Abstract
The peripheral nerve contains diverse cell types that support its proper function and maintenance. We analyzed multiple peripheral nerves using single-nuclei RNA-sequencing (snRNA-seq), which allowed us to circumvent difficulties encountered in analyzing cells with complex morphologies via conventional single cell methods. The resultant mouse peripheral nerve cell atlas highlights a diversity of cell types, including multiple subtypes of Schwann cells (SCs), immune cells, and stromal cells. We identified a distinct myelinating-SC subtype that expresses Cldn14, Adamtsl1, and Pmp2 and preferentially ensheathes motor axons. The number of these motor-associated, Pmp2+ SCs is reduced in both ALS SOD1G93A mouse model and human ALS nerve samples. Our findings reveal the diversity of SCs and other cell types in peripheral nerve and serve as a reference for future studies of nerve biology and disease.
Introduction
Maintaining homeostasis in peripheral nerves is a unique challenge due to the extreme length of the axons, their large surface area, and the substantial energy demands of nerve impulse conduction. In both the PNS and CNS, glia are required for proper signal transduction along axons, and for metabolic support1,2. These glia, called Schwann cells (SCs) are the predominant cell type in peripheral nerve, and have traditionally been classified as either myelinating (mSCs) or non-myelinating (nmSCs), reflecting the elaboration of myelin sheathes around large caliber axons, or the ensheathment of smaller caliber axons into Remak bundles3. Other cellular components of the nerve have received less attention, but nevertheless play important roles in nerve function. Nerve fibroblasts form connective tissue layers that maintain the structural integrity of the nerve, and also play a key role in preserving nerve environment homeostasis4. Interactions between the various components are critical for nerve development and regeneration5,6, and resident immune cells are important in regeneration after nerve injury7. Genetic and anatomic studies of the peripheral nerve in health and disease have provided detailed information regarding many of cells within the nerve, however the variety of cell types of vastly differing morphologies has made characterization of these populations difficult using conventional transcriptional profiling analysis8.
While the localization and geometry of SCs enable these cells to perform indispensable functions, these properties also prevent the use of conventional techniques for single cell transcriptional characterization, because SCs cannot be easily separated from axons. Indeed, recent single-cell RNA-seq (scRNA-seq) experiments in mouse sciatic nerves have found limited numbers of mSCs9–12; and other single-cell RNA-seq experiments have likewise identified very small numbers of SCs in whole organs including heart13 and pancreas14. Since anatomic studies clearly demonstrate that SCs are the predominant cell type within peripheral nerve15, these findings suggest that inconsistent cell capture of SCs in conventional single cell RNA-seq experiments result in their underrepresentation. The recent development of single nuclei RNA-seq (snRNA-seq) has helped eliminate cell capture biases16. Given the complex structure of SCs, we reasoned that a single nuclei RNA-seq approach could be used to analyse transcriptomes of these cells.
In this study, we used snRNA-seq to obtain unbiased cell atlases of four mouse peripheral nerves. This allowed us to identify more than 20 cell types encompassing glial, immune, and stromal populations, as well as to quantify the differences in cellular compositions across nerve types. We use this atlas to demonstrate that Scn7a is a useful marker for nmSCs, and is located in a region containing SNPs that are strongly associated with risk of diabetic neuropathy. We also identified multiple new SC cell types, including a subtype characterized by expression of Pmp2, Adamtsl1 and Cldn14 that preferentially myelinates motor axons. We further showed that these motor-associated SCs are significantly reduced in both an ALS mouse model and in human ALS samples. Finally, to encourage utilization of our data, we built a glial portal (http://milbrandt.wustl.edu/glia-portal/) that allows rich visualization and exploratory data analysis using the SCV infrastructure.
Results
snRNA-seq of sciatic nerve reveals robust SC signatures.
To examine cellular diversity in peripheral nerve, we first obtained single cell expression profiles using whole-cell RNA capture17 technology; however, we found few myelinating Schwann cell (mSC) signatures in our data. This mSC representation was inconsistent with long-standing results from anatomical studies18 and suggested a bias in capturing these cells. A similar capture bias is also apparent in another study reporting scRNA-seq from sciatic nerve9. We determined this was due to issues with the dissociation and capture of the morphologically complex ensheathing glia. We therefore developed a single nuclei extraction protocol that utilizes fluorescence-activated cell sorting (FACS) to isolate GFP-tagged nuclei from dissociated peripheral nerve cells (Fig. 1a, b; Extended Fig. 1a). To fluorescently label nuclei of all cells in the nerve, we mated mice expressing Cre from the ubiquitously expressed ActB promotor (ActB-Cre mice) to mice harbouring a conditional Sun1-GFP allele in the Rosa26 locus that upon Cre excision expresses the Sun1 inner nuclear envelope protein fused to GFP from the CAG promotor, to generate Sun1-GFP/+;ActB-Cre/+ (ActB-Sun1) mice (Fig. 1b). In other experiments, we generated and utilized Sun1-GFP/+;Mpz-Cre/+ (Mpz-Sun1) mice. In these mice, cells that express Mpz at any stage of development, such as those derived from Schwann cell precursors (SCP), will have GFP+ nuclei. The Sun1 mice allowed us to effectively purify nuclei from peripheral nerve that were suitable from generating high-quality single nuclei transcriptional profiles (Fig. 1c,d).
Figure 1. Generation of a single nuclei expression atlas of mouse sciatic nerve.
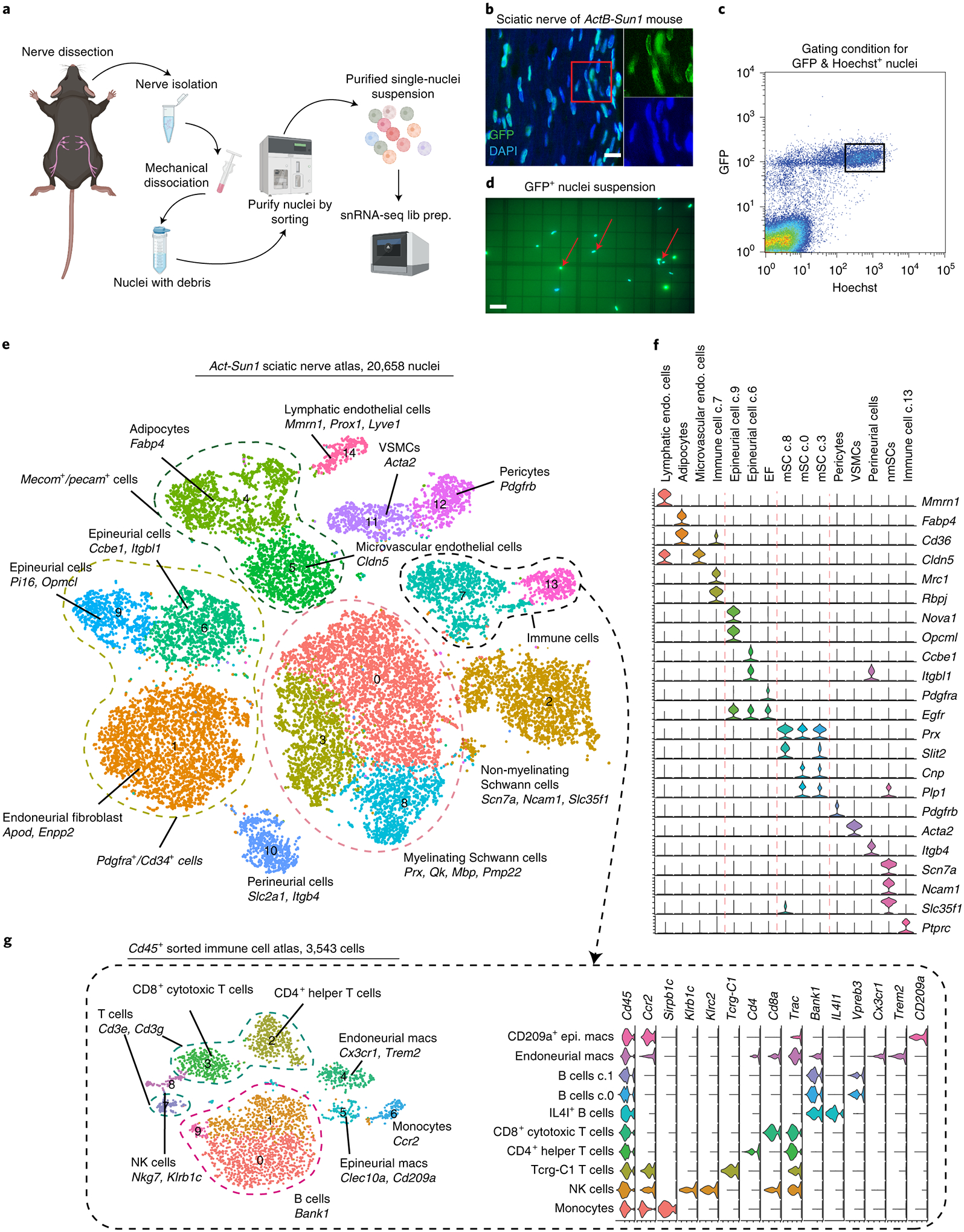
(a) Schematic diagram of the single nuclei isolation and library preparation. (b) Longitudinal section of sciatic nerve of Sun1-GFP/+;ActB-Cre/+ mouse; all nuclei express GFP. (scale bar = 10 um, n=3 biologically independent mice) (c) Gating condition for FACS isolation of GFP and Hoechst positive nuclei; boxed nuclei were selected for scRNA-seq as they represent GFP+ and Hoescht+ population. (d) Fluorescence image of purified GFP-positive single nuclei presented on hemocytometer. (scale bar = 50 um, n=12 biologically independent experiments) (e) Sciatic nerve single nuclei atlas (tSNE plot) with 20,658 high-quality nucleus transcriptional profiles. Distinct cell types are highlighted by different colors. Clusters that share ‘signature’ genes are enclosed in dashed lines, i.e. all mSCs expressed Prx and Mbp, endoneurial/epineurial fibroblasts expressed Pdgfra and Cd34 and immune cells expressed Ptprc (this one missing from figure). (f) Violin plots illustrating different signature genes expressed in 15 clusters. (g) Sciatic nerve immune cell atlas (tSNE plot, left) with 3,543 CD45+ enriched cell transcriptional profiles. Distinct cell types are highlighted with different colors and their signature genes labeled in the violin plot (right).
We performed snRNAseq on the 10X genomics platform using nuclei purified from sciatic nerves (5 sequencing libraries from ActB-Sun1 mice and 2 sequencing libraries from Mpz-Sun1 mice, n=4 mice in each library). As nuclei contain an abundance of intronic pre-mRNA, we also incorporated these intronic reads as we found that intronic and exonic read counts of a specific gene are statistically similar and thus reflect biologically relevant expression (Extended Fig. 1b–f). We derived a sciatic nerve atlas using 20,658 nuclei with an average depth of more than 70,000 reads per nucleus using nerves of ActB-Sun1 mice (Fig. 1e, Supp. Fig. 1). Our initial analysis confirmed that SCs are present in the expected proportions in this dataset (45.1%), and is significantly higher than previously reported percentage, ranging from 3.3%−8%9–12, using single-cell RNA-seq (Supp. Fig. 2). The improvement in mSC representation via analyzing single nuclei provides a more comprehensive view of the cellular components of peripheral nerve and provides an opportunity to assess transcriptional heterogeneity within these cell populations.
Computational data integration using Seurat that adjusts for uncontrolled batch effects enabled clustering and identification of 15 distinct cell types in mouse sciatic nerve (Fig. 1e,f). Among the cells identified, mSCs, marked by expression of Prx, Qk and Mbp, formed the largest population in the atlas (cluster 0, 3 and 8) and constituted 73.5% of all SCs (6,157 nuclei). Interestingly, mSCs were separated into three clusters; Cluster 8, which expressed high levels of Slit2 a chemorepellent factor with strong axon guidance function19, Cluster 0, which expressed high levels of Cnp, the myelin-associated enzyme that catalyses hydrolysis of 2’, 3’-cyclic nucleotides20, and Cluster 3, which expressed moderate levels of Cnp, Plp1 and Slit2. Another large cluster containing 2,218 nuclei, Cluster 2, represented nmSCs, marked by canonical markers such as Ncam1 and Slc35f1, as well as additional markers Scn7a21 (Sodium channel Nax gene) and Csmd1, a complement regulatory protein22. Together, the mSC and nmSC populations represented 40% of the total nuclei in the sciatic nerve. These results are consistent with anatomical studies of peripheral nerve cell composition23, where glia were found to be the predominant cell type.
snRNA-seq uncovers cell type diversity in sciatic nerve.
Our analysis of the sciatic nerve snRNAseq dataset identified a number of additional cell types associated with peripheral nerves. We detected nerve-associated microvascular endothelial cells (Cluster 5: 1,329 nuclei) that express Cldn524, as well as lymphatic endothelial cells (Cluster 14; 331 nuclei) identified by Prox1 and Lyve125 (Fig. 1e,f). In addition to endothelial cells, we also identified vasculature-associated smooth muscle cells (VSMCs, Cluster 11; 659 nuclei), which express Acta2 and support contractility of blood vessels26, and pericytes (Cluster 12; 635 nuclei), which express Pdgfrb and help form the blood-nerve barrier27 (Fig. 1f). Together these vascular cell types constituted 21.7% of cells in sciatic nerve.
