Abstract
T4 RNA ligase 2 catalyses two types of reactions: (i) sealing of a nick structure in double-stranded RNA and (ii) connection of two single-stranded RNA strands. In order to obtain comprehensive views on these two types of reactions and widen the application scope of this RNA ligase, we here systematically analysed the connection of single-stranded RNA strands having different secondary structures. It has been found that the ligation is enormously promoted when a stem of only 4-bp or longer is formed in the 3′-OH side of the joining site. Additional placement of a stem in the 5′-phosphate side further facilitates the ligation. In contrast, perturbation of the stem structures in RNA substrates suppresses the ligation. These results indicate that ligation of two single-stranded RNA strands by T4 RNA ligase 2 is greatly promoted by forming a “nick-like intermediate”. Even the unstable intermediate, formed only temporarily in the solution, is sufficiently effective. By designing the synthetic systems in terms of this finding, short single-stranded RNA rings of versatile sizes, which are otherwise hard to be obtained, are efficiently prepared in high selectivity and yield.
T4 Rnl2 ligates ssRNA via nick-like structures, leading to efficient production of versatile RNA rings for various applications.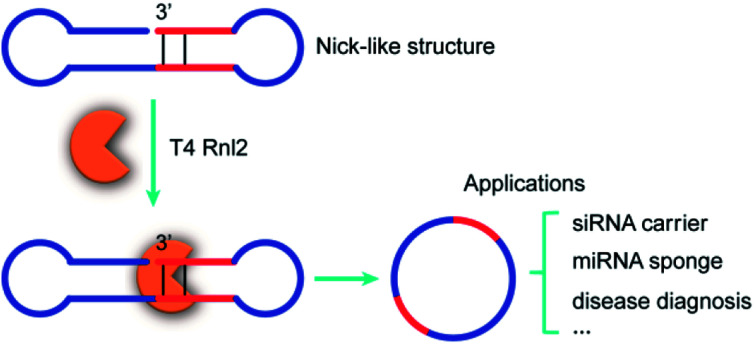
Introduction
The scope of RNA science, either in vivo or in vitro, has been enormously widened and deepened. Many unique biological functions of RNA (e.g., siRNA, miRNA, RNA editing) have been discovered, and completely altered the classical concept of RNA in a central dogma as a simple mediator between DNA and proteins.1–4 Furthermore, promising roles of RNA in nanotechnology and nanomedicine have been envisioned.5–8 One of the key players in these growing fields is RNA ligase which covalently connects two RNA strands (or one RNA strand with one DNA strand) through a phosphodiester linkage. In biological systems, these enzymes participate in repair, splicing, editing, among others. Besides, T4 RNA ligase 2 (Rnl2) and T4 RNA ligase 1 (Rnl1) have been widely used in vitro.9–17 A phosphate at the 5′-end of ssRNA (or ssDNA) is connected with a hydroxyl group at the 3′-end of another ssRNA (not ssDNA). T4 DNA ligase is also available to ligate RNA with DNA. Among these enzymes, Rnl2 is characterized by enormously high intrinsic activity for RNA ligation. In typical cases, this enzyme is more active than Rnl1 by more than 10 folds.12 The reactions by Rnl2 are divided into two categories: (i) seal of nick structure in dsRNA18 and (ii) connection of an ssRNA strand with another ssRNA (or ssDNA) strand. Both reactions are formation of a phosphodiester linkage, but the structures of substrates in them are highly different from each other. For a variety of in vitro applications, the reactions in category (ii) are especially important. For example, two ssRNA strands can be combined to longer ssRNA according to the need. Chimeras of ssRNA and ssDNA are also obtainable. Furthermore, ssRNA rings and ssRNA catenanes, prepared with the use of this enzyme, should exhibit unique properties which cannot be fulfilled by the DNA counterparts.19,20 However, the information on the catalytic function and mechanism of these reactions has been less abundant, preventing still more versatile applications.
The mechanism of Rnl2-mediated nick-seal (category (i)) has been already well established, using the results of X-ray crystallography and others.21,22 There, Rnl2 binds to dsRNA at the nick site, and catalyses the ligation according to Steps 1–3 (the upper part in Scheme 1A). In Step 1, the enzyme reacts with ATP to form a covalent ligase–AMP intermediate in which AMP is linked to Lys 35. Step 2 is the transfer of AMP group from the ligase–AMP intermediate to 5′-phosphate of RNA terminus to produce an adenylated RNA intermediate (AppRNA). The final Step 3 is nucleophilic attack by the 3′-OH of another RNA strand towards the 5′-phosphate group in AppRNA to form a phosphodiester bond at the required position. It should be noted that the detailed dynamics and harmony for Step 2 and Step 3 are rather poorly understood.
