“Everything has an appointed season, and a time for every activity under heaven” (Ecclesiastes 3:1). To better satisfy the demands of living in a cyclical world, organisms from bacteria to humans contain genetically encoded instructions for producing biological time-keeping devices, known as circadian (approximately 24 h) clocks. The first circadian clock gene identified was period (per) in Drosophila (fruit flies) (1), shortly thereafter followed by frequency (frq) in fungi (Neurospora) (2). Later work showed that mammalian clocks are also PER based, having three types (Per1, 2, and 3). PER and FRQ proteins share little amino acid sequence similarity but follow a remarkably similar 24-h life cycle (reviewed in refs. 3–5). Each day is marked by a predictable upswing in the production of newly synthesized PER and FRQ proteins that slowly undergo progressive increases in the addition of negatively charged phosphate groups (a process called phosphorylation) until some 18 to 20 h later when they reach maximally hyperphosphorylated states, a signal that triggers their rapid degradation. This 24-h choreography of phosphorylation/degradation gates when and for how long PER and FRQ function as negative regulators of their own gene expression and that of downstream genes. The integrated circuitry motors negative autoregulatory feedback loops that help amplify daily PER and FRQ rhythms, while also orchestrating molecular oscillations in downstream gene expression that underlie many of the circadian rhythms expressed by life forms. Strikingly, the main kinases catalyzing the slow progressive phosphorylation of PERs and FRQ are members of the casein kinase 1 family (CK1δ/ε in animals and CK1a in Neurospora). Perhaps fittingly, the first mammalian clock mutant identified, termed tau, turned out to be in CK1ε (6). Several human sleep disorders are caused by a mutation in either a PER phosphorylation site or CK1δ (4). Although CK1 is a clock prima donna, other kinases, phosphatases, and numerous regulatory factors modulate the PER/FRQ phosphorylation programs and clock speed, implying the assembly of many moving parts in the cellular workshops of fungi to humans. In PNAS, Marzoll et al. (7) strip down the role of CK1 in generating PER and FRQ phosphorylation programs observed in vivo by reconstituting some of its core attributes in a test tube, a technical breakthrough leading to several unexpected findings.
Acting alone, CK1 goes beyond first impressions of no sequence similarity to drive daily phospho-timing from functionally equivalent modules present in PER and FRQ. The contents within these test tubes also reveal additional biochemical wizardry by maintaining a similar daily rate of progressive phosphorylation even at different temperatures, suggesting this minimal in vitro system is embedded with temperature compensation, one of the holy grails in circadian research and a valuable feature since dawn arises at the same time even if a cold front sweeps in during the night. Surprisingly, the shared timing modules in phospho-timers from bread mold to animals appear to be based on long stretches of disordered regions that also contain a multitude of suboptimal phosphorylation sites. Given this landscape, the binding of CK1 to its substate suffices to drive the slow progressive phosphorylation of suboptimal sites, an overall pace that reliably stretches to a daily time scale despite CK1’s rolling the dice as to which and when any particular site is phosphorylated—not exactly the “quartz crystal” timer described earlier in the cyanobacteria clock (8, 9).
To better appreciate the findings described by Marzoll et al. (7), years of work left unresolved the sufficiency for CK1 family members in the nearly day-long conversion of non- to hyperphosphorylated PER and FRQ, although it is clear that this conversion is necessary for 24-h timing. Casein kinase 1 family members phosphorylate sites that are prephosphorylated at the −3 position with much higher efficiency compared to the suboptimal nonprimed sites (10). There are some 30 to 60 sites phosphorylated on Drosophila and mammalian PERs, whereas FRQ has about 100 (e.g., refs. 11–14). Almost all are exclusively at serine (Ser) and threonine (Thr) residues, consistent with CK1 family members, but also with many other Ser/Thr kinases. The vast majority of phosphorylation sites on PER and FRQ are unlikely to be priming dependent and do not match consensus CK1 sites. Nonetheless, CK1 is rather promiscuous and, importantly, PERs and FRQ have centrally located CK1 binding sites, which could facilitate the ability of CK1 to directly phosphorylate the majority of the available phospho-acceptor sites despite being suboptimal.
In PNAS, Marzoll et al. strip down the role of CK1 in generating PER and FRQ phosphorylation programs observed in vivo by reconstituting some of its core attributes in a test tube, a technical breakthrough leading to several unexpected findings.
Yet, knocking out a single priming site can have strong effects on clock speed. A familial advanced sleep phase syndrome (FASPS) is due to such a priming-site mutation on human Per2 (hPer2), causing the clock to run fast, and affected individuals are sleepy by early afternoon (reviewed in refs. 4 and 15). In this FASPS multisite phospho-module, CK1δ/ε is likely to phosphorylate both the priming site and the nearby downstream sites, but other kinases can also act as priming kinases to stimulate nearby phosphorylation by CK1, such as in the Drosophila PER-short phospho-cluster (12). However, while knocking out a phospho-switch can speed up or slow down the progressive phosphorylation of PERs and FRQ, it does not eliminate the eventual march toward hyperphosphorylation. This leaves unclear how much of the PER and FRQ hypophosphorylation-to-hyperphosphorylation transition observed in vivo is directly due to CK1 kinase activity and dependent on priming.