The immune cells in the peripheral nerve constitute a small percentage of cells (~8.5%, Fig 1e), making it difficult to obtain a comprehensive picture of nerve immune cell heterogeneity from global single cell approaches. To enhance analysis of this limited cell population, we used the global immune marker Cd4528 to FACS purify immune cells from sciatic nerves of 20 mice (Extended Fig. 1g). We performed single-cell RNA-seq on this enriched cell population and obtained a representative peripheral nerve immune cell atlas (Fig. 1g, Supp. Fig. 3; 3,543 cells). We identified 10 distinct immune cell populations, including macrophages as well as B and T lymphocytes, each with subtypes further distinguished by unique expression markers (Fig. 1g). Two distinct macrophage populations could be identified: cluster 4 macrophages express both Cx3cr1 and Trem2 and comprise the resident endoneurial macrophages we previously identified28, while Cluster 5 corresponds to epineurial macrophages36.
A significant portion of the non-glial cells in peripheral nerve are fibroblasts associated with the connective tissue layers of the nerve29. We identified four populations of fibroblasts (Clusters 1, 6, 9, and 10) in our ActB-Sun1GFP sciatic nerve dataset, and together they define the compartments of peripheral nerves – epineurium, perineurium and endoneurium. Epineurial fibroblasts, which surround the outermost layer of the nerve, express Pdgfra and Pcolce29, and were further divided into two populations, with Cluster 9 expressing Opcml and Cluster 6 expressing Pi1630 and Ccbe1 (Figure 1e–f). Ccbe1+ epineurial fibroblasts also expressed Adamts3, which interacts with vascular endothelial growth factor C (Vegfc) and serves as a key regulator of lymphangiogenesis in nerve31 (Extended Fig. 1h). Perineurial fibroblasts (Cluster 10), which ensheath nerve fascicles, were notable for their lack of Pdgfra expression and their expression of markers Itgb4 and Slc2a132 (also known as Glut1). Endoneurial fibroblasts, which reside within the endoneurium, express Pdgfra as well as Cspg4 (also known as NG2) and Enpp233 (Figure 1e).
Previous work showed that endoneurial fibroblasts (EFs) arise from Schwann cell precursor cells (SCPs), thus sharing a developmental lineage with SCs34. In the mouse, SCPs are detectable at E12.5 and express Mpz34. We inspected our snRNAseq dataset derived from peripheral nerve nuclei isolated from Mpz-Sun1 mice (Fig. 2a, b, Extended Fig. 2a and Supp. Fig. 4). We detected EFs in the Mpz-Sun1 atlas (Fig. 2b) and found they express the fibroblast marker Pdgfra as well as the stem cell markers Cd34 and nmSC marker Ngfr (Fig 2c). GO analysis suggests EFs regulate collagen matrix formation and axon development (Extended Fig. 2b–d). We used RNA-FISH to demonstrate co-expression of Pdgfra with the newly identified Ngfr and Cd34 expression in EFs (Fig. 2d–i, Extended Fig. 2e,f). Together, the multiple fibroblast populations responsible for peripheral nerve compartmentalization constitute ~30% of the cells in sciatic nerve.
Figure 2. Endoneurial fibroblasts express non-myelinating Schwann cell markers.
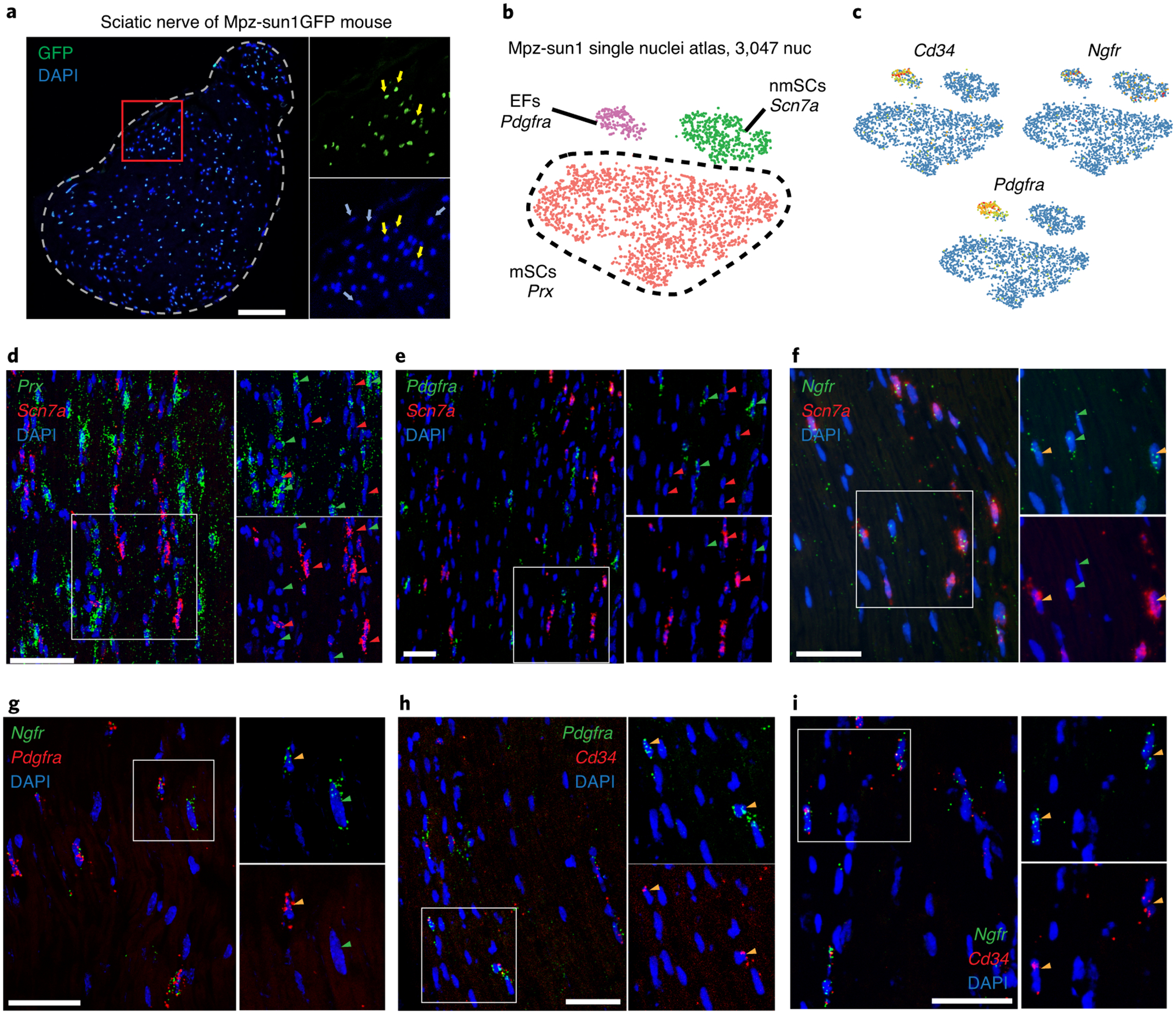
(a) Cross-section of sciatic nerve of Sun1-GFP/+;Mpz-Cre/+ mouse showing only a subpopulation of nuclei express GFP. Yellow arrows indicate GFP+ nuclei and blue arrows indicate GFP- nuclei. (n=3 biologically independent mice) (b) tSNE plot with 3,047 nuclei from Sun1-GFP/+;Mpz-Cre/+ defines three GFP+ cell types – endoneurial fibroblasts, nmSCs and mSCs. (c) Mpz-lineage tSNEs showing expression of Cd34, Ngfr and Pdgfra. Note the expression of Ngfr in both nmSCs and endoneurial fibroblasts. (d-i) RNA-FISH showing expression of Prx, Scn7a, Ngfr, Pdgfra and Cd34; red arrows indicate red gene signal, green arrows indicate green gene signal, orange arrows indicate co-localization of red and green gene signals. Nuclei are stained with DAPI. Scale bar=50 um. (n=3 biologically independent mice for all RNA-FISH experiments) (d, e) Scn7a expression does not overlap with either Prx or Pdgfra. (f, g) Ngfr expression overlaps with subsets of Scn7a+ and Pdgfra+ cells. (h, i) Cd34 expression overlaps with Pdgfra and Ngfr expression.
Unique cellular compositions exist across nerve types.
We performed snRNAseq on additional peripheral nerves to examine the generality of the findings from sciatic nerve. We isolated the pernoneal nerve, a branch of the sciatic that supplies both motor and sensory function to the lower limbs35; sural, a sensory branch of the sciatic that is well studied in models of sensory neuropathy36; and the vagus, a mostly unmyelinated nerve that regulates important autonomic functions37 from ActB-Cre:Sun1-GFP mice, and characterized over 1,500 high quality nuclei from each nerve type (Fig. 3a). After using DeepImpute to impute values for missing genes, Seurat to correct for batch effects, and removing the immune populations, we merged these data with our sciatic nerve dataset using the anchoring approach in Seurat. The combined peripheral nerve atlas, with 22,553 nuclei encompassing 14 clusters of cells, revealed unique differences in cell-type composition across different nerve types (Fig. 3a–c, Supp. Fig. 5 and 6). For instance, whereas most nerves have more glia than fibroblasts, the sural nerve contains more than twice as many fibroblasts (44.2%) as glia (19.2%). This is particularly noticeable for the epineurial fibroblast populations (22.3%), which are 2-fold more abundant in sural compared to other nerves (11.4%). Indeed, the sural nerve has more epineurium than sciatic nerve due to its smaller and more sparsely distributed nerve fascicles38. These structural attributes likely account for the varying sizes of the fibroblast populations we identified.
Figure 3. Single nuclei transcriptional profiling of peroneal, sural and vagus nerves highlights SCN7A as potential DPN risk gene.

(a) UMAP plot of unified peripheral nerve atlas combining data from four peripheral nerves: sciatic (gray), peroneal (red), sural (blue), and vagus (green) nerves. (b) UMAP plot illustrating the 14 cell clusters identified in unified peripheral nerve atlas using annotations consistent with earlier sciatic nerve atlas. Immune cells were not included in this analysis. (c) Quantification of cell type compositions across different peripheral nerves. Glia cells consistently constituted the largest group of cells in all four nerve types. Note the increased proportion of nmSCs in the vagus nerve compared to all other nerves. (d) Genes mutated in peripheral neuropathy were curated from OMIM and their expression in specific cell types was identified using the peripheral nerve atlas. Colored box indicates expression of gene in indicated cell type. Nerve immune cell atlas was used to identify genes expressed in immune cell types. (e) Schematic diagram showing human chromosome 2 locus (164,773,339–168,773,339 (2q24)) with DPN-associated variants identified by GWAS. SCN7A is antisense and immediately upstream of the variant rs13417783. The two other genes uniquely expressed in other peripheral nerve cell types are SLC38A11 and Nostrin (blue arrow). SCN2A is shown in yellow. The directionality of arrow indicates sense or antisense transcription. Expression of genes is shown in UMAP. The two genes Csmd1 and Ntrk3 in dashed box are two additional variants identified in this GWAS study that are also uniquely expressed in nmSCs.
Single cell transcriptional profiling has proven to be a valuable adjunct in identifying disease-relevant cell types in complex diseases such as autism39 and Alzheimer’s disease. We used our peripheral nerve atlas to similarly search peripheral neuropathy datasets. We curated the pathogenic neuropathy variants from OMIM40, and found 20 genes harboring pathogenic variants that are expressed in an unique cell type in the peripheral nerve atlas (Fig. 3d). As expected, most of these are expressed in mSCs or nmSCs. We also examined genome-wide association studies (GWAS) for diabetic neuropathy, a common small-fiber neuropathy, to examine the cellular expression patterns of implicated genes. A cluster of variants at a single locus on chromosome 2q24 was identified as a strong risk factor for diabetic peripheral neuropathy (DPN)41. Sixteen genes lie within 2 MB of the most significant SNP in this locus (rs13417783, chr2:166,773,339). This made it difficult to determine which gene(s) are involved in the disorder (Supp. Fig. 7), however the authors highlighted SCN2A dysregulation as most likely contributor based on its expression in tibial nerve and role as a sodium channel whose dysfunction is linked to epilepsy42. We examined all 16 genes in the locus using our peripheral nerve atlas and found that 3 of them are uniquely expressed in specific cells in peripheral nerve (Fig. 3e). These included Scn7a, which we previously noted is a specific marker of nmSCs, Nostrin that is expressed in microvascular endoethelial cells, and Slc38a11, which is detected in VSMCs. Strikingly, two other disease-associated SNPs, rs11073752 (chr15:88,423,051) and rs13265430 (chr8:4,165,607), from this study are located within NTRK3 and CSMD1 genes, respectively. In the nerve, both of these genes are also specifically expressed in nmSCs (Fig. 3e), cells that enheath the small sensory fibers most affected in DPN43. To confirm these expression patterns, we performed RNA-FISH on sciatic nerves and demonstrated that expression of Csmd1 and Scn7a is completely overlapping (Extended Fig. 2g). The association of nmSCs with small-fiber neuropathy along with our single nuclei expression data (Fig. 1e, f) raises the possibility that the diabetic neuropathy risk variants identified at 2q24 could be influencing SCN7A expression, rather than the SCN2A expression previously reported.
Novel SC subtypes identified by analysis of Mpz+ cell profiles.
We next sought to use our nerve single cell dataset to search for subpopulations of mSCs or nmSCs. We merged the sciatic nerve Act-Sun1 and Mpz-Sun1 snRNA-seq data sets and re-clustered only cells identified as SCs to obtain a more refined view of this population. The sciatic nerve SC-enriched dataset, which contains information from 10,253 nuclei, revealed six distinct subsets of myelinating and non-myelinating SCs (Fig. 4a). The nmSCs, which comprise 26.2% of the glia, were segregated into two clusters, with cluster 4 expressing Scn7a and Csmd1 and cluster 5 expressing additional unique markers including Marcks and Prnp (Fig. 4a). The mSCs, which comprise 73.7% of all glia, were further segregated into four clusters, with cluster 0 expressing high levels of Plp1 and cluster 2 expressing collagen protein Col23a1. The Slit2-high population from global analysis further separated into two clusters, with cluster 1 expressing Lmna and myelination-related transcription factor Egr2, and cluster 3, which is defined by co-expression of the integral membrane protein Cldn14, the secreted protein Adamtsl1, and the myelin protein Pmp2.
