Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has rapidly spread and led to global health crises. COVID-19 causes well-known respiratory failure and gastrointestinal symptoms, such as diarrhea, nausea, and vomiting. Thus, human gastrointestinal cell models are urgently needed for COVID-19 research; however, it is difficult to obtain primary human intestinal cells. In this study, we examined whether human induced pluripotent stem cell (iPSC)-derived small intestinal epithelial cells (iPSC-SIECs) could be used as a SARS-CoV-2 infection model. We observed that iPSC-SIECs, such as absorptive and Paneth cells, were infected with SARS-CoV-2, and remdesivir treatment decreased intracellular SARS-CoV-2 replication in iPSC-SIECs. SARS-CoV-2 infection decreased expression levels of tight junction markers, ZO-3 and CLDN1, and transepithelial electrical resistance (TEER), which evaluates the integrity of tight junction dynamics. In addition, SARS-CoV-2 infection increased expression levels of proinflammatory genes, which are elevated in patients with COVID-19. These findings suggest iPSC-SIECs as a useful in vitro model for elucidating COVID-19 pathology and drug development.
Keywords: COVID-19, SARS-CoV-2, iPSC, Small intestinal epithelial cells, Barrier functions
1. Introduction
The coronavirus disease 2019 (COVID-19), which emerged in Wuhan, China, in November 2019, has rapidly spread and led to global health crises.1 , 2 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a causative and pathogenic virus of COVID-19, with complicated symptoms, including severe pneumonia, causing an urgent demand for the development of efficient drugs and medical treatment.3 , 4
The infectious processes of SARS-CoV-2 are initiated via host receptor recognition, membrane fusion, and viral entry into the target cells. In the host recognition step, SARS-CoV-2 is known to be absorbed by cells via angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), which primes the SARS-CoV-2 spike protein to facilitate viral cellular entry.5 , 6 These receptors are known to be highly expressed in multiple human organs, such as the lung, heart, and small intestine.
COVID-19 causes well-known respiratory disturbances, heart failure, and gastrointestinal symptoms, such as diarrhea, nausea, and vomiting.7 , 8 Thus, digestive organs, including the small intestine, are strongly implicated in the pathogenesis and clinical progression of the disease. Human intestinal cells are essential as in vitro models for COVID-19 research,8 while it is not easy to obtain primary human intestinal cells. In addition, human colon adenocarcinoma-derived Caco-2 cells have been widely used to predict intestinal absorption. However, membrane permeability assays using Caco-2 cells have some limitations; it takes approximately three weeks to form a monolayer membrane and Caco-2 cells have different gene expressions, such as components of tight junctions, compared with human intestinal epithelial cells.
Human-induced pluripotent stem cells (iPSCs) can theoretically differentiate into nearly all cell types in the human body,9 and are expected to be a good source for obtaining intestinal cells. Several groups have successfully developed intestinal cells from human iPSCs using two-dimensional (2D) or three-dimensional (3D) organoid culture methods.10, 11, 12, 13, 14, 15, 16 Recently, several reports indicate SARS-CoV-2 infection with human intestinal organoids.17 , 18 SARS-CoV-2 infection was effectively inhibited by remdesivir and a coronavirus fusion inhibitor EK1.17 Despite these applications of 3D organoids, it has not been fully understood whether SARS-CoV-2 infection affects biological properties, such as permeability, in human iPSC-derived intestinal models.
In this study, we investigated whether human iPSC-derived small intestinal epithelial cells (iPSC-SIECs) could be used as a SARS-CoV-2 infection model. We observed that iPSC-SIECs, including absorptive and Paneth cells, were successfully infected with SARS-CoV-2. Furthermore, SARS-CoV-2 infection decreased transepithelial electrical resistance (TEER) and increased the expression of proinflammatory genes. Remdesivir reversed the decrease in TEER and inhibited the induction of proinflammatory genes. These results suggest that human iPSC-SIECs provide a useful 2D culture model for elucidating intestinal pathology in COVID-19.
2. Materials and methods
2.1. Chemicals
Remdesivir was obtained from Selleck Chemicals (Houston, TX). Penicillin-streptomycin mixture (PS) was obtained from Thermo Fisher Scientific (Waltham, MA, USA). All other reagents were of analytical grade and were obtained from commercial sources.
2.2. Cell culture
TMPRSS2-expressing VeroE6 (JCRB, 1819) cells were obtained from JCRB and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS; Biological Industries, Ashrat, Israel) and 0.05 mg/ml PS at 37 °C in an atmosphere containing 5% CO2. The iPSC-SIECs were purchased from Fujifilm Wako and maintained according to the manufacturer's instructions.19
2.3. SARS-CoV-2 infection
The SARS-CoV-2 strain JPN/TY/WK-52120 was distributed by the National Institute of Infectious Diseases in Japan. After pretreatment with remdesivir (1 μM) for 1 h, the cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 1 for 24 h. After infection, intracellular RNA was extracted using the CellAmp Direct RNA Prep Kit (Takara Bio, Shiga, Japan), according to the manufacturer's instructions. Quantitative real-time PCR was performed using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific), 2019-nCoV RUO Kit (Integrated DNA Technologies, Coralville, Iowa, USA), and 2019-nCoV_N positive control (Integrated DNA Technologies) with a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific).
