Significance
There are five KCNQ channels (KCNQ1–KCNQ5) and they are thought to either homomerize or heteromerize to form tetrameric channels. KCNQ2 and KCNQ3 are highly expressed in the brain and in particular the forebrain, the presumed site of action for many brain disorders. For the last 30 years, the prevailing view is that KCNQ potassium channels in the brain consist of KCNQ2/3 and possibly KCNQ3/5 heteromers. Here, using epitope-tagged knockin mice and split-intein–mediated protein trans-splicing experiments, we demonstrate that KCNQ2 channels form heteromers not only with KCNQ3 but also with KCNQ5 channels. Thus, our findings of unexpected KCNQ subunit composition will shift both our understanding of KCNQ genotype/phenotype relationships as well as drug screening strategies.
Keywords: KCNQ2, KCNQ3, KCNQ5, epilepsy, autism
Abstract
KCNQ2 and KCNQ3 channels are associated with multiple neurodevelopmental disorders and are also therapeutic targets for neurological and neuropsychiatric diseases. For more than two decades, it has been thought that most KCNQ channels in the brain are either KCNQ2/3 or KCNQ3/5 heteromers. Here, we investigated the potential heteromeric compositions of KCNQ2-containing channels. We applied split-intein protein trans-splicing to form KCNQ2/5 tandems and coexpressed these with and without KCNQ3. Unexpectedly, we found that KCNQ2/5 tandems form functional channels independent of KCNQ3 in heterologous cells. Using mass spectrometry, we went on to demonstrate that KCNQ2 associates with KCNQ5 in native channels in the brain, even in the absence of KCNQ3. Additionally, our functional heterologous expression data are consistent with the formation of KCNQ2/3/5 heteromers. Thus, the composition of KCNQ channels is more diverse than has been previously recognized, necessitating a re-examination of the genotype/phenotype relationship of KCNQ2 pathogenic variants.
It is well-accepted that ion channel pathogenic variants elevate the risk for neurodevelopmental and psychiatric disorders (1, 2). Voltage-gated potassium and sodium channels are the ion channels most frequently associated with neurodevelopmental disorders. Members of the KCNQ (Kv7) potassium channel family have been increasingly linked to a spectrum of neurological disorders ranging from self-limiting neonatal epilepsy to developmental and epileptic encephalopathy (DEE) and autism spectrum disorders (ASDs) (1, 3, 4). However, the mechanisms by which KCNQ pathogenic variants lead to the various disorders are not fully understood. KCNQ channels are known to mediate the M-current, an important potassium conductance that controls neuronal firing properties and spike frequency adaptation across the nervous system (5). Due to their emergence as major regulators of brain excitability, KCNQ channels are being therapeutically targeted for multiple diseases including epilepsy (6), stroke (7), tinnitus (8), amyotrophic lateral sclerosis (9), and more recently, Alzheimer’s disease (10).
KCNQ2, KCNQ3, and KCNQ5 exhibit high expression in the brain, particularly the forebrain, the key site of action for many neurodevelopmental disorders including DEE and ASD. It is thought that the five KCNQ channels (KCNQ1–KCNQ5) form either homomeric or heteromeric tetrameric channels. It is important to understand the different combinations of heteromeric KCNQ channels that exist in the brain in order to decipher the genotype and phenotype relationships of pathogenic KCNQ variants and to develop more precise therapeutics for these disorders.
The current dogma is that the majority of native KCNQ channels in the nervous system consist of KCNQ2/3 heteromers (11–13). Various genetic and pharmacological data support this conclusion. For instance, Kcnq2 deletion or expression of Kcnq2 pore mutants reduces the M-current by ∼85% in CA1 pyramidal neurons and eliminates it altogether in superior cervical ganglion neurons (14–16). Likewise, Kcnq3 deletion reduces the M-current by ∼50% (14, 17). Pharmacological data using subunit specific blockers are also consistent with the idea that KCNQ2 and KCNQ3 form heteromers in the adult brain (18, 19). For instance, the native M-current is moderately tetraethylammonium (TEA)-sensitive, an effect mirrored by coexpression of KCNQ2 and KCNQ3 heteromers (12, 20). TEA IC50 strongly depends on the presence of KCNQ2 channels, as KCNQ2 but not KCNQ3 channels, are sensitive to TEA (12, 20).
It is currently assumed that KCNQ5 forms tetrameric channels with KCNQ3, but not KCNQ2, in the brain. This assumption is based on studies in oocytes in which dominant negative KCNQ5 expression reduced currents mediated by KCNQ3, but not KCNQ2 (21). However, a recent study identified some KCNQ2/3/5 heteromeric channels in HEK293T cells expressing all three subunits using STORM super-resolution microscopy (22). Given that many neurons coexpress KCNQ2, KCNQ3, and KCNQ5, such heteromeric channels may also exist in the nervous system. Whether such heteromeric channels are functional has not been studied previously due to technical hurdles. It is difficult to selectively form KCNQ2/3/5 channels in heterologous cells because coexpressing KCNQ2, KCNQ3, and KCNQ5 would lead to the formation of homomeric (KCNQ2, KCNQ3, KCNQ5) or heteromeric KCNQ2/3, KCNQ3/5, or KCNQ2/3/5 channels.
To surmount this obstacle, we used a split-intein protein trans-splicing approach to functionally express KCNQ2/5 tandem channels with and without KCNQ3 in HEK293T cells (23). Surprisingly, we found that KCNQ2/5 tandems can form functional heteromeric channels. To determine whether KCNQ2 indeed interacts with KCNQ5 in the brain, we engineered a mouse line in which the N-terminus of KCNQ2 is tagged with 3xFLAG. Using these mice, we demonstrate that native KCNQ2 partners not only with KCNQ3 but also with KCNQ5, in vivo. Importantly, we show that KCNQ2 interacts with KCNQ5 in Kcnq3 knockout mice, indicating that KCNQ2 can form channels with KCNQ5 independent of KCNQ3. We also provide electrophysiological data suggesting that the previously observed KCNQ2/3/5 heteromeric channels are functional. Together, our work provides insights into KCNQ2 channel composition in the forebrain and introduces an approach for determining the heteromeric composition of ion channels.
Results
KCNQ2/5-Tandems Form Functional Heteromeric Channels.
We reasoned that coexpressing KCNQ2/5 tandems with KCNQ3 would allow us to generate KCNQ2/3/5 heteromers. This is because KCNQ3 readily forms heteromers with either KCNQ2 or KCNQ5 (21, 24, 25). Thus, in principle, we could create heteromers with one KCNQ2, one KCNQ5, and two KCNQ3 subunits.
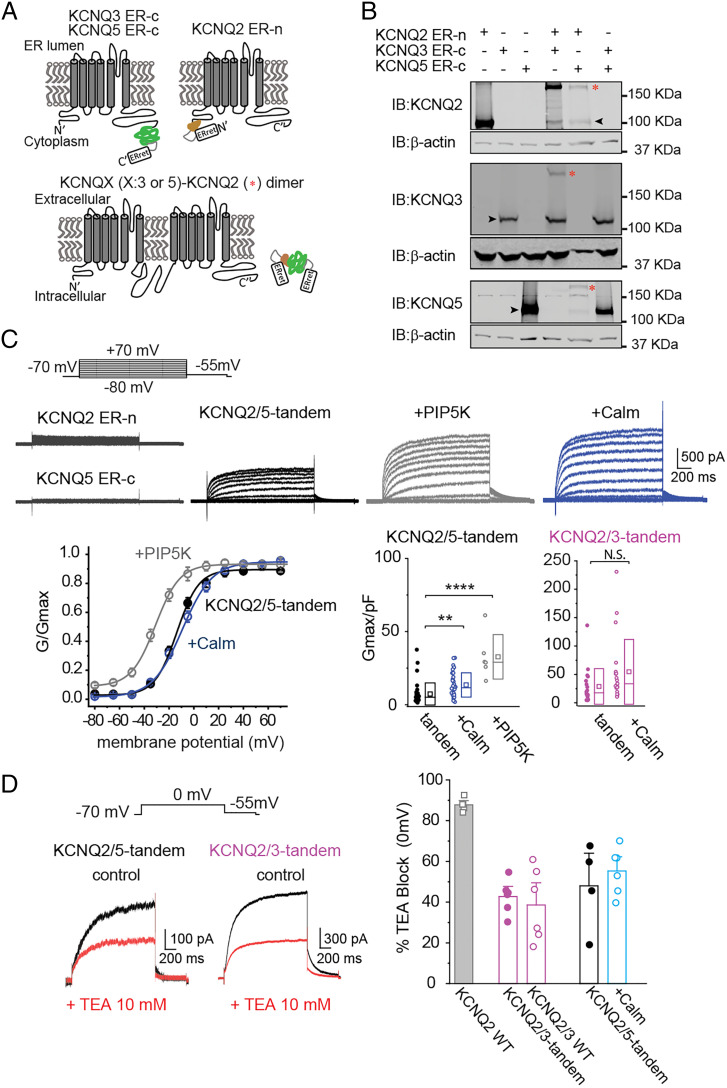
To generate the KCNQ2/5 tandems, we used split-intein protein trans-splicing. In this approach, short protein segments known as split-inteins are incorporated into the targeted proteins. When complementary split-inteins are in close proximity, they rapidly initiate an enzymatic reaction that excises the split-intein segments and forms a peptide bond between the flanking sequences, linking the two target proteins together (Fig. 1A). For our experiments, we incorporated the complementary split-inteins DnaE-n and DnaE-c into the C and N termini of KCNQ5 and KCNQ2, respectively (Fig. 1A) (26). To ensure that KCNQ2 and KCNQ5 do not form homomeric channels on the cell surface, we adopted an approach developed by Ahern and colleagues that utilizes an endoplasmic reticulum (ER) retention motif (27). The ER retention motif was placed downstream of the intein segments, so that it was removed in KCNQ2/5 tandems by the intein excision reaction.