Figure 4. Identification of Schwann cell subpopulations using multiple transcriptional profiling approaches.

Clusters comprising Schwann cells from the ActB-Sun1 and Mpz-Sun1 sciatic nerve atlases were merged to generate a combined Schwann cell atlas. (a) tSNE plot of 10,253 nuclei is shown (middle) with 2 clusters containing cells expressing markers of nmSCs and 4 clusters with expression profiles consistent with mSCs. Markers highlighting subpopulations of nmSCs are shown in top red box (Prnp, Dbi, B2m and Marcks) with sCluster5 exhibiting high expression of these 4 genes. Markers highlighting subpopulations of mSCs are shown in bottom blue box (sCluster0: Plp1; sCluster1: Lmna; sCluster3: Col23a1, Cldn14, Adamtsl1 and Pmp2). (b) Comparison of total nerve RNAseq vs Mpz+ cell ribosome affinity (Ribo-Tag) RNAseq data analysis of sciatic and vagus nerve reveals 3 sets of Mpz+ enriched genes: (i) mSC-enriched genes from sciatic nerve (red), (ii) nmSC-enriched genes from vagus nerve (blue) and (iii) co-enriched genes (green). Each dot represents a gene, and its relative position on the plot indicates the level of enrichment in the ribosome affinity RNAseq (Mpz+ cells) dataset relative to total nerve RNA-seq dataset. (c) Differential expression (DE) analysis comparing vagus and sciatic nerve Mpz+ ribosome affinity RNAseq data reveals signatures of myelinating and non-myelinating specific genes (left). Markers of Schwann cell subtypes derived from snRNAseq sciatic nerve atlas display distinct profiles in mSC vs. nmSC-enriched datasets (right). (d) KEGG-based expression module analysis of SC subpopulations. Cldn14/Adamtsl1/Pmp2+ SCs are enriched for NAD and pyruvate metabolic pathways, as well as axon guidance signaling pathway. (e-h) Expression patterns of Cldn14, Pmp2, Adamtsl1 and Prx were established using RNA-FISH. Red arrows indicate red signal, green arrows indicate green signal, orange arrows indicate co-localization of red and green signals. Both Pmp2 (e) and Adamtsl1 (f) are expressed in subpopulation of mSCs (marked by Prx). Cldn14 (g) and Adamtsl1 (h) expression overlapped extensively with Pmp2. Scale bar=50 um. (n=3 biologically independent mice)
Because single-cell and single-nuclei data are inherently sparse and thus often have limited sensitivity to detect changes in transcript abundance, we leveraged bulk gene expression datasets from mSC and nmSC enriched cell populations in order to contextualize our findings regarding different SC subtypes.For this purpose, we crossed RiboTag mice to Mpz-Cre mice to generate Mpz-RiboTag mice that express epitope-tagged ribosomal protein RPL22HA in cells that expressed MPZ (See Methods). We performed ribosome-affinity purification from nerves of these mice and generated RNA-seq data (Supp. Fig. 8). The majority of glial cells in sciatic nerve are mSCs whereas those in vagus nerve are nmSCs (Fig. 3c). Thus the Mpz-RiboTag approach allows us to identify genes that are differentially expressed in mSC or nmSC by comparing relative expression between these two nerves (Fig. 4b,c). By comparing the SC subpopulation marker genes identified in our snRNAseq analysis with these Mpz-RiboTag datasets (Supp. Table 1), we showed that several of the unique mSC genes, including Slit2, Col23a1, Adamtsl1, Cldn14, and Pmp2, were significantly higher in ribosome-associated transcripts from sciatic nerve (mSC-enriched) (Fig. 4c). In contrast, nmSC markers identified by peripheral nerve snRNAseq, including Scn7a, Prnp, Dbi, and Marcks, were present at high levels in vagus nerve (nmSC-enriched) RiboTag datasets (Fig 4c). This analysis confirms the single nuclei-derived SC transcriptional signatures that we used to identify SC subpopulations.
To examine biological differences across these SC clusters, we defined 298 transcriptional modules based on metabolic and signaling pathways from KEGG database and calculated a module expression score for each nucleus in the SC-atlas. We then derived an aggregated expression z-score for each SC cluster to determine whether a putative SC subpopulation differentially expresses a particular module (Fig. 4d). Using this analysis, we detect profound differences in mSCs vs. nmSCs, with pathways such as cell junction formation and carbon metabolism highly expressed in mSCs and expression of neurotrophin signaling and neuroactive ligand-receptor interactions increased in nmSCs (Supp. Table 2). The Msc-cluster 3 (Pmp2/Cldn14/Adamtsl1+) showed higher scores related to axon guidance (Extended Fig. 3), NAD metabolism and pyruvate metabolism compared to other mSCs (Fig. 4d). Interestingly, motor neurons can readily take up lactate for energy metabolism through coupled-monocarboxylate transporters expressed on both glia and neurons44. In accord, mice with a SC-specific deletion of MCT1 display motor axon defects and disrupted neuromuscular innervation45. This led us to pursue additional experiments to determine whether the SC-cluster 3 could be important in defining a motor nerve environment.
Co-expression of Cldn14, Adamtsl1 and Pmp2 defines a mSC subtype.
While there has been speculation regarding the possibility that SC supopulations could influence the path of regenerating motor vs. sensory axons6,46, previous transcriptome studies were unable to identify motor-specific or sensory-specific SC populations. To confirm mSC-cluster 3 represents a subpopulation of mSCs, we first performed RNA-FISH to examine the expression of Adamtsl1, Cldn14, Pmp2, and the mSC marker Prx. We found that these three transcripts are all detected exclusively in cells expressing the pan-mSC marker Prx47. We also found that Adamtsl1 and Cldn14 are each co-expressed with Pmp2 (Fig. 4e, f), and that Pmp2 as well as Adamtsl1 are expressed in only a fraction of Prx+ cells (Fig. 4g,h), confirming that these cluster3 markers define a subpopulation of mSCs.
To further explore this unique population of mSCs, we performed immunofluorescence (IF) staining on cross sections of sciatic and tibial nerves. We used antibodies against β-tubulin III (β-tub) to highlight axons, MBP to label all mSCs, and PMP2 to highlight the cells corresponding to SC-cluster 3 (Fig. 5a,b). This analysis confirmed that PMP2 is only expressed in a subpopulation of MBP+ mSCs, consistent with previous observations48. In analyzing these images, we noticed that the PMP2+ myelin sheaths are exceedingly thick and morphometric analysis revealed that PMP2+ cells tend to preferentially surround large axons. To confirm these findings, we calculated G-ratios (axon diameter/myelin diameter) of PMP2+ vs. PMP2− SC-associated fibers. This analysis demonstrated that PMP2+ SCs are associated with thicker myelin as noted by significantly smaller G-ratio of PMP2+ myelinated axons (t-test p<0.0001) (Fig. 5c,e). We also found that PMP2+ SCs myelinate larger axons in both sciatic and tibial nerves (Fig. 5d, f; t-test p<0.0001). We next extended our observations to human sciatic nerve samples and again found that PMP2 marks a subpopulation of mSCs. Indeed, PMP2 is expressed similarly in human and mouse nerves, with PMP2+ SCs associated with thickly myelinated, large diameter axons in both species (n=3, Fig. 5g–k, t-test p<0.0001). These immunolabeling and morphometric analyses using defined markers of cluster3 mSCs demonstrate that these cells represent a subpopulation of mSCs present in both mouse and human nerves.
Figure 5. PMP2+ SC subpoulation is associated with thickly myelinated axons.
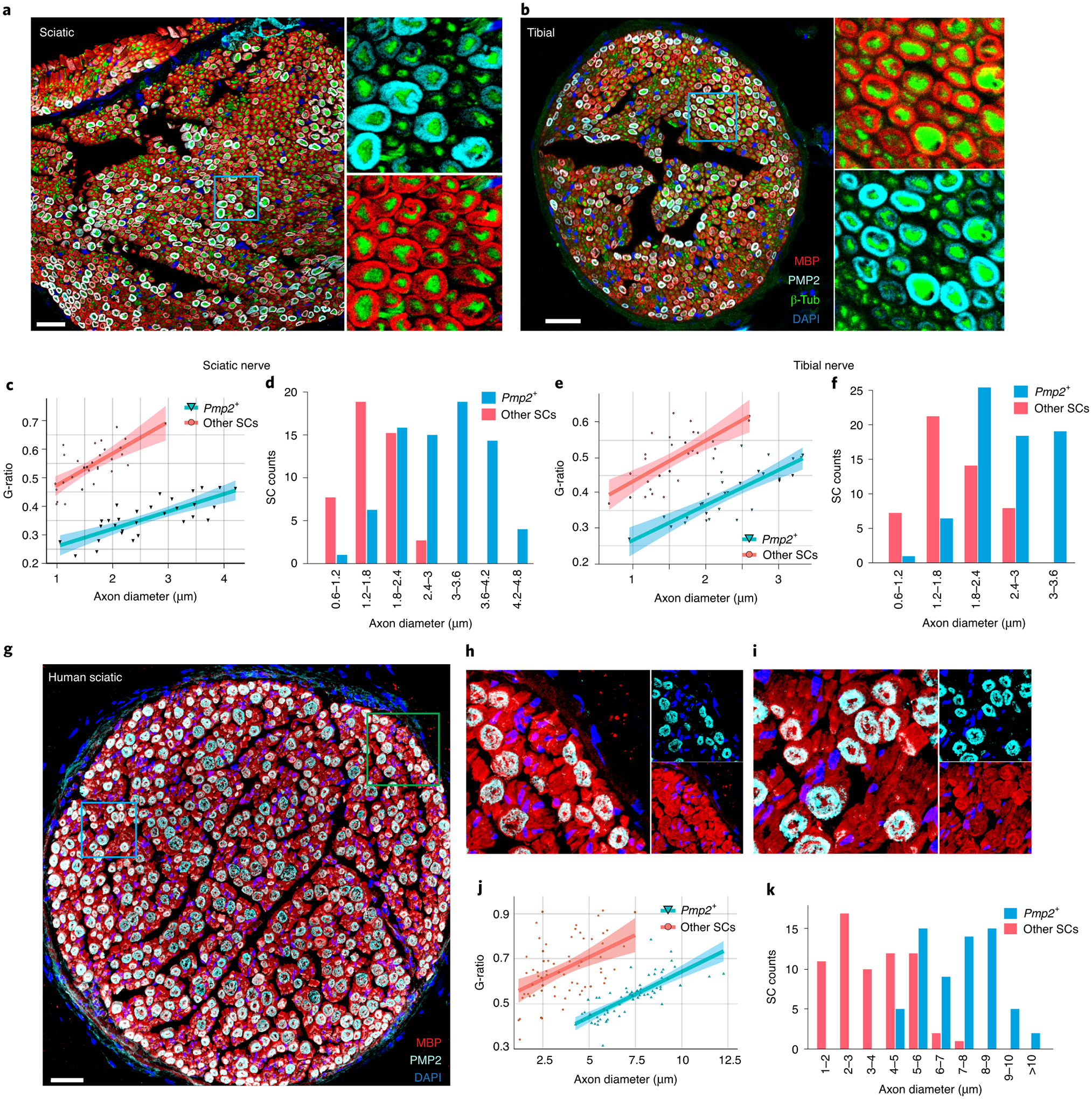
(a,b) Immunostaining of (a) sciatic and (b) tibial nerve cross-sections with antibodies reognizing myelin basic protein (MBP) (marks mSCs), PMP2, β-tubulin (pan-axon marker) and DAPI (scale bar = 30 um, n=3 biologically independent mice). β-tubulin is used for quantification of axon diameter. The color scheme is identical for both sciatic and tibial nerve images, with MBP in red, PMP2 in cyan, β-tubulin in green and DAPI in blue. Note that PMP2 ensheathes only a subpopulation of axons, and the thicker myelin sheaths surrounding PMP2 associated axons are apparent (scale bar = 30 um). (c-f) Pmp2+ SCs are morphologically distinct. (c, e) Scatterplot illustrating the G-ratio vs axon diameter in Pmp2+ SCs vs Pmp2− SCs in (c) sciatic and (e) tibial nerve. The G-ratio distribution of Pmp2+ SCs is significantly smaller (thicker myelin) than that of Pmp2− SCs (n=3 biologically independent mice, two-tailed t-test p<0.0001). 95% confidence level interval is defined by the colored region along the line using a linear model. (d,f) Binned bar graph showing average diameter of axons associated with Pmp2+ SCs is significantly larger than diameter of those associated with Pmp2− SCs in sciatic (d) and tibial (f) nerve (n=3, two-tailed t-test P<0.0001). (g-i) Human sciatic nerve cross-section immunostained for MBP and PMP2 (scale bar = 100 um). Note axons ensheathed by PMP2+ SCs are more thickly myelinated than those myelinated by PMP2− SCs (small panel, right; repeated on 3 independent human sciatic nerves). (j) Scatterplot illustrating the G-ratio vs axon diameter in Pmp2+ SCs vs Pmp2− SCs in human sciatic nerve (n=3 independent human sciatic nerves, two-tailed t-test p<0.0001). 95% confidence level interval is defined by the colored region along the line using a linear model. (k) Binned bar graph showing the average diameter of axons associated with Pmp2+ SCs is significantly larger than diameter of those associated with Pmp2− SCs in human sciatic nerves (n=3 independent human sciatic nerves, two-tailed t-test p<0.0001).
