Significance
Paramutation involves the transfer of a repressive epigenetic mark between silent and active alleles. It is best known from exceptional non-Mendelian inheritance of conspicuous phenotypes in maize but also in other plants and animals. Recent genomic studies, however, indicate that paramutation may be less exceptional. It may be a consequence of wide-cross hybridization and may contribute to quantitative trait variation or unstable phenotypes in crops. Using the sulfurea (sulf) locus in tomato, we demonstrate that a self-reinforcing feedback loop involving DNA- and histone-methyl transferases CHROMOMETHYLTRANSFERASE3 (CMT3) and KRYPTONITE (KYP) is required for paramutation of sulf and that there is a change in chromatin organization. These findings advance the understanding of non-Mendelian inheritance in plants.
Keywords: paramutation, heredity, epigenetic memory, tomato
Abstract
Paramutation involves the transfer of a repressive epigenetic mark from a silent allele to an active homolog and, consequently, non-Mendelian inheritance. In tomato, the sulfurea (sulf) paramutation is associated with a high level of CHG hypermethylation in a region overlapping with the transcription start site (TSS) of the SlTAB2 gene that affects chlorophyll synthesis. The CCG subcontext hypermethylation is under-represented at this region relative to CTG or CAG, implicating the CHROMOMETHYLTRANSFERASE3a (CMT3) in paramutation at this locus. Consistent with this interpretation, loss of CMT3 function leads to loss of the sulf chlorosis, the associated CHG hypermethylation, and paramutation. Loss of KRYPTONITE (KYP) histone methyltransferase function has a similar effect linked to reduced H3K9me2 at the promoter region of SlTAB2 and a shift in higher order chromatin structure at this locus. Mutation of the largest subunit of RNA polymerase V (PolV) in contrast does not affect sulf paramutation. These findings indicate the involvement of a CMT3/KYP–dependent feedback loop rather than the PolV-dependent pathway leading to RNA-directed DNA methylation (RdDM) in the maintenance of paramutation.
Paramutation causes non-Mendelian inheritance in which an epigenetic mark at a silenced paramutagenic allele transfers to and silences an active paramutable homolog (1–3). In plants the DNA is methylated at the paramutated (silenced) sequence region, but this modification is not sufficient to mediate paramutation because many methylated loci do not show paramutation. Early examples of paramutation in maize (1), pea (4), and tomato (5) have striking phenotypes based on pigmentation or gross morphology. However, from genome-wide DNA methylation analyses, there is now evidence that paramutation-like effects may be more widespread (6–8). In some examples, the trigger for paramutation may be hybridization between distantly related varieties or even species (6–8).
Like all heritable epigenetic mutations, paramutation involves separate mechanisms to establish a molecular mark at the paramutable locus and to mediate its maintenance through cycles of cell division and sexual reproduction. Additionally, a defining characteristic of paramutation is the interaction between the participating alleles that, in many models, involves the RNA-directed DNA methylation (RdDM) pathway (1, 2).
RdDM is not, however, exclusive to paramutation. At many genomic loci, it mediates DNA methylation in transposable elements (TEs) and repetitive regions (9). It involves RNA polymerase IV (PolIV) transcription of the target DNA and RNA-dependent RNA polymerase (RDR2) conversion of the transcript into double-stranded RNA. DCL3 trims the double-stranded RNA into 24 nucleotide (nt) fragments and, after unwinding, the single-stranded 24-nt small RNAs (sRNAs) are loaded into Argonaute protein 4 (AGO4). This AGO4 nucleoprotein is then guided by Watson–Crick base pairing to an RNA polymerase V (PolV)–generated scaffold RNA, and it recruits DNA methyltransferases, including DRM2, that catalyze methylation of adjacent DNA cytosines (9).
The RdDM factors identified in paramutation screens in maize include the RDR2 ortholog (Mediator of Paramutation 1 [MOP1]), which is required to maintain and establish paramutation at multiple maize loci, including the classic paramutation example in the booster1 (b1) locus (10). Maize genetic screens have also uncovered multiple PolIV subunits including MOP2/RMR7 (RDP2), MOP3, RMR6, RMR7, RDP1, and RDP2a as affecting paramutation (1). RPD2a has the potential to integrate PolV complexes (11). Consistent with involvement of the RdDM pathway, there may be abundant 24-nt sRNAs and high DNA methylation at the target loci. These 24-nt sRNAs could, in principle, diffuse within the nucleus and mediate the allelic interaction in paramutation.
However, the picture from genetic screens is incomplete. There is no clear evidence for involvement of the PolV subunits, AGO proteins, DNA methyltransferases, or other factors in the downstream part of the RdDM pathway or the 21-nt sRNAs or AGO6 associated with the initial establishment phase (12). Some maize studies also found it difficult to reconcile these findings with an sRNA-based model for paramutation (13). Paramutation at maize purple plant1 (pl1), for example, can be established in mutants that lack sRNAs, including RMR1, an Snf2 protein that affects the stability of nascent transcripts (14). Conversely, mutations in the RMR12 chromodomain helicase DNA-binding 3 (CHD3) protein orthologous to Arabidopsis (Arabidopsis thaliana) PICKLE release the pl1 maize paramutation but in a manner that probably promotes the incorporation of nucleosomes without affecting sRNA accumulation (15). These various findings and observations do not necessarily rule out the involvement of sRNAs or RdDM in paramutation, but they do point to the gaps in current models. Therefore, we are investigating sulfurea (sulf) paramutation in tomato (16). Our reasoning is that sulf paramutation may be mechanistically similar to the maize examples, but the genetic architecture of different plants would provide a new perspective on the factors involved.
The sulf-mediated leaf chlorosis phenotype is due to hypermethylation in a Differentially Methylated Region1 (DMR1) at the 5′ region of SlTAB2 (TAB2), a gene that affects the synthesis of chlorophyll in tomato plants (16). TAB2 is silenced between 0 and 100% in the F1 progeny of sulf × wild-type (WT) plants (17). In crosses between variegated sulf lines and their parental WT lines, paramutation is poorly penetrant in F1 (<12%) (17). In F2, the percentage of variegation among heterozygous also varies, further showing that sulf has incomplete penetrance (5, 17). These differences in paramutation penetrance have been linked with the existence of different sulf epialleles with different degrees of paramutagenicity (5, 17).