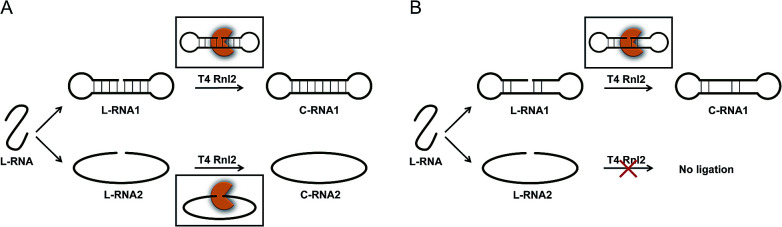
Scheme 1. (A) Usually accepted mechanism for the ligation by Rnl2, and (B) the mechanism proposed here for the connection of two ssRNA strands by Rnl2. In (A), both the nick-sealing (upper part) and the connection of two ssRNAs (lower part) involve the corresponding enzyme–substrate complexes, which are significantly different from each other. In the mechanism proposed in (B), however, the connection of two ssRNA strands also proceeds via “nick-like intermediate” (upper), rather than direct connection of two strands by the enzyme (lower part). The “nick-like intermediate” allows mismatches in both sides. Note that the present figure is never restricted to the intramolecular cyclization to rings and rather also applicable to the ligation of two ssRNA strands to longer ssRNA. “C-RNA” and “L-RNA” refer to circular RNA and linear RNA, respectively.
As compared with category (i), the mechanism for category (ii) has been less understood, except for the ligation involving a helper oligonucleotide which proceeds through a similar mechanism as category (i).15,23 Note that helpers are difficult to design for short RNA substrates. The ligation by Rnl2 in the absence of helper is usually believed to involve direct formation of substrate–enzyme complex from two ssRNA strands and the enzyme (the lower part of Scheme 1A). However, it is unlikely that Rnl2 employs completely different intermediates for each of categories (i) and (ii). Furthermore, remarkable size effect was reported in intramolecular cyclization of ssRNA by Rnl2.24 The reaction occurred only when ssRNA was longer than a critical value (e.g., 15-nt). Independently, we have recently found that the efficiency of intramolecular cyclization of ssRNA by Rnl2 is notably dependent on its length and secondary structure (unpublished results). These results are difficult to understand in terms of the mechanism in which two ssRNA strands simply bind to the enzyme.
On the basis of these results, we have hypothesized that the ligation of two ssRNA strands by Rnl2 also proceeds via a “nick-like structure” (the upper part in Scheme 1B). In this paper, the validity of this hypothesis was confirmed by designing various RNA substrates of different secondary structures and evaluating the efficiency of Rnl2 ligation of these RNAs. It has been found that the stem in the 3′-OH side of joining site enormously promotes the ligation and is essential for the catalysis. Importance of the stem in the 5′-phosphate side is also evidenced. The effects of lengths and positions of these stems are investigated in detail. All the results indicate that the ligation of two RNA strands (or RNA with DNA) by Rnl2 takes advantage of a similar mechanism as the sealing of nick structure in dsRNA. With the use of this finding, a new methodology to synthesize ssRNA rings has been developed in which pseudo nick structure is intentionally formed in the reaction intermediate. Versatile RNA rings, which are otherwise difficult to prepare, can be selectively obtained in high yield.
Materials and methods
Materials
All oligoRNAs used in this study were synthesized by GenScript (Nanjing, China), and the oligoDNAs were purchased from Invitrogen (Shanghai, China). For ligation experiments, a phosphate was enzymatically introduced to the 5′-position of RNA or DNA by using T4 polynucleotide kinase (Thermo Scientific; Pittsburgh, PA, USA). SYBR Gold and RiboLock RNase Inhibitor were also from Thermo Scientific. T4 RNA ligase 2 (Rnl2) and Exonuclease T were purchased from NEW ENGLAND BioLabs (Beijing, China). All other chemicals were from Sigma-Aldrich (St. Louis, MO, USA).
Ligation reactions by Rnl2
Typical reaction system (10 μL) was composed of 1 μM ssRNA (or ssDNA), 4 U Rnl2 and 20 U RiboLock RNase Inhibitor in 1× T4 Rnl2 buffer (50 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 1 mM DTT and 400 μM ATP). Basically, the reaction was carried out at 25 °C for 12 h and quenched by 2 μL of 0.5 M EDTA. When necessary, the cyclic structures of reaction products were confirmed by treating them with Exonuclease T (5 U) at 25 °C for 4 h.
Measurement of Tm values
High resolution melting method (HRM) was used to determine the Tm of RNAs.25 The solutions of RNA (1 μM) were prepared in 1× T4 Rnl2 buffer containing 1× EvaGreen. The mixed oligomer solution (10 μL) was pipetted into 96-well microtiter plates and then transferred to a PikoReal Real-Time PCR instrument (Thermo Scientific, Finland). Fluorescence data were collected over a temperature range of 10–90 °C in 0.1 °C increments. At least three parallel tests were carried out in one plate.
Secondary structure modelling and calculation of Tm values by mfold
Secondary structures of DNA and/or RNA oligos were simulated by Mfold software. Some simple secondary structures are built according to the sequences. The Tm values of DNA/RNA heteroduplexes were also calculated by Mfold Web Server (Website: http://unafold.rna.albany.edu/?q=mfold) using “hybridization of two different strands of DNA or RNA”.26 The salt concentrations were set to be 2 mM Mg2+.