PMP2+ SCs preferentially ensheath motor axons.
Motor axons are generally ChAT+, large diameter axons that are thickly myelinated in order to speed signal propagation. The correlation of Pmp2+ SCs with larger axons that are ensheathed by thick myelin caused us to consider whether these SCs might preferentially myelinate motor axons. We first examined the abundance of Pmp2+ SCs in the nerves included in our peripheral nerve atlas: the sciatic nerve, a mixed nerve containing both motor and sensory axons; the sural nerve, a sensory-specific nerve; and the vagus nerve, an autonomic nerve containing mostly unmyelinated axons. We performed sub-clustering analysis using Seurat (Fig. 6a), and found that Pmp2+ SCs are almost 3-fold enriched in the sciatic nerve compared to sural and vagus nerves (sciatic: 18.7%, sural: 6.6%, vagus: 7.6%; Fig. 6b), suggesting that Pmp2+ SCs could be associated with motor axons. To further examine this idea, we performed immunostaining on the femoral nerve, which is largely composed of motor axons, and sural nerve. We found that Pmp2+ fibers make up a much higher proportion of total myelinated axons in the femoral nerve (motor environment) than in the sural nerve (sensory environment) (Fig. 6c, d).
Figure 6. PMP2+ Schwann cells preferentially ensheath motor axons.
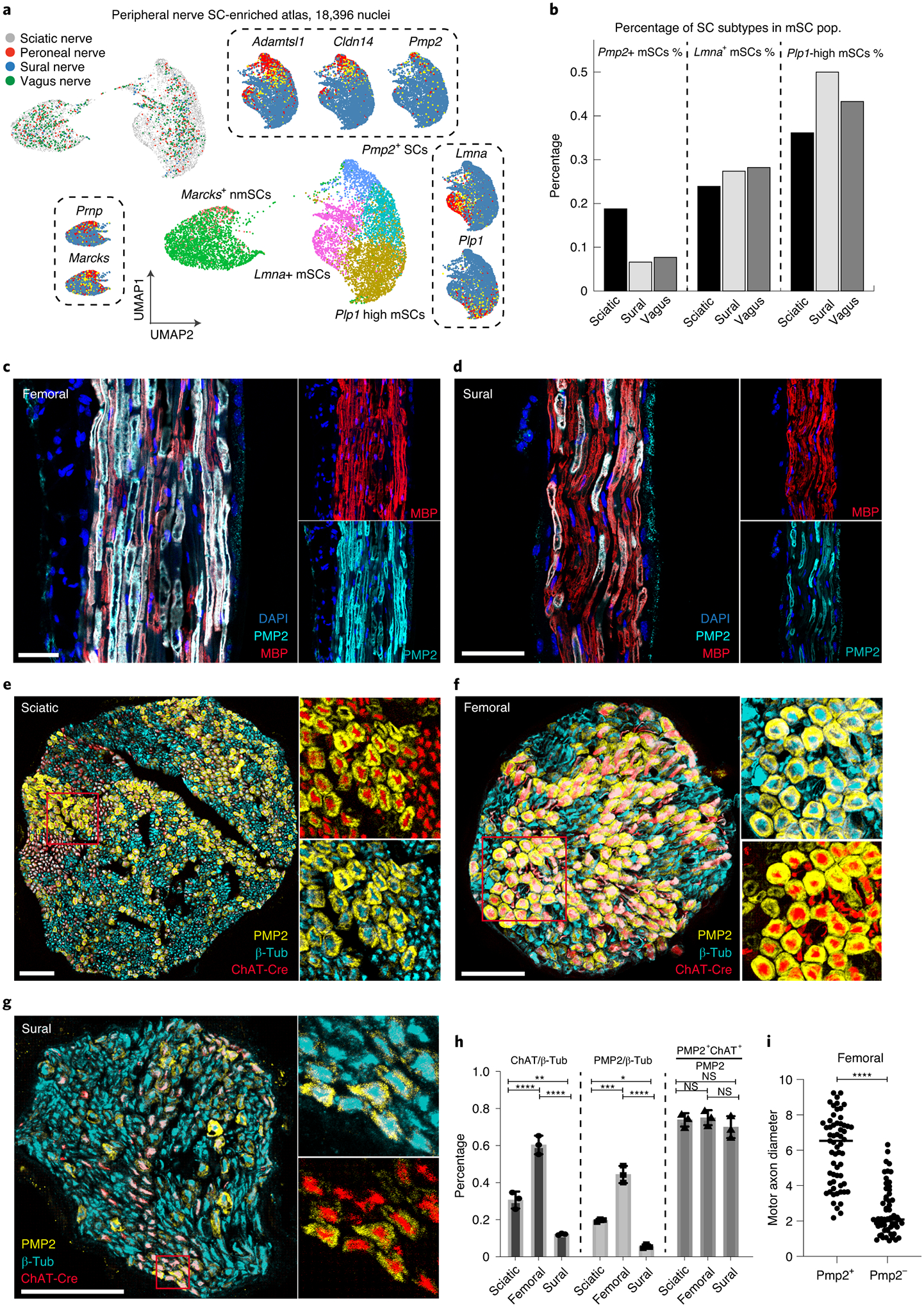
(a, b) SC-specific subclustering of peripheral nerve atlas. (a) UMAP plot of SC populations isolated from the unified peripheral nerve atlas. Identical clustering results are presented with 4 different mSC subtypes and 2 different nmSC subtypes. Marker genes of each SC subtype are also shown in dashed box. (b) Bar plot showing relative proportion of each mSC subpopulation in sciatic, sural and vagus nerves. (c, d) PMP2 (cyan) and MBP (red) immunostaining of longitudinal sections of femoral (motor axon-enriched) and sural (sensory-axon enriched) nerves; DAPI (blue). Overlap of PMP2 and MBP shown in white. (scale bar = 50 um). Note increased number of PMP2+ SCs in femoral vs sural nerves. (n=3 biologically independent mice) (e-g) Immunostaining of cross-sections of indicated peripheral nerves from ChAT-Cre-tdTomato mice (red fluorescence, motor axons) with antibodies for PMP2 (PMP2+ SCs, yellow) and β-tubulin (all axons, cyan). (scale bar = 50 um, n=3 biologically independent mice) (h) Bar charts showing percentages of ChAT axons (ChAT/β-Tub), axons ensheathed by PMP2+ SCs (PMP2/β-Tub), and PMP2+ SCs ensheathing ChAT+ axons (Pmp2+ ChAT+/Pmp2) in each nerve. Note that number of motor axons and PMP2+ SCs is highest in femoral nerve, whereas the sural nerve has the lowest numbers of both ChAT+ axons and PMP2+ SCs. The percentage of PMP2+ SCs associated with motor axons is ~75% in all nerves examined. P-values are as follows: ChAT/β-Tub, sciatic vs. sural (P = 0.0047); PMP2/β-Tub, sciatic vs. femoral (P = 0.0002) and sciatic vs. sural (P = 0.0327). All other P-values are < 0.0001 or not significant (ns). 2-way ANOVA with multiple comparisons. Data are mean +/− SD (n=3 biologically independent mice). (i) Average diameter of ChAT+ axons in femoral nerve ensheathed by PMP2+ SCs vs PMP2−. Unpaired t-test. Data show mean of 20 axons per mouse (n=3 biologically independent mice). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To quantify Pmp2+ SCs in peripheral nerves and illustrate their association with motor axons, we crossed choline acetyl transferase (ChAT)-Cre mice49, which express Cre recombinase primarily in motor neurons, with ROSA-tdTomato (tdT) reporter mice to produce reporter mice that express tdTomato in motor axons (Extended Fig. 4a). We monitored tdT fluorescence in sciatic, femoral, and sural nerves, and found that 30.6%, 60.3%, and 12.1% of axons are ChAT+, respectively (n=3, Fig 6e–g). We saw a similar Pmp2+ SCs distribution in sciatic (19.5%), femoral (44.4%) and sural (5.5%) nerves (Fig. 6h), indicating that Pmp2+ SCs are strongly correlated with ChAT+ axon abundance. We further examined the association of Pmp2+ SCs with ChAT+ axons across these three nerves, and found that, on average, 76% of Pmp2+ SCs ensheath motor axons (Fig. 6h). However, not all ChAT+ axons are ensheathed by Pmp2+ SCs. We found that femoral nerve has the highest percentage of ChAT+ axons ensheathed by Pmp2+ SCs (64.3%, Fig. 6d), whereas ChAT+ axons in sciatic and sural nerves are ensheathed by Pmp2+ SCs at lower rates (45.7% and 28.7%, respectively) (Extended Fig. 4b,c). Additional morphometric analysis of the femoral nerve showed that Pmp2+ SCs preferentially ensheath larger ChAT+ axons while PMP2− ChAT+ axons were of smaller diameter (Fig. 6i). It is possible that these smaller ChAT+ axons represent gamma-motor or parasympathetic axons50. These observations raise the possibility that subtypes of motor neurons with large diameter axons, such as alpha motoneurons, are specifically myelinated by PMP2+ SCs.
Reduced numbers of PMP2+ SCs in SOD1G93A mice and ALS patients.
The association of Pmp2+ SCs with ChAT+ axons led us to hypothesize that they could be affected in disorders of motor neurons. We therefore studied the SOD1G93A mouse model of amyotrophic lateral sclerosis (ALS), which develops a distal to proximal motor impairment and motor neuron death51. We performed immunostaining with ChAT, β-Tub and Pmp2 antibodies to assess the abundance of motor axons and PMP2+ SCs in end-stage P140 SOD1G93A with NeuroScore (NS) 4 and age-matched control mice (Fig. 7a–f, n=3). NeuroScores span from 0–4, with NS0 indicating no symptoms, NS1 exhibiting very early symptoms of ALS (trembling and hindlimb splaying), and NS4 displaying end-stage phenotypes52. In general, we detected a profound reduction in Pmp2+ SCs in all SOD1G93A peripheral nerves examined that was well correlated with the reduction in ChAT+ motor axons (Fig. 7 g–i). As sural nerve contains only 10% motor axons, the preferential depletion of PMP2+ SCs is most apparent in this nerve. This motor-asociated SC subpopulation was dramatically decreased in the SOD1G93A sural nerve, in contrast to the continued presence of other MBP+ SCs (Fig. 7j, Extended Fig. 5). In femoral nerve at P140, we could not accurately count axons due to severe degeneration, however there was clearly greater than 50% axon loss53.
Figure 7. Significant reduction of motor-associated Schwann cells in ALS SOD1G93A mouse model.