Allele-specific DNA methylation analysis at DMR1 in interspecific sulf X Solanum pimpinellifolium F1 hybrids revealed high methylation levels in the CHG context in the de novo paramutated allele (16). In tomato, CHG-methylation is maintained by CHROMOMETHYLTRANSFERASE3a (SlCMT3a, hereafter referred as CMT3) (18). Here, we show that sulf paramutation requires the self-reinforcing loop involving CHG DNA methylation by the DNA methyl transferase CMT3 and H3K9 dimethylation by the histone methyltransferase KRYPTONITE (KYP) at the silenced locus, which correlates with changes in chromosome compartment. Mutation of the largest RNA polV subunit (NRPE1) does not affect sulf. Our findings suggest that the maintenance of sulf paramutation in tomato might be independent of the canonical RdDM pathway and that CHG methylation is required.
Results
CMT3 Maintains the sulf Paramutation.
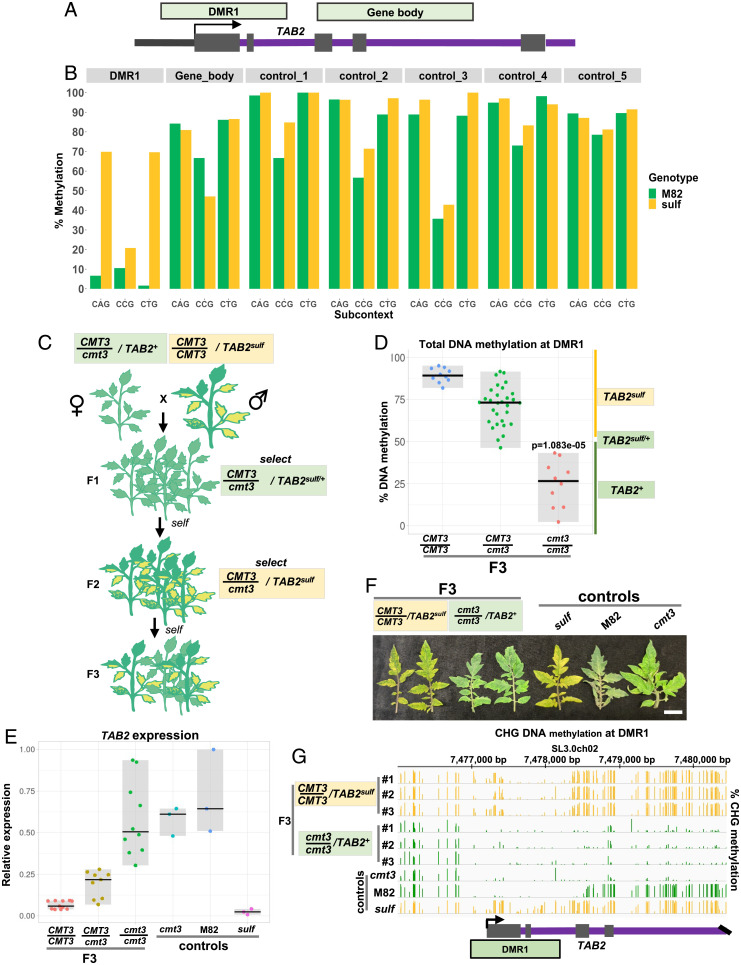
The sulf phenotype results from TAB2 hypermethylation in a DMR1 close to the transcription start site (TSS) (16). The severity of leaf chlorosis is directly proportional to the levels of DMR1 DNA methylation and thus far has not been associated with variation in DNA sequences (16). Here, we classify sulf epialleles based on their correlated phenotype and epigenotype (Table 1) (16). Despite displaying high differential DNA methylation at the CG context (∼50%), DMR1 is unusual in that it has a very high level of CHG hypermethylation (∼60%) (16) consistent with the involvement of CMT3 (18). To test this hypothesis, we assayed methylation at the different CHG subcontexts in DMR1 (Fig. 1A) (16) in leaf tissue excised from plants bearing TAB2+ (WT cv. M82) or TAB2sulf (sulf). CMT3 preferentially methylates CAG and CTG motifs over CCG in multiple plant species, including tomato (18, 19). In TAB2sulf, the percentage of CCG methylation in DMR1 was lower than at CAG and CTG (Fig. 1B), consistent with the involvement of CMT3 in TAB2sulf. The subcontext bias was less pronounced in the gene body of TAB2 than in DMR1 (Fig. 1B). It was also less pronounced in random non-DMR regions of similar size within the pericentric heterochromatin of chromosome 2 (Fig. 1B and SI Appendix, Table S1).
Table 1.
Epiallele classification employed in this study
| Epigenotype | Genotype | DNA Methylation/Phenotype | Paramutated | Identification |
|---|---|---|---|---|
| TAB2+ | M82 DNA sequence | <50% methylation, green | No | Phenotyping |
| McrBC-qPCR | ||||
| TAB2sulf/+ | M82 DNA sequence | 50% methylation, green (e.g., F1s) | No | Phenotyping |
| McrBC-qPCR | ||||
| TAB2sulf | M82 DNA sequence | >50% methylation, variegated leaf chlorosis | Yes | Phenotyping |
| McrBC-qPCR |
Fig. 1.