Fig. 1.
KCNQ2 and KCNQ5 protein trans-splicing leads to functional KCNQ2/5 heteromeric channels. (A) Illustration of the strategy for creating tandem KCNQ2/3 and KCNQ2/5 subunits. The KCNQ2 channel N-terminus was tagged with the DnaE-c fragment, whereas the KCNQ3 and KCNQ5 C-terminus were tagged with the DnaE-n fragment. (B) Coexpression of KCNQ2-ER-n with either KCNQ3-ER-c or KCNQ5 ER-c formed tandems in cells. HEK293T cells transfected in a 1:1 KCNQ2-ER-n: KCNQ3-ER-c or KCNQ2-ER-n: KCNQ5 ER-c ratio led to the formation of a higher molecular band (*) due to linking of the two subunits. Note that the formation is not complete as a substantial amount of monomeric KCNQ3-ER-c and KCNQ5 ER-c remains (black arrow indicates monomers). (C) Top, representative traces from cells expressing KCNQ2-ER-n, KCNQ5-ER-c, or KCNQ2-ER-n and KCNQ5-ER-c together (KCNQ2/5-tandem). Top Center and Right, representative traces from cells expressing KCNQ2-ER-n and KCNQ5-ER-c alone (KCNQ2/5-tandem) or with PIP5K and calmodulin (Calm). Bottom Left, summary graphs showing the conductance-to-voltage relationship of KCNQ2/5-tandem channels alone (V0.5 = −11.1 ± 1.9 mV, n = 23), with PIP5K (V0.5 = −31.5 ± 2.5 mV, n = 6; ****P = 1.4e10−7), or calmodulin (Calm; V0.5 = −8.6 ± 2.2 mV, n = 22; **P = 0.002). Bottom Right, summary graphs of KCNQ2/5-tandem and KCNQ2/3-tandem current densities. Note that coexpressing KCNQ2/5-tandem subunits with either PIP5K or Calm increases their current density. Data are displayed as box plots showing the SD, median (solid line), and mean (square) values. Current densities were calculated from the tail currents recorded at −55 mV following a test pulse. One-way ANOVA, F(22.89), P = 2.2e10−8. Displayed P values were determined using Tukey post hoc test. (D) Left, representative recordings from cells expressing either KCNQ2/5-tandem or KCNQ2/3-tandem channels in the presence or absence of 10 mM TEA at 0 mV. Right, summary graph showing the blocking effect of 10 mM TEA in KCNQ channels with different subunit compositions. Data are displayed as mean ± SEM.
We first validated our split-intein approach using KCNQ2 and KCNQ3, because the properties of these channels are well established. We coexpressed KCNQ2-ER-n with KCNQ3-ER-c, which is KCNQ3 C-terminal tagged with DnaE-n and the ER motif. We confirmed that coexpression of KCNQ2-ER-n and KCNQ3-ER-c led to the formation of KCNQ2/3 tandems using Western blot analysis (Fig. 1B) and that these KCNQ2/3-tandems formed functional potassium channels, albeit with a lower current density than wild-type KCNQ2/3 channels (SI Appendix, Fig. S1 and Table S1). Importantly, we also ran control experiments with noncomplementary split-inteins. Expression of either KCNQ2-ER-n or KCNQ3-ER-c alone did not produce tandems (Fig. 1B), and did not produce KCNQ-like currents (Fig. 1C). Likewise, coexpression of KCNQ3-ER-c with KCNQ5-ER-c did not lead to the formation of KCNQ3/5-tandems (Fig. 1B). Altogether, the KCNQ2/3-tandem data gave us confidence that trans-splicing of KCNQ subunits can lead to functional tandem channels only when complementary split-inteins are present.
Similar to the KCNQ2/3-tandem constructs, coexpression of KCNQ2-ER-n and KCNQ5-ER-c in HEK293T cells led to the formation of KCNQ2/5-tandems as assessed by Western blot analysis (Fig. 1B). Although much of the KCNQ2-ER-n and KCNQ5-ER-c protein was incorporated into tandems, some remained as monomers, similar to what was observed with KCNQ2-ER-n and KCNQ3-ER-c expression (Fig. 1B). Surprisingly, we also detected whole-cell currents in such cells, presumably mediated by heteromeric channels formed by two KCNQ2/5-tandems (Fig. 1C). This was unexpected because KCNQ2 and KCNQ5 are not thought to form functional heteromeric channels (21). In contrast, we did not detect any KCNQ potassium currents when we expressed KCNQ2-ER-n or KCNQ5-ER-c alone (Fig. 1C), consistent with their retention in the ER. Thus, our data suggest that KCNQ2 and KCNQ5 are able to form functional heteromeric channels in heterologous cells.
We next investigated the properties of the observed currents to validate that they are consistent with being mediated by KCNQ2/5-tandem channels. We observed that KCNQ2/5-tandems exhibited a fourfold lower current-density than found when expressing KCNQ2/3-tandems (Fig. 1C and SI Appendix, Table S1). Their lower current density might be due in part to the relatively low opening probability of both KCNQ2 and KCNQ5, which is related to their low affinity for phosphatidylinositol 4,5-bisphosphate (PIP2) binding (28). To test this possibility, we coexpressed KCNQ2-ER-n and KCNQ5-ER-c with phosphatidylinositol 4-phosphate 5-kinase (PIP5K) to increase basal global PIP2 levels. We found that this increased the current density of KCNQ2/5-tandem channels by fourfold and shifted their V0.5 to more hyperpolarized membrane potentials (KCNQ2/5-tandem V0.5 = −11.1 ± 1.9 mV, n = 23; +PIP5K V0.5 = −31.5 ± 2.5 mV, n = 6; Student’s t test, P < 0.0001) consistent with previous work (Fig. 1C and SI Appendix, Table S1) (29). Additionally, coexpression of calmodulin, a KCNQ auxiliary subunit that increases KCNQ2 and KCNQ5 current density, led to an almost twofold increase in KCNQ2/5-tandem currents (Fig. 1C and SI Appendix, Table S1).
Finally, we also investigated the pharmacology of KCNQ2/5-tandems. The pan-KCNQ blockers XE991 and ML252 blocked currents mediated by KCNQ2/5-tandems coexpressed with calmodulin (XE991, 79.2 ± 4.6%, n = 5; ML252, 87.5 ± 3.2%, n = 11, at +55 mV). We also investigated the ability of TEA (10 mM) to block KCNQ2/5-tandem channels, as TEA is commonly used to discriminate between KCNQ2-containing and non-KCNQ2-containing KCNQ channels (20). TEA is a potent KCNQ2 inhibitor (IC50 ∼0.3 mM), but is unable to block homomeric KCNQ3 or KCNQ5 channels (13, 21). The affinity of TEA for KCNQ2 channels, and thus their expected block at 10 mM, is in proportion to the number of KCNQ2 subunits (12, 13, 20). We found that TEA blocked KCNQ2/5-tandems (48.0 ± 10.7% block, n = 4) and KCNQ2/3-tandems (42.7 ± 3.4% block, n = 6) similarly (Fig. 1D), suggesting that both contain two KCNQ2 subunits and TEA cannot distinguish between them. A similar percentage block was seen with KCNQ2/5-tandems expressed with calmodulin (55.3 ± 4.6% block, n = 6) (Fig. 1D). Together, our electrophysiological and pharmacological experiments demonstrate that KCNQ2 and KCNQ5 can form functional heteromeric channels.