Quantitative analysis of ligation
The reaction products were subjected to 12% denaturing polyacrylamide gel electrophoresis (dPAGE, containing 8 M urea), stained by SYBR Gold, and then imaged with a UV Gel imager from BIO-RAD (Hercules, USA). Quantitative analysis of the data was carried out by using Image Lab software. The conversion for the formation of RNA rings were calculated by the following equation: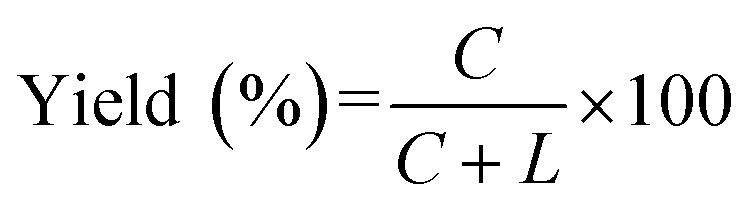 where C and L were the band intensities of ssRNA ring (C-RNA) and remaining linear ssRNA (L-RNA), respectively. The results presented here are the averages of three independent experiments. In the gel electrophoresis of the reaction of L-22d (see Fig. 1A for the sequence), an oligonucleotide complementary (DNA22) to L-22d was added to assist its staining by SYBR Gold (see Fig. S2†), because single-stranded state of L-22d is hard to be stained.
where C and L were the band intensities of ssRNA ring (C-RNA) and remaining linear ssRNA (L-RNA), respectively. The results presented here are the averages of three independent experiments. In the gel electrophoresis of the reaction of L-22d (see Fig. 1A for the sequence), an oligonucleotide complementary (DNA22) to L-22d was added to assist its staining by SYBR Gold (see Fig. S2†), because single-stranded state of L-22d is hard to be stained.
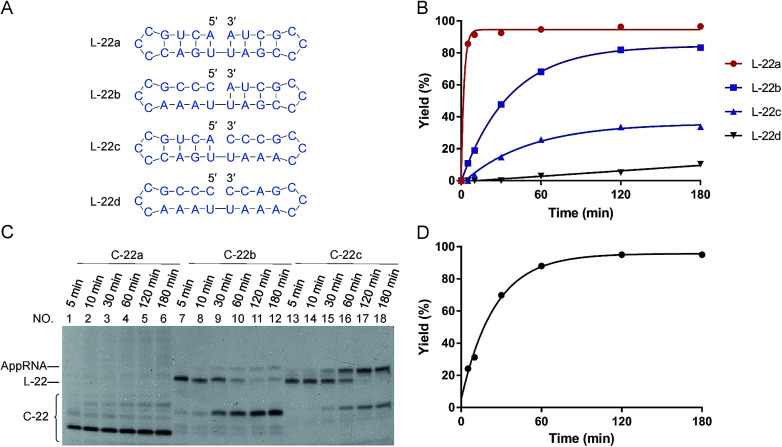
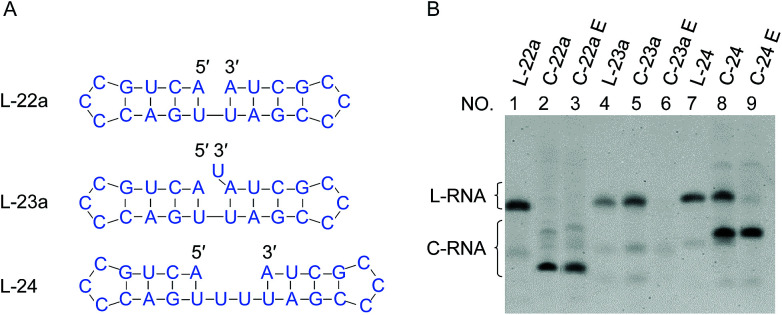
Fig. 1. Effects of “nick-like structure” on RNA cyclization by Rnl2. (A) RNA substrate L-22a having “nick-like structure” (the top) and the relevant structures, which are same as the secondary structures simulated by Mfold software (Fig. S1†). (B) Time-courses for the intramolecular cyclization of L-22a (red circles), L-22b (purple rectangles), L-22c (blue upright triangles) and L-22d (black inverted triangles) in (A). [RNA] = 1 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer at 25 °C. The corresponding 12% denaturing PAGE analyses are presented in (C), except for L-22d (Fig. S2†). L-22 and C-22 refer to 22-nt linear RNA and 22-nt circular RNA, respectively. (D) Time-course for the intramolecular cyclization of L-22a with 0.04 U μL−1 of Rnl2 (the PAGE pattern is shown in Fig. S3†).
Results
Importance of “nick-like structure” for the intramolecular ligation by Rnl2
In Fig. 1, stem structures were introduced near the joining site in RNA substrates, and their roles in Rnl2 ligation were evaluated in terms of the efficiency of intramolecular cyclization. Four kinds of 22-nt RNAs in Fig. 1A were designed, and RNA stems were placed in the 5′-phosphate side and/or in the 3′-OH side (the sequences were listed in ESI Table S1†). The time-courses of cyclization of these RNA substrates, determined by dPAGE (Fig. 1C and S2†), are presented in Fig. 1B. L-22a (the top structure in Fig. 1A), 22-nt linear RNA, involves stable nick structure in which two 4-bp stems existing in the 5′-end and 3′-end, respectively. This RNA was intramolecularly cyclized by Rnl2 to the corresponding 22-membered ring. This reaction was sufficiently fast, and the final yield of RNA ring was close to 100% within 10 min (red line in Fig. 1B). As shown in Fig. 1C (the left-hand part), AppRNA intermediate, formed in Step 2, was rapidly converted to the final ligation product, and never accumulated in the mixture (the particular mechanism was depicted in Fig. S4†). Consistently, the cyclization fairly followed first-order kinetics, as shown in Fig. 1D (the concentration of Rnl2 was decreased here to 0.04 U μL−1 simply for experimental convenience).