2.4. Plaque assay
The plaque assay was performed as previously described.21 Briefly, VeroE6/TMPRSS2 cells were seeded in 12-well plates and incubated with serially diluted cell culture supernatant stock at 24 h post-infection. After 1 h of incubation, the medium was replaced with 1% methylcellulose-containing medium, and the cells were cultured for 72 h. The cells were then fixed with 4% paraformaldehyde and stained with methylene blue. The number of plaques was counted to determine the virus titers.
2.5. Immunocytochemistry
Immunocytochemistry was carried out as previously described.22 Briefly, the cells were fixed, permeabilized, blocked, and incubated with primary antibodies against SARS-CoV-2 nucleocapsid (1:100; GeneTex, Irvine, CA, USA), SARS spike glycoprotein (1:100; Abcam, Cambridge, UK), villin-1 (1:100; Cell Signaling Technology, Danvers, MA, USA), lysozyme (1:100; Thermo Fisher Scientific), mucin-2 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and chr-A (1:100; Santa Cruz Biotechnology) at 4 °C. The cells were then incubated with Alexa 488-conjugated (1:200; Thermo Fisher Scientific) or Alexa 594-conjugated (1:200; Thermo Fisher Scientific) secondary antibodies for 1 h at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole DAPI (Nacalai Tesque, Kyoto, Japan). The cells were mounted in SlowFade (Thermo Fisher Scientific) and examined under a confocal laser-scanning microscope (Nikon A1; Nikon, Tokyo, Japan).
2.6. Measurement of TEER
The TEER of Transwell monolayer cultures was measured using a Millicell ERS-2 volt-ohm meter (Millipore, Bedford, MA, USA), as previously reported.23 TEER values in the absence of cells were used as a blank and subtracted from all cell values.
2.7. Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
RT-qPCR was conducted as previously reported.24 Briefly, total RNA was isolated from iPSCs using TRIzol reagent (Thermo Fisher Scientific). Total RNA of human adult intestine was purchased from BioChain (Hayward, CA, USA). RT-qPCR was performed using a QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA, USA) on an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). Primer sequences are listed in Table 1 . The target transcript levels were normalized to the mRNA levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the ΔΔ Ct method.
Table 1.
PCR primers for RT-qPCR.
| Target gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| ACE2 | CATTGGAGCAAGTGTTGGATCTT | GAGCTAATGCATGCCATTCTCA |
| TMPRSS2 | CAGGAGTGTACGGGAATGTGATGGT | GATTAGCCGTCTGCCCTCATTTGT |
| Villin-1 | CGGAAAGCACCCGTATGGAG | CGTCCACCACGCCTACATAG |
| Lysozyme | CCCTGGTCAGCCTAGCACTC | CCTTGCCCTGGACCGTAACA |
| Mucin-2 | GAGGGCAGAACCCGAAACC | GGCGAAGTTGTAGTCGCAGAG |
| Chr-A | TAAAGGGGATACCGAGGTGATG | TCGGAGTGTCTCAAAACATTCC |
| ZO-1 | CAACATACAGTGACGCTTCACA | CACTATTGACGTTTCCCCACTC |
| ZO-2 | ATGGAAGAGCTGATATGGGAACA | TGCTGAACTGCAAACGAATGAA |
| ZO-3 | GCTTTGGCATTGCGATCTCTG | GATGTGGTCGCCTGTCTGTAG |
| CLDN1 | CCTCCTGGGAGTGATAGCAAT | GGCAACTAAAATAGCCAGACCT |
| CLDN2 | GCCTCTGGATGGAATGTGCC | GCTACCGCCACTCTGTCTTTG |
| IL-1β | CTCGCCAGTGAAATGATGGCT | GTCGGAGATTCGTAGCTGGAT |
| IL-6 | ACTCACCTCTTCAGAACGAATTG | CCATCTTTGGAAGGTTCAGGTTG |
| CCL2 | CAGCCAGATGCAATCAATGCC | TGGAATCCTGAACCCACTTCT |
| CCL3 | AGTTCTCTGCATCACTTGCTG | CGGCTTCGCTTGGTTAGGAA |
| CCL5 | CCAGCAGTCGTCTTTGTCAC | CTCTGGGTTGGCACACACTT |
| CXCL10 | GTGGCATTCAAGGAGTACCTC | TGATGGCCTTCGATTCTGGATT |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
2.8. Statistical analyses
All data are presented as mean ± standard deviation (SD). P values were calculated using a two-sided unpaired Student's t-test. Statistical significance was set at P < 0.05.
