In PNAS, Ren et al. (1) report histone demethylase KDM5B (also called JARID1B or PLU1) as a suppressor of acute myeloid leukemia (AML). AML is one of the most frequently diagnosed adult cancers and is the most common type of acute leukemia (2). It is a heterogeneous group of cancers that arise from clonal expansion and differentiation arrest of myeloid progenitors in the bone marrow. Chromosomal translocation is a hallmark event driving AML initiation. Among different AML subtypes, recurring t(5;11)(q35;15.5) that causes fusion of nuclear receptor–binding SET domain protein 1 (NSD1) to nucleoporin-98 (NUP98) occurs in 16% of pediatric AML (3). NUP98-NSD1–positive AML patients exhibit poor prognosis, with 4-y event-free survival of less than 10% (3), which highlights the urgent demand for new treatment strategies for these patients.
Ren et al. (1) show that UNC1999, a dual inhibitor of the histone H3K27 methyltransferases enhancer of zeste homolog 1/2 (EZH1/2), significantly inhibited the clonogenic growth and induced the differentiation and apoptosis of murine AML cells driven by NUP98-NSD1. Consistently, UNC1999 treatment prolonged the survival of mice injected with these AML cells. These observations are reminiscent of previous studies that revealed the efficacy of the EZH2 inhibitor on AML driven by MLL-AF9, MLL-ENL, MLL-AF10, or MOZ-TIF2 transformation (4, 5). These results support the development of EZH2-targeted therapy to treat AML.
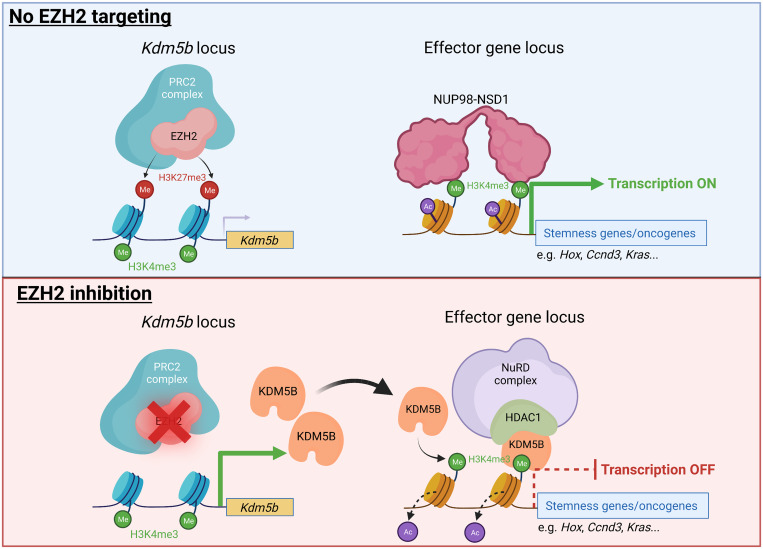
EZH2 is the catalytic subunit of polycomb repressive complex 2 (PRC2). To search for key downstream regulators that become reactivated and account for the antileukemia effect of EZH2 inhibition, Ren et al. (1) compared overlapping transcriptomic changes of AML cells after treatment with EZH2 inhibitors or down-regulation of embryonic ectoderm development (EED), an essential component of PRC2. Kdm5b was identified as one of the 31 consistently induced genes. Chromatin immunoprecipitation sequencing (ChIP-seq) analysis showed that the transcriptional start site of Kdm5b is marked by EZH2, the H3K27me3 repressive mark catalyzed by EZH2, and the H3K4me3 activating mark (1). These suggest that Kdm5b is a direct target of EZH2 poised for transcriptional activation or repression and could have a tumor-suppressive role in AML.
In fact, Ren et al. (1) then demonstrated the tumor suppressor role of Kdm5b in AML through Kdm5b knockdown experiments. Ablation of Kdm5b further enhanced AML clonogenic growth and desensitized AML cells to UNC1999 treatment both in vitro and in vivo. RNA sequencing analyses of AML cells with Kdm5b perturbation consistently showed a negative relationship between KDM5B level and transcriptional activity of genes that promote proliferation and stemness. These findings were consistent with a previous study that indicated the tumor-suppressor role of KDM5B in mixed lineage leukemia (MLL)–rearranged AML (6). Furthermore, Ren et al. (1) showed that low KDM5B expression is significantly correlated with poor survival of human AML patients, reinforcing the notion that KDM5B serves as a tumor suppressor in AML. However, these results contrast with the findings in solid tumors, including melanoma (7, 8) and estrogen receptor–positive breast cancer (9), that indicate the oncogenic and stem-like functions of KDM5B. The mechanisms that dictate the context-dependent roles of KDM5B in cancers remain to be investigated.
NUP98-NSD1 fusion is an oncoprotein mainly because it binds to the regulatory elements of the locus harboring the homeobox (HOX) gene cluster. HOX genes encode a highly conserved family of transcription factors that are required to maintain stemness in both hematopoietic stem cell and AML driven by MLL-AF9 and NUP98-NSD1 (10, 11). Ren et al. (1) performed ChIP-seq to show that KDM5B colocalizes with NUP98-NSD1 to the promoter-proximal sequence of the Hoxa locus as well as genes that either maintain stemness or drive proliferation, such as Sox4, Ccnd3, and Kras. This implies that KDM5B competes with NUP98-NSD1 for binding to the H3K4me3 transcriptional active mark to repress the effector genes that drive AML.
