Significance
DNA damage causes loss of or alterations in genetic information, resulting in cell death or mutations. Ionizing radiations produce local, multiple DNA damage sites called clustered DNA damage. In this study, a complete protocol was established to analyze the damage complexity of clustered DNA damage, wherein damage-containing genomic DNA fragments were selectively concentrated via pulldown, and clustered DNA damage was visualized by atomic force microscopy. It was found that X-rays and Fe ion beams caused clustered DNA damage. Fe ion beams also produced clustered DNA damage with high complexity. Fe ion beam–induced complex DNA double-strand breaks (DSBs) containing one or more base lesion(s) near the DSB end were refractory to repair, implying their lethal effects.
Keywords: clustered DNA damage, atomic force microscopy, ionizing radiation, DNA repair
Abstract
Clustered DNA damage is related to the biological effects of ionizing radiation. However, its precise yield and complexity (i.e., number of lesions per damaged site) in vivo remain unknown. To better understand the consequences of clustered DNA damage, a method was established to evaluate its yield and complexity in irradiated cells by atomic force microscopy. This was achieved by isolating and concentrating damaged DNA fragments from purified genomic DNA. It was found that X-rays and Fe ion beams caused clustered DNA damage in human TK6 cells, whereas Fenton's reagents did it less efficiently, highlighting clustered DNA damage as a signature of ionizing radiation. Moreover, Fe ion beams produced clustered DNA damage with high complexity. Remarkably, Fe ion beam–induced complex DNA double-strand breaks (DSBs) containing one or more base lesion(s) near the DSB end were refractory to repair, implying the lethal effect of complex DSBs.
Ionizing radiation produces various types of DNA damage: base damage, apurinic/apyrimidinic (AP) sites, DNA single-strand breaks (SSBs), DNA double-strand breaks (DSBs), and DNA–protein crosslinks (DPCs) (1–4). These DNA lesions may lead to biological effects such as cell death, mutations, and ultimately cancers. It is of note that endogenous DNA lesions induced by the Fenton reaction of hydrogen peroxide in cells are chemically indistinguishable from the individual lesions induced by ionizing radiation (5, 6). However, even though there is no significant difference in the type and the total amount of damage produced by ionizing radiation and the Fenton reaction, the biological effects differ significantly between these agents (7, 8). From these observations, it is considered that one of the signatures of ionizing radiation is DNA damage clustering, while endogenous and exogenous chemical agents generate mainly isolated DNA damage (9, 10). Simulation studies on the radiation track structure also suggest that ionizing radiation frequently induces clustered DNA damage and that its yield and damage complexity (i.e., the number of lesions per damaged site) relative to isolated lesions increase with increasing linear energy transfer (LET) of the radiation (11–14). Ionizing radiation produces simple clustered DNA damage (damage complexity = 2) and complex clustered DNA damage (damage complexity ≥ 3). Simple clustered DNA damage includes the simple base damage cluster (BDC) that contains two vicinal base and/or AP lesions and a simple DSB that contains no associated base/AP lesions near the DSB end. Complex clustered DNA damage include the complex BDC that contains more than three vicinal base and/or AP lesions and a complex DSB that contains one or more base and/or AP lesion(s) near the DSB end. Currently, much attention is paid to complex BDCs and complex DSBs to understand the biological effect of ionizing radiation.
Clustered DNA damage is believed to be mainly responsible for the detrimental effects of ionizing radiation, because the repair of clustered DNA damage is often compromised (15–21). In vitro studies using chemically synthesized defined DNA lesions have revealed that clustered DNA damage with a certain configuration is refractory to repair. In the case of BDCs, the repair efficacy is governed by the separation between the lesions and the type of lesions within a cluster. Furthermore, there is hierarchy on the damage excision that depends on the type of lesions. Compromised repair of lesions in a cluster is further supported by the findings in vivo, in which replication inhibition or mutation induction is readily observed depending on the configuration of the lesions in a cluster. In the case of complex DSBs, their rejoining is very often retarded in vitro (22–26), and when they are rejoined, the excision of base damage from the termini likely follows the rejoining (15, 24–26).
Many studies have analyzed the yield of radiation-induced BDCs in vivo. The incision of bistranded base or AP lesions by DNA glycosylases with an AP lyase activity or AP endonucleases greatly facilitated the detection of simple BDCs, since these were converted to DSBs, allowing quantitative measurements by gel electrophoresis (16, 27–30). From these studies, it was estimated that BDCs were formed several times more frequently than DSBs upon the exposure of cells to low-LET radiations. More recently, the localization of DNA damage was estimated using fluorescence resonance energy transfer between fluorescent dyes that were labeled to AP sites (31–33). The in vivo processing of radiation-induced BDCs has also been examined and demonstrated to be inefficient. For instance, the processing of bistranded AP clusters in γ-irradiated human cells was much slower than that of DSBs and took over 14 d (34). Similarly, clustered DNA damage generated by irradiation of human monocytes with Fe ion beams was repaired over 4 to 5 d, while DSBs were repaired within 1 to 2 d (35). Interestingly, a transient increase of the amount of DSBs is often observed after irradiation and is attributed to the conversion of bistranded BDCs to DSBs by attempted base excision repair (36–38). In contrast to the case of BDCs, very few pieces of information have been accumulated on the in vivo yield of complex DSBs. In previous studies using immunofluorescent staining and microscopy, complex DSBs were indirectly detected by observing the colocalization of DSB foci (53BP1 or γ-H2AX) and base damage/AP site foci (XRCC1, hOGG1, or APE1) in cells (39–41). However, the sizes of observed foci encompassed a few megabase pairs (21). Thus, whether the colocalization of the foci is indeed due to the clustered DNA damage generated within one to two helical turns of DNA remains to be clarified (20). The inefficient repair of complex DSBs is inferred from the observations that DSBs induced by high-LET radiations are repaired much slower than those induced by low-LET radiations (42, 43), although alternative mechanisms, such as clustering of DSBs, could account for their inefficient repair (44–47).
Despite extensive studies, the information on the structural complexity of radiation-induced clustered DNA damage and its influence on repair and biological consequences in vivo still remains limited. The conventional method using gel electrophoresis in the detection of BDCs only reveals that at least one base/AP lesion on each DNA strand is present in close proximity and is unable to determine whether there are additional lesions nearby. In addition, to date, the extent of damage complexity around the DSB end and how the complexity affects the repair process in vivo have hardly been elucidated. To investigate the yield and complexity of clustered DNA damage, we have recently developed a method to directly observe the cluster DNA damage in plasmid DNA irradiated in vitro (48). By labeling DNA damage sites with an aldehyde reactive probe (ARP) containing a biotin moiety and subsequently with streptavidin, we enabled direct visualization of damage sites with atomic force microscopy (AFM). We showed that high-LET Fe ion beams produced more cluster DNA damage than low-LET X-rays and that the complexity of clustered DNA damage was higher with Fe ion beams than with X-rays. The proportion of the types of X-ray-induced clustered DNA damage observed with AFM was qualitatively consistent with that calculated by the simulation of radiation track structure (49).
In the present study, we have extended our in vitro study (48) to in vivo. The human TK6 cells were irradiated with ionizing radiation (X-rays and Fe ion beams) or treated with the Fenton’s reagents and the genomic DNA was isolated from cells. The damage-containing DNA fragments were selectively concentrated by pulldown with the streptavidin magnetic beads and analyzed for clustered DNA damage using AFM.
Results
Outline of DNA Damage Analysis.
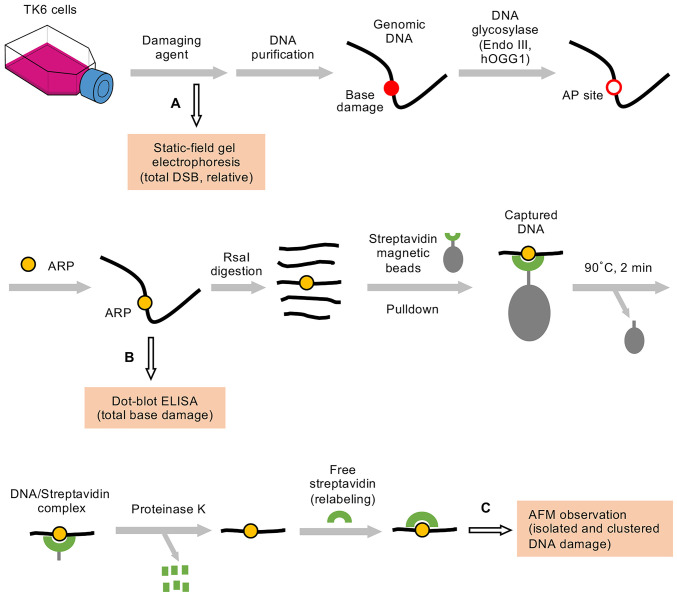
Fig. 1 shows the outline of DNA damage analysis in this study. TK6 cells were treated with ionizing radiation or Fenton’s reagents, and the genomic DNA was purified from the cells by CsCl density gradient centrifugation (18, 19). The purified DNA was treated with DNA glycosylases (hOGG1 and Endo III) to convert base lesions to AP sites. hOGG1 excises oxidative purine damage to form AP sites (a mixture intact and nicked forms), while Endo III excises oxidative pyrimidine damage to form nicked AP sites (50). Ionizing radiation and Fenton’s reagents produced AP sites directly together with base damage. In this study, these AP sites were also considered as base damage, since AP sites generated by DNA glycosylases and DNA-damaging agents were indistinguishable in the present method of DNA damage detection with AFM (Fig. 1). The AP sites were labeled with ARP containing a biotin tag. The total DNA damage was quantified by dot-blot enzyme-linked immunosorbent assay (ELISA) using horseradish peroxidase (HRP)–biotin antibodies (20). Alternatively, the DNA was digested with RsaI to reduce its size to around 1.1 kbp. DNA fragments containing ARP (i.e., damage) were selectively concentrated by pulldown with streptavidin magnetic beads, and clustered DNA damage was analyzed using AFM.