In L-22b (the second structure from the top in Fig. 1A), a 4-bp RNA stem is formed in the 3′-OH side of joining site, whereas its 5′-phosphate side is left single-stranded and unstructured. This RNA was intramolecularly cyclized more slowly than L-22a, but the yield attained around 80% at 180 min (compare purple line with red line in Fig. 1B). However, when a 4-bp stem exists only in the 5′-phosphate side of joining site and its 3′-side remains single-stranded (L-22c), the cyclization was significantly slower (blue line). Furthermore, notable amount of AppRNA intermediate was accumulated (the right-hand part of Fig. 1C), and the yield of RNA ring was very low (around 30%). When no stem exists either in the 5′-phosphate side or in the 3′-OH side (L-22d), the cyclization hardly proceeded (black line). All these results showed that the stem in the 3′-OH side of joining site was essential for the ligation by Rnl2, whereas the stem in the 5′-phosphate side further promoted the reaction.
Length of the neighbouring duplex part required for efficient intermolecular ligation by Rnl2
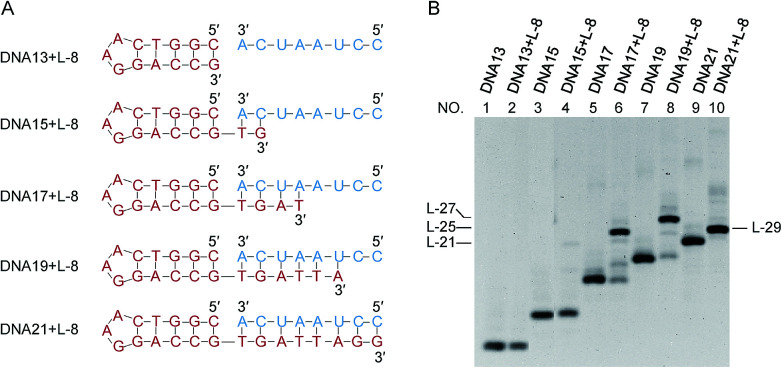
It has been confirmed by Fig. 1 that the stems next to the joining site are necessary for the efficient ligation by Rnl2. In Fig. 2 and 3, the lengths of these duplex parts, required for sufficient promotion of the ligation, are quantitatively determined. In order to obtain direct information on the connection of two strands by Rnl2, the ligation was achieved intermolecularly, and unnecessary perturbation of the assay by ring-size effects and other factors in intramolecular cyclization is minimized. As the component in the 5′-phosphate side of ligation, DNA strand was used simply because DNA-5′-phosphate/3′-OH-RNA pairs are satisfactorily ligated by Rnl2 16 and the usage of DNA instead of RNA is believed not to affect the results much (the DNA sequences are listed in Table S2†). In Fig. 2, the length of the duplex in 3′-OH side of joining site (a DNA/RNA heteroduplex) was systematically increased from 0- to 8-bp, by incorporating the corresponding sequence to the 3′-end of DNA. When there existed no complementary part between the DNA and the RNA (DNA13 + L-8 in the bottom of (A)), the ligation never occurred (lane 2 in (B)). Apparently, intermolecular ligation of ssRNA and ssDNA by Rnl2 is too inefficient under the conditions employed. With the complementary part of 2-bp, the yield was also marginal (lane 4). Only when DNA17, DNA19 and DNA21 were used and the complementary parts were 4-, 6- and 8-bp, respectively, the ligation proceeded to notable extents (lanes 6, 8 and 10). The ligation efficiency monotonously increased with increasing length of the DNA/RNA heteroduplex, and was almost 100% with the 8-bp stem (lane 10). It can be concluded that the length of duplex part in the 3′-OH side must be 4-bp or longer. The ligation satisfactorily proceeds even with only 4- or 6-bp duplex part. It is noteworthy that only one G·C pair is present in the 4- or 6-bp duplex and all others are A·U (or A·T) pairs. Thus, the duplex is very unstable under the ligation conditions.
Fig. 2. Dependence of the intermolecular ligation efficiency on the length of complementary part in the 3′-OH side (DNA/RNA heteroduplex) of the nick. On the 5′-phosphate side of the nick, a DNA/DNA duplex is formed. (A) The sequences of RNA (blue) and DNA (red) were used to systematically change the length of stem in the 3′-OH side; (B) denaturing PAGE (12%) of the products by Rnl2 at 25 °C for 12 h. Lanes 1, 3, 5, 7 and 9: only DNA13, DNA15, DNA17, DNA19 and DNA21, respectively. Lanes 2, 4, 6, 8 and 10: Rnl2 ligation of L-8 in the presence of DNA13, DNA15, DNA17, DNA19 and DNA21, respectively. [RNA] = 2 μM, [DNA] = 1 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer. The migration rate of L-29 was faster than L-27 because it involves a very stable stem-loop structure which is very difficult to be denatured.