3. Results
3.1. SARS-CoV-2 infection in human iPSC- SIECs
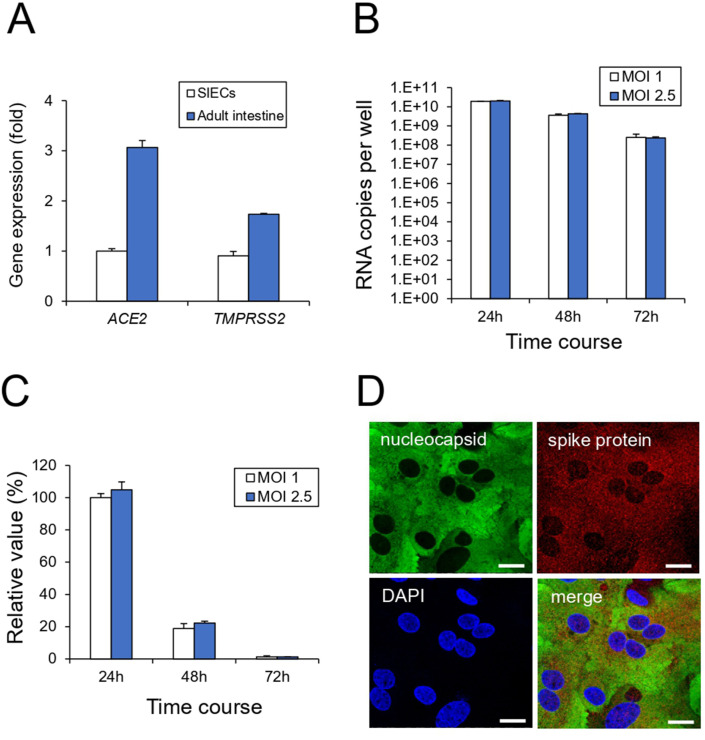
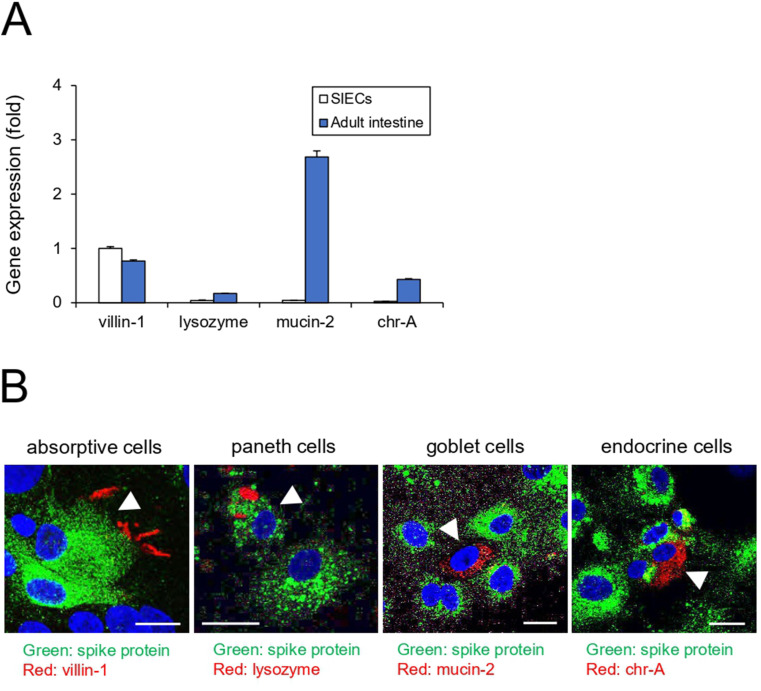
First, we examined whether iPSC-SIECs were infected with SARS-CoV-2. The entry of SARS-CoV-2 into target cells is initiated by the binding of the spike protein to ACE2.6 The spike protein is cleaved by the TMPRSS2 serine protease and triggers viral entry into the target cell. RT-qPCR analysis revealed that iPSC-SIECs expressed ACE2 and TMPRSS2 to a similar extent as in the adult intestine (Fig. 1 A). We determined the optimal infection conditions for human iPSC-SIECs. As shown in Fig. 1B, iPSC-SIECs were infected with SARS-CoV-2 at an MOI of 1 or 2.5 at different time courses (24–72 h). We observed that the intracellular SARS-CoV-2 copy number was the highest after 24 h of infection (Fig. 1B and C). Since there was no significant difference in copy number even if the MOI was raised above 1, we infected iPSC-SIECs with SARS-CoV-2 at an MOI of 1 for 24 h. The immunocytochemical analysis confirmed that iPSC-SIECs expressed SARS-CoV-2 protein (Fig. 1D). In addition, RT-qPCR analysis revealed that iPSC-SIECs mainly expressed villin-1 (absorptive cell marker)-positive cells at a level comparable to that of the adult intestinal tissue (Fig. 2 A). We further performed co-immunostaining for viral proteins with intestinal cell type-specific markers. Viral proteins were detected in villin-1 (absorptive cell marker) and lysozyme (Paneth cell marker)-positive cells, where lysozyme-positive intracellular granules did not contain viral proteins (Fig. 2B). Conversely, mucin-2 (a goblet cell marker) and chr-A (an endocrine cell marker) were not co-immunostained with viral proteins (Fig. 2B). These findings suggest that SARS-CoV-2 infects iPSC-SIECs, mainly absorptive and Paneth cells.