Fig. 1.
Outline of DNA damage analysis. TK6 cells were treated with ionizing radiation or Fenton’s reagents. The total DSBs (possibility a mixture of simple and complex DSBs) that were directly produced in the genomic DNA by ionizing radiation or Fenton’s reagents were analyzed by static-field gel electrophoresis (path A). Alternatively, the genomic DNA was purified from the cells by CsCl density gradient centrifugation. The purified DNA was treated with DNA glycosylases (hOGG1 and Endo III) to convert base lesions to AP sites. Ionizing radiation and Fenton’s reagents produced AP sites directly together with base damage. The AP sites were included in the base damage in this study, since the AP sites generated by the DNA glycosylases and DNA-damaging agents cannot be distinguished in the present method of DNA damage detection with AFM. The AP sites were labeled with ARP containing a biotin tag. The total DNA damage (as base damage) was quantified by dot-blot ELISA using HRP-biotin antibodies (path B). Alternatively, DNA was digested with RsaI to reduce its size to around 1.1 kbp. DNA fragments with ARP (i.e., damage) were selectively concentrated by pulldown with streptavidin magnetic beads. ARP-containing DNA/streptavidin complexes (not free DNA) were released from the beads by a brief heat treatment (90 °C, 2 min) and digested with proteinase K. The ARP in DNA was fully relabeled with free streptavidin, and DNA damage was observed with AFM (path C).
Quantification of Total Base Damage by Dot-Blot ELISA.
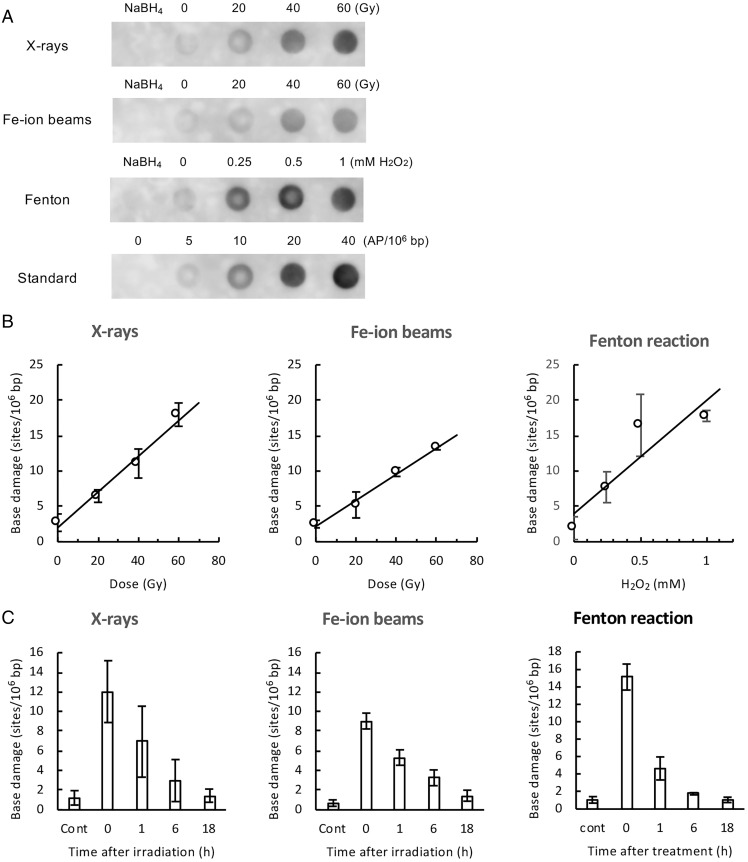
The total base damage produced by ionizing radiation or the Fenton reaction in the genomic DNA of TK6 cells was quantified by dot-blot ELISA (Fig. 2A), as described in Materials and Methods. The amount of base damage increased with increasing dose of X-rays or Fe ion beams (Fig. 2B). Similarly, treatment with Fenton’s reagents resulted in an increase in the amount of base damage in a concentration-dependent manner of hydrogen peroxide (Fig. 2B). The assay revealed that the genomic DNA of TK6 cells contained about 2.4 endogenous base damage per 106 bp (Table 1). According to the slopes of the dose–response plots (Fig. 2B), X-rays and Fe ion beams produced about 0.25 and 0.18 base damage per 106 bp per Gy, respectively (Table 1). The yield of total base damage with Fe ion beams was lower than that with X-rays. This is consistent with previous observations, in which the in vivo yield of DNA damage generally decreases with increasing LET of radiation (11, 36). The Fenton reaction induced about 16 base damage per 106 bp per mM hydrogen peroxide per h (Fig. 2B and Table 1). The amount of total base damage produced by both X-rays, Fe ion beams, and Fenton’s reagents decreased with postincubation time, and the damage was mostly repaired at 18 h (Fig. 2C).
Fig. 2.
Quantification of total base damage by dot-blot ELISA. The total base damage in the treated TK6 cells was quantified by dot-blot ELISA, as described in Materials and Methods (Fig. 1, path B). Purified genomic DNA was treated with Endo III and hOGG1, and the resulting aldehydic groups in DNA were labeled with ARP. ARP-labeled DNA (without RsaI digestion) was dot blotted on a membrane, and ARP labels (i.e., damage sites) in DNA were detected with HRP-biotin antibodies and ECL reagents. The amount of base damage present in DNA was calibrated from the signal intensity of DNA standards containing known amounts of AP sites. (A) Typical dot-blot ELISA images (chemiluminescence signals) of DNA obtained from cells treated with indicated doses of X-rays, Fe ion beams, and Fenton’s reagents. NaBH4 indicates NaBH4-treated DNA, showing nonspecific background signals of the assay. Standard denotes DNA samples containing zero to 40 AP sites per 106 bp, which were used for the calibration of chemiluminescence signals. (B) The amount of total base damage immediately after the indicated treatment as a function of the dose (Gy) of X-rays and Fe ion beams or the concentrations (mM) of hydrogen peroxide in Fenton reactions. (C) Changes in the amount of total base damage during postincubation. Cells were treated with X-rays/Fe ion beams (40 Gy) or Fenton’s reagents (1 mM hydrogen peroxide) and were incubated for up to 18 h. Base damage was quantified by dot-blot ELISA. “Cont” indicates untreated control cells. The data points were the average values of three independent experiments, while error bars indicate the SDs in B and C.
Table 1.
Yields of DNA damage in TK6 cells treated with DNA-damaging agents
| Method of analysis | Damage | Yield (sites/106 bp) | |||
|---|---|---|---|---|---|
| Endogenous | X-rays | Fe ion | Fenton | ||
| (/Gy) | (/Gy) | (/mM H2O2/h) | |||
| AFM | Isolated base damage | 2.579 | 0.330 | 0.134 | 21.17 |
| Simple BDC | 0.018 | 0.028 | 0.055 | 0.23 | |
| Complex BDC | ND* | 0.004 | 0.011 | ND* | |
| Complex DSB | 0.056 | 0.033 | 0.021 | 1.97 | |
| (Total clustered damage)† | (0.074) | (0.065) | (0.087) | (2.20) | |
| Total | 2.653 | 0.395 | 0.221 | 23.37 | |
| Dot-blot ELISA | Total | 2.376 | 0.252 | 0.184 | 15.99 |
*ND: not detected.
†Total clustered damage = Simple BDC + Complex BDC + Complex DSB.
AFM Observation of DNA Damage in Genomic DNA.