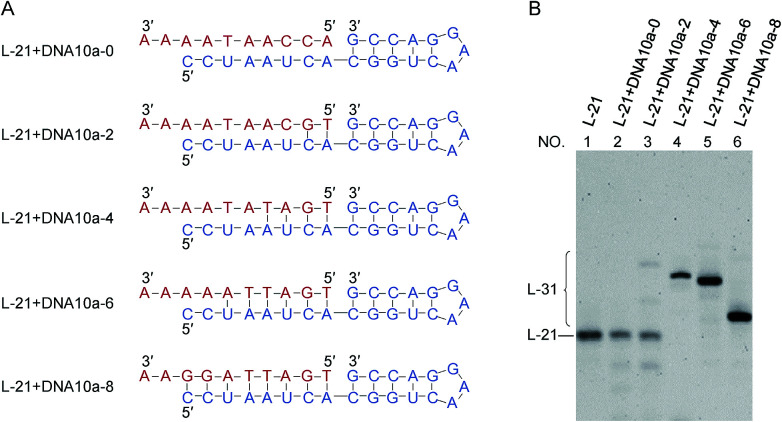
Fig. 3. Dependence of the intermolecular ligation efficiency on the length of complementary part (DNA/RNA heteroduplex) on the 5′-phosphate side of the nick. On the 3′-OH side of the nick, RNA/RNA duplex is formed. (A) The sequences of RNA (blue) and DNA (red). DNA10a-8 is a 10-nt linear DNA whose complementary part with the 5′-phosphate side of L-21 is 8-nt. (B) Denaturing PAGE (12%) for the ligation at 25 °C for 12 h. Lane 1, L-21 only. Lanes 2, 3, 4, 5 and 6: intermolecular ligation of L-21 with DNA10a-0, DNA10a-2, DNA10a-4, DNA10a-6 and DNA10a-8 by Rnl2, respectively. [RNA] = 1 μM, [DNA] = 2 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer.
The effects of duplex length (DNA/RNA heteroduplex) in the 5′-phosphate side of joining site were examined in Fig. 3 (as well as in Fig. S5†). For example, DNA10a-8 is a 10-nt linear DNA, in which the length of complementary part with the 5′-phosphate side of L-21 is 8-nt. When the complementary sequences between RNA (blue) and DNA (red) were 4-, 6- and 8-bp (lanes 4, 5 and 6), the ligation successfully produced RNA/DNA chimera almost completely. With shorter stems of 0- and 2-bp lengths, however, the ligation hardly proceeded (lanes 2 and 3). The stem in the 5′-phosphate side must be also 4-bp or longer. Again, the ligation for 4- or 6-bp duplex is relatively efficient although the corresponding duplex is never stable under the ligation conditions.
Effects of alteration of the “nick-like structure”
In Fig. 4, “nick-like structure” in L-22a was altered to clarify whether an exact “nick-like structure” is necessary for efficient ligation or not. Under the conditions employed, the cyclization of L-22a proceeded smoothly (the yield > 90% as shown in lane 2 in Fig. 4B). When a overlapping U was introduced to the 3′-end of the RNA (L-23a), the yield of cyclization was decreased to 20% (lane 5), demonstrating that the disturbance of nick structure affected the ligation greatly. The overlapping U should form a U·U mismatch when the stem on the 5′-side is open. It is also possible that in L-23a sequence, the 3′ terminal U residue might form a UAU base triple, which would inhibit ligation.27 With the addition of a U–U gap into the counter strand (L-24), the yield of cyclization decreased to 65% (lane 8). Apparently, these structural changes notably suppressed the ligation, but the effects were never fatal. Even with these modified “nick-like structures”, the ligation occurred with reasonable efficiency.
Fig. 4. Effects of overlap or gap structure on the ligation by Rnl2. (A) Structural illustration of RNAs. (B) Denaturing PAGE (12%) for the ligation at 25 °C for 12 h. Lanes 1, 4 and 7: L-22a (nick), L-23a (overlap) and L-24 (2-nt gap) only, respectively. Lanes 2, 5 and 8: Rnl2 ligation of L-22a, L-23a and L-24 by Rnl2, respectively. The cyclic structures of the products in lanes 2, 5 and 8 were confirmed by their intactness against Exonuclease T in lanes 3, 6 and 9, respectively. [RNA] = 1 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer.