Marzoll et al. (7) take the biochemist approach, aiming to reconstitute progressive phosphorylation of PER and FRQ with a daily time scale using purified components mixed in a test tube. The degree of overall FRQ and PER phosphorylation was monitored by electrophoretic mobility; the more phosphorylation, the less movement through a solid gel matrix. Incredibly, simply mixing purified FRQ and CK1a in the presence of an ATP-generating source led to the slow transition of nonphosphorylated FRQ to maximally hyperphosphorylated over a 24-h incubation period. Similar success was obtained by adding purified CK1δ to mammalian cell-free extracts containing PER2. Since the identity of all the priming-dependent sites on PERs and FRQ are not known, Marzoll et al. (7) relied on a neat trick to ask if non–priming-dependent phosphorylation is sufficient for CK1 to generate the in vitro progressive phosphorylation with a daily time scale. Earlier work showed that CK1ε with the tau mutation (R178C) strongly attenuates preference at primed sites (the positively charged R residue increases affinity for primed sites) with little effect on the rate at non–priming-dependent sites (in a sense, all CK1-tau target sites are treated as suboptimal) (see ref. 15). Surprisingly, progressive phosphorylation of PER2 and FRQ with wild-type pattern and daily time scale did not seem to miss a beat when the experiments were performed with validated priming-defective tau mimetics, CK1a (Ck1a-R181Q) and CK1δ (R178Q). Similar global phosphorylation kinetics for PER and FRQ were observed at 4° and 20 °C (unfortunately, PER and FRQ were unstable at more physiological temperatures). Removing key parts of the CK1 binding sites found on PER or FRQ abolished mobility shifts (indicating no or very little phosphorylation).
Moving to more in vivo settings, overexpressing CK1a-R181Q in Neurospora and CK1δ-R178Q in mammalian cells led to much faster clocks even when compared to similar manipulations with the cognate wild-type versions, supporting an in vivo role for CK1-mediated suboptimal phosphorylation in setting clock period. In addition, it suggests that priming-dependent phosphorylation (and its proximal phospho-switches) largely function to stretch out the overall rate of CK1-mediated suboptimal phosphorylation. Overexpressing Neurospora CK1a and CK1a-R181Q in mammalian cells sped up clocks to the same extent as that observed with the CK1δ versions, suggesting CK1 binding motifs on PERs and FRQ are interchangeable. Overall, Marzoll et al. (7) demonstrate that despite all the complexity linked to daily phospho-timing in fungal to human clocks, 24-h timing is mainly based on the slow filling of suboptimal phosphorylation sites by CK1 once it binds to the phospho-timer operating in that system (e.g., PER and FRQ) (Fig. 1).
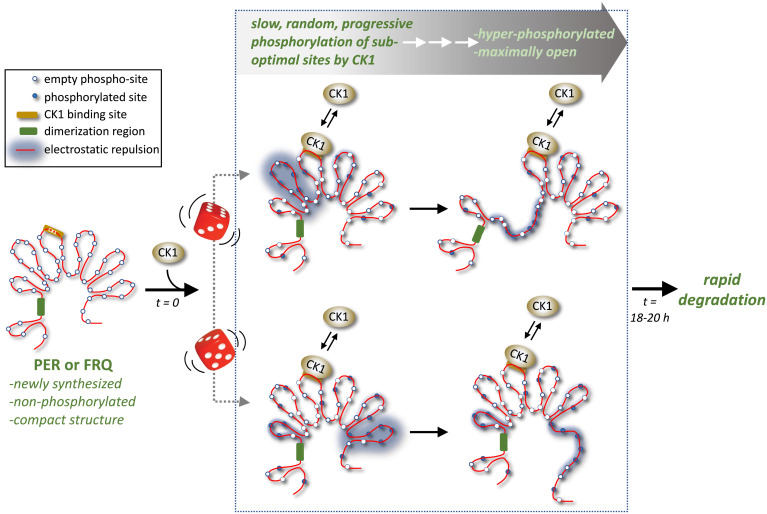
Fig. 1.
Casein kinase 1 works with functionally similar modules on PER and FRQ to drive their slow conversion from compact nonphosphorylated forms to extended hyperphosphorylated forms over a daily time scale, despite randomness in pathways to hyperphosphorylation (the beginnings of two random paths are indicated by the dice). Conserved modules include long stretches of highly disordered regions attached to a centrally located CK1 binding site. Increased phosphorylation by CK1 leads to electrostatic repulsion that force more open conformations, eventually facilitating the rapid degradation of PER and FRQ, which helps set the daily timing for the next round of PER and FRQ synthesis (not shown). Adapted from ref. 7.