CMT3 maintains sulf. (A) Diagram illustrates TAB2 locus and relative DMR1 and gene body positions. These regions were used for analysis in Fig. 1B (refer to SI Appendix, Table S1 for precise coordinates). (B) Mean % DNA methylation in leaf tissue at CHG subcontext in DMR1, TAB2 gene body, and five random control regions within SL3.0ch02. M82: S. lycopersicum cv. M82 TAB2+ (green), n = 2; and sulf: S. lycopersicum cv. Lukullus TAB2sulf (yellow), n = 2. Coordinates are listed in SI Appendix, Table S1. (C) Diagram illustrates crossing scheme used to generate F3 populations (F3 pedigree). (D) Jittered dots depict % of DNA methylation at DMR1 in individual plants determined by McrBC-qPCR. Plants denote F3 siblings–4-wk-old leaf tissue. The summary of the data is shown as the horizontal line indicating the median. Gray boxes illustrate the data range. The × axis refers to CMT3 genotypes. CMT3/CMT3 TAB2sulf, n = 10; CMT3/cmt3 TAB2sulf, n = 30; cmt3/cmt3 TAB2+, n = 10. P value cmt3/cmt3 versus CMT3/CMT3 was calculated employing a Mann-Whitney–Wilcoxon test. (E) Jittered dots depict relative TAB2 expression in individual plants (4-wk-old leaf tissue) normalized to the geometric mean of the expression of two reference genes (SI Appendix, Table S3). The summary of the data is shown as horizontal line indicating the median. Gray boxes illustrate the data range. F3 plants: CMT3/CMT3 TAB2sulf, n = 11; CMT3/cmt3 TAB2sulf, n = 8; cmt3/cmt3 TAB2+, n = 9. Controls: M82- (S. lycopersicum cv. M82) CMT3/CMT3 TAB2+, n = 3; sulf- (S. lycopersicum cv. Lukullus) CMT3/CMT3 TAB2sulf, n = 3; cmt3- cmt3/cmt3 TAB2+, n = 3. (F) Young leaves from 6-mo-old plants. White bar, 2 cm.1E. (G) TAB2 Integrative Genomics Viewer (IGV) screenshot of bisulfite sequencing data exhibiting CHG DNA methylation (4-wk-old leaf tissue), range [0 to 100]; F3 plants and controls: same as in Fig. 1E. Green tracks refer to green leaf phenotype, and yellow tracks refer to plants that display chlorosis. (A and C–G) Yellow boxes refer to plants displaying sulf chlorosis, and green boxes refer to green plants.
To further test this possibility, we took advantage of the plants bearing CRISPR-mediated deletion in the SlCMT3a gene—cmt3a (referred to as cmt3) (18). The heterozygous CMT3/cmt3 was crossed to CMT3/CMT3 plants with the TAB2sulf epiallele (Fig. 1C). In the F1 generation, we selected plants heterozygous for cmt3 and, from McrBC-qPCR (16), for the TAB2sulf/+ epialleles characteristic of the epiheterozygous state at DMR1 (Fig. 1C and SI Appendix, Fig. S1A). The epiheterozygous state is characteristic of a sulf allele with low paramutation penetrance. Some sulf epialleles have strong F1 penetration, whereas others, such as the one used here, only display paramutation symptoms in F2 (17). Because pure sulf homozygotes are lethal, it is likely that F2 chlorotic TAB2sulf plants underwent paramutation (5) during early growth of TAB2sulf/+.
In the F2 generation, only CMT3 homozygotes or heterozygotes displayed the TAB2sulf phenotype and epigenotype (>50% methylation by McrBC-qPCR) (SI Appendix, Fig. S1B). The outcome was the same in reciprocal crosses in which the paternal plants carried cmt3 and the maternal CMT3/CMT3 plants carried the TAB2sulf epiallele (SI Appendix, Fig. S1 C and D). From this result, we conclude that DMR1 hypermethylation and CMT3 are required for establishment or maintenance of sulf paramutation. There was a similar genetic dependence on CMT3 for the TAB2sulf epiallele in the F3 progeny of CMT3/cmt3 F2 plants TAB2sulf (Fig. 1D). As in the F2, the homozygous cmt3 F3 plants were green and had lower DMR1 DNA methylation levels than their CMT3 siblings (Fig. 1D), indicative of the epiallele TAB2+. This observation suggests that CMT3 is required to maintain sulf paramutation that established in the F2.
Further support for a link between sulf chlorosis, DNA methylation, and CMT3 level comes from the analysis of DMR1 DNA methylation in the heterozygous CMT3/cmt3 F2 and F3 plants. The distribution of DMR1 DNA methylation in these plants was skewed toward lower values than in CMT3 homozygotes (SI Appendix, Fig. S1B and Fig. 1D), and in the F2, a smaller proportion of plants were TAB2sulf and chlorosis in CMT3/cmt3 (∼17%) than in CMT3/CMT3 (∼24%) (SI Appendix, Fig. S1B). Many of the green F2 CMT3/cmt3 heterozygous plants may have inherited TAB2sulf and TAB2sulf/+ but with incomplete maintenance of TAB2sulf and reversion to TAB2+ due to reduced dosage of CMT3.
Consistent with the low DNA methylation levels of DMR1 in the F3 cmt3 backgrounds, TAB2 expression in these plants was higher than in the F3 CMT3 homozygous and heterozygous siblings (Fig. 1E), although with more variation than in CMT3 or cmt3 lines with TAB2+ that had not been crossed with sulf (Fig. 1E). The homozygous cmt3 plants lacked sulf chlorosis, whereas their F3 CMT3 siblings were chlorotic (Fig. 1F and SI Appendix, Fig. S2).
More detailed analysis of the F3 DNA methylation pattern by bisulfite sequencing analysis confirmed that, compared to M82, the CHG DNA methylation in sulf plants bearing the TAB2sulf allele was higher in DMR1 extending from 212 bp in the 5′ upstream region into intron 2 at 852 bp downstream of the TSS (16) (Fig. 1G). In the cmt3 TAB2+ F3 progeny, the hyper CHG DMR1 was lost (Fig. 1G). There was also CHG hypomethylation of the transcribed DNA but no indication that this gene body DNA methylation influenced paramutation (Fig. 1G). From these patterns, we conclude that sulf is associated with CMT3-mediated CHG hypermethylation of DMR1.
In the CG context, the sulf hyper DMR was restricted to the DMR1 region, but the association with sulf was weaker than with CHG (SI Appendix, Fig. S3). The degree of hyper CG methylation was markedly lower in the cmt3 TAB2+ F3 progeny than in the CMT3 TAB2sulf siblings (SI Appendix, Fig. S3). The CHH context did not exhibit significant hypermethylation in the TAB2 DNA of sulf plants (SI Appendix, Fig. S3). There was a region of CHH hypomethylation on the upstream side of DMR1 (SI Appendix, Fig. S3) corresponding to a Differentially Methylated Region2 (DMR2) (16), but it was not correlated with the sulf phenotype in the F3 progeny: The hypomethylation was lost to a varying extent irrespective of whether the plants were CMT3 and TAB2sulf or cmt3 and TAB2+ (SI Appendix, Fig. S3). These results indicate that CHG methylation patterns, but not CG or CHH, primarily govern the sulf phenotype.