Suppression of the formation of “nick-like structure” by complementary DNA as competitive inhibitor
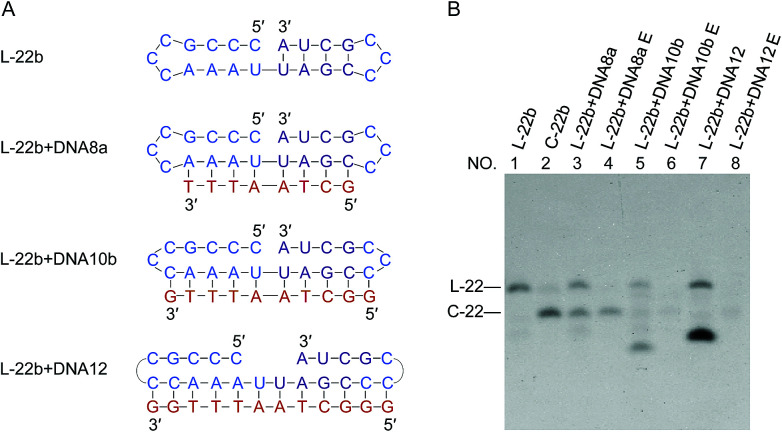
If the ligation proceeds through a “nick-like structure” as we propose, its ligation efficiency should be decreased by suppressing its formation. As the competitive inhibitors on the formation of nick-like structure of L-22b, oligoDNAs (8-, 10- and 12-nt) binding to the stem region of this RNA were added. In Fig. 5, when the oligoDNA was complementary with the RNA in 8-nt length (DNA8a), the cyclization was retarded by 50% (compare lane 3 with lane 2). With the use of longer complementary oligoDNAs (10- and 12-nt), the cyclization was further suppressed to extremely low yields (lanes 5 and 7, respectively). As expected, the Tm values of the corresponding DNA/RNA duplexes increased in this order, which are 23, 40 and 51 °C, respectively. Note that no intermolecular ligation occurred to form longer RNA strands, indicating that the low flexibility of duplex structure was not the main reason of low efficiency of intramolecular cyclization. Significant roles of “nick-like structure” in the ligation by Rnl2 are further supported.
Fig. 5. Inhibition of intramolecular ligation by Rnl2 through the suppression of formation of nick-like structure of L-22b by complementary oligoDNAs. (A) L-22b substrate and the oligoDNAs used as the competitive inhibitors. (B) Denaturing PAGE (12%) for the ligation at 25 °C for 12 h. Lane 1, L-22b. Lane 2, L-22b treated with Rnl2. Lanes 3, 5 and 7: Rnl2 ligation of L-22b in the presence of DNA8a, DNA10b and DNA12, respectively. Lanes 4, 6 and 8: Exonuclease T treatment of the products in lanes 3, 5 and 7, respectively. The bands in the bottoms of lanes 5 and 7 are due to DNA10b and DNA12, respectively. [RNA] = 1 μM, [DNA] = 5 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer.
Enzymatic cyclization of short RNAs involving short stems
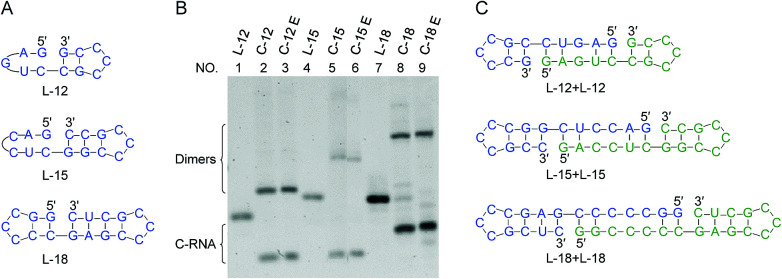
In Fig. 6, RNAs of rather short sizes (12-, 15- and 18-nt) were ligated by Rnl2. The sizes of loop were also decreased. In all of the three RNAs investigated, the intramolecular cyclization to the 12-, 15- and 18-nt rings successfully occurred as expected (the lowest bands in lanes 2, 5 and 8). Very interestingly, even for L-12 with 2-bp stems on both sides of the nick, the ligation satisfactorily occurred (lane 2). Apparently, Rnl2 allows very short duplex part for substrate-binding, although it prefers longer duplex for still more efficient ligation. Note that, in this 12-nt RNA, there is only one nucleotide in the loop of the 5′-side, which makes the stem formation further difficult. With the increase of stem length in the 3′-side, the yield of monomeric rings (not dimeric rings) also increased. In these systems involving short ssRNAs, two RNA molecules were concurrently dimerized, and the corresponding dimeric rings (24-, 30- and 36-nt long) were also formed (the upper bands in these lanes). The formation of the 24-nt long ring was consistent with the previous report.24 The dimeric cyclization was more dominant, when the size of RNA was smaller (lane 2, 12-nt; lane 5, 15-nt; lane 8, 18-nt). The formation of monomeric ring from shorter RNA is less efficient due to larger steric hindrance, and this factor is more explicitly released by the formation of dimeric rings. The yield of cyclic dimer was 60% for 12-nt RNA (L-12), while the yield of intramolecular cyclization was 40%. At higher temperature (37 °C), the yield of dimeric cyclization decreased greatly, and the monomeric ring was dominantly produced (see Fig. S6†).
Fig. 6. Size effects on the ligation by Rnl2. (A) Short RNAs having various nick-like structures. (B) Denaturing PAGE (12%) for the ligation at 25 °C for 12 h. Lanes 1, 4 and 7: L-12, L-15 and L-18, respectively. Lanes 2, 5 and 8: Rnl2 cyclization of L-12, L-15 and L-18, respectively. Lanes 3, 6 and 9: Exonuclease T treatment of the products in lanes 2, 5 and 8, respectively. [RNA] = 1 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer. In (C), the plausible conformations leading to dimeric cyclization are presented.