The amount of DNA damage produced in irradiated cells is expected to be low due to the high radical-scavenging environment of the cell interior. According to our previous study, the yield of DNA damage in cells was 18-fold lower than that in isolated DNA upon irradiation with the same dose of γ-rays (50). Therefore, a selective enrichment of damage-containing DNA can facilitate the AFM analysis of DNA damage produced in cells.
In this study, DNA fragments containing ARP (i.e., damage) were selectively concentrated by pulldown with streptavidin magnetic beads. The conditions to concentrate the biotin-labeled DNA were preliminarily investigated using a model DNA containing biotin-dT (SI Appendix, SI Text I and Fig. S1). The results showed that the biotin-labeled DNA was recovered about 16-fold more efficiently than that without the label. It was also revealed that DNA was released from the magnetic beads as biotin-labeled DNA/streptavidin complexes (i.e., not free DNA) upon brief heat treatment (90 °C for 2 min). For the genomic DNA of TK6 cells, DNA fragments containing ARP were similarly concentrated with streptavidin magnetic beads, as illustrated in Fig. 1. The amount of DNA recovered by pulldown was 3 to 4% of the input DNA (SI Appendix, SI Text II). According to the results of the quantitation of total base damage (Fig. 2B), the genomic DNA of treated cells contained the following number of base lesions per 106 bp: 6 to 18 (X-rays with 20 to 60 Gy), 5 to 13 (Fe ion beams with 20 to 60 Gy), and 8 to 18 (Fenton reaction with 0.25 to 1 mM hydrogen peroxide). These numbers are equivalent to those of damage-containing DNA fragments. Moreover, 106 bp DNA comprised 909 fragments of 1.1 kbp DNA (average size of DNA used for pulldown). Thus, with 106 bp DNA, the theoretical recovery (percentage) of damage-containing DNA fragments with the magnetic beads was calculated as follows: 100 × (number of damage-containing DNA fragments/total 909 DNA fragments). The calculated recovery (percentage) was 0.7 to 2.0% (X-rays), 0.6 to 1.4% (Fe ion beams), and 0.9 to 2.0% (Fenton reaction). The actual recovered DNA by pulldown was 3 to 4% of the input DNA. Although there were some differences between the theoretical percentages (0.6 to 2.0%) and actual ones (3 to 4%), both values were not significantly different from each other. Therefore, the pulldown of DNA with the magnetic beads enriched the damage-containing DNA fragments effectively (ca. 25- to 33-fold enrichment based on the 3 to 4% DNA recovery), facilitating the AFM analysis of DNA damage.
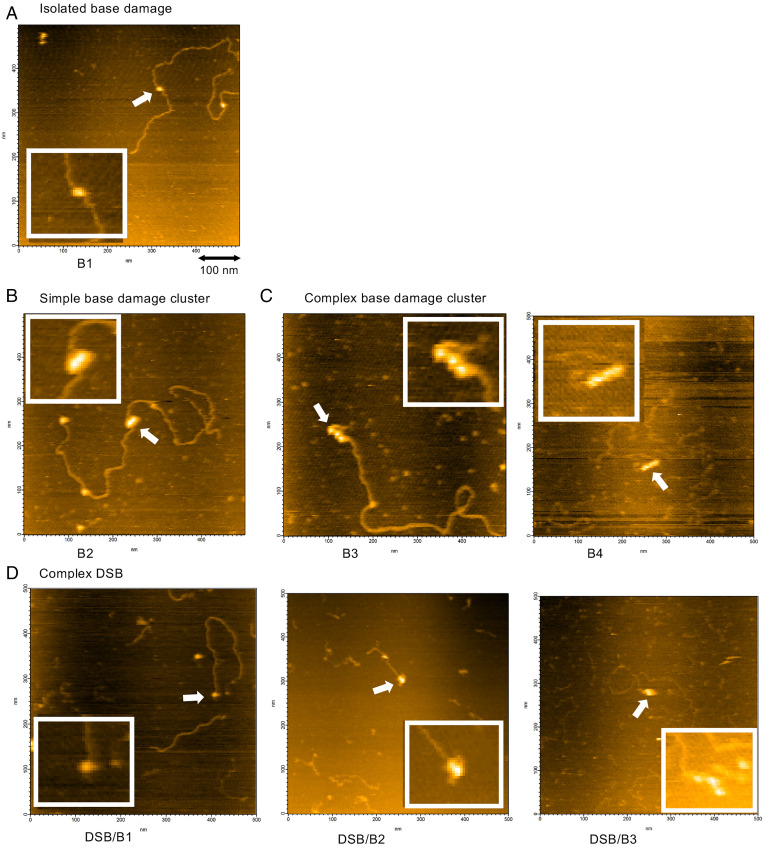
Ionizing radiation produces simple and complex clustered DNA damage. The AFM analysis of the genomic DNA from irradiated TK6 cells revealed isolated base damage (B1), simple BDCs (B2), complex BDCs (B3 and B4, among others), and complex DSBs (DSB/B1 and DSB/B2, among others). The number suffix to B indicates the number of base/AP lesions in the clustered damage. The prefix of “DSB/” to B indicates that base/AP lesion(s) are present near the DSB end. SI Appendix, Fig. S3 summarizes the configuration of DNA lesions, abbreviation, and damage complexity that were observed in this study. It was noted that simple DSBs that contain no associated lesions near the DSB end were not included in the present AFM analysis of DNA damage, since simple DSBs were present in every 1.1 kbp and were exclusively produced by RsaI digestion and not by DNA-damaging agents. Fig. 3 shows the typical DNA lesions observed in the genomic DNA of TK6 cells irradiated with Fe ion beams. Clustered DNA damage with damage complexity of up to 5 (DSB/B3) is shown. Although less frequently observed, two adjacent BDCs were also produced by Fe ion beams (SI Appendix, Fig. S4). As in our previous study (48), we used a slightly modified definition of clustered DNA damage in the present study. The conventional definition is a site containing two or more lesions within 10 to 20 bp, while our definition is a site containing two or more lesions within 37 bp. In our previous study that used a DNA model for clustered damage (48), two base lesions three or eight bases apart were observed as partially resolved streptavidin molecules in AFM imaging when these were tagged with streptavidin. Furthermore, the apparent diameter of a streptavidin molecule in AFM imaging was ∼12.5 nm, corresponding to the apparent DNA length of 37 bases. Therefore, in our AFM imaging, two base lesions with an interlesion distance up to 37 bases are all observed as partially resolved streptavidin molecules regardless of the interlesion distance. Therefore, a tentative interlesion distance threshold of 37 bp was introduced to distinguish isolated and clustered DNA damage, and it was considered that two or more streptavidin tags in contact with each other represent clustered DNA damage, whereas those with baseline separation represent isolated DNA damage.
Fig. 3.
AFM images of genomic DNA damage in TK6 cells irradiated with Fe ion beams. Representative images of genomic DNA damage produced by irradiation of TK6 cells with Fe ion beams (40 Gy) are shown. (A) Isolated base damage. (B) Simple BDC containing two vicinal base lesions (B2). (C) Complex BDCs containing three vicinal base lesions (B3, Left) and four vicinal base lesions (B4, Right). (D) Complex DSBs containing one base lesion (DSB/B1, Left), two base lesions (DSB/B2, Middle), and three base lesions (DSB/B3, Right) near the DSB end. The box within each image shows the enlarged view of the region indicated by the arrow. For the abbreviations of the types of clustered DNA damage, refer to SI Appendix, Fig. S3.
Types and Yields of Genomic DNA Damage Induced by X-rays, Fe Ion Beams, and Fenton Reactions.
