Contaminants in Marker-Gene and Metagenomics Sequencing Data (2019). https://bioconductor.org/packages/decontam/. Accessed 1 September 2020.
- 63.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Ondov B. D., Bergman N. H., Phillippy A. M., Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12, 385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wood D. E., Lu J., Langmead B., Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huerta-Cepas J., et al. , Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell A. L., et al. , InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jurtz V. I., Villarroel J., Lund O., Voldby Larsen M., Nielsen M., MetaPhinder—Identifying bacteriophage sequences in metagenomic data sets. PLoS One 11, e0163111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Meijenfeldt F. A. B., Arkhipova K., Cambuy D. D., Coutinho F. H., Dutilh B. E., Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ren J., et al. , Identifying viruses from metagenomic data using deep learning. Quant. Biol. 8, 64–77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang H. B., et al. , Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol. 37, 632–639 (2019). [DOI] [PubMed]
- 74.Tisza M. J., Belford A. K., Domínguez-Huerta G., Bolduc B., Buck C. B., Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus Evol. 7, veaa100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997 (Accessed 16 March 2013).
- 76.Deboutte W., et al. , Honey-bee-associated prokaryotic viral communities reveal wide viral diversity and a profound metabolic coding potential. Proc. Natl. Acad. Sci. U.S.A. 117, 10511–10519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falony G., et al. , Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Tito R. Y., et al. , Brief Report: Dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheumatol. 69, 114–121 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Amaral-Zettler L. A., McCliment E. A., Ducklow H. W., Huse S. M., A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4, e6372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wickham H., et al. , ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics (R package version 2, 2019). https://CRAN.R-project.org/package=ggplot2. Accessed 1 September 2020.
- 81.Guy L., Kultima J. R., Andersson S. G. E., genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McMurdie P. J., Holmes S., Jordan G., Chamberlain S., Package ‘phyloseq’: Handling and Analysis of High-Throughput Microbiome Census Data (2014). https://rdrr.io/bioc/phyloseq/. Accessed 1 September 2020.
- 83.Dinno A., dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums (2017). https://CRAN.R-project.org/package=dunn.test. Accessed 1 September 2020.
- 84.Oksanen J., et al. , vegan: Community Ecology Package (2019). https://cran.r-project.org/web/packages/vegan/index.html. Accessed 1 September 2020.
- 85.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T., ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Virome-sequencing data and metadata used in this study have been deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA693793/) after removal of all human reads (using the Bowtie2 sensitive mode, mapping to reference genome hg38). The code to perform the analyses and create figures starting from the ASV abundance table has been made available at GitHub (https://github.com/Matthijnssenslab/BabyGutVirome/). Previously published data were used for this work (https://doi.org/10.1128/mbio.01857-21).