Fig. 1.
SARS-CoV-2 infection in human iPSC-SIECs. (A) The expression levels of ACE2 and TMPRSS2 in iPSC-SIECs and adult intestines were examined by RT-qPCR. (B) After the cells were infected with SARS-CoV-2 (MOI = 1, 2.5) for indicated time courses, the intracellular viral copy number was determined by RT-qPCR. (C) The relative value of SARS-CoV-2 copy number was indicated with the infectious condition at MOI = 1 for 24 h as 100%. (D) After SARS-CoV-2 infection (MOI = 1) for 24 h, the cells were stained with antibodies against SARS-CoV-2 nucleocapsid and SARS spike glycoprotein. Nuclei were counterstained with DAPI. Bar = 20 μm. Data are represented as mean ± standard deviation (SD; n = 3).
Fig. 2.
Cell-type-specific incorporation of SARS-CoV-2 in human iPSC-SIECs. (A) The expression levels of cell-type-specific markers in SIECs and the adult intestine were examined by RT-qPCR. (B) After SARS-CoV-2 infection (MOI = 1) for 24 h, the cells were stained with antibodies against cell-type specific markers and SARS spike glycoprotein. Nuclei were counterstained with DAPI. Bar = 20 μm. Data are represented as mean ± standard deviation (SD; n = 3).
3.2. Effect of remdesivir on SARS-CoV-2 infection in iPSC-SIECs
To examine whether antiviral drugs can be evaluated in iPSC-SIECs, we investigated the effect of remdesivir on SARS-CoV-2 infection. Remdesivir, a nucleotide analog developed for the treatment of Ebola virus, has been used as a pharmacological tool to inhibit viral RNA synthesis and clinical treatment against COVID-19.25 Treatment with remdesivir (1 μM) at 1 h pre-infection decreased the intracellular SARS-CoV-2 copy number by 99% (Fig. 3 A). The plaque assay indicated that the SARS-CoV-2 viral titer was reduced by remdesivir in the cells (Fig. 3B). These findings suggest that human iPSC-SIECs may be useful for the evaluation of COVID-19 drugs.
Fig. 3.
Effect of remdesivir on SARS-CoV-2 infection in human iPSC-SIECs. (A) Cells were treated with remdesivir (1 μM) 1 h before SARS-CoV-2 infection (MOI = 1) for 24 h. The intracellular viral copy number was determined by RT-qPCR. Left panel: SARS-CoV-2 RNA copies per well. Right panel: Normalized value of SARS-CoV-2 RNA copies against vehicle control as 100%. (B) After the cells were treated with remdesivir and infected with SARS-CoV-2, viral titers in the culture supernatants were determined by plaque assay in Vero E6 cells. Data are represented as mean ± SD (n = 3). ∗P < 0.05.