The cmt3 mutants lose DMR1 hypermethylation and sulf chlorosis, but in principle, they could retain other epigenetic marks with the potential to reestablish TAB2sulf in a CMT3 background. To assess this possibility, we backcrossed an F2 cmt3/cmt3 TAB2+ plant with one of the highest levels of methylation (27%) (SI Appendix, Fig. S3B) with an M82 TAB2+ allele (SI Appendix, Fig. S4A) and quantified DNA methylation at DMR1 using McrBC-qPCR in the BC1 and two generations of selfed progeny (SI Appendix, Fig. S4A). None of these plants had DMR1 DNA methylation levels higher than 40% (SI Appendix, Fig. S4 B–D), and plants were not chlorotic (SI Appendix, Fig. S4E). Bisulfite sequencing confirmed lower levels of CHG methylation at DMR1 in backcrossed plants than in sulf controls (SI Appendix, Fig. S5), suggesting that sulf memory is CMT3 dependent and that RdDM is not sufficient to maintain sulf.
NRPE1 and sulf Paramutation.
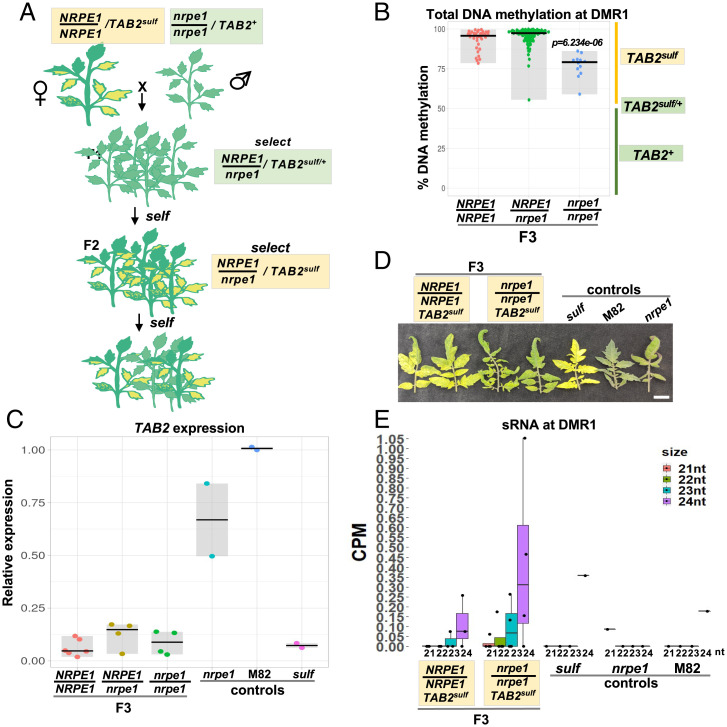
To assess a possible role of an RdDM cofactor, we crossed sulf plants with a mutation in an RdDM component DNA-directed RNA NRPE1 (Fig. 2A) (19) and then selfed F1 plants that were epiheterozygous (TAB2sulf/+) (Fig. 2A and SI Appendix, Fig. S6A) for two generations. We were unable to obtain progeny with a maternal nrpe1 mutation; however, the parent of origin does not affect sulf paramutagenicity (20), and the lack of reciprocal crosses would not compromise our interpretation.
Fig. 2.
sulf maintenance remains unaffected in nrpe1 mutants. (A) Diagram illustrates crossing scheme used to obtain F3 populations (F3 pedigree). (B) Jittered dots depict % DNA methylation of individual plants at DMR1 determined by McrBC-qPCR. Plants denote F3 siblings–4-wk-old leaf tissue. The summary of the data is shown as horizontal line indicating the median. Gray boxes illustrate the data range. The × axis refers to NRPE1 genotypes. NRPE1/NRPE1 TAB2sulf, n = 35; NRPE1/nrpe1 TAB2sulf, n = 94; nrpe1/nrpe1 TAB2sulf, n = 12. P value nrpe1/nrpe1 versus NRPE1/NRPE1 was calculated employing a Mann-Whitney–Wilcoxon test. (C) Jittered dots depict relative TAB2 expression in individual plants (4-wk-old leaf tissue) normalized to the geometric mean of the expression of two reference genes (SI Appendix, Table S3). The summary of the data is shown as horizontal line indicating the median. Gray boxes illustrate the data range. F3 plants: NRPE1/NRPE1 TAB2sulf, n = 6; NRPE1/nrpe1 TAB2sulf, n = 4; nrpe1/nrpe1 TAB2sulf, n = 4. Controls: M82- (S. lycopersicum cv. M82) NRPE1/NRPE1 TAB2+, n = 2; sulf- (S. lycopersicum cv. Lukullus) NRPE1/NRPE1 TAB2sulf, n = 2; nrpe1-nrpe1/nrpe1 TAB2,+ n = 2. (D) Young leaves from 6-mo-old plants. White bar, 2 cm. (E) DMR1 sRNA size distribution in counts per million (CPM) in F3 plants and controls (4-wk-old leaf tissue). Genotypes are the same as in Fig. 2C. Jittered dots represent individual plants. The summary of the data is shown as horizontal line indicating the median. Errors bars represent the SD. Genotypes are the same as in Fig. 2C. (A–E) Yellow boxes refer to plants displaying sulf chlorosis, and green boxes refer to green plants.
The nrpe1 mutant has a deletion of two codons in the NRPE1 coding sequence, reduced 24-nt sRNAs, and hypomethylation at RdDM loci (19). In the F2 and F3 progeny, the DMR1 DNA methylation profiles were similar in all NRPE1 genotypes (Fig. 2B and SI Appendix, Fig. S6B). In the F2, the mean level was close to 50% with a distribution from 0 to 100% (SI Appendix, Fig. S6B). In the F3, all of the plants had the TAB2sulf epiallele since DMR1 methylation was above 50% in all individuals regardless of their NRPE1 genotype (Fig. 2B). On average, the F3 nrpe1 plants had 20% lower DMR1 DNA methylation levels than their NRPE1 F3 siblings (Fig. 2B), but this reduction did not suppress silencing of TAB2 expression (Fig. 2C) or the sulf chlorosis (Fig. 2D and SI Appendix, Fig. S7). From these data, we conclude that reduced NRPE1 function has a minor effect on DMR1 DNA methylation but that PolV is not an essential cofactor of sulf silencing.