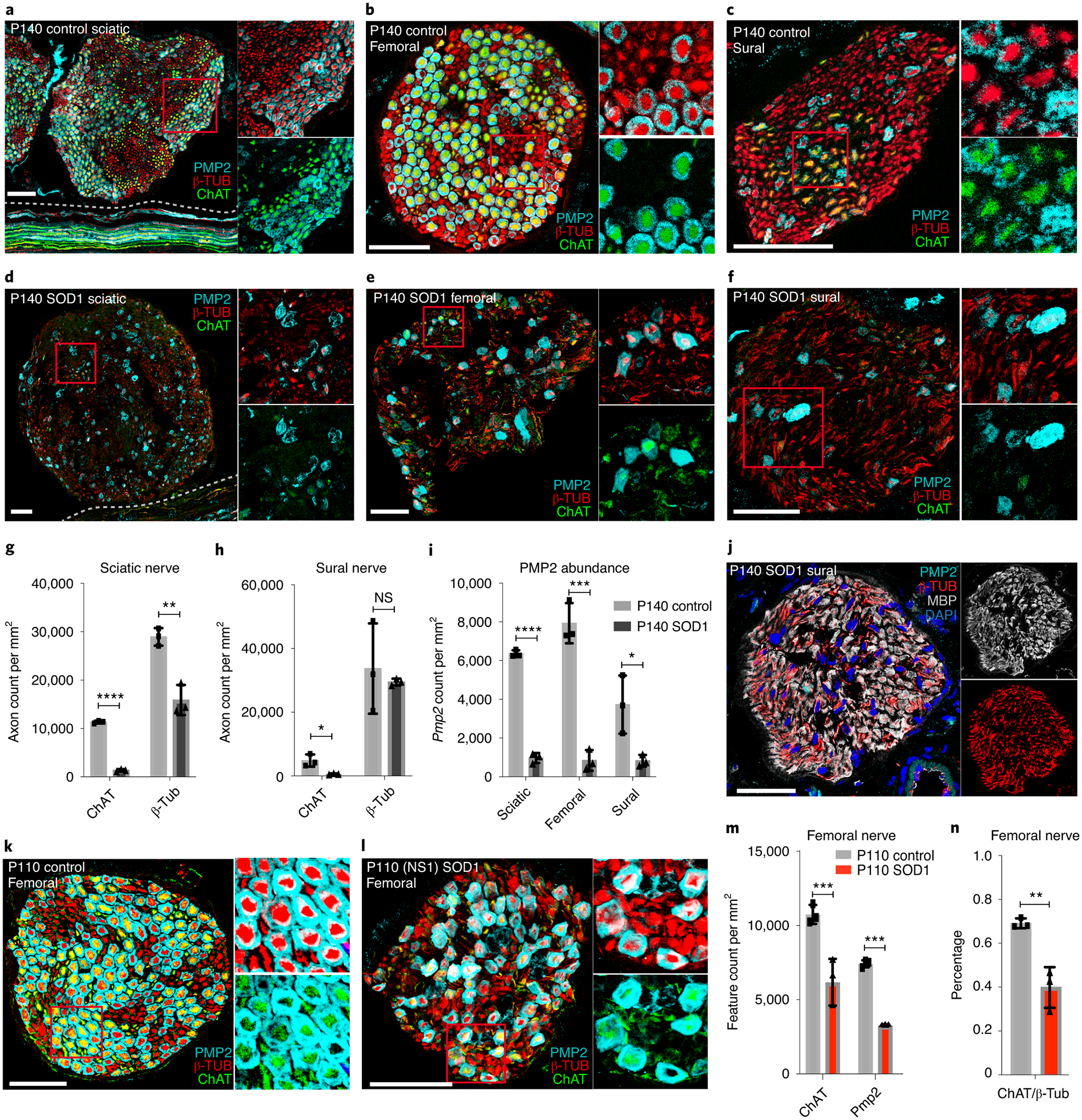
(a-f) Immunostaining of PMP2 (cyan), β-TUB (red) and ChAT (green) on cross-sections of sciatic (a, d), sural (b, e), and femoral (c, f) nerves from P140 NS4 ALS SOD1G93A and age-matched control mice. (scale bar = 50 um) (g, h). Quantification of motor axons (ChAT) vs. total axons (β-TUB) in sciatic (g) and sural (h) nerves of P140 NS4 SOD1G93A and age-matched control mice (n=3 biologically independent mice). Note that axons in NS4 ALS SOD1G93A femoral nerve are severely degenerated and could not be reliably quantified. Multiple unpaired t-tests (n=3). (g) Sciatic ChAT (P < 0.0001), β-Tub (P = 0.0033); (h) Sural ChAT (P = 0.0199), β-Tub (P = 0.6359). (i) Comparison of PMP2+ SCs in sciatic, femoral and sural nerves between P140 NS4 ALS SOD1G93A and age-matched control mice. Multiple unpaired t-tests (n=3). Sciatic (P < 0.0001), femoral (P = 0.0005), sural (P = 0.0302). (j) Immunostaining of PMP2 (cyan), β-TUB (red) and MBP (white) on cross-section of sural nerve from P140 NS4 ALS SOD1 G93A mouse. Note that Pmp2+ SCs are depleted while other SCs remain (marked by MBP) (scale bar = 50 um, n=3 biologically independent mice). (k,l) Immunostaining of PMP2 (cyan), β-TUB (red) and ChAT (green) on cross-sections of femoral nerves from P110 NS1 ALS SOD1G93A and age-matched control mice. (scale bar = 50um). (m) Quantification of motor axons (ChAT+) vs PMP2+ SCs in femoral nerves between P110 NS1 ALS SODG93A and age-matched control mice. 2 way ANOVA with multiple comparisons (n=3). ChAT (P = 0.0004); Pmp2 (P = 0.0007) (n) Bar charts showing percentages of ChAT+ axons in femoral nerves of P110 NS1 ALS SODG93A and age-matched control mice. 2 way ANOVA with multiple comparisons (n=3). ChAT/-tub (P = 0.0016). Data are mean +/− SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To examine the femoral nerves before extensive axon degeneration occurs, we performed additional experiments on younger mice (P110–120) when the ALS SOD1G93A mice had early stage disease [NeuroScore=1 (NS1)]. We observed early signs of axon degeneration in femoral nerves from these mice (Fig. 7k, l). We found a 42.4% reduction of ChAT+ axons and a concordant 55.9% reduction in Pmp2+ SCs compared to age-matched controls (n=3, Fig. 7m, n, t-test p<0.001). These results are consistent with the idea that Pmp2+ SCs are preferentially associated with motor axons including those that are vulnerable in ALS.
To extend our observations to human disease, we obtained samples of sciatic nerves from 3 ALS patients as well as 3 controls with no peripheral neuropathy reported. The ALS samples were from individuals aged 61 to 77 years old, with ALSFRS decline ranging from −3.02 to −5.7 (Supp. Table 3). We identified a substantial difference in the number of Pmp2+ SCs in the ALS samples. While there is variability regarding the intensity of staining across fascicles in controls and ALS samples, we never observed depletion of Pmp2+ fibers in control fascicles (Fig. 8a–c), whereas this was common in the ALS samples (Fig. 8 d–f). While the mechanisms underlying the “dying-back” phenomenon in both SOD1G93A transgenic mice and ALS patients are unclear, our observations indicate that maintenance of PMP2+ motor-associated SCs, or the expression of PMP2 itself, is decreased in ALS. This could be due to a primary effect on these SCs, or perhaps, it reflects the loss of contact between these SCs and motor axons.
Figure 8. Pmp2-depleted fascicles identified in sciatic nerve of ALS patients.

(a-c) Immunostaining of cross-sections of human sciatic nerves with no peripheral neuropathy reported. Only a sub-population of SCs expressed PMP2 across the three samples. (d-f) Immunostaining of cross-section of human ALS sciatic nerve. PMP2+ staining is vastly decreased in all three ALS samples. PMP2 (green), MBP (red), DAPI (blue). Scale bar = 100um.
Discussion
Schwann cell heterogeneity has been a topic of interest ever since the discovery of anatomically distinguishable myelinating and non-myelinating cells. Yet profiling SCs has been immensely difficult due to their complex morphologies, even with the scRNA-seq approach. In this study, we devised a peripheral nerve-specific nuclei isolation protocal and present a peripheral nerve atlas that covers four different peripheral nerves. The cellular composition of the peripheral nerve is complex, with over 20 different cell types identified. Furthermore, the abundance of each cell type varies across different nerves depending on their biological functions – motor, sensory or autonomic. More importantly, our transcriptional profiling study provides the first high resolution identification of SCs, revealing novel markers and six new SC subtypes. The nmSCs could be separated into two subtypes based on Marcks expression. The mSC population contained more subtypes, including a Lmna+ group that was associated with Tnf, and Tgfb signaling, and a Cldn14+ Adamtsl1+ Pmp2+ group associated with NAD and pyruvate metabolism. These observations confirm the existence of SC subtypes extending beyond conventional myelinating and non-myelinating SCs and suggest that there is functional divergence within different SC subtypes.
PMP2 is an abundant, primarily peripheral myelin protein that plays a role in intracellular trafficking of lipids. PMP2 missense mutations are causative for a dominant demyelinating neuropathy54,55. While Pmp2 knock-out mice have a surprisingly mild phenotype48, transgenic mice expressing the disease-causing PMP2 (I43N) mutation exhibit a significant reduction of motor NCV at 5 months of age, suggesting a toxic gain of function consistent with the dominant transmission of the disorder54. Pmp2 appears to spontaneously induce the formation of myelin-like membrane multilayers56, which could be related to the observation that Pmp2+ SCs ensheath larger axons with thicker myelin (smaller G-ratio).
Along with Pmp2, RNA-FISH experiments showed that the motor-associated SCs co-express Cldn14, a component of tight junctions, and Adamtsl1, a thrombospondin-domain matrix protein that serves as an axon guidance cue in C. elegans57. More importantly, we found that PMP2 marks a SC population in human sciatic nerves with similar features to those in mouse. Through the transcriptional modules, we identified unique metabolic and signaling signatures of the motor-associated SCs, such as the pyruvate and NAD metabolism pathway. Lactate shuttling has long been shown to play a key role in the metabolic support of axons in the CNS by both astrocytes and oligodendrocytes through monocarboxylate transporters58 (MCTs), and disruption of MCT1 affects the maintenance of motor end-plate innervation in PNS45. Future studies of the metabolic activity of these motor-associated SCs, especially in regard to pyruvate/lactate metabolism, could reveal important insights regarding their role in supporting motor axons.
Numerous experiments have demonstrated that motor axons preferentially regenerate along motor-specific pathways59, leading to the hypothesis that SC subtypes define specific microenvironments for motor or sensory nerves. We noted that PMP2+ vs. PMP2- ChAT+ axons, particularly in the sciatic nerve, are clustered together and appear to reside in ‘sectors’. A similar ‘sectoring’ pattern in the sciatic was detected by injecting a retrograde dye into the lateral gastrocnemius muscle in mice60. Although this sectoring pattern is not clearly observed in our human nerve analysis, this could be due to either anatomical differences29 or biopsy location. Taken together with the size differential of PMP2+ axons, their spatial arrangement and transcriptional signatures, these data suggest that motor-associated SCs may be associated with ChAT+ axons that preferentially innervate fast-twitch, glycolytic motor muscle fibers.
The ALS SOD1G93A model is a motor neuropathy caused by death of motor neurons61. The strong reduction of motor-associated SCs in SOD1G93A mice agrees with the motor neuron vulnerability in ALS, and suggests the maintenance of PMP2+ motor-SC identity is dependent on contact with motor axons. Differential motor neuron vulnerability and resistance in ALS has long been identified62, In the SOD1G93A mouse model, muscles innervated by a high percentage of fast-twitch type II fibers, such as the gastrocnemius, are affected earlier than muscles innervated by slow-twitch fibers63. While we have shown motor-associated SCs are preferentially depleted at end-stage of ALS, future studies of these PMP2+ SCs in cranial nerves, where differential susceptibility in ALS is prominent in patients, could reveal important insights on differential motor neuron vulnerability in this disorder.
In summary, the existence of SC subtypes has long been speculated but the methods to identify them have been technically challenging due to the complex, close apposition of the glia cells with the axons they ensheath. We have made our dataset readily accessible through our glia portal (http://milbrandt.wustl.edu/glia-portal/), and anticipate it will stimulate future discoveries and serve as a resource for peripheral neuropathy studies.
Materials & Methods
Experimental animals.
Mouse care and experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Washington University in St. Louis under the protocols 20170154 and 20170030. Mice were kept on a 12-hour light dark cycle and received food and water ad libitum. The following strains were used:
Sun1-sfGFP: B6.129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J
Actb-Cre: B6N.FVB-Tmem163Tg(ACTB-cre)2Mrt/CjDswJ
Mpz-Cre: B6N.FVB-Tg(Mpz-cre)26Mes/J
Ribotag: Rpl22tm1.1Psam
Ngfr-GFP: Tg(Ngfr-EGFP)QU6Gsat
C57BL/6NTac
B6SJL-Tg(SOD1*G93A)1Gur/J
FVB.Cg-Tg(SOD1*G93A)1Gur/J
Unless stated otherwise, both female and male mice between the ages of 8 and 12 weeks were used in these experiments.
Nuclei isolation and single nuclei RNA-seq.
Nerves were isolated from reporter mice as indicated below. For sciatic nerve snRNA-seq libraries, 2 male and 2 female mice were pooled for each experiment. For vagus nerve snRNA-seq libraries, were prepared from a pool of 9 male and 9 female mice. For peroneal and sural nerve snRNA-seq libraries, 5 male and 5 female mice were pooled for each experiment. Upon dissection, the nerves were immediately transferred to digestion media containing 0.3% collagenase IV (Sigma Aldrich C5138), 0.04% hyaluronidase (Sigma Aldrich H3506) and 0.04% DNase I (Sigma Aldrich DN25) for 10–15 min at 37°C. The cells were filtered through 70 um cell strainers and washed twice with cold 5% PBS containing 0.1% BSA, RNaseIn and SuperaseIn. Cells were mechanically dissociated using a Dounce homognizer on ice; media for mechanical dissociation was prepared according to Krishnaswami’s protocol. The nuclei and cellular debris were filtered through 40 um cell strainers and washed with cold 5% PBS containing 0.1% BSA, RNaseIn (Ambion AM2682) and SuperaseIn (Invitrogen AM2694) and the debris was removed from the nuclei by immediate FACS sorting. The purified nuclei were used to prepare single nuclei RNA-seq libraries following manufacturer’s instructions (10X Genomics). A total of 11 single nuclei libraries were prepared: 5 libraries from sciatic nerves from Sun1-GFP/+:ActB-Cre/+ mice (v2 chemistry); 2 libraries from sciatic nerves from Sun1-GFP/+:Mpz-Cre/+ mice (v3 chemistry); 2 libraries from vagus nerves from Sun1-GFP/+:ActB-Cre/+ mice (v2 chemistry); 1 library from peroneal nerves dissected from Sun1-GFP/+:ActB-Cre/+ mice (v2 chemistry); 1 library from sural nerves dissected from Sun1-GFP/+:ActB-Cre/+ mice (v2 chemistry).
Immune cell isolation and generation of sciatic nerve single-cell immune cell atlas.
Cell sorting was following ImmGen protocols. In brief, sciatic nerves from 20 mice were pooled, incubated with gentle shaking for 15 min in digestion media containing 0.3% collagenase IV (Sigma Aldrich C5138), 0.04% hyaluronidase (Sigma Aldrich H3506), and 0.04% DNAse I (Sigma Aldrich DN25). Cells were then washed with 5% PBS containing 0.1% BSA with RNaseIn and SuperaseIn, filtered through 70 uM cell strainers and sorted twice into 5uL TCL buffer containing 5% PBS/BSA with RNase inhibitors using the CD45 antibody (Biolegend cat #103101, dilution 1:200). Cd45+ cells were loaded to one lane of the 10X Chip for preparation of one single-cell library. The library preparation was performed according to the manufacturer’s instructions. Flow analysis was performed using FlowJo (version 10.7.1)
Sequencing and single nuclei RNA-seq analysis.