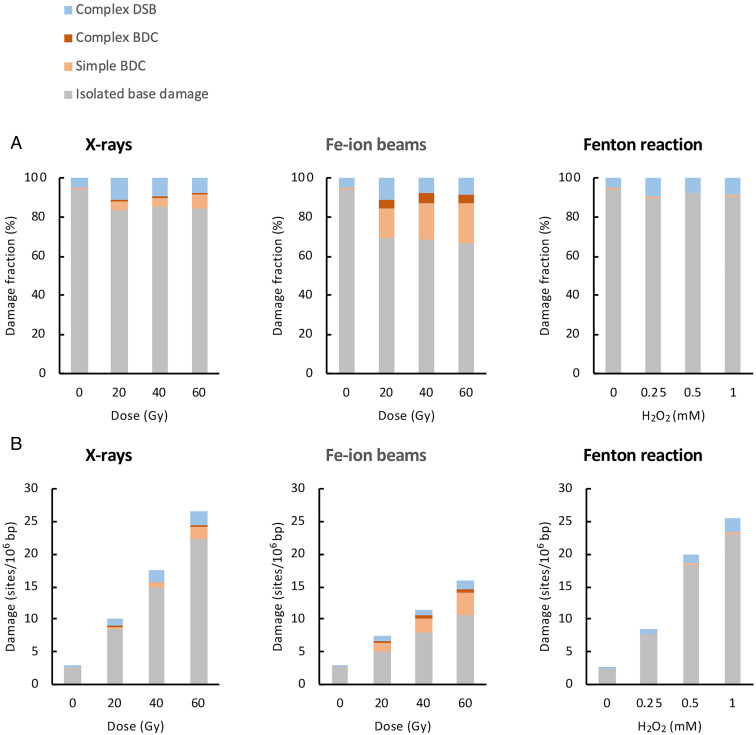
The genomic DNA damage in treated TK6 cells was analyzed with AFM. The DNA damage spectra for individual data points were determined by analyzing 300 DNA fragments containing DNA damage. The percentage of each type of DNA damage relative to the total damage (i.e., DNA damage spectra) was calculated and is shown in Fig. 4A, including the data for isolated base damage, simple and complex BDCs, and complex DSBs but not those for simple DSBs (AFM Observation of DNA Damage in Genomic DNA). The average percentages of isolated base damage, simple BDCs, complex BDCs, and complex DSBs were 84.7%, 5.2%, 0.7%, and 9.4%, respectively, for X-rays (20 to 60 Gy) and 68.0%, 18.3%, 4.4%, and 9.2%, respectively, for Fe ion beams (20 to 60 Gy). Accordingly, these data clearly demonstrate that densely ionizing Fe ion beams (LET = 200 keV/µm) produce clustered DNA damage in cells more frequently than sparsely ionizing X-rays (LET = 10 keV/µm). In the case of Fenton reactions (0.25 to 1 mM H2O2), the average percentages of isolated base damage, simple BDCs, complex BDCs, and complex DSBs were 90.8%, 0.9%, 0%, 8.3%, respectively, indicating that the major DNA damage in Fenton reactions was isolated base damage. This is consistent with previously reported results (8). For the formation of complex DSBs by Fenton reactions, refer below.
Fig. 4.
Spectra and yields of genomic DNA damage in treated TK6 cells. TK6 cells were treated with X-rays, Fe ion beams, and Fenton’s reagents, and the genomic DNA damage immediately after the treatments was analyzed by AFM. (A) Spectra of genomic DNA damage. The type of DNA damage for individual doses were determined by analyzing 300 damage-containing DNA fragments. The percentage of each type of DNA damage relative to the total damage (i.e., DNA damage spectra) is plotted for each dose. (B) Yields of genomic DNA damage. The yield of each type of DNA damage (sites/106 bp) was estimated from the DNA damage spectra (A) and percentages of recovered biotin-containing model DNA (SI Appendix, SI Text I and Fig. S1C) and damage-containing genomic DNA (SI Appendix, SI Text II) by pulldown with the magnetic beads. The dose–response plots of the yields of isolated base damage, simple and complex BDCs, and complex DSBs are shown as cumulative bar charts for each damaging agent.
The yield of each type of DNA damage (sites/106 bp) was estimated from the DNA damage spectra determined by AFM (Fig. 4A) and the percentages of recovered biotin-containing model DNA (SI Appendix, Fig. S1) and damage-containing genomic DNA (SI Appendix, Fig. S2) by pulldown with magnetic beads. The details of estimated yields of DNA damage are described in SI Appendix, SI Text III. The dose–response plots of the amounts of isolated base damage, simple and complex BDCs, and complex DSBs are shown in Fig. 4B. The rates of the formation of these lesions (sites/106 bp/Gy or sites/106 bp/mM H2O2/h) were evaluated from the slopes of the dose–response plots and are summarized in Table 1. For each damaging agent, the rate of the formation of total DNA damage determined by AFM agreed fairly well with that determined by dot-blot ELISA (Table 1), although these two methods used different measurements. This corroborates the validity of the DNA damage data obtained by AFM. The ratio of the formation rates of total clustered damage (simple and complex BDCs + complex DSBs) to isolated base damage was 1:1.5 (Fe ion beams), 1:5.1 (X-rays), and 1:9.6 (Fenton reaction), indicating that Fe ion beams produced clustered damage most efficiently. These ratios are comparable to those obtained when plasmid DNA was treated with the same agents in vitro (48) and are qualitatively consistent with the results of previous simulation studies (11–14, 49). Among the clustered damage, Fe ion beams produced simple and complex BDCs more efficiently than X-rays, while X-rays somehow produced complex DSBs slightly more efficiently than Fe ion beams (Table 1).
Complex DSBs were commonly observed for ionizing radiation (X-rays and Fe ion beams) and Fenton’s reagents. These were also observed as endogenous damage in untreated control cells (Fig. 4A), but their amounts were very low (Fig. 4B). Although the exact reason why Fenton’s reagents produced detectable complex DSBs was unknown, it was reported that the hydrogen peroxide used in the Fenton reaction induced the clustering of oxidative DNA damage and/or AP sites in the genomes of TK6 and chicken DT40 cells (15, 26). However, further studies are necessary to clarify the mechanism.
Repair of DNA Damage in Cells Induced by X-rays and Fe Ion Beams.
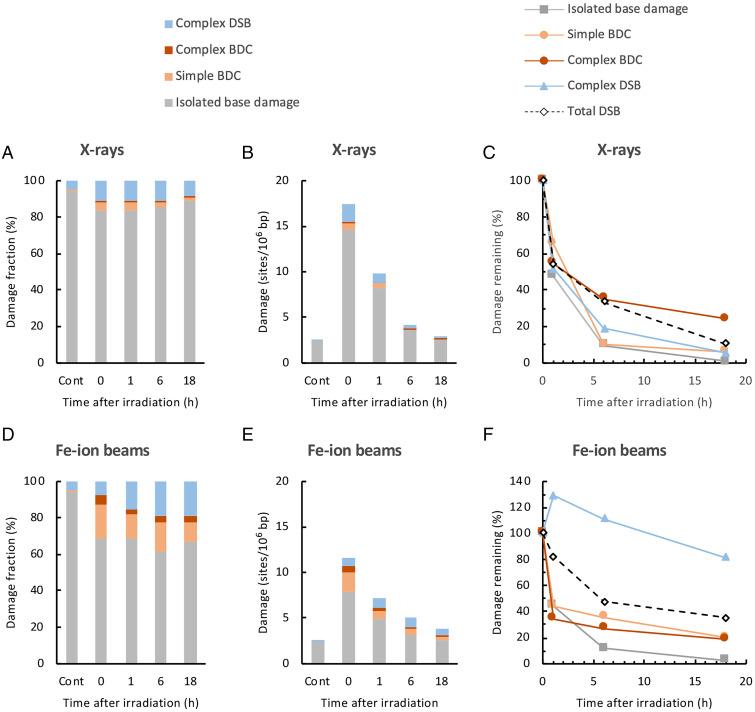
The repair of radiation-induced DNA damage in cells was studied. TK6 cells were irradiated with X-rays or Fe ion beams (40 Gy) and were incubated for up to 18 h. The genomic DNA was isolated at 0, 1, 6, and 18 h after irradiation, and the DNA damage spectra and amount of each type of DNA damage (sites/106 bp) were determined, as described in Types and Yields of Genomic DNA Damage Induced by X-rays, Fe Ion Beams, and Fenton Reactions and Fig. 4 A and B. In addition, the relative changes in total DSBs were measured by static-field gel electrophoresis for comparison (Fig. 5 C and F).
Fig. 5.
Repair of X-ray- and Fe ion beam–induced DNA damage in TK6 cells. TK6 cells were irradiated with 40 Gy of X-rays or Fe ion beams and incubated for up to 18 h. The genomic DNA from the cells was isolated after 0, 1, 6, and 18 h of irradiation, and DNA damage was analyzed by AFM. (A) Changes in DNA damage spectra for X-rays. The types of DNA damage were determined by analyzing 300 damage-containing DNA fragments. The percentage of each type of DNA damage relative to the total damage is shown as a cumulative bar chart. “Cont” indicates untreated control cells. (B) Changes in the amounts of DNA damage for X-rays. The amount of each type of DNA damage (sites/106 bp) was estimated as described in Fig. 4B. The amounts of isolated base damage, simple and complex BDCs, and complex DSBs are shown as a cumulative bar chart. (C) Time courses of the repair of each type of X-ray-induced DNA damage. The percentages of the amount of remaining damage relative to that immediately after irradiation (0 h) were calculated for each type of damage and are plotted against time after irradiation. The amounts of endogenous damage in untreated control cells were subtracted in the calculation of the data in C. The total DSBs were separately analyzed by static-field gel electrophoresis. (D) Changes in the DNA damage spectra. (E) Amounts of DNA damage for Fe ion beams. (F) Time courses of the repair of each type of Fe ion beam–induced DNA damage. The data were obtained as described in A through C for X-rays.