Efficient production of target ssRNA ring by designing ssRNA substrate which involves a pseudo nick structure
When we attempt to prepare ssRNA ring from linear ssRNA by enzymatic ligation, the ligation efficiency is often too low for practical purposes. With the use of the present finding, however, this problem can be successfully solved. One may design a to-be-ligated ssRNA substrate sequence by having the ligation site within the dsRNA regions that may form in the desired RNA ring product.
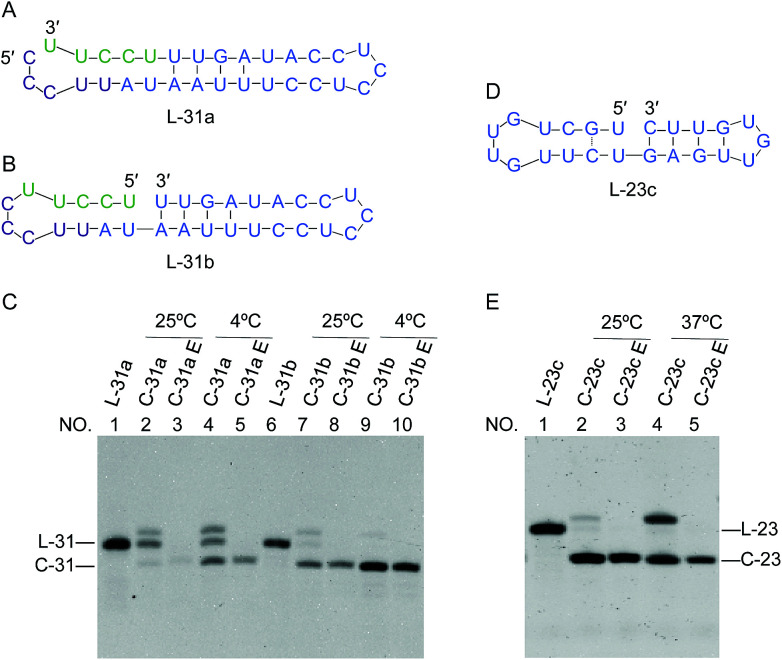
In Fig. 7, two 31-nt long RNAs (L-31a and L-31b) were treated by Rnl2. Both of these ssRNAs are the precursors of the same 31-nt ssRNA ring and converted to the C-31 ring upon the cyclization. Among them, L-31a has no stable nick-like structure near the joining site (Fig. 7A). When this RNA was treated by Rnl2 at 25 °C, C-31 was obtained only in a minimal amount (the faint band below the band of L-31a in lane 2; the upper band is for AppRNA intermediate). Even at 4 °C (lane 4), the formation of C-31 was inefficient (41% yield). With the use of another precursor L-31b which has a 4-bp double-stranded portion (containing 1 G·U base pair) in the 3′-side of the joining site, however, the intramolecular ligation to the ring was far more efficient. In lane 7, this RNA was treated with Rnl2 at 25 °C under the same conditions as described above. Significantly, C-31 was efficiently formed in 76% yield. At 4 °C (lane 9), the yield of C-31 was still higher and almost 100%, since the formation of pseudo nick structure is more favourable. Note that L-31b has exactly the same sequence as L-31a, and the difference is only the presence of pseudo-nick structure near the joining site or its absence. This finding was further extended to the preparation of ssRNA rings which are directly useful for practical applications. It was previously reported that small interfering activity of RNA is greatly promoted by cyclizing one of the two siRNA strands, mainly because the ring structure is more resistant against in vivo enzymatic degradation.20 Accordingly, we attempted to synthesize a ring from 23-nt RNA, which composes siRNA to reduce hypothalamic AGRP.28 Its sequence is 5′-CUGAGUUGUGUUCUGCUGUUGTT-3′. The corresponding 23-nt RNA ring was hypothetically cut at various sites, and the secondary structure of each RNA was investigated. It was found that L-23c (in Fig. 7D) takes the most stable pseudo nick structure. There exist four base-pairs in the 3′-side of the joining site (containing 2 G·U) and one G·C pair in the 5′-side. Exactly as designed, when this RNA was treated by Rnl2, the targeted 23-nt ssRNA ring was successfully obtained almost in 100% selectivity (lanes 2 and 4 in Fig. 7E). Practical importance and availability of the present “pseudo nick strategy” have been further conclusive.
Fig. 7. Circularization of two substrates with the same sequence but with different ends. (A) L-31a without nick-like structure; (B) L-31b with nick-like structure. The nucleotides in the ligation site in L-31a (A) are colored in green and purple to show the difference from L-31b (B) more clearly. (C) Denaturing PAGE patterns (12%) for the ligation of L-31a and L-31b. L-31 and C-31 refer to 31-nt linear RNA and circular RNA, respectively. In (E), 23-nt RNA ring as eminent siRNA was prepared by designing the most appropriate precursor L-23c (in (D)) which has a nick-like structure. [RNA] = 1 μM and [Rnl2] = 0.4 U μL−1 in 1× buffer. The reaction time was 12 h at 25 °C and 4 °C, or 2 h at 37 °C.