KDM5B is a large multidomain protein with an AT rich interaction domain (ARID) domain for DNA binding, a jumonji C (JmjC) domain that confers the histone demethylase activity to erase H3K4me3, and three plant homeodomain (PHD) fingers. In order to determine the role of different domains in KDM5B to suppress AML, Ren et al. (1) ectopically expressed the wild-type (WT) and a series of KDM5B mutants in the NUP98-NSD1+ AML model. Surprisingly, all domains, except the catalytic JmjC domain, abolished the antileukemic activity of KDM5B both in vitro and in vivo. Ren et al. (1) further performed ChIP-seq to show that the WT and catalytic inactive mutant of KDM5B not only substantially overlap globally with each other to promoter-proximal sequences, but also locally to the similar set of stemness and proliferation genes that are also bound by NUP98-NSD1. Such a high similarity of chromatin localization patterns between these two forms of KDM5B also supports the catalytic-independent role of KDM5B to suppress AML.
Ren et al. (1) conclude the study by showing that both WT and catalytic-inactive KDM5B interact with the histone deacetylase HDAC1, which is a component of the transcriptional repressive nucleosome remodeling and deacetylase (NuRD) complex. Although the authors did not further examine the importance of the NuRD complex in AML, these observations are consistent with two previous studies in breast cancer cells, which showed that KDM5B interacts with the NuRD complex to repress the expression of genes that drive proliferation, migration, and angiogenesis (12, 13). Li et al. (12) showed that histone lysine demethylase KDM1A (also called LSD1) also interacts with the KDM5B/NuRD complex to suppress gene transcription. However, the epigenetic regulatory roles of these two histone demethylases within the same repressor complex were not further investigated. On the other hand, Klein et al. (13) showed that the reader function of the PHD1 domain toward unmethylated H3K4 is critical for KDM5B to inhibit migration.
A recent study published by our group also unveiled the noncatalytic role of KDM5B in regulating gene expression and melanoma progression (8). When KDM5B was depleted, retroelements became actively transcribed to induce cytosolic RNA- and DNA-sensing pathways, which in turn, trigger type I interferon signaling to mount antitumor immune responses. However, inhibitors of KDM5 demethylases were unable to induce type I interferon responses, suggesting that the oncogenic activity of KDM5B in melanoma is independent of its demethylase activity (8). Consistently, both WT and catalytic-inactive KDM5B were able to restore tumorigenicity of Kdm5b knockout melanoma cells (8). Furthermore, we found that KDM5B recruits the H3K9 methyltransferase SETDB1 to repress the expression of retroelements (8). The demethylase activity–independent function of KDM5 proteins is not limited to human cancers and has previously been elucidated in Drosophila (14). However, the contextual mechanisms that determine KDM5B to execute either its histone demethylase activity or chromatin reader/scaffolding function remained to be identified.
The work by Ren et al. (1) shows a prominent antileukemic effect when Kdm5b is induced. Previous studies in our group showed that two pan-KDM5 inhibitors CPI-48 and KDM5-C70 induced KDM5B protein level while the demethylase activity was suppressed (15). Although the mechanism of this up-regulation remains to be determined, these observations suggest that KDM5B induction by pan-KDM5 inhibitors can be repurposed to inhibit AML.
In summary, EZH2 inhibition elicits an antileukemic effect in AML driven by NUP98-NSD1 and MLL-rearranged transformation. Mechanistically, epigenetic repression of Kdm5b transcription by EZH2 is important to prevent KDM5B from interacting with the NuRD complex to condense chromatin surrounding the set of effector genes driving AML progression (Fig. 1). Ren et al. (1) identified the tumor suppressor role of KDM5B in AML and highlighted the importance of the noncanonical scaffolding function of KDM5B in epigenetic regulation of its target genes.
Fig. 1.
The molecular basis of KDM5B activation to suppress AML. (Upper) In AML cells, KDM5B is normally suppressed by the transcriptional repressive H3K27me3 mark at its promoter-proximal sequence. NUP98-NSD1 fusion oncoprotein drives the expression of genes that are required to maintain stemness and oncogenic features of AML. (Lower) EZH2 inhibition leads to reactivation of KDM5B transcription. KDM5B binds to the H3K4me3 at the regulatory sequence of stemness genes/oncogenes, replaces NUP98-NSD1, and recruits the NuRD complex to deacetylate the histone and shut down these genes.
Acknowledgments
This work is supported by NIH Grants R01CA237586 (to Q.Y.) and P50CA121974 (to Q.Y.). The figure is created with https://biorender.com/.
Footnotes
The authors declare no competing interest.
See companion article, “A PRC2–Kdm5b axis sustains tumorigenicity of acute myeloid leukemia,” 10.1073/pnas.2122940119.
References
- 1.Ren Z., et al. , A PRC2–Kdm5b axis sustains tumorigenicity of acute myeloid leukemia. Proc. Natl. Acad. Sci. U.S.A. 119, 10.1073/pnas.2122940119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shallis R. M., Wang R., Davidoff A., Ma X., Zeidan A. M., Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 36, 70–87 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Hollink I. H., et al. , NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 118, 3645–3656 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Xu B., et al. , Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 125, 346–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita S., et al. , Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia 32, 855–864 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Wong S. H., et al. , The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell 28, 198–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roesch A., et al. , A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S. M., et al. , KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 598, 682–687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinohara K., et al. , KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 34, 939–953.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krivtsov A. V., et al. , Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Wang G. G., Cai L., Pasillas M. P., Kamps M. P., NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 9, 804–812 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Li Q., et al. , Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 71, 6899–6908 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Klein B. J., et al. , The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6, 325–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Secombe J., The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep. 13, 2219–2231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., et al. , KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 16, e2006134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]