3.3. Effect of SARS-CoV-2 infection on the intestinal epithelial integrity
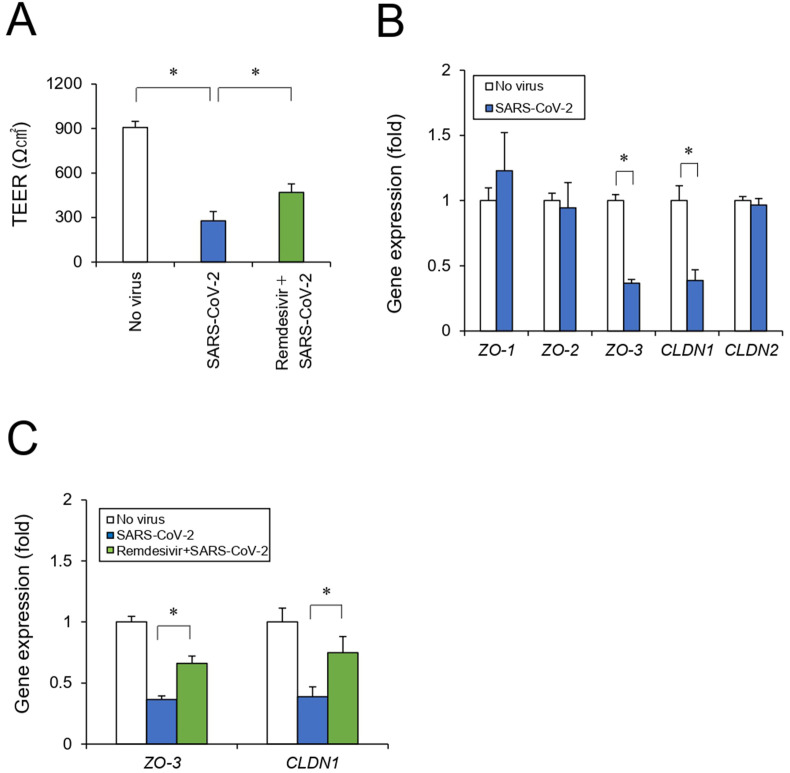
To investigate the effects of SARS-CoV-2 infection on intestinal function, we focused on the intestinal epithelial barrier. We measured an epithelial barrier indicator after iPSC-SIECs were seeded in a 24-well Transwell plate to generate epithelial monolayers. The SARS-CoV-2 infection resulted in a 67% reduction in the TEER. Remdesivir treatment partially reversed this decrease in TEER levels (Fig. 4 A). In the intestinal mucosa, the epithelial barrier is mainly formed by a 3D structure of monolayer cells with intercellular tight junctions (TJ).26 Reportedly, SARS-CoV-2 infection causes airway epithelial damage, including TEER decrease, with disruption of ZO expression.27 Thus, to clarify the mechanism underlying intestinal barrier disruption by SARS-CoV-2, we examined the expression of representative tight junction marker genes (ZO-1, ZO-2, ZO-3, CLDN1, and CLDN2) of intestinal epithelium.28, 29, 30 We observed that SARS-CoV-2 infection decreased the expression of several genes, such as ZO-3 and CLDN1, which were reversed by remdesivir treatment (Fig. 4B and C), suggesting that SARS-CoV-2 disrupts tight junction marker expression and intestinal epithelial barrier integrity.
Fig. 4.
Effect of SARS-CoV-2 infection on the intestinal epithelial barrier. (A) Cells were seeded in Transwell chambers. After the cells were treated with remdesivir and infected with SARS-CoV-2 for 24 h, the TEER values across monolayers were measured. (B) After the cells were infected with SARS-CoV-2 for 24 h, tight junction marker genes (ZO-1, ZO-2, ZO-3, CLDN1, CLDN2) were analyzed by RT-qPCR. (C) After the cells were treated with remdesivir and infected with SARS-CoV-2 for 24 h, tight junction marker genes (ZO-3, CLDN1) were analyzed by RT-qPCR. Data are represented as mean ± SD (n = 3). ∗P < 0.05.
3.4. Effect of SARS-CoV-2 infection on inflammatory responses
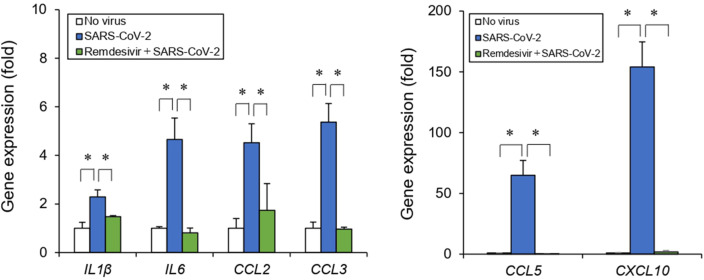
Clinical studies have reported that plasma cytokines and chemokines, such as IL-1β, IL-6, CCL2, CCL3, CCL5, and CXCL10, increase in patients with COVID-19 and may result in an excessive inflammatory response and subsequent cytokine storm.31, 32, 33, 34 We examined these inflammatory responses to investigate further the effect of SARS-CoV-2 infection on the intestinal epithelial barrier. As shown in Fig. 5 , SARS-CoV-2 infection increased the expression of proinflammatory genes such as IL-1β, IL-6, CCL2, CCL3, CCL5, and CXCL10. In addition, remdesivir treatment reduced the induction of these cytokines and chemokines (Fig. 5). These results suggest that SARS-CoV-2 infection induces an inflammatory response in the intestinal epithelium.
Fig. 5.
Effect of SARS-CoV-2 infection on inflammatory responses. Cells were treated with remdesivir (1 μM) 1 h before SARS-CoV-2 infection (MOI = 1) for 24 h. Inflammatory response genes (IL-1β, IL-6, CCL2, CCL3, CCL5, and CXCL10) were analyzed by RT-qPCR. Data are represented as mean ± SD (n = 3). ∗P < 0.05.