The 24-nt sRNAs constitute a hallmark of RdDM activity (9). Given the nonessential role of NRPE1 in maintaining sulf, we reassessed sRNAs at the TAB2 locus employing sRNA sequencing. On average, 24-nt sRNAs accumulated more in TAB2sulf than in TAB2+ at DMR1 (16) but at a low and variable level (SI Appendix, Fig. S8A). Gene body sRNAs showed a more consistent up-regulation in sulf plants (SI Appendix, Fig. S8B).
Whole-genome sRNA-sequencing data revealed that 13% of all sRNA loci were affected by NRPE1 (19), but in F3 NRPE1 and F3 nrpe1 plants, the low levels of DMR1 24-nt sRNA were not significantly different (Fig. 2E). Similarly, in CMT3 and cmt3 F2 plants of the crosses with TAB2sulf, the DMR1 sRNA of all 21- to 24-nt size classes was unaffected by genotype or sulf epigenotype (SI Appendix, Fig. S9 A–C). There was a reduction of gene body sRNAs in cmt3 genotypes with TAB2+ (SI Appendix, Fig. S9D), but this effect did not correlate with sulf chlorosis: DNA methylation was abundant in plants with TAB2+ epigenotype (Fig. 1G and SI Appendix, Fig. S3). From these sRNA data, it is therefore unlikely that DMR1 or gene body sRNAs play a role in sulf maintenance.
KYP and H3K9me2 Are Required to Maintain the sulf Paramutation.
In Arabidopsis, a self-reinforcing loop model involving H3K9me2, a repressive histone modification, can account for maintenance of CHG methylation by CMT3. In this model, KYP family proteins bind CHG DNA methylation and methylate histone H3K9 tails. In turn, CMT3 binds H3K9me2 and increases the existing level of CHG DNA methylation (21, 22). Given the signature of CMT3-dependent DNA methylation at TAB2, the CMT3/KYP reinforcement model predicts higher levels of H3K9me2 in sulf than in WT tissues at DMR1.
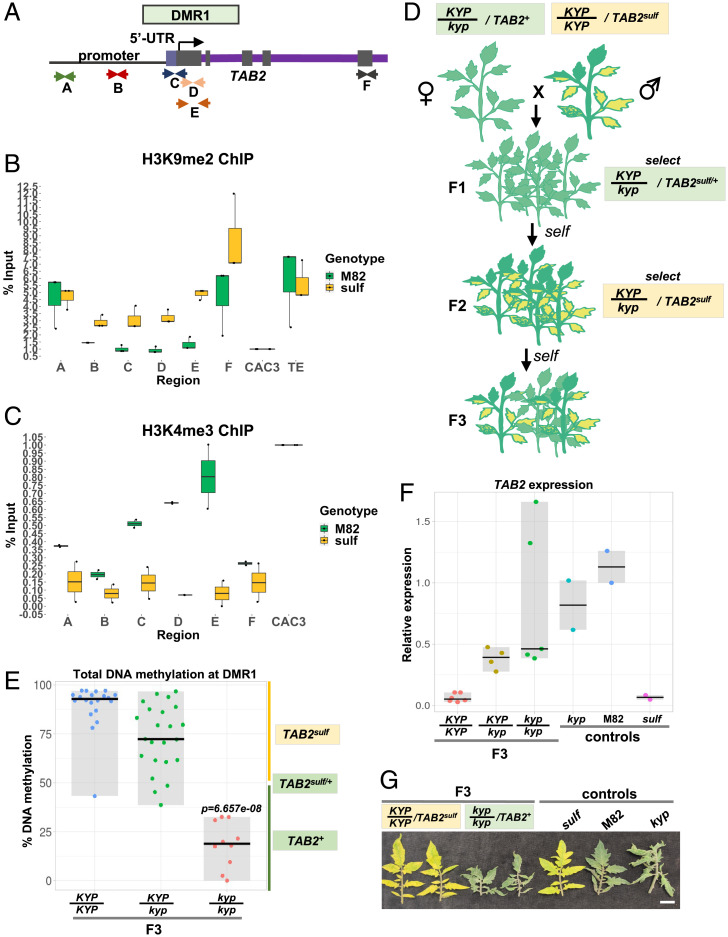
To test this prediction, we carried out H3K9me2 chromatin immunoprecipitation (ChIP) followed by quantitative real-time PCR (ChIP-qPCR) with oligonucleotide pairs A–F spanning different regions of the TAB2 locus (Fig. 3A) in M82 and sulf plants bearing TAB2+ and TAB2sulf epigenotypes, respectively. We normalized the input using an unrelated reference locus. This approach allowed us to determine the relative H3K9me2 abundance but not the proportion of modified nucleosomes. Consistent with our prediction, the H3K9me2 enrichment was higher in TAB2sulf than in TAB2+ at the DMR1 overlapping regions C, D, and E, including the TAB2 TSS, and also at the promoter region B (Fig. 3B). In contrast, we detected no or smaller H3K9me2 differences between TAB2+ and TAB2sulf at regions outside DMR1, including A and F and an unrelated TE (Fig. 3B). From these results, we conclude that the link between CMT3 and sulf is associated with increased levels of H3K9me2 at DMR1.
Fig. 3.