KCNQ2/5-Tandems Likely Form Heteromeric Channels with KCNQ3.
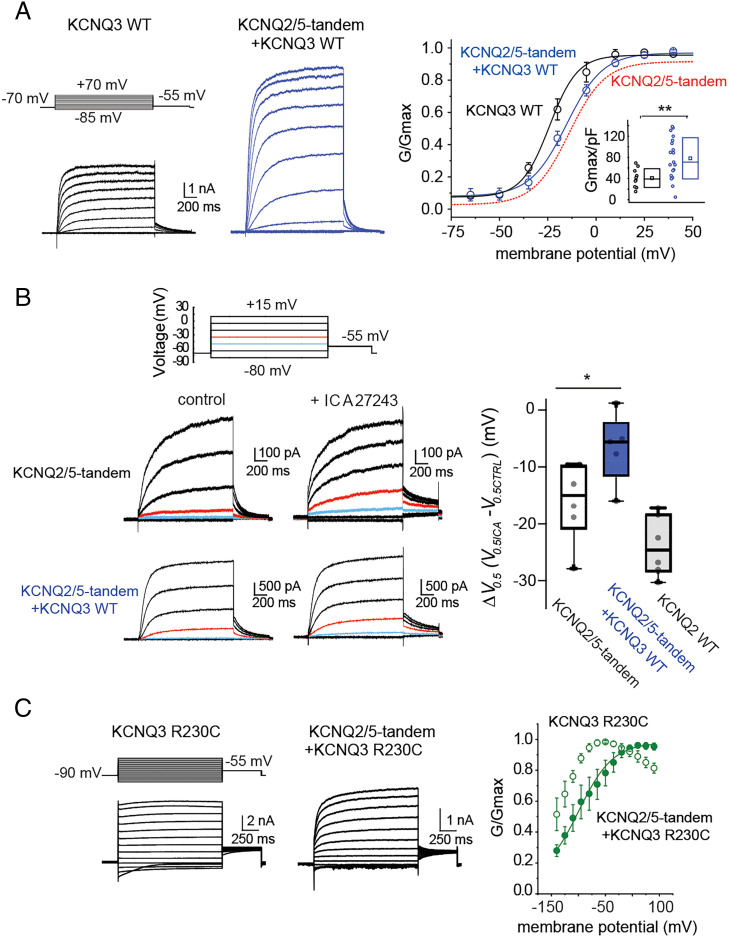
Using super-resolution microscopy, Shapiro and colleagues suggested that KCNQ2/3/5 heteromeric channels may exist when all three subunits are coexpressed in heterologous cells (22). However, the presence of such channels was not validated functionally. We therefore tested whether our KCNQ2/5-tandems might form functional heteromeric channels with KCNQ3. Coexpressing KCNQ2/5-tandems (KCNQ2/5-tandem V0.5 = −11.1 ± 1.9 mV, n = 23) with wild-type KCNQ3 channels boosted the KCNQ2/5-tandem current density by six- to eightfold (Fig. 2A and SI Appendix, Table S1) and shifted their V0.5 away from the KCNQ3 V0.5 (KCNQ3 V0.5 = −21.7 ± 3.1 mV, n = 11; KCNQ2/5-tandem+KCNQ3 V0.5 = −14.4 ± 1.7 mV, n = 18; Student’s t test P < 0.05) indicating the formation of a new channel complex with unique biophysical properties. Heteromeric KCNQ2/3 channels and KCNQ3/5 channels have higher current densities than KCNQ2 and KCNQ5 homomers because KCNQ3 has a high PIP2 affinity that increases their opening probability. Additionally, the presence of KCNQ3 is thought to support increased cell surface trafficking. Thus, the increased current-density seen when expressing KCNQ2/5-tandems with KCNQ3 is consistent with the formation of KCNQ2/5-tandem/KCNQ3 heteromeric channels.
Fig. 2.
Properties of KCNQ2/3/5 heteromeric channels. (A) Left, representative recordings from cells expressing either KCNQ2/5-tandem subunits or KCNQ2/5-tandem subunits along with KCNQ3. Right, summary graphs comparing the conductance-to-voltage relationship of KCNQ2/5-tandem channels (the curve fit from Fig. 1C is illustrated for comparison), KCNQ3 channels (V0.5 = −21.7 ± 3.2 mV, n = 11), and KCNQ2/5-tandem/KCNQ3 channels (V0.5 = −14.4 ± 1.7 mV, n = 18). Note that coexpression of KCNQ2/5-tandem subunits with KCNQ3 led to a current-to-voltage relationship similar to one obtained for KCNQ2/5-tandem channels. Data are displayed as mean ± SEM. Inset, summary graph comparing KCNQ3 to KCNQ2/5-tandem/KCNQ3 current densities. Significance was determined using the Mann–Whitney U test (**P = 0.0063). (B) Left, representative recordings from cells expressing either KCNQ2/5-tandem subunits or KCNQ2/5-tandem subunits with KCNQ3 in the presence or absence of 10 µM ICA27243. Right, summary graph showing that ICA27243 leads to a smaller shift in the V0.5 of KCNQ2/5-tandem/KCNQ3 (n = 5) channels compared to KCNQ2/5-tandem channels (n = 6). Statistical significance was determined using the Mann–Whitney U test (*P = 0.036). For illustration purposes, we also show the effect of 10 µM ICA27243 to unlinked KCNQ2 channels (n = 6). (C) Representative traces from HEK293T cells expressing either KCNQ3R230C subunits or KCNQ2/5-tandem and KCNQ3R230C subunits. Right, summary graph showing that coexpression of KCNQ2/5-tandem subunits with KCNQ3R230C leads to a right shifted conductance-to-voltage relationship (KCNQ3R230C V0.5 was not determined as KCNQ3R230C channels were constitutive open and could not be fitted with a Boltzmann function, n = 4; KCNQ2/5-tandem/KCNQ3R230C, V0.5 = −85 ± 5.8 mV, n = 6). Data are displayed as mean ± SEM.
To corroborate this interpretation, we investigated the pharmacology of currents produced by coexpression of KCNQ2/5-tandems with KCNQ3. If heteromeric KCNQ2/5-tandem/KCNQ3 channels were formed, they would have one KCNQ2, one KCNQ5 and two KCNQ3 subunits, and thus be resistant to TEA block at 10 mM. Indeed, we found that currents resulting from coexpression of KCNQ2/5-tandems with KCNQ3 were only weakly blocked by TEA (13.0 ± 3.0% block, n = 6) (12), unlike its effect on KCNQ2/5-tandem channels (Fig. 1D). We also investigated the effect of ICA27243 (10 µM), a specific KCNQ2 activator that induces a shift in the KCNQ2 V0.5 (Fig. 2B) in proportion to the number of available KCNQ2 voltage sensors in each tetramer (30). Application of ICA27243 to KCNQ2/5-tandem channels, containing two KCNQ2 subunits, led to a −16 mV shift in V0.5, whereas the application to cells expressing KCNQ2/5-tandems and KCNQ3 led to only a −6 mV shift (Fig. 2B). The smaller V0.5 shift in the cells additionally expressing KCNQ3 suggests that heteromeric KCNQ2/5-tandem/KCNQ3 channels, containing only one KCNQ2 subunit, are present.
Finally, we also investigated the possibility of heteromeric channels using the KCNQ3R230C pathogenic variant, which shifts the voltage activation of KCNQ3 to strongly hyperpolarized membrane potentials, transforming the channels from voltage-activated to “leak”-like channels (Fig. 2C) (31). Coexpression of the KCNQ2/5-tandem with KCNQ3R230C shifted the voltage activation to more depolarized values than KCNQ3R230C alone and introduced a time-dependent component in the activation (Fig. 2C). The voltage activation could be fitted with a one-component Boltzmann function suggesting that there is predominantly one population of KCNQ channels that are distinct from the KCNQ3R230C homomers and KCNQ2/5-tandem heteromers. Most likely these are KCNQ2/5-tandem/KCNQ3 heteromeric channels. Together, our data support the earlier microscopy finding (22) suggesting that such heteromeric channels can form in heterologous cells and that such channels are functional. However, our data cannot completely rule out a mixed population of KCNQ2/5-tetramers and KCNQ3 homomers, because KCNQ3 homomers can produce currents in HEK293T cells (Fig. 2A and SI Appendix, Table S1), unlike Xenopus laevis oocytes (11).
Native KCNQ2/5 Heteromers Are Present in the Brain.