All 12 libraries were sequenced by using Illumina HiSeq2500 or NovaSeq6000 according to manufacturer’s instructions. The reads were then aligned to mouse genome mm10 using Cell Ranger v3.0.2 from 10X Genomics. As nuclei contain mostly pre-mRNA with intron retention, over 40% of the mapped reads were at introns. We therefore incorporated both intronic and exonic reads during the analysis and then determined the UMI counts for each gene using PySam (v15). The first QC step involved the removal of nuclei with less than 500 genes and genes seen in 10 or fewer nuclei, the second QC step involved the removal of nuclei with high mitochondrial content (>5%). The expression matrix of individual library was then normalized and scaled based on the UMI counts and mitochondria percentage. Highly variable genes were identified by using FindVariableGenes function in Seurat64 and subjected to downstream analysis. For sciatic nerve atlas generated from Sun1-GFP/+;ActB-Cre/+ mice as well as peripheral nerve atlas that combined three nerve types, the batch effect was corrected by using DeepImpute65 and Seurat SCTransform. For Mpz-nuclei atlas generated from Sun1-GFP/+;Mpz-Cre/+ mice and vagus nerve atlas generated from Sun1-GFP/+;ActB-Cre/+ mice, the batch effect was corrected by SCTransform. For peroneal nerve atlas generated from Sun1-GFP/+;ActB-Cre/+ mice, only one library was generated and hence principal component analysis (PCA) was performed and PCA dimensions were used for downstream analysis. As the third QC step, we calculated the ratio of variance and removed nuclei with inconsistent lower dimensional information, which were likely to be caused by doublets or nuclei rupture during the nuclei capture step in creating the single nucleus libraries. Clustering analysis was then performed by using shared nearest neighbor (SNN) modularity optimization approach with at least 100 random starts and 100 iterations. Data visualization was performed using t-SNE. For cell trajectory analysis, the normalized matrix was imported to Monocle and cell trajectory analysis was performed. The analyses were done by using customized R code, Seurat (v3.2.3), DeepImpute (Apr2020), Monocle66 (v2.9.0) and Python3.7. To evaluate the potential functions of a cell cluster of interest, we calculated the expression scores of 298 mouse transcriptional modules (Entry ID: T01002) defined in the KEGG67 database (https://www.genome.jp/kegg/). Specifically, we first calculated the expression module score using the AddModuleScore function in Seurat at single-nuclei level. We then calculated the average expression module score per cell clusters and derived the aggregated z-score for comparison across modules by using the scale module in R. Data visualization was performed using Morpheus from Broad Institute.
Sciatic and vagus Ribo-tag RNA extraction.
The protocol was modified based on Sanz et al68. Specifically, we generated Mpz-Cre:Ribo-Tag mice that, upon the expression of Mpz, cells would express HA-tagged ribosomal protein RPL22. The mRNA of Mpz-expressing cells from the nerve was then isolated by immunoprecipitation of polyribosomes with anti-HA and subjected to RNA-seq. Both direct and indirect pull down approaches were tested and indirect pull down was selected because of higher yields. Both sciatic and vagus nerves were dissected from Mpz-Cre:RiboTag mice and immediately placed into PBS with RNaseIn and SuperaseIn on ice. The epineurium of sciatic nerve was also carefully removed by forceps before placing the remaining nerves into PBS. Upon dissection, the nerves were transferred to dissociation media with TL liberase, RNaseIn and SuperaseIn for enzymatic dissociation for 15min at 37°C. The cells were pelleted by spinning at 1500rpm for 5 min, homogenized using Dounce homogenizer in Polysome buffer and then spin at 10,000g for 10min. Supernatant was then collected, with HA antibody-coupled magnetic beads added to the cleared supernatant. After an overnight incubation at 4°C, the magnetic beads that contained immunoadsorbed polysomes were washed with a high salt buffer. mRNA was then extracted using the Qiagen RNA extraction kit according to manufacturer’s protocol.
Sciatic and vagus bulk RNA extraction.
Sciatic and vagus nerves were harvested from C57/B6 mice and immediately placed into PBS with RNaseIn and SuperaseIn on ice, the epineurium of sciatic nerve was carefully removed by forceps. The nerves were then transferred to dissociation media with TL liberase, RNaseIn and SuperaseIn for enzymatic dissociation for 15min at 37°C. The cells were then pelleted by spinning at 1500rpm for 5 mins, homogenized using Dounce homogenizer and then centrifuged at 10,000g for 10min. Supernatant was then collected and mRNA extraction was performed using Qiagen RNA extraction kit according to manufacturer’s protocol.
Ribo-tag and bulk RNA sequencing and analysis.
Samples were prepared according to Illumina library kit manufacturer’s protocol, indexed, pooled, and sequenced on an Illumina HiSeq. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq (v1.8) software and a custom python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with STAR version 2.5.1a1 (Parameters: -genomeDir Ensembl_GRCh38.76_genome.fa --genomeLoad NoSharedMemory --outSAMmapqUnique 60 --outSAMunmapped Within KeepPairs --outFilterIntronMotifs RemoveNoncanonicalUnannotated -outSAMstrandField intronMotif --runThreadN 8 --outStd BAM_Unsorted --outSAMtype BAM Unsorted --alignTranscriptsPerReadNmax 100000 --outFilterMismatchNoverLmax 0.1 --sjdbGTFfile Ensembl_GRCh38.76_genes.gtf > genome_accepted_hits.bam). Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.6-p5. Isoform expression of known Ensembl transcripts was estimated with Salmon version 0.8.2. Sequencing performance was assessed for the total number of aligned reads, total number of uniquely aligned reads, and features detected. The ribosomal fraction, known junction saturation, and read distribution over known gene models were quantified with RSeQC version 2.6.2. All gene counts were then imported into the R (3.2.3) /Bioconductor (3.2) package EdgeR and TMM normalization size factors were calculated to adjust for samples for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one sample greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma. Weighted likelihoods based on the observed mean-variance relationship of every gene and sample were then calculated for all samples with the voomWithQualityWeights. The performance of all genes was assessed with plots of the residual standard deviation of every gene to their average log-count with a robustly fitted trend line of the residuals. Differential expression analysis was then performed to analyze for differences between conditions and the results were filtered for only those genes with Benjamini-Hochberg false-discovery rate adjusted p-values less than or equal to 0.05.
Intercellular communication analysis on the peripheral nerve atlas.
Cell-cell interaction was predicted by a method similar to that described by Ximerakis et al70. A cell communication interactome was created by collecting known protein-protein interactions between receptor, ligand and extracellular matrix (ECM) proteins, based on information from Baderlab’s CellCellInteractions interaction database (http://baderlab.org/CellCellInteractions). Subpopulation of SCs, immune cells and epineurial fibroblasts were considered as a whole respectively. To predict cell-cell interactions, the ligand-receptor interaction datasets were filtered for genes that were differentially expressed within the cell group (Seurat FindMarkers function with the default Wilcox Rank Sum test; p-value < 1e-40, fold-change > 0.5 for each cell type). We used CCInx R package (v0.5.1) for testing the marker genes of each cell type against the databases. With this, the node and edge information was given, with each node denoting a gene and each edge connecting two genes. We filtered the edges by focusing on intercellular communication, intracellular edges were removed for this analysis; The number of combinations was then calculated, structured and the circos plot data was generated. We also performed Monte Carlo simulation by randomly permutating the genes connecting by each edge repeated for 100,000 times, and determine the p-value of two cell types having intercellular communication by chance using Python3.7. The circos plot was generated using circos-0.69–6.
Comparative analysis on peripheral neuropathy GWAS.
Peripheral neuropathy GWAS data were downloaded from GWAS Central and the significant variants were curated from both GWAS Central and the publications. Variants were represented by rsIDs, and the coordinates of each rsID were identified on the human genome GRCh38.p7 by using vcftools (v.0.1.17). The coordinates were converted to BED format by vcf2bed (v.2.4.39) and the closest gene were identified by using closest-features (v2.4.39). The rsIDs were also categorized into genic or intergenic variants, and the associated genes were crosschecked with the publications. We then identified the corresponding mouse homolog by using the NCBI homologene database and Jackson’s laboratory homolog database. For genic variants there was no conflict observed, and for intergenic variants, manual curation was performed with the showcase of rs13417783 shown in Supp. Fig. 7. We further determined if the gene is expressed in the peripheral nerve atlas, and if it is, whether it is a signature gene of certain cell types (differential gene expression analysis through Seurat FindMarkers function with the default Wilcox Rank Sum test; p-value < 1e-40, fold-change > 0.5 for each cell type). The data were visualized using Python (v3.7) and Morpheus from Broad Institute (https://software.broadinstitute.org/morpheus/), cell-types without disease-associated genes were not included. Enrichment analysis was performed using binomial test through R binom.test.
Comparative analysis on hereditary peripheral neuropathy conditions and neurological disorders with peripheral neuropathy phenotype.
A total of 360 hereditary peripheral neuropathy genes and neurological disorder genes were curated from OMIM (Supp. Table 4) and categorized into 4 categories based on the Mayo Clinic Laboratory classification. We identified the corresponding mouse homolog by using the NCBI homologene database and the Jackson Laboratory homolog database. We further refined the analysis by locating genes that are signature genes within the peripheral nerve atlas (differential gene expression analysis through Seurat FindMarkers function with the default Wilcox Rank Sum test (p-value < 1e-40, fold-change > 0.5 for each cell-type). A total of 41 genes identified, and 20 genes are visualized as they are responsible for either hereditary peripheral neuropathy or neurological disorders with peripheral neuropathy phenotypes. The data were visualized using Python (v3.7) and Morpheus from Broad Institute. Enrichment analysis was performed using binomial test through R binom.test.
RNA-FISH.
C57/B6 mice were perfused with fresh PBS, the nerves were harvested, immediately embedded into OCT, and flash frozen at −80C. 10 micron sections were prepared from all nerve OCT blocks. To prepare the slides for ThermoFisher viewRNA protocol, slides were fixed in 4% paraformaldehyde at 4°C overnight, dehydrated at 50%, 70% and 100% ethanol for 10 min each, baked at 60°C for 30 min and rehydrated in RNA-grade PBS. The remaining steps were performed according to manufacturer’s protocol. All RNA-FISH probes are listed in Supp. Table 5.
IHC.
For whole mount imaging, mouse nerve samples were harvested and immediately fixed in 4% PFA for 1 hour and then transferred to 40% sucrose overnight. Samples were washed in PBS, blocked, stained, and imaged. For frozen sections, samples were harvested, stored in PFA/sucrose, and embedded into OCT. 10 micron sections were prepared, blocked in 1% BSA, stained, and imaged. Antibodies to the following proteins were used: anti-GFP (Invitrogen A21311, dilution 1:200), PMP2(ProteinTech 12717–1-AP, dilution 1:100), MBP (Temecula California MAB386 82–87, dilution 1:500), β-tubulin (Aves lab TUJ, dilution 1:500) and ChAT (EMD Millipore AB144P, dilution 1:100). Depending on the dataset and experimental design, different statistical methods (e.g. t-test and ANOVA) were used as stated in the figure legends. Two-tailed tests were applied if not indicated otherwise. All imaging analyses were performed blinded without the knowledge of nerve type, genotype of mice or clinical report of patients.
Statistics and reproducibility
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to or larger than those reported in previous publications13, 19–21. The experiments were not randomized as all our experiments were performed using either mouse or human tissue, experimental groups were determined by mouse genotype or human clinical report. Data were analyzed using Prism 9 (GraphPad). Significance was considered when P < 0.05. Depending on the dataset and experimental design, different statistical methods (e.g. t-test and ANOVA) were used. Two-tailed tests were applied if not indicated otherwise. Data distribution from imaging analysis was assumed to be normal but this was not formally tested. For bulk RNA sequencing studies, 3 libraries were prepared across three independent experiments. Imaging studies were performed with at least three replicates per experiment unless stated otherwise. Animal experiments were repeated as stated by the N number. Animals were age-matched, sex-matched and littermate conditions were fulfilled. No animals or data points were excluded from the analyses.
Data availability
A total of 24 files, including 18 raw sequencing reads and 6 processed data such as expression matrices and RDS files for single-cell and single nuclei atlases, has been depositied onto the GEO repository under the reference series ID GSE182099. The Mpz-RiboTag and bulk RNA-seq data can be accessed through the sub-series ID GSE181858 and the single nuclei data can be obtained through GSE182098. The glia portal can also be accessed through the link – http://milbrandt.wustl.edu/glia-portal/.
Code availability
The source code for data analysis and visualization in this study will be available online at https://github.com/aldrinyim/PNS-glia
Extended Data
Extended Data Fig. 1. Generation and analysis of sciatic nerve and immune cell atlases.