KYP maintains sulf. (A) Diagram represents the relative oligonucleotide position (arrows) spanning the TAB2 locus used for ChIP-qPCR experiments in Fig. 3B. The different letters represent the different TAB2 locus regions. (B) Box plot depicts H3K9me2 enrichment per % input normalized to CAC3 reference locus determined by Chip-qPCR. Jittered dots represent different biological replicates. M82- S. lycopersicum cv. M82 TAB2+ (green), n = 3; sulf- S. lycopersicum cv. Lukullus TAB2sulf (yellow), n = 3. The summary of the data is shown as horizontal line indicating the median of biological replicates. Error bars represent the SD. TE refers to T135 retrotransposon element. (C) Box plot depicts H3K4me3 enrichment per % input normalized to CAC3 reference locus determined by Chip-qPCR. Jittered dots represent different biological replicates. M82- S. lycopersicum cv. M82 TAB2+ (green), n = 2; sulf- S. lycopersicum cv. Lukullus TAB2sulf (yellow), n = 2. The summary of the data is shown as horizontal line indicating the median of biological replicates. Error bars represent the SD. (D) Diagram illustrates crossing scheme used to obtain F3 populations (F3 pedigree). (E) Jittered dots depict % DNA methylation at DMR1 individual plants determined by McrBC-qPCR. Plants denote F3 siblings. The summary of the data is shown as horizontal line indicating the median. Gray boxes illustrate the data range. The × axis refers to KYP genotypes. F3 plants: KYP/KYP TAB2sulf, n = 20; KYP/kyp TAB2sulf, n = 23; kyp/kyp TAB2+, n = 10. P value kyp/kyp versus CMT3/CMT3 was calculated employing a Mann-Whitney–Wilcoxon test. (F) Jittered dots depict relative TAB2 expression in individual plants normalized using the geometric mean of the expression values for two reference genes (SI Appendix, Table S3). The summary of the data is shown as horizontal line indicating the median. Gray boxes illustrate the data range. F3 plants: KYP/KYP TAB2sulf, n = 6; F3 plants: KYP/kyp TAB2sulf, n = 4; kyp/kyp TAB2+, n = 5. Controls: M82- (S. lycopersicum cv. M82) KYP/KYP TAB2+, n = 2; sulf- (S. lycopersicum cv. Lukullus) KYP/KYP TAB2sulf, n = 2; kyp- kyp/kyp TAB2+, n = 2. (G) Young leaves excised from 6-mo-old plants. White bar, 2 cm. (A–G) Yellow boxes refer to plants displaying sulf chlorosis, and green boxes refer to green plants. UTR, untranslated region.
In M82 leaves bearing TAB2+, the H3K9me2 levels were lower close to the TSS than in more distal regions (Fig. 3B). Such low levels are exceptional in pericentromeric heterochromatin (where TAB2 resides) (18), and it is likely that these low H3K9me2 levels enable transcription of TAB2. Consistent with this interpretation, there were high levels of the active transcription mark H3K4me3 close to TAB2 TSS (Fig. 3C) detected by ChIP-qPCR in M82 plants at regions C, D, and E. The association between histone modifications with sulf silencing is reinforced by reduced levels of the H3K4me3 mark at these sites at TAB2sulf (Fig. 3C).
To further test the involvement of H3K9me2, we crossed sulf with a mutant bearing CRISPR-Cas9–mediated deletion at the gene encoding for the Solanum lycopersicum KYP (18) (Fig. 3D). The crossing strategy was parallel to that used for cmt3, and in the F2 and F3 progeny of the F1 KYP/kyp epiheterozygous TAB2sulf/+ (SI Appendix, Fig. S10A), the kyp/kyp plants all had less than 50% DMR1 DNA methylation (SI Appendix, Fig. S10B and Fig. 3E). Accordingly, TAB2 expression in F3 kyp plants was higher than in the KYP siblings (Fig. 3F), and none displayed symptoms of chlorosis (Fig. 3G and SI Appendix, Fig. S11). Only progeny that carried an active KYP allele displayed TAB2sulf epigenotype, sulf phenotypes, and low TAB2 expression (Fig. 3 E and F), as observed in the original TAB2sulf parent. The sulf paramutation in tomato, therefore, is associated with high levels of CHG DNA methylation and H3K9me2 at DMR1 that would be expected to influence chromatin organization.
Chromatin Architecture Changes in sulf.
To test this hypothesis, we carried out chromatin conformation capture (Hi-C) analysis in TAB2sulf and TAB2+. Despite having distinct leaf colors, M82 and sulf plants showed highly similar Hi-C maps, suggesting that there were no intensive genome rearrangements or changes in chromatin contacts in sulf plants bearing TAB2sulf epialleles (SI Appendix, Fig. S12 A and B). Principal component analysis (PCA) of the contact matrix of chromosome 2 revealed A/B compartments that correlated to the delineation of euchromatin/heterochromatin across this chromosome (SI Appendix, Fig. S13 A and B). This A/B compartment partition was broadly similar in the two epigenotypes, and in all the samples, the genomic region containing TAB2 belonged to the heterochromatic B compartment (SI Appendix, Fig. S13C). However, there was a change in the PCA eigenvalue of this genomic region in all the sulf replicates, indicating a change in chromatin interaction between this region and the A/B compartments. By examining the interaction strength between this TAB2-containing region and the A compartment, we found that it was significantly weaker in the TAB2sulf plants than in the TAB2+ plants (SI Appendix, Fig. S13D), indicating a general shift away from euchromatic regions.
Discussion
We confirm here that sulf paramutation is associated with DMR1 that overlaps the TSS of TAB2 (16) (Fig. 1G and SI Appendix, Fig. S3), and we have shown additionally the involvement of CMT3/KYP (Figs. 1 and 3) and an associated change in chromatin organization (SI Appendix, Fig. S13). CMT3/KYP have not been previously associated with endogenous gene paramutation, although in Arabidopsis, a LUC paramutation transgene relies on multiple proteins including MET1 and CMT3/KYP (23). The involvement of CMT3 in maize is difficult to test because loss of function in the ZMET2 and ZMET5 homologs is not compatible with plant viability (24). However, the CHG subcontext methylation of B’, a highly paramutagenic maize allele at the b1 locus, has the CMT3 signatures (SI Appendix, Fig. S14). This pattern is consistent with ZMET2 and ZMET5 playing a role in the maintenance of b1 paramutation in maize. In addition, there are high H3K9me2 levels correlated with b1 and p1 paramutagenic alleles (25, 26). As with the subcontext data, this feature is consistent with a CMT3/KYP self-reinforcing loop in paramutation in maize as well as tomato.
The dynamics of sulf paramutation vary depending on the sulf epiallele used as parent. Some sulf epialleles transfer the repressive state in the F1, but with the sulf epialleles used here, the transfer likely occurs in the F2 (17). The sulf epiallele was transmitted and maintained in the CMT3/CMT3 homozygous F1 or CMT3/cmt3 heterozygous F1 and then progressively transferred between TAB2 alleles in the CMT3/CMT3 or CMT3/cmt3 F2 but not in the cmt3/cmt3 homozygous F2. While we cannot rule out that F2 plants with the sulf phenotype had inherited two silent alleles of TAB2, it is known that TAB2sulf homozygotes die at the seedling stage (5); therefore, a more likely scenario is that they germinated as TAB2sulf/+ and then progressed to TAB2sulf through paramutation shortly after germination.