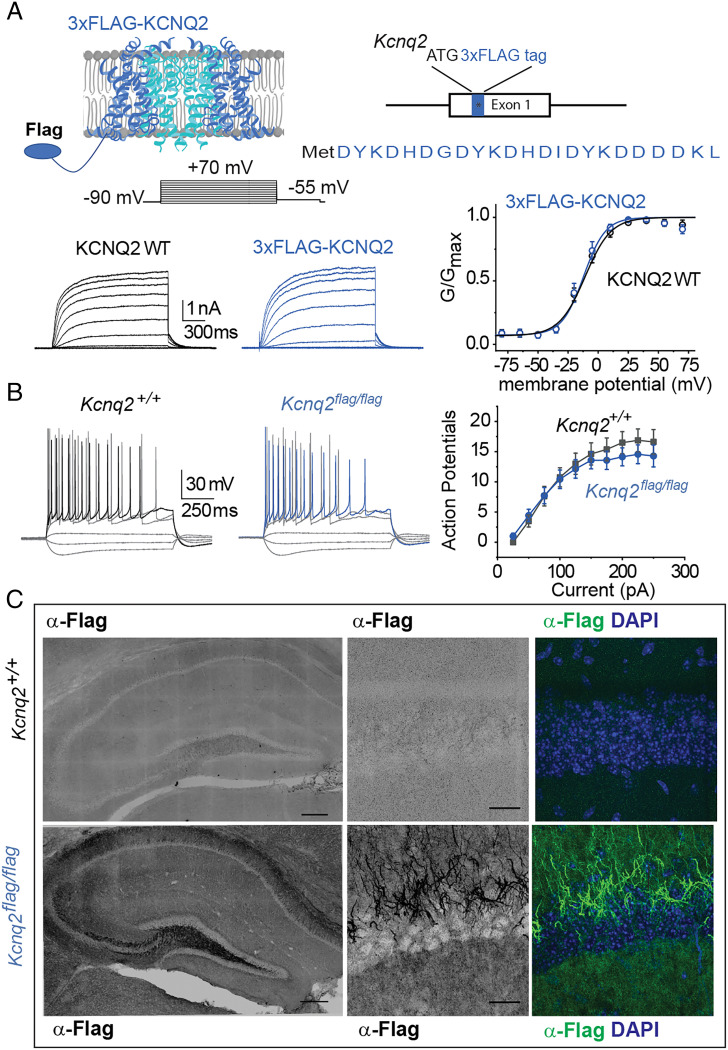
Our data for HEK293T cells demonstrates that KCNQ2 can form heteromeric channels with either KCNQ3 or KCNQ5 as well as KCNQ2/5/3 heteromers. Do such KCNQ2 complexes exist natively in brain tissue? We addressed this question using coimmunoprecipitation of native KCNQ complexes from the forebrain, a region with enriched KCNQ2, KCNQ3, and KCNQ5 expression. To ensure antibody specificity and increase the likelihood of detecting low-abundance KCNQ2 channel complexes, we first generated an N-terminus epitope-FLAGed Kcnq2 knockin mouse line (Fig. 3A).
Fig. 3.
Development and characterization of Kcnq2 epitope-tagged mice. (A) Top, illustration showing the location of the epitope tag on the KCNQ2 channels and Kcnq2 gene. Bottom Left, representative recordings from cells expressing either wild-type KCNQ2 channels or 3XFLAG-tagged KCNQ2 channels. Bottom Right, summary graph showing that introduction of the 3XFLAG epitope does not change the properties of the KCNQ2 channels (wild-type V0.5= −13.7 ± 2.8 mV, n = 4; 3XFLAG-KCNQ2 V0.5 = −11.9 ± 2.6 mV, n = 5). Data are displayed as mean ± SEM (B) Left, representative recordings from either control CA1 pyramidal neurons or neurons expressing the 3XFLAG-tagged KCNQ2 channels. The membrane potential was held at −65 mV, maintained with small DC current injections. Right, summary graph showing that introduction of the 3XFLAG epitope does not change the firing properties of CA1 pyramidal neurons (Kcnq2+/+ n = 8, 2 mice; Kcnq2flag/flag n = 7; 2 mice). Data are displayed as mean ± SEM (C) Coronal sections showing localization of the 3XFLAG-tagged KCNQ2 channels in axons. Slices were stained with either DAPI (blue) or an anti-FLAG M2 antibody.
Prior work has shown that incorporating tags into the KCNQ2 N-terminus does not interfere with the channel’s biophysical properties or its ability to traffic to the plasma membrane (32). First, we examined the properties of tagged vs. untagged KCNQ2 channels with electrophysiology when expressed in HEK293T cells, and we found no significant differences (Fig. 3A). We next examined CA1 pyramidal neuron excitability in Kcnq2flag/flag knockin mice and controls. As shown in Fig. 3B, the firing properties of CA1 pyramidal neurons from wild-type and Kcnq2flag/flag mice were similar, with similar numbers of action potentials fired in response to a series of depolarizing steps (Fig. 3B). Importantly, knockin mice carrying the KCNQ2 3XFLAG epitope were viable, with no evidence of increased lethality or behavioral seizures, which occur in animals with reduced KCNQ2 channel activity (14, 33, 34). Additionally, births followed a Mendelian distribution when heterozygous Kcnq2flag/+ mice were bred together (18 wild-type: 25.7%; 32 heterozygous: 45.7%; 20 homozygous: 28.6%), consistent with normal KCNQ2 function.
We next examined the selectivity of KCNQ2 protein detection in brain slices using anti-FLAG immunohistochemistry. Fig. 3C shows an example of anti-FLAG labeling of coronal brain sections. Antibody labeling was evident across the hippocampus of Kcnq2flag/flag knockin mice, showing a wide distribution consistent with known KCNQ2 channel localization (Fig. 3C). Within the hippocampus, anti-FLAG staining primarily occurred in axons and fibers based on the staining of mossy fibers and finger-like staining in CA1, indicating axon initial segment localization (Fig. 3C). This is in agreement with known KCNQ2 subcellular distribution. Importantly, no staining was observed in control littermates (Fig. 3C).
Having validated the utility of the Kcnq2flag/flag mouse line in visualizing KCNQ2 channels in the brain, we next determined the composition of KCNQ2 channel complexes using mass spectrometry on affinity-purified KCNQ2 complexes. We first solubilized protein complexes from the cortex of Kcnq2flag/flag mice with either CHAPS or Nonidet P-40 lysis buffer in order to preserve protein–protein interactions. We then affinity-purified KCNQ2 protein complexes using the monoclonal anti-FLAG M2 antibody and subjected the entire pulldown to analysis via high resolution mass spectrometry.