4. Discussion
In this study, we demonstrated SARS-CoV-2 infection by using iPSC-SIECs. We observed that absorptive and Paneth cells in the iPSC-SIECs were infected with SARS-CoV-2. In addition, we observed that SARS-CoV-2 infection decreased TEER and increased the expression of proinflammatory genes. However, remdesivir treatment recovered the decrease in TEER and induction of proinflammatory genes by SARS-CoV-2.
We observed that SARS-CoV-2 infection was observed in the absorptive and Paneth cells (lysozyme-positive intracellular granules did not contain viral proteins) but not in the goblet and endocrine cells. Although Krüger reported that SARS-CoV-2 infects Paneth and endocrine cells but not goblet cells by co-staining analysis using intestinal organoids,17 we were unable to detect viruses in endocrine cells, which secrete multiple hormones, such as chr-A.35 Goblet cells are also known to synthesize and secrete mucus.35 Thus, we consider the possibility that SARS-CoV-2 is less likely to infect secretory cell types. However, further studies are required to determine the characteristics of cell types that are susceptible to SARS-CoV-2.
SARS-CoV-2 infection caused a reduction in the TEER value via ZO-3 and CLDN1 downregulation. Since disruption of ZO-3 or CLDN1 expression has been reported to impair tight junction assembly and paracellular flux regulation,36 , 37 both genes are considered to play a crucial role in the maintenance of intestinal barrier integrity. Epithelial barrier impairment increases the risk of various intestinal diseases, including inflammatory bowel diseases, which cause nausea, vomiting, and diarrhea.38 Thus, this barrier injury of the intestinal mucosa by viral infection could be a causative factor of COVID-19-induced gastrointestinal symptoms.
Furthermore, we observed that SARS-CoV-2 infection increased the expression of proinflammatory genes in SIECs. Increased levels of cytokines have often been reported in patients with severe COVID-19.31, 32, 33, 34 Autopsy reports of fatal COVID-19 patients have also indicated widespread systemic inflammation involving the gastrointestinal tract.39 Proinflammatory cytokines are generally known to have a drastic effect on TJ expression, which significantly affects the epithelial barrier. For example, IL-1β exposure to intestinal epithelial Caco-2 cells reportedly disrupts the integration of TJ via the NF-κB pathway and reduces the TEER value.40 In the case of the human airway epithelium, SARS-CoV-2 infection is reported to distort ZO expression, causing barrier dysfunction (TEER decrease),27 presumably caused by inflammatory cytokine production. Since IL-1β was upregulated in SARS-CoV-2 infected SIECs, TJ regulatory mechanisms may mediate viral infection-induced barrier impairment. The intestinal epithelial inflammatory response is known to regulate immune cells against pathogens. In the gut mucosal environment, CCL2, CCL3, CCL5, and CXCL10 are involved in the recruitment, activation, and migration of various immune cells (T lymphocytes, macrophages, monocytes, NK cells, and immature dendritic cells).41 , 42 In addition, IL-6 has been shown to cause TEER disruption via negative TJ regulator CLDN2 in Caco-2 cells.43 In contrast, serum levels of IL-6 has not been linked with gastrointestinal symptom in patient with COVID-19.44 Although increased fecal CCL-28 was observed in the COVID-19 patients with diarrhea,45 CCL-28 has not been correlated with SARS-CoV2 infection or gastrointestinal symptoms.44 Increased IL-23 level has been reported in the stools of the COVID-19 patients.46 RT-qPCR analysis revealed that IL-23 was below the limit of detection in human iPSC-SIECs with or without SARS-CoV2 infection (data not shown). The upregulation of various cytokines is considered an intestinal defensive response after viral infection. Future studies should investigate the pathophysiological role of these inflammatory responses to SARS-CoV-2 infection in human organs in detail, including the digestive tract.
We also observed that remdesivir suppressed SARS-CoV-2 infection and recovered the SARS-CoV-2-induced barrier damage and inflammatory responses. There has been reported the clinical characteristics of COVID-19 patients with gastrointestinal symptoms, including nausea, vomiting and diarrhea.7 , 8 , 31 In this study, we indicated that these gut injuries were possible to be caused by mucosal barrier dysfunction and inflammatory responses after SARS-CoV-2 infection. Our results indicated the recovery of intestinal function by remdesivir; clinical data using remdesivir are expected to analyze in COVID-19 patients with gastrointestinal symptoms. In consistent with these findings, clinical research has been reported that remdesivir prevents cytokine storm in COVID-19 patients,47 which suggest that remdesivir further improves gastrointestinal symptoms of the patients. However, antiviral drugs, including remdesivir, widely used in the treatment of COVID-19 patients have been known to contain gastrointestinal side effects, such as nausea, vomiting and diarrhea.48 , 49 Thus, further research of prediction and alleviation of drug-induced side effects should be also progressed using human iPSC-SIECs.