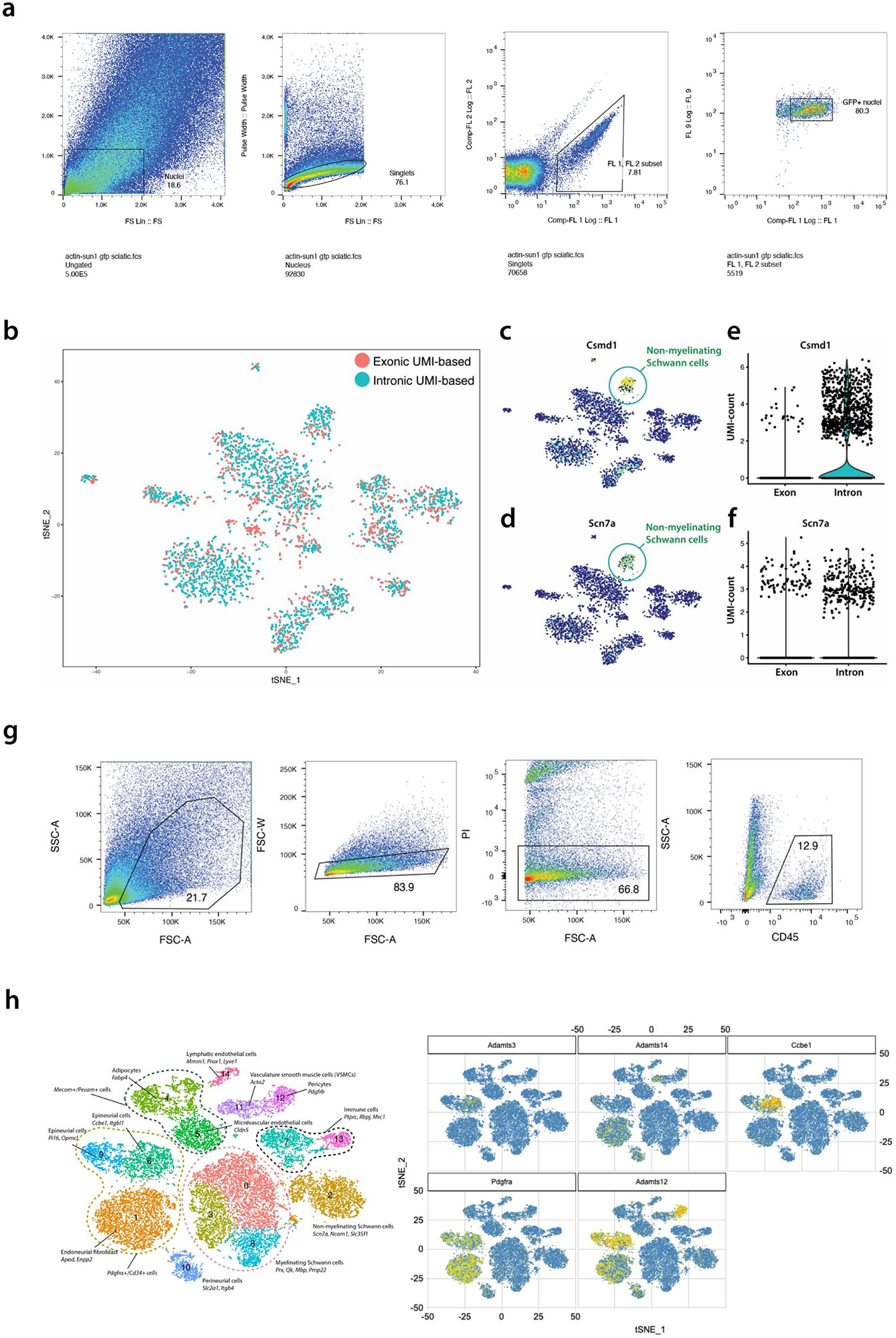
(a) Gating strategy for sorting GFP+ nuclei from Sun1-GFP/+:Act-Cre/+ sciatic nerves. Nuclei were isolated from single cell suspensions containing pooled sciatic nerves from at least 5 Sun1-GFP/+:Act-Cre/+ mice. Purified nuclei were selected based on GFP and Hoechst expression. (b-f) Intronic reads contain biological information. Single nuclei atlases were generated by using either exonic or intronic reads, and CCA analysis was performed to align the two data sets. (b) The two atlases showed high level of concordance. c-f, Contribution of intronic reads to signature genes in nmSCs. (c) Csmd1 and (d) Scn7a were found to mark nmSCs. (e) Over 90% of the UMIs were from intronic reads for Csmd1. (f) Approximately 60% of the UMIs were from intronic reads for Scn7a. (g) Gating strategy for sorting CD45+ cells from sciatic nerves. Sciatic nerves were pooled from 20 Cx3cr1-GFP/+ mice and prepared for FACS sorting. Cells were stained with Propidium Iodide (PI) to select for viable cells, and with CD45 antibody to select for CD45+ population. (h) Expression of lymphangiogenesis-related genes in epineurial fibroblasts. Adamts3, Adamts14, Ccbe1, Pdgfra and Adamts12 expression in the ActB-Sun1 sciatic nerve atlas.
Extended Data Fig. 2. Generation and analysis of Mpz-enriched single nuclei atlas.
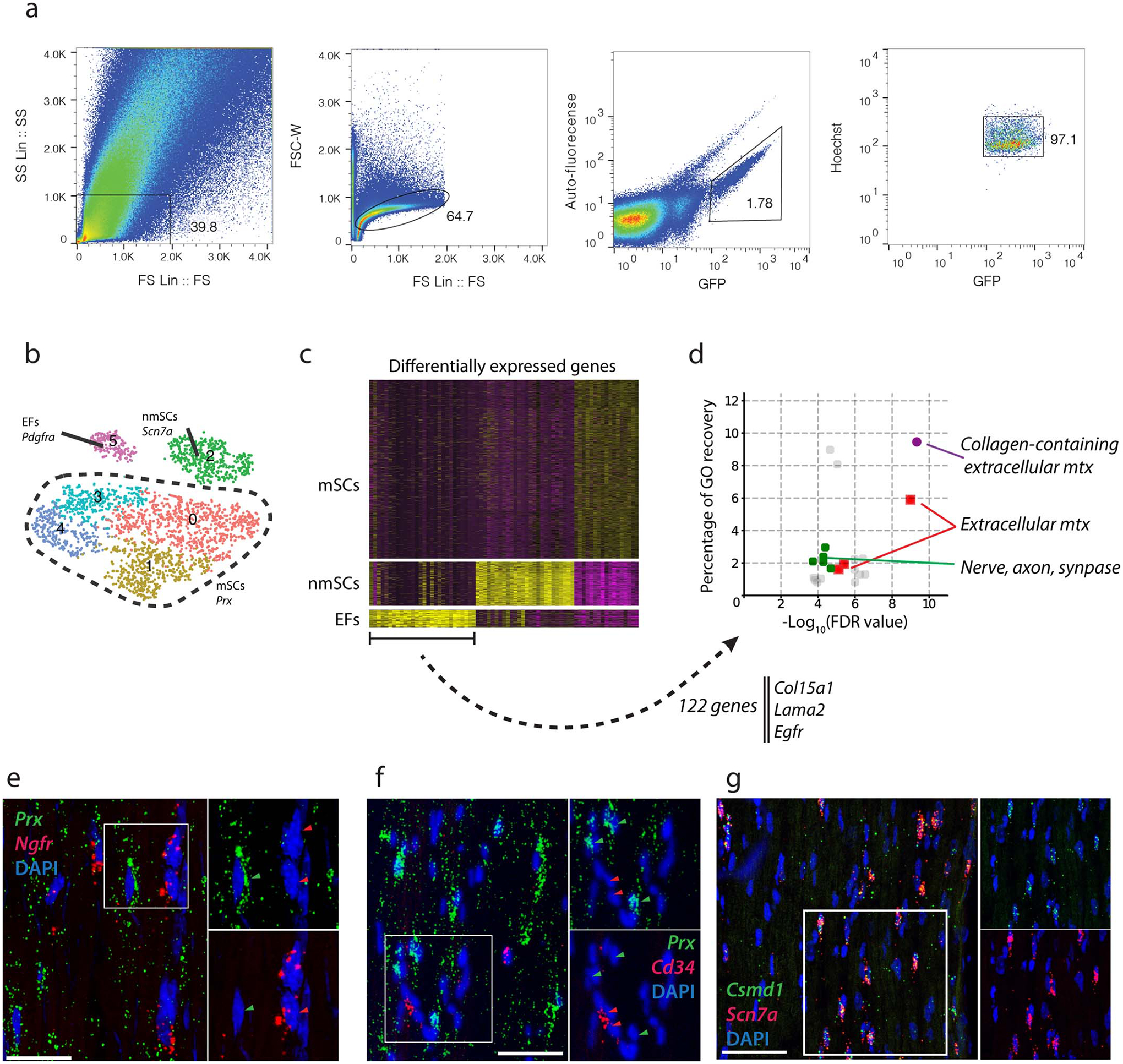
(a) Gating strategy for sorting GFP+ nuclei from Sun1-GFP/+:Mpz-Cre/+ mice. Nuclei were isolated from single cell suspensions containing pooled sciatic nerves from at least 5 Sun1-GFP/+:Act-Cre/+ mice. Purified nuclei were selected based on GFP and Hoechst expression. (b) tSNE plot with 3,047 nuclei from Sun1-GFP/+;Mpz-Cre/+ defines three GFP+ cell types – endoneurial fibroblasts, nmSCs and mSCs. (c) Differential expression analysis from Mpz-Sun1 atlas shows signature gene expression that defines endoneurial fibroblasts, including Col15a1, Lama2 and Egfr. Each column represents a gene and each row represents a nucleus, genes with high expression level are shown in yellow and low expression in purple. (d) GO analysis showing that endoneurial fibroblast markers (122 genes) are enriched for components related to collagen-containing extracellular matrix, extracellular matrix and formation of axon, neurons and synapse, percentage of GO recovery is plotted against the false discovery rate (FDR) and selected GO terms are highlighted in color. (e,f) RNA-FISH analysis of Prx, Ngfr, and Cd34 expression. (scale bar=50um) e, Representative expression of Prx and Ngfr, indicating that myelinating Schwann cells do not express Ngfr. f, Representative expression of Prx and Cd34, indicating that myelinating Schwann cells do not express Cd34. (g) RNA-FISH analysis of Csmd1 and Scn7a. Representative overlapping expression of Csmd1 (green) and Scn7a (red) in adult sciatic nerves (scale bar=50um).
Extended Data Fig. 3. Pathway-based transcriptional module analysis.
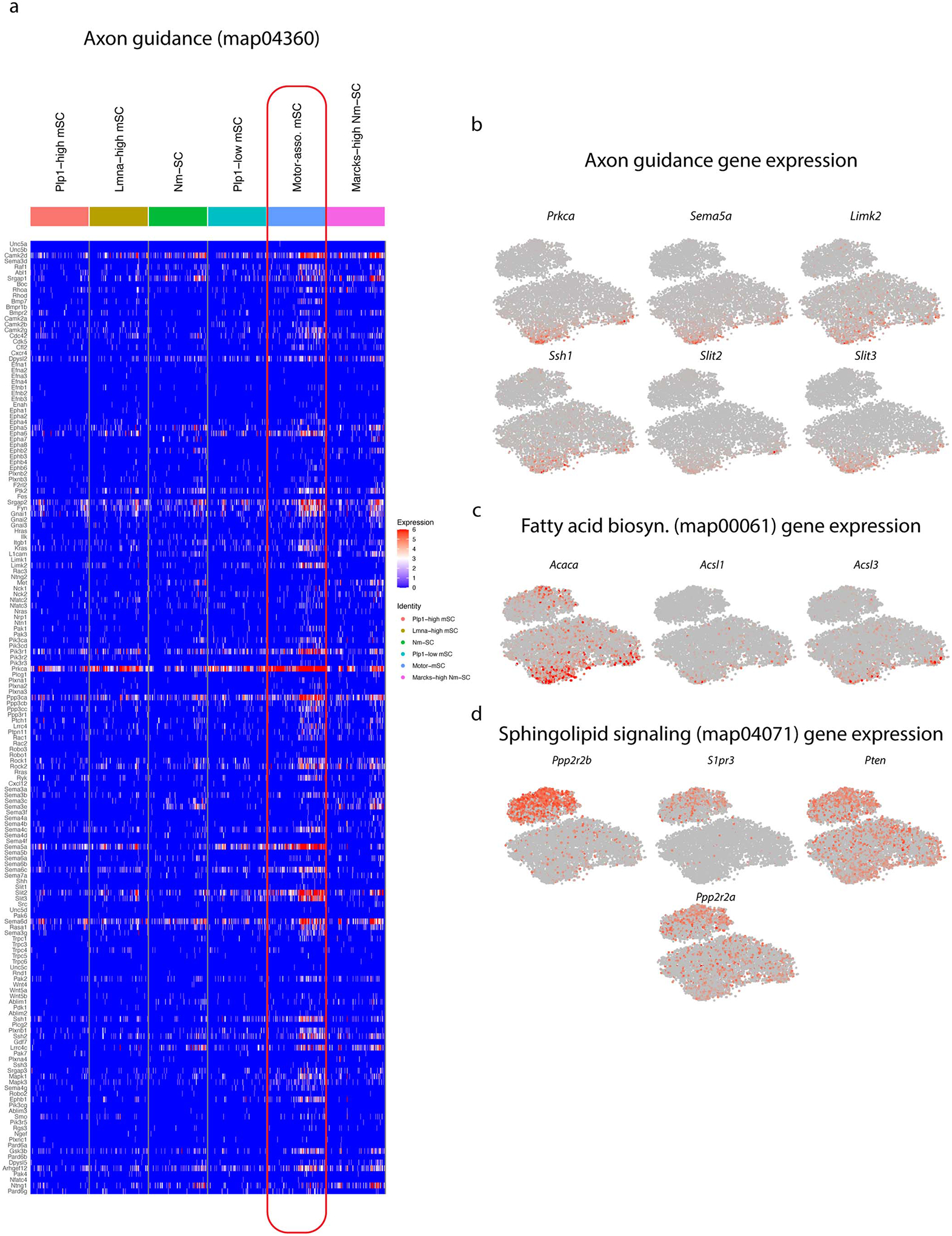
(a) Expression of all genes in the axon guidance pathway (map04360) defined by the KEGG database are shown in here, with red indicating high expression and blue indicate no expression. Many of the genes are more abundantly expressed in the Pmp2+ mSCs. (b) tSNE plot showing the expression of genes in the axon guidance pathway. (c) tSNE plot showing the expression of genes in fatty acid biosynthesis pathway (map00061). (d) tSNE plot showing the expression of genes in the sphingolipid signaling pathway (map04071).
Extended Data Fig. 4. Pmp2+ Schwann cells preferentially ensheath Chat+ axons.