Here and elsewhere, Brunner and coworkers speculate that the progressive phosphorylation of PER and FRQ by CK1 might be linked to a shared landscape (Fig. 1). Both PER and FRQ have a centrally located CK1 binding site that is flanked on both sides by long stretches of potentially intrinsically disordered regions (IDRs). Long stretches of IDRs collapse to form compact structures and contain an overabundance of disorder-promoting amino acids, such as Ser and Thr. Marzoll et al. (7) note that the putative IDRs in FRQ and PER are about the same length (∼1,000 amino acids) and contain similar proportions of Ser (∼14%) and Thr (∼6 to 7%), marrying disordered regions to potential influence from multisite phosphorylation. Addition of negatively charged phosphates to this landscape by CK1 would increase electrostatic repulsion, pushing more open conformations of PER and FRQ, presumably facilitating degradation of hyperphosphorylated isoforms (12, 16). Just like grains of sand are not expected to funnel in the same order every time, yet hourglasses are reliable timers, so it seems CK1 can synchronize a population of PER and FRQ proteins to follow a similar overall rate in progressive phosphorylation despite the enormous randomness in which and when a specific suboptimal site is filled on any individual PER or FRQ molecule (Fig. 1; portrayed by the roll of the dice).
A very different model emerged from the first circadian phospho-timing generated in a test tube, which used the cyanobacterial clock system that is based on three interacting proteins, KaiA, KaiB, and KaiC. In a landmark study, Kondo and coworkers showed that simply mixing these three proteins in the presence of ATP led to a temperature-compensated 24-h cycle in the multisite phosphorylation of KaiC that persisted for days (17). The mechanism is unambigously engineered whereby the two phospho-sites (yes, there are only two in this system) follow precise instructions as to the order of which site gets phosphorylated and when. If there are little cross-over lessons in comparing clock mechanisms in cyanobacteria and those in eukaryotes, where is the presumed Rosetta stone of “quartz” timing in eukaryotic clocks so elegantly described for cyanobacteria (9)?
This brings me to perhaps the key insight from Marzoll et al. (7). A timing system that is based on the functional strength of many interchangeable individuals but not any specific individual (or pathway) is rather resistant to mutation and misregulation. A solid framework ensures the wheel always turns with an ∼24-h pace but in its simplicity and lack of rigidity offers flexibility and adaptability that because of its preeminence snaps back to core attributes, minimizing long-term distortions. Indeed, if the basic progressive phosphorylation that ensures a daily time scale is also largely temperature insensitive or relies on a simple thermal balance in the efficiency of phosphorylation at suboptimal sites by CK1, this would add another considerable foundational stability and argue against the need for multiple precisely counterbalanced thermal rates at each step of clock progression. Moreover, the aforementioned negative feedback loops add stability to the overall phase and period of the timing mechanism, namely, by linking the degradation of hyperphosphorylated PER and FRQ to the start of the next round of newly synthesized PER and FRQ. This sets in motion a similarly timed biochemical fate as its predecessors (Fig. 1). If provided with the address for PER and FRQ, CK1 is up to the task because it is a workhorse kinase that will steadily march through phosphorylating each suboptimal site with the same indifference. For now, it is gratifying to see old rivals like PER and FRQ come together to tell a compelling story of shared timing modules that diverged some 1.5 billion y ago but remained loyal to the same kinase. More secrets in the blueprints used to build protein-based biochemical oscillators with 24-h periodicities await to be discovered.
Footnotes
The author declares no competing interest.
See companion article, “Casein kinase 1 and disordered clock proteins form functionally equivalent, phospho-based circadian modules in fungi and mammals,” 10.1073/pnas.2118286119.
References
- 1.Konopka R. J., Benzer S., Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldman J. F., Hoyle M. N., Isolation of circadian clock mutants of Neurospora crassa. Genetics 75, 605–613 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunlap J. C., Loros J. J., Making time: Conservation of biological clocks from fungi to animals. Microbiol. Spectr., 10.1128/microbiolspec.FUNK-0039-2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirano A., Fu Y. H., Ptáček L. J., The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 23, 1053–1060 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Young M. W., Kay S. A., Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–715 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Ralph M. R., Menaker M., A mutation of the circadian system in golden hamsters. Science 241, 1225–1227 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Marzoll D., et al. , Casein kinase 1 and disordered clock proteins form functionally equivalent, phospho-based circadian modules in fungi and mammals. Proc. Natl. Acad. Sci. U.S.A. 119, 10.1073/pnas.2118286119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T., A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb. Symp. Quant. Biol. 72, 47–55 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Rosbash M., The implications of multiple circadian clock origins. PLoS Biol. 7, e62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong J. K., Virshup D. M., Casein kinase 1: Complexity in the family. Int. J. Biochem. Cell Biol. 43, 465–469 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Baker C. L., Kettenbach A. N., Loros J. J., Gerber S. A., Dunlap J. C., Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell 34, 354–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu J. C., Ko H. W., Edery I., NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145, 357–370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garbe D. S., et al. , Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet. 9, e1003749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanselow K., et al. , Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20, 2660–2672 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narasimamurthy R., Virshup D. M., The phosphorylation switch that regulates ticking of the circadian clock. Mol. Cell 81, 1133–1146 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Querfurth C., et al. , Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol. Cell 43, 713–722 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Nakajima M., et al. , Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). [DOI] [PubMed] [Google Scholar]