In conclusion, we have demonstrated that human iPSC-SIECs are permissive to SARS-CoV-2 infection and could be used to evaluate the effectiveness of remdesivir against SARS-CoV-2-induced intestinal impairment. Thus, human iPSC-SIECs may be useful as 2D models for drug development.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (#21H02634 to Y. K.), the Research on Regulatory Harmonization and Evaluation of Pharmaceuticals, Medical Devices, Regenerative and Cellular Therapy Products, Gene Therapy Products, and Cosmetics from Japan Agency for Medical Research and Development, AMED (JP21mk0101189 to Y. K.), Emerging and Re-emerging Infectious Diseases, AMED (JP20fk0108518 to Y. K.), and a grant from the Smoking Research Foundation (Y. K.).
Footnotes
Peer review under responsibility of Japanese Pharmacological Society.
References
- 1.Wang M., Cao R., Zhang L., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettit S.D., Jerome K.R., Rouquié D., et al. 'All In': a pragmatic framework for COVID-19 testing and action on a global scale. EMBO Mol Med. 2020;12(6):e12634. doi: 10.15252/emmm.202012634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Moore M.J., Vasilieva N., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghafoor K., Ahmed A., Abbas M. Fulminant myocarditis with ST elevation and cardiogenic shock in a SARS-CoV-2 patient. Cureus. 2021;13(7):e16149. doi: 10.7759/cureus.16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galanopoulos M., Gkeros F., Doukatas A., et al. COVID-19 pandemic: pathophysiology and manifestations from the gastrointestinal tract. World J Gastroenterol. 2020;26(31):4579–4588. doi: 10.3748/wjg.v26.i31.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K., Kong Y., Zhang M., Xie F., Liu P., Xu S. Differentiation of pluripotent stem cells for regenerative medicine. Biochem Biophys Res Commun. 2016;471(1):1–4. doi: 10.1016/j.bbrc.2016.01.182. [DOI] [PubMed] [Google Scholar]
- 10.Clinton J. Directed differentiation of gastrointestinal epithelial organoids using ATCC CELLMATRIX basement membrane from multiple human ATCC iPSC lines. AP notes. 2016;26:1–8. [Google Scholar]
- 11.McCracken K.W., Howell J.C., Wells J.M., Spence J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6(12):1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negoro R., Takayama K., Nagamoto Y., Sakurai F., Tachibana M., Mizuguchi H. Modeling of drug-mediated CYP3A4 induction by using human iPS cell-derived enterocyte-like cells. Biochem Biophys Res Commun. 2016;472(4):631–636. doi: 10.1016/j.bbrc.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Ogaki S., Morooka M., Otera K., Kume S. A cost-effective system for differentiation of intestinal epithelium from human induced pluripotent stem cells. Sci Rep. 2015;5:17297. doi: 10.1038/srep17297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onozato D., Yamashita M., Nakanishi A., et al. Generation of intestinal organoids suitable for pharmacokinetic studies from human induced pluripotent stem cells. Drug Metab Dispos. 2018;46(11):1572–1580. doi: 10.1124/dmd.118.080374. [DOI] [PubMed] [Google Scholar]
- 15.Spence J.R., Mayhew C.N., Rankin S.A., et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchida H., Machida M., Miura T., et al. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight. 2017;2(1):e86492. doi: 10.1172/jci.insight.86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krüger J., Groß R., Conzelmann C., et al. Drug inhibition of SARS-CoV-2 replication in human pluripotent stem cell-derived intestinal organoids. Cell Mol Gastroenterol Hepatol. 2021;11(4):935–948. doi: 10.1016/j.jcmgh.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y., Duan X., Yang L., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589(7841):270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabeya T., Mima S., Imakura Y., et al. Pharmacokinetic functions of human induced pluripotent stem cell-derived small intestinal epithelial cells. Drug Metabol Pharmacokinet. 2020;35(4):374–382. doi: 10.1016/j.dmpk.2020.04.334. [DOI] [PubMed] [Google Scholar]
- 20.Pezzotti G., Boschetto F., Ohgitani E., et al. Raman molecular fingerprints of SARS-CoV-2 British variant and the concept of Raman barcode. Adv Sci. 2022;9(3):e2103287. doi: 10.1002/advs.202103287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baer A., Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. JoVE. 2014;93:e52065. doi: 10.3791/52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji K., Yamada S., Hirai K., Asakura H., Kanda Y. Development of alveolar and airway cells from human iPS cells: toward SARS-CoV-2 research and drug toxicity testing. J Toxicol Sci. 2021;46(9):425–435. doi: 10.2131/jts.46.425. [DOI] [PubMed] [Google Scholar]
- 23.Yamada S., Kanda Y. Retinoic acid promotes barrier functions in human iPSC-derived intestinal epithelial monolayers. J Pharmacol Sci. 2019;140(4):337–344. doi: 10.1016/j.jphs.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Yanagida S., Satsuka A., Hayashi S., Ono A., Kanda Y. Chronic cardiotoxicity assessment of BMS-986094, a guanosine nucleotide analogue, using human iPS cell-derived cardiomyocytes. J Toxicol Sci. 2021;46(8):359–369. doi: 10.2131/jts.46.359. [DOI] [PubMed] [Google Scholar]