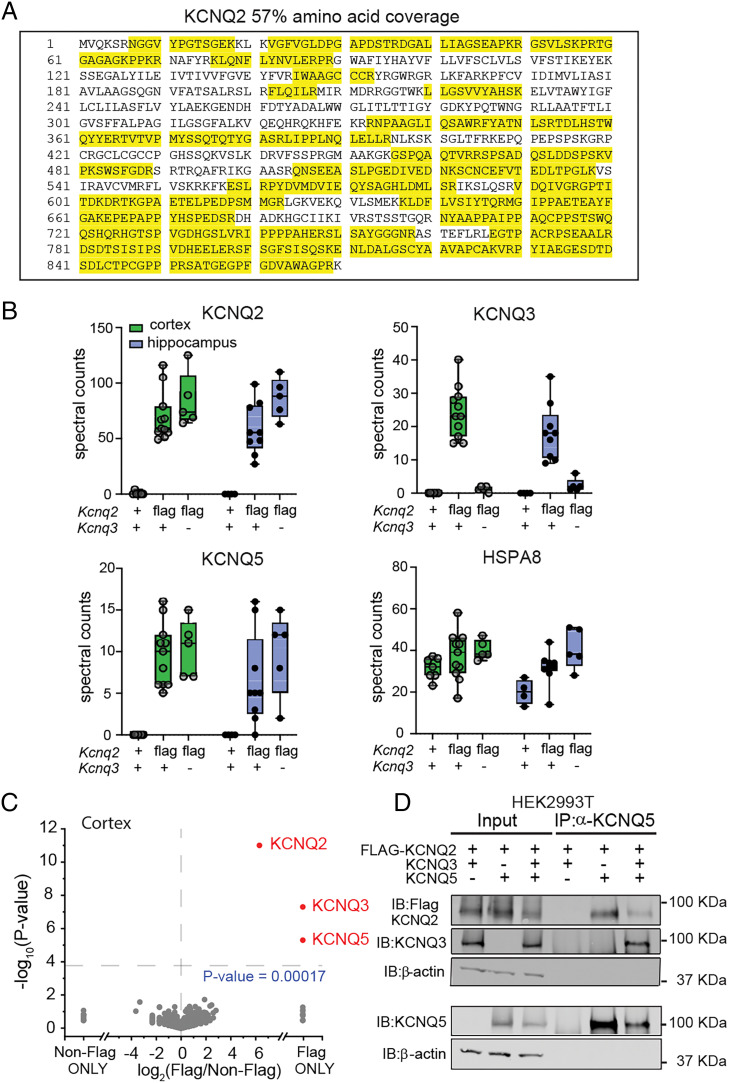
To identify the proteins that were found in the KCNQ2 protein complexes, we implemented bottom-up proteomic methodologies. We digested the anti-FLAG M2-bound proteins using trypsin and then separated and mass analyzed the generated peptides using nanoflow ultra-high-performance liquid chromatography coupled to tandem mass spectrometry. Label-free quantitation was achieved by MaxQuant using tandem mass spectrometry (MS/MS) spectral counting. Spectral counting has been shown to correlate with overall protein relative abundance when using data-dependent acquisition methods like those used in our study (35, 36). For head-to-head comparisons of identified proteins across pulldown conditions, we directly compared unnormalized spectral counts. The identification of many KCNQ2-specific peptides that span the entire primary sequence from cortical Kcnq2flag/flag immunoprecipitation pulldowns demonstrates the effectiveness and specificity of our methods (Fig. 4 A and B). In each pulldown, we identified roughly 50 or more spectral counts for KCNQ2 in Kcnq2flag/flag mouse tissue that covered 36 to 60% of the KCNQ2 protein (mean 48.5 ± 2.1% sequence coverage, n = 7). In addition to KCNQ2, our proteomic analysis yielded significant spectral counts from numerous unique peptide sequences from KCNQ3 (25.1 ± 2.8% sequence coverage, n = 7) and KCNQ5 (11.3 ± 1.4% sequence coverage, n = 7) in the cortex (Fig. 4B and SI Appendix, Fig. S2) and the hippocampus (Fig. 4B). As expected, negligible spectral counts from KCNQ2, KCNQ3, or KCNQ5 were identified from anti-FLAG affinity purification of control mice. We carried out a two-tailed t test analysis comparing cortical KCNQ2-associated proteins isolated from Kcnq2flag/flag mice and controls, and this revealed that KCNQ2, KCNQ3, and KCNQ5 were uniquely enriched in the Kcnq2flag/flag samples (Fig. 4C). Interestingly, although peptide spectral counts from hundreds of other proteins such as HSPA8 were also detected in the anti-FLAG pulldowns, these proteins showed no differential abundance patterns when compared to control pulldowns which suggests they nonspecifically bind the M2 antibody or magnetic bead solid support (Fig. 4 B and C). This strongly suggests that the KCNQ2, KCNQ3, and KCNQ5 proteins bind directly rather than through adaptor or scaffolding proteins.
Fig. 4.
KCNQ2 and KCNQ5 form a complex in the brain. (A) Representative sequence coverage of KCNQ2 channels from anti-FLAG (M2 antibody) immunoprecipitated proteins followed by mass spectrometry analysis. Yellow indicates recovered peptides. (B) Total nonnormalized spectral counts of KCNQ2, KCNQ3, and KCNQ5 channels identified in Kcnq2+/+;Kcnq3+/+ (cortex, n = 7; hippocampus, n = 4), Kcnq2flag/flagKcnq3+/+ (cortex, n = 11; hippocampus, n = 8), and Kcnq2flag/flag;Kcnq3−/− (cortex, n = 5; hippocampus, n = 5). Data are displayed as box and whisker plots. (C) Volcano plot comparing calculated log2 fold change using unnormalized spectral count values of different cortical proteins identified in Kcnq2+/+;Kcnq3+/+ (n = 7) and Kcnq2flag/flag;Kcnq3+/+ (n = 7) mice. (D) Immunoblot showing immunoprecipitated proteins from HEK293T cells expressing various 3XFLAG-KCNQ2, KCNQ3, and KCNQ5 channel combinations. Immunoprecipitation was performed using an anti-KCNQ5 antibody. n indicates number of mice.
These data are consistent with the known presence of KCNQ2/3 channels in the brain. It also indicates the existence of channels complexes containing both KCNQ2 and KCNQ5 in the brain. However, these data do not distinguish between the presence of KCNQ2/5 and KCNQ2/3/5 heteromeric channels. We reasoned that if all KCNQ2/5 channels additionally contain KCNQ3, then ablating KCNQ3 would prevent the formation of KCNQ2/3/5 heteromeric complexes. We therefore repeated our mass spectrometry experiments in Kcnq2flag/flag mice that are devoid of KCNQ3 channels. To do this, we crossed our Kcnq2flag/flag mice with our previously developed constitutive Kcnq3−/− mouse line to obtain Kcnq2flag/flag;Kcnq3−/− mice (29, 37). As expected, affinity-purified KCNQ2 complexes from these mice did not lead to the detection of KCNQ3 peptides via mass spectrometry (Fig. 4B). Surprisingly, we found that affinity-purified KCNQ2 complexes from Kcnq2flag/flag;Kcnq3−/− mice generated a similar number of KCNQ5 spectral counts in the cortex and the hippocampus as in mice with intact KCNQ3 levels (Fig. 4B). This suggests that in the brain, KCNQ2 can indeed form a channel complex with KCNQ5 directly, independent of KCNQ3.
To strengthen this conclusion, we expressed 3XFLAG-KCNQ2, KCNQ3, and KCNQ5 in HEK293T cells, and performed coimmunoprecipitation and Western blotting. As expected, anti-FLAG, anti-KCNQ3, and anti-KCNQ5 antibodies detected each protein specifically in protein extracts from cells transfected with different combinations of these proteins. Actin was used as a loading control. We then immunoprecipitated KCNQ5 protein with the anti-KCNQ5 antibody and found that KCNQ2 was copurified, both with and without KCNQ3 coexpression (Fig. 4D). Additionally, we also detected KCNQ5 protein when the immunoprecipitation occurred with anti-FLAG KCNQ2 antibodies, both with and without KCNQ3 coexpression (SI Appendix, Fig. S2B). Together, these experiments align with our mass spectrometry data and indicate that KCNQ2 and KCNQ5 interact directly in both the brain and heterologous mammalian cells. Thus, the diversity of KCNQ2 channel subunit stoichiometry in the brain is likely much richer than previously assumed.
Discussion
In current models, the majority of native KCNQ2 channels in the central nervous system are in a complex comprising two KCNQ2 subunits and two KCNQ3 subunits. Accordingly, KCNQ2 and KCNQ3 genotype/phenotype studies and ongoing drug-screening strategies to identify therapeutic compounds utilize KCNQ2/3 heteromers. Using protein trans-splicing and our newly developed epitope-tagged Kcnq2flag/flag knockin mice, we have shown that KCNQ5 forms a complex with KCNQ2 in heterologous cells and in the brain. It is currently assumed that KCNQ5 associates with KCNQ3, but not with KCNQ2. Our data challenge this notion, demonstrating that KCNQ2 channels can form a complex with KCNQ5 channels independent of the presence of KCNQ3 channels. Moving forward, our findings require a reconsideration of drug-screening strategies seeking to ameliorate KCNQ-related neurological disorders, as well as a re-examination of the mechanisms underlying the genotype/phenotype relationship of KCNQ2 and KCNQ5 pathogenic variants in humans.
KCNQ2 and KCNQ5 Interact.
The use of trans-splicing allowed us to form KCNQ2/5 heteromers and, upon coexpression with KCNQ3, KCNQ2/3/5 heteromers. To our knowledge, this study presents the first expression and description of KCNQ2/5 and KCNQ2/3/5 heteromeric channels. In heterologous cells, we found that the heteromeric complex is poorly blocked by TEA, and the ability of ICA27243 to activate these heteromeric channels is diminished compared with KCNQ2/5, in agreement with expectations for channels containing only one KCNQ2 subunit (12, 30). Importantly, we also showed that TEA cannot distinguish between KCNQ2/3 and KCNQ2/5 heteromers, raising the possibility that some previously recorded somatic M-currents might be due to KCNQ2/5 heteromers.
Our data also support the idea that KCNQ2 and KCNQ5 interact in the brain. This potentially explains why we previously found that Kcnq2 knockout mice have decreased KCNQ5 protein levels (14). Such decreases in protein levels between interacting proteins have been previously reported in potassium channels (38), but were not expected since KCNQ2 and KCNQ5 were not thought to interact.