(a) Validation of ChAT-Cre-tdT expression specificity. Representative expression of PMP2 (yellow) and ChAT-Ab (white) in ChAT-Cre-tdT (red) sciatic nerves. The two panels on the right have shown almost complete overlap between the antibody (white) and tdT reporter (red) for ChAT (scale bar = 50um, n=3 biologically independent mice). (b) ChAT+ axons are mostly ensheathed by Pmp2+ SCs. Representative imaging showing the expression of MBP (blue) and PMP2 (green) in ChAT-tdT (red) sciatic nerve. (scale bar = 50um, n=3 biologically independent mice). (c) Quantification of percentage of ChAT axons ensheathed by a Pmp2+ SCs in three different nerve types. 2-way anova with multiple comparisons. Sciatic vs Sural: P=0.0159, Sciatic vs Femoral: P= 0.0456, Sural vs Femoral: P=0.0009. Data are mean +/− SD (n=3 biologically independent mice).
Extended Data Fig. 5. SOD1G93A mice show reduced PMP2 expression in SCs.

Representative imaging of PMP2+ SCs (yellow), b-TUB+ axons (cyan) and MBP+ SCs (gray) in sciatic, femoral, and sural nerves in SOD1G93A mice. Note that femoral nerve is severely degenerated as shown by MBP staining. Sciatic nerve remains relatively intact with depletion of PMP2, while sural nerve has intact axons with MBP+ SCs ensheathing them (scale bar = 50um, n=3 biologically independent mice).
Supplementary Material
Supplementary Table 1 Schwann Cell enriched atlas Differential expression analysis
Supplementary Table 2 KEGG-expression-module-analysis
Supplementary Table 4 Hereditary Peripheral Neuropathic Genes
Supplementary Table 5 List of RNA FISH probes
Acknowledgements
The authors thank all our collaborators at Washington University School of Medicine, St. Louis (WUSM) for their advice and discussion. We thank Richard Head, Ruteja Barve, Paul Cliften at GTAC@MGI and McDonnell Genome Institute for discussion and assistance with RNA sequencing. The authors are grateful for assistance from Gwendalyn Randolph’s laboratory and Bernd Zinselmeyer for confocal imaging and use of equipment (supported by NIH R01DK119147 and R37AI049653 to G.R.). The authors thank Mark Shabsovich, Tao Shen and Collin Kreple for ALS SOD1G93A mice, Maggie Ireland for curating patient samples (supported by NIH R01NS078398 to T.M.), and Robert Schmidt for curating clinical pathology report and many helpful discussions. The authors thank Rachel McClarney and Cassidy Menendez for the experimental assistance, and members of the Milbrandt and DiAntonio labs for their helpful comments and discussion. This project was supported by NIH grants RF1MH117070 and RO1GM123203 to R.D.M. and R01NS105645 and R01AG013730 to J.M.
Footnotes
Competing interests
J.M. is a co-founder, consultant, and shareholder of Disarm Therapeutics, a wholly owned subsidiary of Eli Lilly & Company. All other authors declare no financial interests.
References
- 1.Fünfschilling U et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature (2012) doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nave KA Myelination and support of axonal integrity by glia. Nature (2010) doi: 10.1038/nature09614. [DOI] [PubMed] [Google Scholar]
- 3.Monk KR, Feltri ML & Taveggia C New insights on schwann cell development. Glia 63, 1376–1393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richard L et al. Endoneurial fibroblast-like cells. Journal of Neuropathology and Experimental Neurology (2012) doi: 10.1097/NEN.0b013e318270a941. [DOI] [PubMed] [Google Scholar]
- 5.Brushart TME Preferential Reinnervation of Motor Nerves by Regenerating Motor Axons. The Journal of neuroscience 8, 1026–1031 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madison RD, Sofroniew MV & Robinson GA Schwann cell influence on motor neuron regeneration accuracy. Neuroscience 163, 213–221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguilar SV et al. ImmGen at 15. Nature Immunology 21, 700–703 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Jesuraj NJ et al. Differential gene expression in motor and sensory Schwann cells in the rat femoral nerve. Journal of Neuroscience Research 90, 96–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr MJ et al. Mesenchymal Precursor Cells in Adult Nerves Contribute to Mammalian Tissue Repair and Regeneration. Cell Stem Cell 24, 240–256.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Toma JS et al. Peripheral Nerve Single Cell Analysis Identifies Mesenchymal Ligands that Promote Axonal Growth. eneuro (2020) doi: 10.1523/eneuro.0066-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolbert J et al. Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proceedings of the National Academy of Sciences of the United States of America (2020) doi: 10.1073/pnas.1912139117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber D et al. Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas ( SNAT ). 1–28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skelly DA et al. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Reports (2018) doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 14.Baron M et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Systems (2016) doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Causey G & Barton AA The cellular content of the endoneurium of peripheral nerve. Brain (1959) doi: 10.1093/brain/82.4.594. [DOI] [PubMed] [Google Scholar]
- 16.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macosko EZ et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters A & Muir AR The relationship between axons and Schwann Cells during development of peripheral nerves in the rat. Quarterly Journal of Experimental Physiology and Cognate Medical Sciences 44, 117–130 (1959). [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Teng HL & Huang ZH Repulsive migration of schwann cells induced by slit-2 through Ca2+-dependent RhoA-Myosin signaling. GLIA (2013) doi: 10.1002/glia.22464. [DOI] [PubMed] [Google Scholar]
- 20.Yoshino JE, Dinneen MP, Sprinkle TJ & DeVries GH Localization of 2′,3′-cyclic nucleotide 3′-phosphodiesterase on cultured Schwann cells. Brain Research (1985) doi: 10.1016/0006-8993(85)90316-6. [DOI] [PubMed] [Google Scholar]
- 21.García-Villegas R, López-Álvarez LE, Arni S, Rosenbaum T & Morales MA Identification and functional characterization of the promoter of the mouse sodium-activated sodium channel Nax gene (Scn7a). Journal of Neuroscience Research (2009) doi: 10.1002/jnr.22069. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Martínez J, Azcona LJ, Bergareche A, Martí-Massó JF & Paisán-Ruiz C Whole-exome sequencing associates novel CSMD1 gene mutations with familial Parkinson disease. Neurology: Genetics (2017) doi: 10.1212/NXG.0000000000000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friede RL & Samorajski T Relation between the number of myelin lamellae and axon circumference in fibers of vagus and sciatic nerves of mice. Journal of Comparative Neurology 130, 223–231 (1967). [DOI] [PubMed] [Google Scholar]
- 24.Morita K, Sasaki H, Furuse M & Tsukita S Endothelial claudin: Claudin-5/TMVCF constitutes tight junction strands in endothelial cells. Journal of Cell Biology (1999) doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliver G Lymphatic vasculature development. Nature Reviews Immunology (2004) doi: 10.1038/nri1258. [DOI] [PubMed] [Google Scholar]
- 26.Wendling O, Bornert JM, Chambon P & Metzger D Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis (2009) doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan L, Chow BW & Gu C Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nature Reviews Neuroscience (2020) doi: 10.1038/s41583-020-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang PL et al. Peripheral nerve resident macrophages share tissue-specific programming and features of activated microglia. Nature communications 11, 2552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunderland S The connective tissues of peripheral nerves. Brain (1965) doi: 10.1093/brain/88.4.841. [DOI] [PubMed] [Google Scholar]
- 30.Singhmar P et al. The fibroblast-derived protein PI16 controls neuropathic pain. Proceedings of the National Academy of Sciences of the United States of America (2020) doi: 10.1073/pnas.1913444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeltsch M et al. CCBE1 enhances lymphangiogenesis via a disintegrin and metalloprotease with thrombospondin motifs-3-mediated vascular endothelial growth factor-C activation. Circulation (2014) doi: 10.1161/CIRCULATIONAHA.113.002779. [DOI] [PubMed] [Google Scholar]
- 32.Hirose T et al. Immunohistochemical demonstration of EMA/Glut1-positive perineurial cells and CD34-positive fibroblastic cells in peripheral nerve sheath tumors. Modern Pathology (2003) doi: 10.1097/01.MP.0000062654.83617.B7. [DOI] [PubMed] [Google Scholar]
- 33.Richard L, Védrenne N, Vallat JM & Funalot B Characterization of Endoneurial Fibroblast-like Cells from Human and Rat Peripheral Nerves. Journal of Histochemistry and Cytochemistry 62, 424–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jessen KR & Mirsky R The origin and development of glial cells in peripheral nerves. Nature Reviews Neuroscience (2005) doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 35.Baima J & Krivickas L Evaluation and treatment of peroneal neuropathy. Current Reviews in Musculoskeletal Medicine (2008) doi: 10.1007/s12178-008-9023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herrmann DN, Griffin JW, Hauer P, Cornblath DR & McArthur JC Epidermal nerve fiber density and sural nerve morphometry in peripheral neuropathies. Neurology (1999) doi: 10.1212/wnl.53.8.1634. [DOI] [PubMed] [Google Scholar]
- 37.Yuan H & Silberstein SD Vagus Nerve and Vagus Nerve Stimulation, a Comprehensive Review: Part i. Headache (2016) doi: 10.1111/head.12647. [DOI] [PubMed] [Google Scholar]
- 38.Zilic L et al. An anatomical study of porcine peripheral nerve and its potential use in nerve tissue engineering. Journal of Anatomy 227, 302–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velmeshev D et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF & Hamosh A OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an Online catalog of human genes and genetic disorders. Nucleic Acids Research (2015) doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Y et al. A genetic locus on chromosome 2q24 predicting peripheral neuropathy risk in type 2 diabetes: Results from the ACCORD and BARI 2D studies. Diabetes (2019) doi: 10.2337/db19-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders SJ et al. Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends in Neurosciences 41, 442–456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldman EL, Nave KA, Jensen TS & Bennett DLH New Horizons in Diabetic Neuropathy: Mechanisms, Bioenergetics, and Pain. Neuron 93, 1296–1313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandoorne T, de Bock K & van den Bosch L Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathologica 135, 489–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouçanova F et al. Disrupted function of lactate transporter MCT1, but not MCT4, in Schwann cells affects the maintenance of motor end-plate innervation. Glia 69, 124–136 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Hoke A Schwann Cells Express Motor and Sensory Phenotypes That Regulate Axon Regeneration. Journal of Neuroscience 26, 9646–9655 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman DL, Fabrizi C, Gillespie CS & Brophy PJ Specific disruption of a Schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron (2001) doi: 10.1016/S0896-6273(01)00327-0. [DOI] [PubMed] [Google Scholar]
- 48.Zenker J et al. A role of peripheral myelin protein 2 in lipid homeostasis of myelinating Schwann cells. GLIA (2014) doi: 10.1002/glia.22696. [DOI] [PubMed] [Google Scholar]
- 49.Rossi J et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metabolism (2011) doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blum JA et al. Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nature Neuroscience 24, 572–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guttenplan KA et al. Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nature Communications (2020) doi: 10.1038/s41467-020-17514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatzipetros T et al. A quick phenotypic neurological scoring system for evaluating disease progression in the SOD1-G93A mouse model of ALS. Journal of Visualized Experiments 2015, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gould TW et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. Journal of Neuroscience (2006) doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong Y. Bin et al. A Mutation in PMP2 Causes Dominant Demyelinating Charcot-Marie-Tooth Neuropathy. PLoS Genetics 12, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Motley WW et al. De novo PMP2 mutations in families with type 1 Charcot-Marie-Tooth disease. Brain 139, 1649–1656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruskamo S et al. Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice. Journal of Biological Chemistry (2020) doi: 10.1074/jbc.RA120.013087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seetharaman A et al. MADD-4 Is a Secreted Cue Required for Midline-Oriented Guidance in Caenorhabditis elegans. Developmental Cell 21, 669–680 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Domènech-Estévez E et al. Distribution of monocarboxylate transporters in the peripheral nervous system suggests putative roles in lactate shuttling and myelination. Journal of Neuroscience (2015) doi: 10.1523/JNEUROSCI.3534-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madison RD, Archibald SJ & Brushart TM Reinnervation accuracy of the rat femoral nerve by motor and sensory neurons. The Journal of neuroscience 16, 5698–703 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Llewellyn ME, Thompson KR, Deisseroth K & Delp SL Orderly recruitment of motor units under optical control in vivo. Nature Medicine 16, 1161–1165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nijssen J, Comley LH & Hedlund E Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathologica (2017) doi: 10.1007/s00401-017-1708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hegedus J, Putman CT & Gordon T Time course of preferential motor unit loss in the SOD1G93A mouse model of amyotrophic lateral sclerosis. Neurobiology of Disease 28, 154–164 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Frey D et al. Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. Journal of Neuroscience 20, 2534–2542 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nature Biotechnology (2015) doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arisdakessian C, Poirion O, Yunits B, Zhu X & Garmire LX DeepImpute: An accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biology (2019) doi: 10.1186/s13059-019-1837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Goto S, Sato Y, Furumichi M & Tanabe M KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research 40, D109–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanz E et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America (2009) doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanz E et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America 106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ximerakis M et al. Single-cell transcriptomic profiling of the aging mouse brain. Nature Neuroscience (2019) doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Schwann Cell enriched atlas Differential expression analysis
Supplementary Table 2 KEGG-expression-module-analysis
Supplementary Table 4 Hereditary Peripheral Neuropathic Genes
Supplementary Table 5 List of RNA FISH probes
Data Availability Statement
A total of 24 files, including 18 raw sequencing reads and 6 processed data such as expression matrices and RDS files for single-cell and single nuclei atlases, has been depositied onto the GEO repository under the reference series ID GSE182099. The Mpz-RiboTag and bulk RNA-seq data can be accessed through the sub-series ID GSE181858 and the single nuclei data can be obtained through GSE182098. The glia portal can also be accessed through the link – http://milbrandt.wustl.edu/glia-portal/.