- 25.Eastman R.T., Roth J.S., Brimacombe K.R., et al. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia M.A., Nelson W.J., Chavez N. Cell-cell junctions organize structural and signaling networks. Cold Spring Harbor Perspect Biol. 2018;10(4):a029181. doi: 10.1101/cshperspect.a029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao S., Ning K., Kuz C.A., Vorhies K., Yan Z., Qiu J. Long-term modeling of SARS-CoV-2 infection of in vitro cultured polarized human airway epithelium. mBio. 2020;11(6) doi: 10.1128/mBio.02852-20. e02852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer zum Büschenfelde D., Tauber R., Huber O. TFF3-peptide increases transepithelial resistance in epithelial cells by modulating claudin-1 and -2 expression. Peptides. 2006;27(12):3383–3390. doi: 10.1016/j.peptides.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147(6):1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Itallie C.M., Tietgens A.J., Anderson J.M. Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol Biol Cell. 2017;28(4):524–534. doi: 10.1091/mbc.E16-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fajgenbaum D.C., June C.H. Cytokine storm. N Engl J Med. 2020;383(23):2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas C., Wong P., Klein J., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith N., Goncalves P., Charbit B., et al. Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat Immunol. 2021;22(11):1428–1439. doi: 10.1038/s41590-021-01028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali A., Tan H., Kaiko G.E. Role of the intestinal epithelium and its interaction with the microbiota in food allergy. Front Immunol. 2020;11:604054. doi: 10.3389/fimmu.2020.604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak Y.K., Vikström E., Magnusson K.E., Vécsey-Semjén B., Colque-Navarro P., Möllby R. The Staphylococcus aureus alpha-toxin perturbs the barrier function in Caco-2 epithelial cell monolayers by altering junctional integrity. Infect Immun. 2012;80(5):1670–1680. doi: 10.1128/IAI.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W.Y., Wang M., Zhang J., Barve S.S., McClain C.J., Joshi-Barve S. Acrolein disrupts tight junction proteins and causes endoplasmic reticulum stress-mediated epithelial cell death leading to intestinal barrier dysfunction and permeability. Am J Pathol. 2017;187(12):2686–2697. doi: 10.1016/j.ajpath.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rios-Arce N.D., Collins F.L., Schepper J.D., et al. Epithelial barrier function in gut-bone signaling. Adv Exp Med Biol. 2017;1033:151–183. doi: 10.1007/978-3-319-66653-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurink B., Roos E., Radonic T., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Sadi R.M., Ma T.Y. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178(7):4641–4649. doi: 10.4049/jimmunol.178.7.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmerman N.P., Vongsa R.A., Wendt M.K., Dwinell M.B. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14(7):1000–1011. doi: 10.1002/ibd.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulkarni N., Pathak M., Lal G. Role of chemokine receptors and intestinal epithelial cells in the mucosal inflammation and tolerance. J Leukoc Biol. 2017;101(2):377–394. doi: 10.1189/jlb.1RU0716-327R. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki T., Yoshinaga N., Tanabe S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J Biol Chem. 2011;286(36):31263–31271. doi: 10.1074/jbc.M111.238147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livanos A.E., Jha D., Cossarini F., et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160(7):2435–2450. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y., Jiang X., Wang X., et al. CCL28 mucosal expression in SARS-CoV-2-infected patients with diarrhea in relation to disease severity. J Infect. 2021;82(1):e19–e21. doi: 10.1016/j.jinf.2020.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Britton G.J., Chen-Liaw A., Cossarini F., et al. Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19. Sci Rep. 2021;11(1):13308. doi: 10.1038/s41598-021-92740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kivrak A., Ulaş B., Kivrak H. A comparative analysis for anti-viral drugs: their efficiency against SARS-CoV-2. Int Immunopharm. 2021;90:107232. doi: 10.1016/j.intimp.2020.107232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grein J., Ohmagari N., Shin D., et al. Compassionate use of remdesivir for patients with severe covid-19. N Engl J Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottlieb R.L., Vaca C.E., Paredes R., et al. Early remdesivir to prevent progression to severe covid-19 in outpatients. N Engl J Med. 2022;386(4):305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]