Where might native KCNQ2/5 heteromeric channels be found? Although there is a wealth of information about KCNQ2 and KCNQ3 channel localization in the brain, little is known about KCNQ5 channels. For instance, it is currently unknown whether KCNQ5 channels localize at the AIS, a site that is highly enriched in KCNQ2 and KCNQ3 channels (39). Recent work has identified KCNQ2, KCNQ3, and KCNQ5 channels in the dendrites and spine heads of prefrontal cortical neurons (40), suggesting that these regions might be sites of interaction between different KCNQ subunits. Other work has suggested that KCNQ5 and KCNQ2 are both found in synaptic terminals (41, 42). KCNQ5 exhibits high expression levels in interneurons (43), some of which are devoid of KCNQ3 (44), but not KCNQ2. Thus, KCNQ2 and KCNQ5 might form heteromeric channels in these cell populations. The extent to which these different KCNQ combinations contribute to the native M-current in different cell types is currently unclear. Further work using KCNQ knockout mice is needed to address this question.
Are KCNQ3 and KCNQ5 the Only KCNQ2 Transmembrane Binding Partners in the Brain?
Recent work has shown that KCNQ2 channels can also interact with calmodulin (45), solute carriers (46), and single transmembrane auxiliary proteins (47) in neurons. Although we did not detect such proteins in the anti-FLAG KCNQ2Flag immunoprecipitate, our work does not exclude this possibility. In our study, we used high-stringency conditions to eliminate proteins that bind relatively weakly to KCNQ2. Doing so allowed us to have negligible detection of KCNQ peptides in control non-epitope-tagged mice using mass spectrometry. The detection of both KCNQ3 and KCNQ5 under such conditions suggests they form tightly bound complexes. However, these high-stringency conditions also meant that we may have missed proteins with low affinity associations with KCNQ2. Such proteins might alter the properties of KCNQ2 channels in the cortex and hippocampus. Further, our study focused on the forebrains of mature mice. KCNQ2 channels express early in development and across the central and peripheral nervous system, and thus it is possible that the KCNQ2 interactome differs from region to region and/or changes as the animal ages. Future studies using our epitope-tagged Kcnq2 mice can examine these dynamic properties of the KCNQ2 interactome.
Epitope-Tagged Kcnq2 Knockin Mice.
In this study, we developed and validated a KCNQ2 FLAG-tagged knockin mouse line. These mice are viable and do not exhibit any obvious behavioral abnormalities. Our mice support previous work showing that introducing tags to the KCNQ2 N-terminus does not affect the properties of KCNQ2 channels (32). Our mice have three FLAG epitopes, which allowed us to perform KCNQ2 localization studies and identify low-abundance complexes. Thus, KCNQ2 localization using a FLAG antibody might allow detection of KCNQ2 protein even in heterozygous mice. This may be important for investigators studying mice carrying Kcnq2 pathogenic variants. Some Kcnq2 variants identified in patients with KCNQ2 encephalopathy lead to a redistribution of KCNQ2 channels from the axon to the soma (48), but it is unknown whether the wild type, the variant, or both mislocalize. It should be straightforward to localize these channels using immunohistochemistry on mice containing Kcnq2flag and the Kcnq2 variant. An additional use of Kcnq2flag/flag mice would be for investigations of the phosphorylation state of KCNQ2 channels under different experimental conditions, such as conditions relevant to seizures, alcohol consumption, stress, or learning and memory paradigms. KCNQ2 channels have multiple phosphorylation sites that may have profound effects on their properties and subsequently on neuronal physiology (49). Thus, Kcnq2flag/flag mice would, in principle, allow for a comprehensive analysis of KCNQ2 phosphorylation sites in vivo.
Conclusion
In summary, we have determined the properties of KCNQ2/5 and KCNQ2/3/5 heteromers. Additionally, we developed a Kcnq2 epitope-tagged mouse as a tool for studying the properties of KCNQ2 channels in the brain. Using this mouse line, we demonstrated that KCNQ2 channels can directly interact with KCNQ5 in the brain without KCNQ3, challenging the current models of KCNQ heteromer formation. We propose that neurons have a flexible KCNQ2 channel stoichiometry determined by KCNQ2, KCNQ3, and KCNQ5 levels and their subcompartmental localization.
Materials and Methods
All unique reagents generated in this study are available from the lead contact without restriction. All experiments were performed according to the guidelines described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Connecticut.
Mice.
Kcnq2flag/flag mice were generated in a C57BL/6J mouse strain background using CRISPR/Cas9 (sg: 5′-GGTGCCTGCGGCGCCCTATC GGG-3′). Mice were generated and validated by the University of Rochester transgenic facility. Constitutive Kcnq3 knockout mice have been described before (29, 50). Mice were housed in ventilated cages with controlled temperature (22–23 °C) and a 14 h:10 h light:dark cycle. Mice had ad libitum access to chow and water. Mice were genotyped using PCR.
Constructs.
KCNQ2, KCNQ3, and KCNQ5 ERret-intein plasmids were developed in house. We generated the constructs as described previously by Lueck et al (27). Briefly, we inserted the DnaE-n sequence (CLSYETEILTVEYGLLPIGKIVEKRIECTVYSVDNNGNIYTQPVAQWHDRGEQEVFEYCLEDGSLIRATKDHKFMTVDGQMLPIDEIFERELDLMRVDNLPN), a linker (PINQPANTHERPR), and an ER retention/retrieval motif (italic) (LLDALTLASSRGPLRKRSVAVAKAKPKFSISPDSLSPRKKFQstop) at the C-terminus of KCNQ3 and KCNQ5 channels. For the Kcnq2 cDNA plasmid, we removed the start codon and added a linker (ARSMEAS) to the KCNQ2 N-terminus as well as an ER retention/retrieval motif (MLLDALTLASSRGPLRKRSVAVAKAKPKFSISPDSLS) and the DnaE-c sequence (PRIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASNCFN). To introduce the additional amino acids to KCNQ channels, DNA fragments including partial KCNQ sequences were synthesized (Integrated DNA Technologies, Inc.) and ligated using existing restriction enzyme sites. We note that the synthesized template was optimized for synthesis by reducing the amount of GC content but not changing amino acid sequence of KCNQ channels, verified later using Sanger sequencing. The calmodulin plasmid was a gift from Naoto Hoshi (University of California, Irvine, CA). All other constructs used in this study have been described previously (51).
Electrophysiological Recordings.
Postnatal (P) day P15 to P17 mice were anesthetized using isoflurane (Baxter Healthcare). After ensuring mice were fully anesthetized, we euthanized them by decapitation. Brains were quickly dissected out of the skull, cerebella were then removed and the remaining brain placed in an ice-cold cutting solution consisting of: 26 mM NaHCO3, 210 mM sucrose, 10 mM glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, and 7 mM MgCl2. Horizontal slices 300 µm thick were prepared and housed in a holding chamber containing artificial cerebrospinal fluid (aCSF) consisting of the following: 125 mM NaCl, 26 mM NaHCO3, 2.5 mM KCl, 1 mM NaH2PO4, 1.3 mM MgCl2, 2.5 mM CaCl2, and 12 mM d-glucose. Slices were recovered at 35 °C for 30 min and then were left at room temperature (∼22 °C) for ≥1 h before current clamp electrophysiological recordings. Both cutting and aCSF solutions were saturated with 95% O2/5% CO2. Recordings were done at room temperature.
Whole-cell recordings were obtained using electrodes pulled from thin-walled borosilicate glass capillaries (World Precision Instruments, Sarasota, FL) that had resistances of 3–5 MΩ when filled with recording solution. CA1 pyramidal neurons from the CA1 area were visually identified using infrared differential interference contrast optics (Olympus BX51WI, Olympus). Glass electrodes (resistance 3–5 MΩ) were filled with intracellular solution containing the following (in mM): 130 potassium methylsulfate, 10 KCl, 5 Tris-phosphocreatine, 10 Hepes, 4 NaCl, 4 Mg2ATP, and 0.4 Na4GTP. The pH was adjusted to 7.2–7.3 with KOH. The extracellular solution contained the following (in mM): 125 NaCl, 26 NaHCO3, 2.5 KCl, 1 NaH2PO4, 1.3 MgCl2, 2.5 CaCl2, and 12 glucose saturated with 95% O2 and 5% CO2. NBQX (4 μM), D-AP5 (10 μM), and picrotoxin (100 μM; Abcam) were included in the extracellular medium to block AMPA-, NMDA-, and GABA-mediated synaptic transmission, respectively. For all current-clamp experiments membrane potential was kept between −60 and −65 mV by applying a small DC current. Bridge balance was engaged throughout the duration of the recordings. For the voltage-clamp experiments with HEK293T cells, the extracellular and intracellular solutions were as follows: Extracellular (in mM): 144 NaCl, 2.5 KCl, 2.25 CaCl2, 1.2 MgCl2, 10 Hepes, and 22 D-glucose with a pH of 7.2 adjusted with NaOH and with an osmolarity of 300–306 mOsm; Intracellular (in mM): 132 K-gluconate, 10 KCl, 4 Mg*ATP, 20 Hepes, and 1 EGTA*KOH with a pH 7.2–7.3 adjusted with KOH, with an osmolarity 290–300 mOSm HEK293T cells were held at −70 mV. Data were acquired using a Multiclamp 700B amplifier (Molecular Devices) and digitized using the Digidata 1440A system (Molecular Devices). Data were low-pass filtered at 2–10 kHz and sampled at 10–50 kHz.