Fig. 5 A and B show the changes in the DNA damage spectra and amount of each type of DNA damage, respectively, with time after irradiation with X-rays. Based on the data in Fig. 5B and those for total DSBs, the percentages of the amount of remaining damage relative to that immediately after irradiation (0 h) were calculated for each type of damage and are plotted against the postincubation time in Fig. 5C. The values of endogenous damage for untreated cells were subtracted in the calculation of data shown in Fig. 5C. With X-rays, isolated base damage was repaired most efficiently, and 99% of the damage was repaired after 18 h. Simple BDCs and complex DSBs were also repaired. Complex BDCs were repaired, but the repair rate was somewhat lower than that of other types of clustered damage (simple BDCs and complex DSBs) (Fig. 5C). The repair efficiency of complex DSBs determined with AFM was virtually comparable to that of total DSBs determined by static-field gel electrophoresis (Fig. 5C).
Fig. 5 D–F show the changes in the DNA damage spectra, amount of each type of DNA damage, and percentage of the amount of remaining damage relative to that immediately after irradiation, respectively, with time after irradiation with Fe ion beams. With Fe ion beams, isolated base damage was repaired efficiently (Fig. 5F), similar to the case for X-rays. Simple and complex BDCs were also repaired, but their repair rates were somewhat lower than those of isolated base damage. The repair of complex DSBs induced by Fe ion beams were markedly retarded (Fig. 5F). Moreover, 81% of initially formed complex DSBs remained unrepaired after 18 h of incubation, while other types of damage were mostly repaired (unrepaired fraction was 2 to 20%). Interestingly, the amount of complex DSBs increased after 1 h of incubation; then, it decreased slowly (Fig. 5 E and F). A possible mechanism for this observation is discussed (Discussion). Similar to complex DSBs, the total DSBs were repaired by Fe ion beams more slowly than those by X-rays (Fig. 5F). In addition, 36% of the total DSBs remained unrepaired at 18 h. The retardation of the repair of Fe-induced complex DSBs may partly account for the impairment of the repair of the total DSBs. However, their amount did not increase after 1 h of incubation.
Complexity and Repairability of Fe Ion Beam–Induced Clustered DNA Damage.
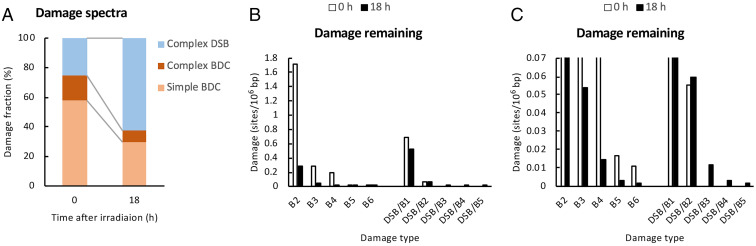
Fe ion beams produced clustered DNA damage with high efficiencies (Fig. 4 A and B). In addition, complex DSBs induced by Fe ion beams remained in the genome for up to 18 h after irradiation (Fig. 5F). To obtain further insights into the nature of clustered DNA damage induced by Fe ion beams, the numbers of DNA fragments containing clustered DNA damage for AFM analysis was increased to 531 and 636 after 0 h and 18 h of irradiation (40 Gy), respectively.
Fig. 6A shows the spectra of clustered DNA damage at 0 and 18 h after irradiation with Fe ion beams. Immediately after irradiation (0 h), simple/complex BDCs and complex DSBs accounted for 75% and 25% of the total clustered damage, respectively. However, after 18 h of incubation, the spectra of clustered DNA damage changed dramatically, and the fractions of simple/complex clustered base damage and complex DSBs were reversed to 37% and 63%, respectively. The observed changes in the damage spectra after incubation indicates that simple/complex BDCs and complex DSBs induced by Fe ion beams exhibit distinct repairabilities in cells.
Fig. 6.
Complexity and repairability of Fe ion beam–induced clustered DNA damage. TK6 cells were irradiated with Fe ion beams (40 Gy) and incubated for 18 h. Clustered DNA damage after 0 and 18 h of irradiation was analyzed by AFM. In AFM analysis, large numbers of DNA fragments containing clustered DNA damage (531 for 0 h and 636 for 18 h) were analyzed to obtain a detailed information about the damage spectra and repairability. (A) Changes in the spectra of clustered DNA damage after repair incubation. The percentages of BDCs (simple/complex) and complex DSBs relative to the total clustered damage are shown for 0 and 18 h. (B) Changes in the amount (sites/106 bp) of each type of clustered DNA damage after repair incubation for 18 h. The amount of each type of clustered DNA damage was calculated as described in Fig. 4B and is plotted against the type of clustered damage for 0 and 18 h. For abbreviations of the type of clustered DNA damage, refer to SI Appendix, Fig. S3. (C) Expanded view of the graph in B. The y-axis region between 0 and 0.07 in B was magnified to show the amounts of clustered DNA damage with very low amounts.
Fig. 6B shows the amount of each type of clustered DNA damage (sites/106 bp) at 0 h and 18 h after irradiation with Fe ion beams. The amounts of DNA damage were calculated, as described in SI Appendix, SI Text III. The data for 0 and 18 h show that Fe ion beams produced BDCs and complex DSBs with a damage complexity of up to 6 (B6) and 7 (DSB/B5), respectively. Immediately after irradiation (0 h), simple BDCs (B2) were the major clustered damage. The amounts of other types of complex BDCs and complex DSBs decreased with increasing damage complexity. After 18 h of incubation, the amounts of all types of BDCs (B2 through B6) significantly decreased (8 to 23% relative to 0 h), suggesting a complexity-independent efficient repair of BDCs in cells. Complex DSB/B1-type DSBs were the major clustered damage remaining after 18 h of incubation. Remarkably, the amounts of complex DSBs with damage complexities of 3 (DSB/B1) and 4 (DSB/B2) were virtually unchanged relative to 0 h (76% and 106%, respectively). Furthermore, complex DSBs with higher complexities of 5 to 7 were concurrently scored in AFM analysis at 18 h (Fig. 6C). Complex DSBs with higher complexities of 5 to 7 (DSB/B3, DSB/B4, and DSB/B5) were detected in the samples at 18 h (636 observed DNA fragments) but not at 0 h (531 observed DNA fragments), since the populations of these types of complex DSBs in the samples increased due to the concurrent decrease in the population of complex BDCs. Taken together, the observed complex DSBs with a damage complexity of up to 7 might be refractory to repair and remain for a long time in cells.
The cells were irradiated with a relatively high dose of Fe ion beams (40 Gy) during the analysis of the repair of clustered DNA damage (Fig. 6). The formation of excessive amount of DNA damage in the genome caused by the high radiation dose might affect the repair of clustered DNA damage. Therefore, the cells were irradiated with low-dose Fe ion beams (5 Gy), and the repair of clustered DNA damage was analyzed (SI Appendix, Fig. S5). Although the number of observed clustered DNA damage was not high owing to the low dose (90 and 87 clustered damages after 0 h and 18 h of irradiation, respectively), the repair of complex DSBs but not simple/complex BCDs was retarded. Accordingly, the result with 5 Gy was generally consistent with that with 40 Gy.
Discussion
Detection of Clustered DNA Damage in Genomic DNA.
In this study, a method was established to analyze the radiation-induced clustered DNA damage in the genomic DNA of cells (Fig. 1). The keys to a successful analysis are twofold: enrichment of damage-containing DNA fragments by pulldown with magnetic beads and a direct observation of streptavidin-tagged damage sites with AFM. Using this method, the clustered DNA damage produced in TK6 cells was analyzed. The results showed that X-rays and Fe ion beams generated various types of clustered DNA damage in vivo, including simple/complex BDCs and complex DSBs, in a dose-dependent manner (Fig. 4). Densely ionizing Fe ion beams induced clustered DNA damage more efficiently than sparsely ionizing X-rays. Fenton’s reagents induced clustered DNA damage less efficiently. These observations are consistent with those obtained in our previous work, in which plasmid DNA was treated with the same damaging agents in vitro (48).
The analysis of the damage complexity of clustered DNA damage induced by Fe ion beams revealed that damage complexities were as high as 6 (B6) for complex BDCs and 7 (DSB/B5) for complex DSBs (Fig. 6C). In irradiated cells, clustered DNA damage, including BDCs, was originally detected and quantified using the method developed by Sutherland et al. (51, 52). However, their method could not determine the damage complexity of the clusters since all types of clustered DNA damage were ultimately detected as DSBs with damage complexity = 2. Therefore, to the best of our knowledge, the present work experimentally detects and quantify highly complex clustered DNA damage (B3 through B6 and DSB/B1 through DSB/B5) in the genomic DNA of irradiated cells. The level of complexity of in vivo clustered DNA damage observed in this study was comparable to that observed in in vitro plasmid DNA irradiated with Fe ion beams (48).