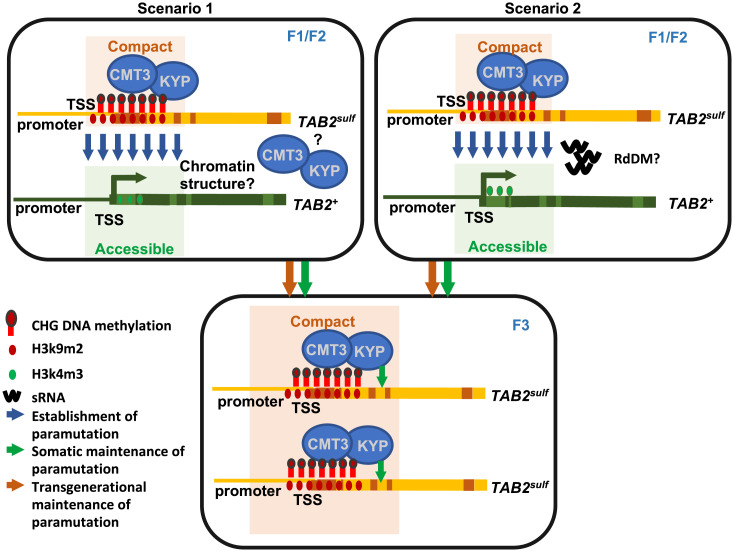
One scenario is that CMT3/KYP is required to make the TAB2+ chromatin sensitive to receive the paramutation silencing signal from a silenced allele (Fig. 4, Scenario 1). Alternatively, the paramutagenic allele (TAB2sulf) silences the paramutable allele (TAB2+) in the cmt3 homozygote, as it would do so in the CMT3 genotype; however, the silencing is not maintained, as CMT3 is needed to either complete the sulf silencing process or to allow the repressed state to persist (Fig. 4, Scenario 2). In the former scenario, the role of CMT3 would be in establishment, and in the latter, it would be in maintenance of paramutation (Fig. 4).
Fig. 4.
Model to explain the establishment (F1 or F2) and maintenance of the sulf paramutation in tomato. Scenarios 1 and 2 represent alternative hypotheses to explain the establishment of the sulf paramutation, which are not mutually exclusive. In Scenario 1, the establishment of paramutation requires a CMT3/KYP–dependent compact chromatin conformation, which in turn facilitates the exchange of epigenetic marks. In Scenario 2, the initial round of DNA methylation at TAB2 is initiated by RdDM and sRNAs. We show that CMT3/KYP is required either to complete silencing or/and maintain TAB2sulf in subsequent cell divisions and across generations. Yellow/orange boxes represent the silenced paramutagenic epiallele TAB2sulf, which exists in a compact chromatin conformation. Green boxes represent the active paramutable TAB2+ epiallele in which chromatin exists in a less compact structure and therefore remains more accessible to the transcription machinery.
The involvement of KYP and the changes to the chromatin modifications and organization of TAB2 in sulf are consistent with all of these hypotheses. However, the finding that CMT3 is absolutely required to maintain sulf epigenetic memory (SI Appendix, Figs. S4 and S5) implies a role of CMT3/KYP and the associated change in chromatin architecture in maintenance. This finding contrasts with demonstration that paramutated states persist through backcrosses with maize RdDM mutants (10). The various CMT3 hypotheses are not, however, mutually incompatible because TAB2 methylation could be lost directly in cmt3 gametes in F2 plants or during F3 early development in cmt3/cmt3 plants.
The accepted paradigm for establishment of paramutation invokes sRNAs in the communication between interacting alleles. In our sulf system, however, there is a very low level of (24-nt) sRNA accumulation at TAB2sulf (SI Appendix, Fig. S8) (16), and loss or reduced function of PolV does not lead to loss of sulf silencing (Fig. 2). These findings are not easy to reconcile with a simple role of RdDM in the maintenance of sulf paramutation as previously described (16), although we cannot rule it out conclusively in terms of establishment (Fig. 4). It could also be that there is residual PolV function in the mutant line and that sulf sRNA is abundant at specific developmental stages when communication between alleles takes place. Although unlikely, we cannot also exclude that in nrpe1 pollen (used here to introgress nrpe1 in the sulf background) sRNA populations are less affected than maternal sRNAs (27), thereby decreasing the effect of NPRE1 loss in the sulf phenotype.
Even in maize, however, the 24-nt sRNA role in paramutation also remains unclear. For instance, RMR1 and RMR7 are not required for paramutation establishment at Pl1-Rhoades despite affecting 24-nt sRNA accumulation (13, 14). In addition, paramutation is affected by genetic backgrounds with normal 24-nt sRNA levels (15). Furthermore, sRNA accumulation is not sufficient to direct paramutation at b1, indicating that other factors must be involved (28).
In summary, our results indicate that models of paramutation should accommodate the involvement of CMT3/KYP and changes in chromatin structure in at least the maintenance/memory phase if not in establishment (Fig. 4). These models should be open to the possibility that there could be locus-specific and/or species-specific mechanisms of paramutation and that there could be communication between alleles by mechanisms other than through sRNA and RdDM. Further understanding of chromatin organization and the 4D genome will also be necessary to understand how paramutation, uniquely among epigenetic phenomena, enables communication between alleles.
Materials and Methods
Plant Growth.
Plants were germinated and grown in F2 compost and transferred to John Innes 2 compost 3 wk after germination. Plants were grown in 16-h light (22 °C) and 8-h dark (18 °C) cycles with 70% humidity and light intensity 300 µmol l× m−2 × s−1 photosynthetically active radiation (PAR). Extended plant methods and genotyping details are provided in the SI Appendix.
McrBC Digestion.
Genomic DNA was isolated from 100 mg leaf tissue using DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The DNA samples used for McrBC digestion were the same as the samples used for genotyping. McrBC digestion was performed as described in (29). Details about qPCR are provided in the SI Appendix.