Current and voltage‐clamp experiments were analyzed using Clampfit (Molecular Devices) and Origin 2019b (OriginLab Corp., Northampton, MA) software programs. Tail current density-to-voltage relationships were calculated by measuring tail peak currents in response to different applied test voltage steps followed by normalization to cell capacitance (pA/pF). As the tail currents were measured at the same voltage (i.e., the same driving force Vm-Ek), they reflected changes in conductance. For all G-V measurements we used the tail current, as only KCNQ-expressing HEK293T cells show prominent tail currents (52). G-V curves were fit to a Boltzmann equation where V0.5 is the potential of half‐maximal activation and k is the slope. All Boltzmann equations were fit using Origin 2019b software built-in algorithms.
Immunohistochemistry.
P25-P30 mice were deeply anesthetized with isoflurane, and then transcardially perfused with ice-cold 1× phosphate buffered saline (PBS), followed by 2% paraformaldehyde (PFA) in ice-cold PBS (pH 6). Brains were removed and placed in cold 2% PFA (pH 6), stored for 1 h at 4 °C, and then transferred to 30% wt/vol sucrose and incubated overnight at 4 °C. Brains were snap-frozen using isopentane on dry ice and then embedded in optimal cutting temperature compound (Tissue‐Tek) and stored at -80 °C until ready for sectioning. Coronal sections (40-60 μm) were taken using a Leica CM3050S cryostat and collected in 6-well or 24-well plates. For FLAG‐tag immunohistochemistry, sections were incubated in blocking solution (5% normal goat serum, 0.5% Triton X‐100 in PBS) for 2 h at room temperature, followed by an overnight incubation, at 4 °C, in mouse anti‐FLAG antibody (F3165, Sigma) diluted 1:1,000 in blocking solution. Following three washes in PBS, slices were incubated for 2 h at room temperature in either an Alexa 488- or 647-conjugated goat anti‐mouse secondary antibody (1:500, Invitrogen). Slices were then washed three more times in 1× PBS and then mounted and covered with ProLong Gold mounting medium with DAPI (Invitrogen, P36931). Prepared slides were stored at 4 °C until they were ready to be imaged. Images were taken using a Leica SP8 confocal.
Co‐Immunoprecipitation and Western Blotting.
Cortex and hippocampus were dissected and homogenized with an electric homogenizer in homogenization buffer (320 mM sucrose, 5 mM Na phosphate pH 7.4, 100 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride [PMSF] protease inhibitor). Brain homogenate was added to an equal volume of 2× lysis buffer (final concentration; 25 mM Tris⋅HCl pH 7.0, 150 mM sodium chloride, 1.5% CHAPS or 1.5% Nonidet P-40) and incubated on ice for 30 min. Following this incubation, insoluble material was pelleted by centrifugation at 16,100 × g for 10 min at 4 °C. The supernatant was incubated with primary antibodies (Sigma, mouse anti-FLAG, catalog #3165) and Dynabeads protein G (Invitrogen, catalog #1004D) overnight at 4 °C. The beads were washed 5 times in wash buffer (25 mM Tris⋅HCl pH 7.0, 450 mM sodium chloride) for mass spectrometry.
Transfected HEK293T (Lipofectamine 3000, Invitrogen, L3000015) cells were washed 3 times in PBS and incubated in lysis buffer (25 mM Tris⋅HCl pH7.0, 150 mM sodium chloride, 1.5% CHAPS, 1 mM PMSF) on ice for 30 min. Following this incubation, cell lysate was centrifuged at 16,100 × g for 10 min at 4 °C, and supernatant was collected for immunoprecipitation. Primary antibody, either mouse anti-FLAG (Sigma) or rabbit anti-KCNQ5 (Millipore, catalog #ABN1372), and Dynabeads protein G (Invitrogen) were added to the supernatant and incubated overnight at 4 °C. The beads were washed 3 times in wash buffer (25 mM Tris⋅HCl pH 7.0, 300 mM sodium chloride) and eluted in 4× sodium dodecyl sulfate sample buffer for Western blot analysis.
Shotgun Proteomics Analysis via Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry.
We performed all experiments using 1- to 2-mo-old male and female mice. Immunoprecipitated proteins were isolated on magnetic beads, washed in detergent-free IP buffer, and resuspended to a total volume of 100 µL using 0.1 M ammonium bicarbonate in water. The protein and bead slurry was reduced and alkylated at 37 °C using 5 mM dithiothreitol and 10 mM iodoacetamide in 0.1 M ammonium bicarbonate for 1.5 h uncovered and 45 min in the dark, respectively. Proteins were digested on-beads using trypsin (Promega sequencing grade modified porcine trypsin, part number V5113) at a 1:20 enzyme:protein ratio at 37 °C for 16 h while rotating at 700 rpm in a Thermo Scientific Thermal Mixer. Peptides were removed with the supernatant, acidified to pH 3 with concentrated formic acid and desalted with Pierce C18 Peptide Desalting Spin Columns (Thermo Scientific part number 89851) using the manufacturer’s instructions. Eluted and desalted peptides were dried to completion using a Labconco Speedvac Concentrator, resuspended in 0.1% formic acid in water, and frozen at −20 °C until shotgun proteomics analysis via ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis.
Peptides were quantified using a Thermo Scientific Nanodrop OneC Spectrophotometer and diluted using 0.1% formic acid to a final concentration of 2 µg/µL. For each injection, 1 µL was directly loaded onto a Waters nanoEase m/z Peptide UPLC BEH C18 analytical column (75 µm × 25 cm) using a Dionex Ultimate 3000 RSLCnano ultra-high performance liquid chromatography system connected to a Thermo Scientific Q Exactive HF mass spectrometer. A 300 nL/min, 2-h linear reversed phase gradient was used to elute peptides directly into the mass spectrometer inlet via nanoflow electrospray ionization through a PicoTip emitter (New Objective). A Top 15 data-dependent acquisition method was utilized to generate UPLC-MS/MS data to enable peptide sequencing via higher energy collisional dissociation.
Peptide and protein identification with label-free quantification was achieved using MaxQuant v1.6.1.0 searches against the Flag-KCNQ sequence plus the Uniprot Mus musculus database (UP000000589, accessed May 16, 2017) (53). All results were filtered to a 1% false discovery rate at the protein and peptide-spectrum match levels. Variable modifications included oxidized Met, protein N-terminal acetylation, peptide N-terminal Gln to pyroGlu, and phosphorylation of Ser, Thr, and Tyr. Carbamidomethylation on Cys was selected as a fixed modification and a 5 amino acid per peptide minimum was used. All proteomics results were visualized and analyzed further using Scaffold Q+S v.5 (Proteome Software).
Data Analysis and Statistics.
Experimental details on the number of samples used in each individual experiment and statistical analysis are specified in the figure legends or in the text. Statistical analyses were performed using Origin 2019b and Scaffold Q+S v5. Summary data are expressed as X-Y plots, bar graphs, or box plots. For analysis, we used Mann–Whitney nonparametric unpaired, Student’s t test, or one-way ANOVA tests, followed by Tukey post hoc test. Values of P < 0.05 were considered statistically significant. For the volcano plots, we calculated P values and adjusted the significance based on the Benjamini–Hochberg false discovery rate (FDR) correction (FDR <0.05).
Supplementary Material
Acknowledgments
This work was supported by NIH (R01NS101596 to A.V.T. and R21NS118262 to A.V.T. and H.S.). We thank members of the A.V.T. laboratory and Dr. Menuz for discussions and Rebecca Oramas for developing the illustration in Fig. 3A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117640119/-/DCSupplemental.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PDX028142 (54).