Notably, the yields of total DNA damage determined by AFM agreed fairly well to those determined with dot-blot ELISA (Table 1), corroborating the validity of DNA damage data obtained with AFM. In addition, there are reports on the measurements of DSBs in human cells irradiated with X/γ-rays and Fe ion beams, in which DSBs were analyzed by pulse-field gel electrophoresis (PFGE) (30, 53, 54). Our estimations of the yield of complex DSBs by AFM is compared with the reported values (SI Appendix, Table S1). With both X-rays and Fe ion beams, the yields of complex DSBs obtained in the present study are virtually comparable to those of total DSBs reported previously. Complex DSBs observed in the present study constitute part of total DSBs that comprise simple DSBs and complex DSBs, yet our estimations of complex DSBs were somewhat larger than the reported values for total DSBs. As one DSB causes two DSB ends, the yield of complex DSBs observed in this study could be, at most, twice as large of the yield of DSBs. It is also likely that the relatively large yield of radiation-induced complex DSBs could have resulted from an enhanced sensitivity of the AFM method over PFGE, as AFM could detect DSB ends even from clustered DSBs that result in short DNA fragments.
The dose to generate the amount of DNA damage comparable to that of endogenous DNA damage can be estimated from the rate of damage formation. Regarding total clustered damage (simple and complex BDCs + complex DSB), 1.1 Gy of X-rays and 0.85 Gy of Fe ion beams generate the amount of DNA damage comparable to that of endogenous DNA damage (Table 1). These doses were comparable to the lethal doses for 37% survival (D37) of TK6 cells irradiated with X-rays and Fe ion beams (1.9 and 0.56 Gy, respectively) (SI Appendix, Fig. S6). This highlights the importance of clustered damage in the lethal effect of ionizing radiation. In contrast, regarding isolated base damage, 7.8 Gy of X-rays and 19.2 Gy of Fe ion beams generate the amount of DNA damage comparable to that of endogenous DNA damage. These doses far exceed LD37 values of TK6 cells.
Repair of Clustered DNA Damage in Cells.
We also analyzed the repair of radiation-induced clustered DNA damage in TK6 cells. With X-ray-irradiated cells, both isolated base damage and all types of observed clustered damage (simple/complex BDCs and complex DBSs) were repaired (Fig. 5C). With Fe ion beam–irradiated cells, isolated base damage, simple/complex BDCs were also repaired as was the case for X-ray-irradiated cells. Remarkably, the repair of complex DSBs was markedly retarded, and 81% of the initially formed complex DSBs remained unrepaired after 18 h of irradiation (Fig. 5F). The major complex DSBs remaining after 18 h was DSB/B1 (Fig. 6B). Complex DSBs with higher complexities (DSB/B2-DSB/B5) also remained after 18 h (Fig. 6C). Thus, complex DSBs induced by Fe ion beams were refractory to repair, whereas simple/complex BDCs are not particularly refractory to repair (Fig. 6 and SI Appendix, Fig. S5). Fe ion beams exhibit highly detrimental effects on cells relative to X-rays. The LD37 values of Fe ion beams and X-rays used in the present study was 0.56 and 1.9 Gy, respectively (SI Appendix, Fig. S6). Accordingly, the extent of unrepairable complex DSBs parallels the level of lethality, potentially accounting for the high biological effects of Fe ion beams. Complex DSBs were proposed to be one of the primary DNA lesions responsible for the lethal effects of ionizing radiation (23–25). However, complex DSBs may not be the only etiology of the high biological effects of Fe ion beams. Chromosome rearrangements, such as translocations and inversions, are indicated in cell killing by high-LET radiations (55, 56). These are formed by the rejoining DSB ends, and such damage cannot be detected by the present AFM imaging. Moreover, high-LET radiations produce multiple DSBs in close proximity (clustered DSBs) that are also suggested in the high biological effects of high-LET radiations (reviewed in refs. 57, 58). The present method of DNA damage analysis cannot distinguish isolated DSBs from clustered ones. Notably, the retardation of the repair of complex DSBs were not observed with X-ray-irradiated cells for unknown reasons (Fig. 5C). Elucidation of the critical difference between complex DSBs induced by Fe ion beams and those by X-rays is important subject of future studies to account for the distinct repairability of complex DSBs.
The amount of complex DSBs transiently increased during 1 h after irradiation; then, it decreased slowly when cells were irradiated with Fe ion beams (Fig. 5 E and F). During 1 h after irradiation, simple/complex BDCs were repaired efficiently. This suggests that during this time, simple and/or complex BDCs might be cleaved by DNA glycosylases/AP endonucleases in cells, raising the yield of complex DSBs over prompt complex DSBs formed directly by irradiation (Fig. 5F). In contrast, this transient increase in the yield of complex DSBs was not observed when cells were irradiated with X-rays (Fig. 5 B and C). There are two possible reasons for this observation. First, a previous study showed that a certain amount of enzymatic DSBs is formed due to an attempted repair of bistranded clustered base damage after the γ-irradiation of TK6 cells (38). However, the doses used in previous (3 Gy) (38) and present (40 Gy) studies are quite different. Accordingly, the formation of a high level of prompt DSBs with a high dose of X-rays (40 Gy) might completely mask the subtle formation of complex DSBs by attempted repair. Second, the amount of simple/complex BDCs repaired in 1 h after irradiation with X-rays (Fig. 5 B and C) was lower than that irradiated with Fe ion beams (Fig. 5 E and F). This led to a lower yield of enzymatic DSBs and obscured formation of complex DSBs. In any case, further studies are necessary to clarify the distinct variations in the amount of complex DSBs after irradiations with Fe ion beams and X-rays.
In summary, we have shown that X-rays and Fe ion beams produce various types of clustered DNA damage in vivo, including simple/complex BDCs and complex DSBs and that densely ionizing Fe ion beams induce clustered DNA damage more efficiently than sparsely ionizing X-rays. The highest damage complexity observed with Fe ion beams was 6 (B6) for complex BDCs and 7 (DSB/B5) for complex DSBs. Complex DSBs induced by Fe ion beams are refractory to repair and might be related to the highly lethal effect of Fe ion beams. Our analysis makes an important step toward extrapolating the biological relevance of clustered DNA damage to the killing of tumor cells by radiotherapy and chemotherapy and to the risk of cancer in nontumor cells.
Materials and Methods
Cell Culture.
In this study, the human lymphoblastoid TK6 cell line was used. The cell line is one of standard mammalian cell lines used in genotoxicity tests. The cells were maintained in an Roswell Park Memorial Institute (RPMI) medium 1640 (Nacalai) supplemented with 5% heat-inactivated horse serum (Life Technologies). The cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 (59, 60).
Cell Treatments.
X-rays and Fe ion beams were generated, as described previously (48). The dose rate and dose-averaged LET of X-rays were 3.7 Gy/min and around 10 keV/μm, respectively. The dose rate was determined by the Fricke dosimeter (61), whereas the LET was estimated from the published data (62). The dose rate and dose-averaged LET of Fe ion beams were 12 Gy/min and ∼200 keV/μm, respectively. The dose rate was determined according to the model of Kanai et al. (63, 64), whereas the calculation of LET was reported previously (65). TK6 cells were cultured in an RPMI medium 1640 supplemented with 10% horse serum. Immediately before irradiation, cells in midlogarithmic phase were collected, suspended in a 50-mL culture medium, and placed in a 70-mL canted-neck cell culture flask with a vent cap (Corning). The vent cap allowed for gas exchange between the inside and outside of the flask. The culture flask with cells was cooled with ice (ca. 4 °C) and irradiated with X-rays or Fe ion beams (0 to 60 Gy) under aerobic conditions. The cell culture medium remained cool after irradiation, although the flask containing cells was exposed to air during irradiation (5.4 to 16.2 min for X-rays and 1.6 to 5.0 min for Fe ion beams). For repair assays, the irradiated cells were further incubated in the culture media at 37 °C for up to 18 h. For the Fenton reactions, the cells in midlogarithmic phase were incubated with 0 to 1 mM H2O2 and 0.3 mM Fe(NH4)2(SO4)2/ethylenediaminetetraacetic acid (EDTA) in an RPMI medium 1640 supplemented with 10% horse serum at 37 °C for 2 h.
Purification of Genomic DNA.
After the treatment with ionizing radiation (X-rays and Fe ion beams) or Fenton’s reagents, the cells were collected by centrifugation. The genomic DNA was isolated from the cells using CsCl density gradient ultracentrifugation, as described previously (66, 67).
Enrichment of Damage-Containing Genomic DNA Fragments for AFM Imaging.