Expression Analysis.
Total RNA isolation was performed with the Direct-zol RNA Miniprep (Zymo Research) and TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For reverse-transcription qPCR analyses, 1 μg of total RNA was reverse transcribed using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions with random hexamer primers. Details about qPCR are provided in the SI Appendix.
ChIP.
Chromatin extraction was carried out as described in (18) using 1 g of leaf tissue of 4-wk-old plants. Chromatin was fragmented between 200 and 600 bp in a Covaris E220 evolution (duty cycle: 20%, peak intensity: 140, cycles of burst: 200, time: 3 min). Immunoprecipitation was carried out as described in (18) with Anti-Histone H3 (di methyl K9) antibody–ChIP Grade (Abcam, ab1220) and Anti-Histone H3 (tri methyl K4) antibody–ChIP Grade (Abcam, ab8580). The material was reverse cross-linked by adding NaCl to a final concentration of 200 mM and incubating at 65 °C overnight. This was followed by a 30-min treatment with 1 μL RNase A (Thermo Fisher Scientific) at 37 °C and a 90-min treatment with 1.5 µL Proteinase K (Thermo Fisher Scientific) at 65 °C. DNA was purified using the MinElute Kit (Qiagen) according to manufacturer’s instructions and eluted in 35 µL. Details about qPCR are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Melanie Steer for the invaluable horticultural support. We thank Pawel Baster, James Barlow, Neuza Duarte, and Jean-Francois Popoff for technical support. We also thank Christoph Lambing and Paul Fransz for the effort of troubleshooting fluorescent in situ hybridization (FISH) experiments, which weren’t successful and could not be included in this manuscript. Finally, we thank all the colleagues who supported this study through scientific discussions: Jake Harris, Ian Henderson, Hajk-Georg Drost, Sara Lopez-Gomollon, Hadi Putra, and Natasha Elina. This study was supported by Biotechnology and Biological Sciences Research Council Grant No. RG87392.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112240119/-/DCSupplemental.
Data Availability
Sequencing data have been deposited in Array Express (accession Nos. E-MTAB-10556, E-MTAB-10557, E-MTAB-10565, E-MTAB-10568, and E-MTAB-10574). All other study data are included in the article and/or SI Appendix.
Previously published data were used for this work (DOI: 10.1093/jxb/erw096 BioProject SRP066362).
References
- 1.Hollick J. B., Paramutation and related phenomena in diverse species. Nat. Rev. Genet. 18, 5–23 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Pilu R., Paramutation phenomena in plants. Semin. Cell Dev. Biol. 44, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Hövel I., Pearson N. A., Stam M., Cis-acting determinants of paramutation. Semin. Cell Dev. Biol. 44, 22–32 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Ellew C. P., The genetics of “rogues” among culinary peas (Pisum sativum). Proc. R. Soc. London. Ser. B, Contain. Pap. a Biol. Character 91, 186–195 (1920). [Google Scholar]
- 5.Hagemann R., [Somatic conversion in Lycopersicon esculentum Mill] [Article in German]. Z. Vererbungsl. 89, 587–613 (1958). [PubMed] [Google Scholar]
- 6.Regulski M., et al. , The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23, 1651–1662 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouil Q., Baulcombe D. C., BMC genomics paramutation-like features of multiple natural epialleles in tomato. BMC Genomics 19, 203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves I. K., Groszmann M., Wang A., Peacock W. J., Dennis E. S., Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. U. S. A. 111, 2017–2022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Dorweiler J. E., et al. , Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12, 2101–2118 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stonaker J. L., Lim J. P., Erhard K. F. Jr., Hollick J. B., Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 5, e1000706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuthikattu S., et al. , The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbour J. E. R., et al. , Required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell 24, 1761–1775 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale C. J., Stonaker J. L., Gross S. M., Hollick J. B., A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 5, e275 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deans N. C., Giacopelli B. J., Hollick J. B., Locus-specific paramutation in Zea mays is maintained by a PICKLE-like chromodomain helicase DNA-binding 3 protein controlling development and male gametophyte function. PLoS Genet. 16, e1009243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouil Q., Novák O., Baulcombe D. C., SLTAB2 is the paramutated SULFUREA locus in tomato. J. Exp. Bot. 67, 2655–2664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagemann R., Somatische Konversion (Paramutation) am sulfurea Locus von Lycopersicon esculentum Mill. IV. Die genotypische Bestimmung der Konversionshäufigkeit [Article in German]. Theor. Appl. Genet. 39, 295–305 (1969). [DOI] [PubMed] [Google Scholar]
- 18.Wang Z., Baulcombe D. C., Transposon age and non-CG methylation. Nat. Commun. 11, 1221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouil Q., Baulcombe D. C., DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12, e1006526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemann R., Somatische Konversion am sulfurea Locus von Lycopersicon esculentum Mill. II. Weitere Beweise für die somatische Konversion. Die Kult. 14, 171–200 (1966). [Google Scholar]
- 21.Lindroth A. M., et al. , Requirement of chromomethylase3 for maintenance of CpXpG methylation. Science 292, 2077–2080 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E., Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Zheng Z., et al. , Involvement of multiple gene-silencing pathways in a paramutation-like phenomenon in Arabidopsis. Cell Rep. 11, 1160–1167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., et al. , Genetic perturbation of the maize methylome. Plant Cell 26, 4602–4616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekhon R. S., Wang P. H., Sidorenko L., Chandler V. L., Chopra S., Maize unstable factor for orange1 is required for maintaining silencing associated with paramutation at the pericarp color1 and booster1 loci. PLoS Genet. 8, e1002980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haring M., et al. , The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 63, 366–378 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Lu J., Zhang C., Baulcombe D. C., Chen Z. J., Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 109, 5529–5534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arteaga-Vazquez M., et al. , RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc. Natl. Acad. Sci. U.S.A. 107, 12986–12991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bond D. M., Baulcombe D. C., Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 917–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in Array Express (accession Nos. E-MTAB-10556, E-MTAB-10557, E-MTAB-10565, E-MTAB-10568, and E-MTAB-10574). All other study data are included in the article and/or SI Appendix.
Previously published data were used for this work (DOI: 10.1093/jxb/erw096 BioProject SRP066362).