References
- 1.Satterstrom F. K., et al. ; Autism Sequencing Consortium; iPSYCH-Broad Consortium, Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbrici P., Camerino D. C., Tricarico D., Major channels involved in neuropsychiatric disorders and therapeutic perspectives. Front. Genet. 4, 76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nappi P., et al. , Epileptic channelopathies caused by neuronal Kv7 (KCNQ) channel dysfunction. Pflugers Arch. 472, 881–898 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Sands T. T., et al. , Autism and developmental disability caused by KCNQ3 gain-of-function variants. Ann. Neurol. 86, 181–192 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Delmas P., Brown D. A., Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat. Rev. Neurosci. 6, 850–862 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Gunthorpe M. J., Large C. H., Sankar R., The mechanism of action of retigabine (ezogabine), a first-in-class K+ channel opener for the treatment of epilepsy. Epilepsia 53, 412–424 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Bierbower S. M., Choveau F. S., Lechleiter J. D., Shapiro M. S., Augmentation of M-type (KCNQ) potassium channels as a novel strategy to reduce stroke-induced brain injury. J. Neurosci. 35, 2101–2111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S., Choi V., Tzounopoulos T., Pathogenic plasticity of Kv7.2/3 channel activity is essential for the induction of tinnitus. Proc. Natl. Acad. Sci. U.S.A. 110, 9980–9985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wainger B. J., et al. , Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 7, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee S. H., et al. , APP family regulates neuronal excitability and synaptic plasticity but not neuronal survival. Neuron 108, 676–690.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H. S., et al. , KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Shapiro M. S., et al. , Reconstitution of muscarinic modulation of the KCNQ2/KCNQ3 K(+) channels that underlie the neuronal M current. J. Neurosci. 20, 1710–1721 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadley J. K., et al. , Differential tetraethylammonium sensitivity of KCNQ1-4 potassium channels. Br. J. Pharmacol. 129, 413–415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soh H., Pant R., LoTurco J. J., Tzingounis A. V., Conditional deletions of epilepsy-associated KCNQ2 and KCNQ3 channels from cerebral cortex cause differential effects on neuronal excitability. J. Neurosci. 34, 5311–5321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins J., Passmore G. M., Abogadie F. C., Reilly J. M., Brown D. A., Effects of KCNQ2 gene truncation on M-type Kv7 potassium currents. PLoS One 8, e71809 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters H. C., Hu H., Pongs O., Storm J. F., Isbrandt D., Conditional transgenic suppression of M channels in mouse brain reveals functions in neuronal excitability, resonance and behavior. Nat. Neurosci. 8, 51–60 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Gao X., et al. , Place fields of single spikes in hippocampus involve Kcnq3 channel-dependent entrainment of complex spike bursts. Nat. Commun. 12, 4801 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springer K., Varghese N., Tzingounis A. V., Flexible stoichiometry: Implications for KCNQ2- and KCNQ3-associated neurodevelopmental disorders. Dev. Neurosci. 43, 191–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dirkx N., Miceli F., Taglialatela M., Weckhuysen S., The role of Kv7.2 in neurodevelopment: Insights and gaps in our understanding. Front. Physiol. 11, 570588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadley J. K., et al. , Stoichiometry of expressed KCNQ2/KCNQ3 potassium channels and subunit composition of native ganglionic M channels deduced from block by tetraethylammonium. J. Neurosci. 23, 5012–5019 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroeder B. C., Hechenberger M., Weinreich F., Kubisch C., Jentsch T. J., KCNQ5, a novel potassium channel broadly expressed in brain, mediates M-type currents. J. Biol. Chem. 275, 24089–24095 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Zhang J., Carver C. M., Choveau F. S., Shapiro M. S., Clustering and functional coupling of diverse ion channels and signaling proteins revealed by super-resolution STORM microscopy in neurons. Neuron 92, 461–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanda A., Nasker S. S., Mehra A., Panda S., Nayak S., Inteins in science: Evolution to application. Microorganisms 8, E2004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lerche C., et al. , Molecular cloning and functional expression of KCNQ5, a potassium channel subunit that may contribute to neuronal M-current diversity. J. Biol. Chem. 275, 22395–22400 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Wickenden A. D., Zou A., Wagoner P. K., Jegla T., Characterization of KCNQ5/Q3 potassium channels expressed in mammalian cells. Br. J. Pharmacol. 132, 381–384 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanyam P., et al. , Manipulating L-type calcium channels in cardiomyocytes using split-intein protein transsplicing. Proc. Natl. Acad. Sci. U.S.A. 110, 15461–15466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueck J. D., et al. , Atomic mutagenesis in ion channels with engineered stoichiometry. eLife 5, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Gamper N., Hilgemann D. W., Shapiro M. S., Regulation of Kv7 (KCNQ) K+ channel open probability by phosphatidylinositol 4,5-bisphosphate. J. Neurosci. 25, 9825–9835 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K. S., Duignan K. M., Hawryluk J. M., Soh H., Tzingounis A. V., The voltage activation of cortical KCNQ channels depends on global PIP2 levels. Biophys. J. 110, 1089–1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Maghera J., Lamothe S. M., Marco E. J., Kurata H. T., Heteromeric assembly of truncated neuronal Kv7 channels: Implications for neurologic disease and pharmacotherapy. Mol. Pharmacol. 98, 192–202 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Miceli F., et al. , Early-onset epileptic encephalopathy caused by gain-of-function mutations in the voltage sensor of Kv7.2 and Kv7.3 potassium channel subunits. J. Neurosci. 35, 3782–3793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez-Posada J. C., et al. , A pore residue of the KCNQ3 potassium M-channel subunit controls surface expression. J. Neurosci. 30, 9316–9323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E. C., et al. , Heterozygous loss of epilepsy gene KCNQ2 alters social, repetitive and exploratory behaviors. Genes Brain Behav. 19, e12599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aiba I., Noebels J. L., Kcnq2/Kv7.2 controls the threshold and bi-hemispheric symmetry of cortical spreading depolarization. Brain 144, 2863–2878 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., Sadygov R. G., Yates J. R. III, A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Kirmiz M., Vierra N. C., Palacio S., Trimmer J. S., Identification of VAPA and VAPB as Kv2 channel-interacting proteins defining endoplasmic reticulum-plasma membrane junctions in mammalian brain neurons. J. Neurosci. 38, 7562–7584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varghese N., Lauritano A., Taglialatela M., Tzingounis A. V., KCNQ3 is the principal target of retigabine in CA1 and subicular excitatory neurons. J. Neurophysiol. 125, 1440–1449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norris A. J., Foeger N. C., Nerbonne J. M., Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J. Neurosci. 30, 13644–13655 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kole M. H., Cooper E. C., Axonal Kv7.2/7.3 channels: Caught in the act. Channels (Austin) 8, 288–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galvin V. C., et al. , Muscarinic M1 receptors modulate working memory performance and activity via KCNQ potassium channels in the primate prefrontal cortex. Neuron 106, 649–661.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang H., Trussell L. O., KCNQ5 channels control resting properties and release probability of a synapse. Nat. Neurosci. 14, 840–847 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martire M., et al. , M channels containing KCNQ2 subunits modulate norepinephrine, aspartate, and GABA release from hippocampal nerve terminals. J. Neurosci. 24, 592–597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fidzinski P., et al. , KCNQ5 K(+) channels control hippocampal synaptic inhibition and fast network oscillations. Nat. Commun. 6, 6254 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Goff K. M., Goldberg E. M., Vasoactive intestinal peptide-expressing interneurons are impaired in a mouse model of Dravet syndrome. eLife 8, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung H. J., Role of calmodulin in neuronal Kv7/KCNQ potassium channels and epilepsy. Front. Biol. 9, 205–215 (2014). [Google Scholar]
- 46.Manville R. W., Abbott G. W., Teamwork: Ion channels and transporters join forces in the brain. Neuropharmacology 161, 107601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hessler S., et al. , β-Secretase BACE1 regulates hippocampal and reconstituted M-currents in a β-subunit-like fashion. J. Neurosci. 35, 3298–3311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abidi A., et al. , A recurrent KCNQ2 pore mutation causing early onset epileptic encephalopathy has a moderate effect on M current but alters subcellular localization of Kv7 channels. Neurobiol. Dis. 80, 80–92 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Greene D. L., Hoshi N., Modulation of Kv7 channels and excitability in the brain. Cell. Mol. Life Sci. 74, 495–508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou B., Varghese N., Soh H., Santaniello S., Tzingounis A. V., Loss of KCNQ2 or KCNQ3 leads to multifocal time-varying activity in the neonatal forebrain ex vivo. eNeuro 8, ENEURO.0024-21.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalappa B. I., et al. , Potent KCNQ2/3-specific channel activator suppresses in vivo epileptic activity and prevents the development of tinnitus. J. Neurosci. 35, 8829–8842 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suh B. C., Hille B., Electrostatic interaction of internal Mg2+ with membrane PIP2 Seen with KCNQ K+ channels. J. Gen. Physiol. 130, 241–256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PDX028142 (54).