The genomic DNA (30 µg) purified from TK6 cells was treated with DNA glycosylases (Endo III and hOGG1) to convert base lesions to AP sites, and the aldehydic group of intact AP sites and 3′-nicked AP sites generated by the DNA glycosylases was labeled with ARP (Dojindo), as described previously (48, 68, 69). ARP-labeled DNA was partially digested with RsaI (10 units, New England Biolabs) to reduce the fragment size of DNA to around 1.1 kbp. The reduction of DNA sizes eliminated the overlapping of long DNA fibers in AFM imaging. This also enabled the selective enrichment of damage-containing DNA fragments by pulldown with streptavidin magnetic beads. Both facilitated the damage analysis with AFM.
The ARP-labeled DNA fragments were selectively concentrated using a Dynabeads kilobaseBINDER kit (Invitrogen). The conditions to concentrate the biotin-labeled DNA with the magnetic beads were preliminarily investigated using the model DNA, as described in SI Appendix, SI Text I and Fig. S1. For the enrichment of damage-containing genomic DNA, the ARP-labeled DNA digested by RsaI (SI Appendix, Fig. S2) was prepared from TK6 cells. The DNA (5 µg) in 50 µL of 10 mM Tris⋅HCl (pH 8.0) and 1 mM EDTA was mixed with 50 µL of binding solution included in the kit (Bottle 1, Invitrogen) for a DNA concentration of 5 µg/100 µL (total volume, 100 µL). Then, 20 µL of Dynabeads M-280 streptavidin (Invitrogen) from the original stock was initialized, following the method provided by the supplier; it was resuspended in 50 µL of the binding solution and was added to the DNA solution (5 µg DNA in a total volume of 150 µL). The sample in a tube was incubated at room temperature for 12 h on a roller to keep the beads in the suspension. The beads were washed twice with 200 μL of washing solution contained in the kit (Bottle 2: 10 mM Tris⋅HCl [pH 7.5], 1 mM EDTA, and 2.0 M NaCl). The beads were recovered and resuspended in 100 µL of 10 mM EDTA (pH 8.2). The solution was briefly heated at 90 °C for 2 min. This resulted in the release of ARP-labeled DNA/streptavidin complex from the beads, as demonstrated in the model DNA (SI Appendix, SI Text I and Fig. S1). The released DNA retained the double-stranded structure. The amount of genomic DNA recovered by the pulldown with the magnetic beads was 3 to 4% of the input DNA (SI Appendix, SI Text II). The supernatant was recovered and mixed with proteinase K (final concentration of 500 ng/µL, Wako). The sample was incubated overnight at room temperature to digest the streptavidin. DNA was recovered by ethanol precipitation and resuspended in 55 µL of Milli-Q water. The sample was mixed with 5 µg streptavidin (New England Biolabs) to fully label the damage sites in DNA (total volume, 60 µL). It should be noted that it was unlikely for all ARP-modified damage sites (particularly those in clustered damage) in DNA to be captured by streptavidin tethered to the surface of the magnetic beads, since the spacing of lesions in clustered DNA damage sites was much narrower than that of streptavidin molecules on the bead surface. Thus, it was essential to relabel the damage sites with free streptavidin to completely label them. The sample was incubated at room temperature for 12 h. Free streptavidin was removed by Chroma SPIN TE200 column, Clontech). DNA damage was observed with AFM, as described previously (48).
Quantification of the Total Base Damage Based on Dot-Blot ELISA.
The total amount of base damage in the treated TK6 cells was quantified by ARP labeling of damage sites and dot-blot ELISA, as described previously with some modifications (50). Briefly, the genomic DNA was purified from the cells. DNA was treated with Endo III and hOGG1, and aldehydic groups in DNA were labeled with ARP, as described previously (48, 50). ARP-labeled DNA (without RsaI digestion) was dot blotted on a membrane, and ARP labels (i.e., damage sites) in DNA were detected with anti-biotin antibodies conjugated with HRP-biotin antibodies (Dojindo) and enhanced chemiluminescence (ECL) reagents (BioRad). Chemiluminescence was quantified with the ChemiDoc Touch MP imaging system (BioRad). To evaluate the nonspecific background signal, NaBH4-treated DNA was also assayed in parallel (67, 70, 71). NaBH4 reduced the aldehydic groups of AP sites to alcohol, which has no reactivity to ARP. Standard DNA with known amounts of AP sites was prepared by heat-acid depurination of NaBH4-treated DNA. A linear relationship was obtained between the number of AP sites and the chemiluminescence intensity for the DNA standards containing 0 to 40 AP sites per 106 bp.
Analysis of DSBs by Static-Field Gel Electrophoresis.
The DSBs produced in the treated TK6 cells were also analyzed as the total DSBs by static-field gel electrophoresis, as reported previously (67, 70, 71). The fraction (percentage) of DNA released from the plug relative to the total DNA (i.e., released and retained DNA) was used as a measure of DSBs.
Supplementary Material
Acknowledgments
We thank Drs. Masaki Unno (Ibaraki University), Yoshiyuki Tanaka (Tokushima Bunri University), and Yoshikazu Hattori (Tokushima Bunri University) for kindly providing the hOGG1 plasmid. The authors also thank Satomi Kayamura (National Institutes for Quantum and Radiological Science and Technology (QST, Kansai) for her technical assistance. This research was supported by Research Project with Heavy Ions at QST–Heavy Ion Medical Accelerator in Chiba. This work was supported by Grant-in-Aid for Scientific Research (KAKENHI) (21K12248 to T.N., 16H02959 to N.S., and 18H03374 to H.I.) from the Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119132119/-/DCSupplemental.
Data Availability
Raw data and DNA damage videos are available upon request. All study other data are included in the article and/or SI Appendix.
References
- 1.Ward J. F., DNA damage produced by ionizing radiation in mammalian cells: Identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 35, 95–125 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Téoule R., Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 51, 573–589 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Cadet J., Ravanat J. L., TavernaPorro M., Menoni H., Angelov D., Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 327, 5–15 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Nakano T., Xu X., Salem A. M. H., Shoulkamy M. I., Ide H., Radiation-induced DNA-protein cross-links: Mechanisms and biological significance. Free Radic. Biol. Med. 107, 136–145 (2017). [DOI] [PubMed] [Google Scholar]
- 5.von Sonntag C., Free-Radical-Induced DNA Damage and Its Repair (Springer, 2006). [Google Scholar]
- 6.Wallace S. S., Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 33, 1–14 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Bradley M. O., Erickson L. C., Comparison of the effects of hydrogen peroxide and X-ray irradiation on toxicity, mutation, and DNA damage/repair in mammalian cells (V-79). Biochim. Biophys. Acta 654, 135–141 (1981). [DOI] [PubMed] [Google Scholar]
- 8.Ward J. F., Blakely W. F., Joner E. I., Mammalian cells are not killed by DNA single-strand breaks caused by hydroxyl radicals from hydrogen peroxide. Radiat. Res. 103, 383–392 (1985). [PubMed] [Google Scholar]
- 9.Goodhead D. T., Initial events in the cellular effects of ionizing radiations: Clustered damage in DNA. Int. J. Radiat. Biol. 65, 7–17 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Ward J. F., The complexity of DNA damage: Relevance to biological consequences. Int. J. Radiat. Biol. 66, 427–432 (1994). [DOI] [PubMed] [Google Scholar]
- 11.Nikjoo H., O’Neill P., Wilson W. E., Goodhead D. T., Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 156, 577–583 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Semenenko V. A., Stewart R. D., A fast Monte Carlo algorithm to simulate the spectrum of DNA damages formed by ionizing radiation. Radiat. Res. 161, 451–457 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Watanabe R., Rahmanian S., Nikjoo H., Spectrum of radiation-induced clustered non-DSB damage – A Monte Carlo track structure modeling and calculations. Radiat. Res. 183, 525–540 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Fulford J., Nikjoo H., Goodhead D. T., O’Neill P., Yields of SSB and DSB induced in DNA by Al(K) ultrasoft X-rays and alpha-particles: Comparison of experimental and simulated yields. Int. J. Radiat. Biol. 77, 1053–1066 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Sharma V., et al. , Oxidative stress at low levels can induce clustered DNA lesions leading to NHEJ mediated mutations. Oncotarget 7, 25377–25390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terato H., Ide H., Clustered DNA damage induced by heavy ion particles. Biol. Sci. Space 18, 206–215 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Shikazono N., Noguchi M., Fujii K., Urushibara A., Yokoya A., The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J. Radiat. Res. (Tokyo) 50, 27–36 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Eccles L. J., O’Neill P., Lomax M. E., Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. 711, 134–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sage E., Harrison L., Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res. 711, 123–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sage E., Shikazono N., Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 107, 125–135 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Mavragani I. V., et al. , Complex DNA damage: A route to radiation-induced genomic instability and carcinogenesis. Cancers (Basel) 9, 91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou R. Z., et al. , Tolerance for 8-oxoguanine but not thymine glycol in alignment-based gap filling of partially complementary double-strand break ends by DNA polymerase lambda in human nuclear extracts. Nucleic Acids Res. 36, 2895–2905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta K., Purkayastha S., Neumann R. D., Pastwa E., Winters T. A., Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat. Res. 175, 97–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almohaini M., et al. , Nonhomologous end joining of complex DNA double-strand breaks with proximal thymine glycol and interplay with base excision repair. DNA Repair (Amst.) 41, 16–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobbs T. A., Palmer P., Maniou Z., Lomax M. E., O’Neill P., Interplay of two major repair pathways in the processing of complex double-strand DNA breaks. DNA Repair (Amst.) 7, 1372–1383 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Chastain P. D. II, et al. , Abasic sites preferentially form at regions undergoing DNA replication. FASEB J. 24, 3674–3680 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulston M., Fulford J., Jenner T., de Lara C., O’Neill P., Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 30, 3464–3472 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland B. M., Bennett P. V., Sutherland J. C., Laval J., Clustered DNA damages induced by x rays in human cells. Radiat. Res. 157, 611–616 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Hada M., Georgakilas A. G., Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. (Tokyo) 49, 203–210 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Tsao D., et al. , Induction and processing of oxidative clustered DNA lesions in 56Fe-ion-irradiated human monocytes. Radiat. Res. 168, 87–97 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Akamatsu K., Shikazono N., Saito T., Localization estimation of ionizing radiation-induced abasic sites in DNA in the solid state using fluorescence resonance energy transfer. Radiat. Res. 183, 105–113 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Akamatsu K., Shikazono N., Saito T., New method for estimating clustering of DNA lesions induced by physical/chemical mutagens using fluorescence anisotropy. Anal. Biochem. 536, 78–89 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Akamatsu K., Shikazono N., Saito T., Fluorescence anisotropy study of radiation-induced DNA damage clustering based on FRET. Anal. Bioanal. Chem. 413, 1185–1192 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Georgakilas A. G., Bennett P. V., Wilson D. M. III, Sutherland B. M., Processing of bistranded abasic DNA clusters in gamma-irradiated human hematopoietic cells. Nucleic Acids Res. 32, 5609–5620 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gollapalle E., et al. , Detection of oxidative clustered DNA lesions in X-irradiated mouse skin tissues and human MCF-7 breast cancer cells. Radiat. Res. 167, 207–216 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Tokuyama Y., Furusawa Y., Ide H., Yasui A., Terato H., Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Radiat. Res. (Tokyo) 56, 446–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gulston M., de Lara C., Jenner T., Davis E., O’Neill P., Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 32, 1602–1609 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang N., Chaudhry M. A., Wallace S. S., Base excision repair by hNTH1 and hOGG1: A two edged sword in the processing of DNA damage in gamma-irradiated human cells. DNA Repair (Amst.) 5, 43–51 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Asaithamby A., Hu B., Chen D. J., Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proc. Natl. Acad. Sci. U.S.A. 108, 8293–8298 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asaithamby A., Chen D. J., Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 711, 87–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikitaki Z., et al. , Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 50 (sup1), S64–S78 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Moore S., Stanley F. K., Goodarzi A. A., The repair of environmentally relevant DNA double strand breaks caused by high linear energy transfer irradiation – No simple task. DNA Repair (Amst.) 17, 64–73 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Okayasu R., Repair of DNA damage induced by accelerated heavy ions – A mini review. Int. J. Cancer 130, 991–1000 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Iliakis G., Mladenov E., Mladenova V., Necessities in the processing of DNA double strand breaks and their effects on genomic instability and cancer. Cancers (Basel) 11, 1671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schipler A., et al. , Chromosome thripsis by DNA double strand break clusters causes enhanced cell lethality, chromosomal translocations and 53BP1-recruitment. Nucleic Acids Res. 44, 7673–7690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickoloff J. A., Sharma N., Taylor L., Clustered DNA double-strand breaks: Biological effects and relevance to cancer radiotherapy. Genes (Basel) 11, 99 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Wang X., Zhang P., Wang Y., The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst.) 7, 725–733 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Xu X., et al. , Direct observation of damage clustering in irradiated DNA with atomic force microscopy. Nucleic Acids Res. 48, e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuya Y., et al. , A simplified cluster analysis of electron track structure for estimating complex DNA damage yields. Int. J. Mol. Sci. 21, 1701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohsin Ali M., et al. , Detection of endonuclease III- and 8-oxoguanine glycosylase-sensitive base modifications in gamma-irradiated DNA and cells by the aldehyde reactive probe (ARP) assay. J. Radiat. Res. (Tokyo) 45, 229–237 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Sutherland B. M., Bennett P. V., Sidorkina O., Laval J., Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 97, 103–108 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutherland B. M., Georgakilas A. G., Bennett P. V., Laval J., Sutherland J. C., Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat. Res. 531, 93–107 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Prise K. M., et al. , A review of DSB induction data for varying quality radiations. Int. J. Radiat. Biol. 74, 173–184 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Belli M., et al. , DNA fragmentation induced in human fibroblasts by accelerated (56)fe ions of differing energies. Radiat. Res. 165, 713–720 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Pouget J. P., Mather S. J., General aspects of the cellular response to low- and high-LET radiation. Eur. J. Nucl. Med. 28, 541–561 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Cucinotta F. A., Durante M., Cancer risk from exposure to galactic cosmic rays: Implications for space exploration by human beings. Lancet Oncol. 7, 431–435 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Schipler A., Iliakis G., DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 41, 7589–7605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mladenova V., Mladenov E., Iliakis G., Novel biological approaches for testing the contributions of single DSBs and DSB clusters to the biological effects of high LET radiation. Front. Oncol. 6, 163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuda M., et al. , Tyrosyl-DNA phosphodiesterase 2 (TDP2) repairs topoisomerase 1 DNA-protein crosslinks and 3′-blocking lesions in the absence of tyrosyl-DNA phosphodiesterase 1 (TDP1). DNA Repair (Amst.) 91–92, 102849 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Tsuda M., Kitamasu K., Hosokawa S., Nakano T., Ide H., Repair of trapped topoisomerase II covalent cleavage complexes: Novel proteasome-independent mechanisms. Nucleosides Nucleotides Nucleic Acids 39, 170–184 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Fricke F. S., Hart E. J., “Chemical dosimetry” in Radiation Dosimetry, Attix F. H., Roesch W. C., Eds. (Academic Press, New York, ed. 2, 1966), pp. 167–239. [Google Scholar]
- 62.Wyckoff H. O., et al. , International Commission on Radiation Units and Measurements (ICRU), Linear Energy Transfer (ICRU Publications, Bethesda, MD, 1970). [Google Scholar]
- 63.Kanai T., et al. , Irradiation of mixed beam and design of spread-out Bragg peak for heavy-ion radiotherapy. Radiat. Res. 147, 78–85 (1997). [PubMed] [Google Scholar]
- 64.Kanai T., et al. , Biophysical characteristics of HIMAC clinical irradiation system for heavy-ion radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 44, 201–210 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Hirayama R., et al. , Determination of the relative biological effectiveness and oxygen enhancement ratio for micronuclei formation using high-LET radiation in solid tumor cells: An in vitro and in vivo study. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793, 41–47 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Shoulkamy M. I., et al. , Detection of DNA-protein crosslinks (DPCs) by novel direct fluorescence labeling methods: Distinct stabilities of aldehyde and radiation-induced DPCs. Nucleic Acids Res. 40, e143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakano T., et al. , Induction of DNA-protein cross-links by ionizing radiation and their elimination from the genome. Mutat. Res. 771, 45–50 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Kubo K., Ide H., Wallace S. S., Kow Y. W., A novel, sensitive, and specific assay for abasic sites, the most commonly produced DNA lesion. Biochemistry 31, 3703–3708 (1992). [DOI] [PubMed] [Google Scholar]
- 69.Ide H., et al. , Synthesis and damage specificity of a novel probe for the detection of abasic sites in DNA. Biochemistry 32, 8276–8283 (1993). [DOI] [PubMed] [Google Scholar]
- 70.Hirayama R., Furusawa Y., Fukawa T., Ando K., Repair kinetics of DNA-DSB induced by X-rays or carbon ions under oxic and hypoxic conditions. J. Radiat. Res. (Tokyo) 46, 325–332 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Hirayama R., et al. , Induction of DNA DSB and its rejoining in clamped and non-clamped tumours after exposure to carbon ion beams in comparison to X rays. Radiat. Prot. Dosimetry 143, 508–512 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and DNA damage videos are available upon request. All study other data are included in the article and/or SI Appendix.