Discussion
For intramolecular cyclization of ssRNAs to rings by Rnl2, a stem structure in the 3′-OH side of joining site is crucially important (Fig. 1). Another stem in the 5′-phosphate side of joining site further facilitates the ligation. Cooperation of these two stems is evident. The stem in the 5′-phosphate side is necessary to precisely place the terminal 5′-phosphate of RNA (or DNA) near the Rnl2 and allow efficient acceptance of an adenyl group from the ligase–AMP intermediate (Steps 1 and 2). Consistently, these three steps proceed smoothly for L-22a having stems on both 3′-OH side and 5′-phosphate side, and the ligation is completed without the accumulation of AppRNA intermediate. Inhibition by a overlap or a gap introduced to a nick structure (Fig. 4) is probably associated with deterioration of Step 2 and/or Step 3. The ligation was not completely inhibited because the hairpin structure is not very stable and dynamically repeats open-close process. Even in this case, we believe only when the “nick-like structure” forms, the ligation can be carried out, and in our researches, the Tm values of stem structures derived from RNAs are mostly higher than the reaction temperature (25 °C), which ensure the stability of double-stranded structures of RNAs (Fig. S7†).
The intermolecular ligation by Rnl2 also proceeds via a “nick-like intermediate”. The requirements of stems in both 3′- and 5′-sides are clearly evidenced by Fig. 2 and 3. The stem in the 3′-OH side of joining site (3′-end of one RNA strand) provides the Rnl2-binding site.21 It is noteworthy that, for sufficient intermolecular ligation, these stems do not have to be concretely stable under the reaction conditions. Even 4-bp stem successfully promotes the ligation (Fig. 2). In addition to classical Watson–Crick base-pairings, non-Watson–Crick pairings are available to form these stems. The ligation is successful, even when the duplex part in the 3′-OH side is composed of 1 A·U + 4 G·U, and the one in the 5′-phosphate side is from 1 G·C + 2 C·U (Fig. S8†). Compared with the binding of Rnl2 to unstructured RNA, the entropy loss upon the binding of the enzyme to stem-like structure should be smaller, and the formation of enzyme–substrate complex is greatly facilitated. This substrate binding by Rnl2 in turn stabilizes otherwise unstable stem structure. Furthermore, the position of the 5′-terminal phosphate in the second strand of RNA (or DNA) is precisely regulated by the stem in the 3′-OH side so that it accepts the adenylyl group from the covalent ligase–adenylate intermediate (Step 2). Furthermore, the adenylated phosphate at the 5′-end is appropriately oriented for the nucleophilic attack by the 3′-OH (Step 3). Single-stranded RNA is sufficiently flexible to form these pseudo-stems in the course of molecular movements, and thus a “nick-like structure” is easily taken for a variety of the sequences of RNA (and DNA) in the solutions.
Remarkable substrate-specificity of Rnl2 reaction is interpretable in terms of the mechanism involving “nick-like intermediate”. For example, product distribution (intramolecular cyclization vs. intermolecular ligation) strongly depends on the size of ssRNA (Fig. 6 and S9†). As shown previously, no products (either monomeric or dimeric) were formed, when a 9-mer ssRNA was treated with this enzyme.24 From 12-mer ssRNA, however, both monomeric circle and dimeric product were obtained. From 15-mer ssRNA, monomeric circle was formed together with small amount of dimeric product. With the use of 18-mer ssRNA, only monomeric circle was produced. Due to increasing strain, the stability of the “nick-like intermediate” for the formation of monomeric circle decreased in the following order; 18-mer > 15-mer > 12-mer ≫ 9-mer. On the other hand, the pseudo-rings of the intermediates for the dimerization were much larger, and thus their stability was less dependent on the size of ssRNA. Accordingly, the contribution of the dimerization in the whole reaction decreased with increasing size of ssRNA, and monomeric circle were dominantly produced for 18-mer ssRNA. With the smallest ssRNA (9-mer), even the “nick-like intermediate” leading to the dimeric product was too unstable. If the reactions do not involve the “nick-like intermediates”, these drastic dependencies of the product distribution on ring size can never be expected. Our results also confirm that Rnl2 shows a very high ligase activity, when the ligation system is designed to form a “nick-like structure”. Successful production of the ring of 23-nt siRNA in Fig. 7 is one of the typical applications of this finding.
DNA/RNA can be also ligated efficiently, as long as the 3′-side of the nick is RNA. On the other hand, another RNA ligase Rnl1 is less efficient for nick-ligation. T4 DNA ligase is very active, but requires at least 5–6 bp duplex part on each side of the DNA nick.29 Furthermore, the activity of this ligase is highly sensitive to mismatches. Thus, Rnl2 may be the most convenient ligase which successfully tolerates both mismatches and short substrate-binding part, and is applicable to ubiquitous purposes. Detailed mechanism of RNA ligation involving the formation of pseudo nick structures is depicted in Fig. S4.†
All these arguments fairly agree with previous analysis on Rnl2. First, according to X-ray crystallography on the complex between Rnl2 and nick-bearing dsRNA, the protein primarily interacts with the dsRNA at the 3′-OH side of the nicking site.21 The interactions with the stem in the 5′-phosphate side are marginal. Secondarily, both the rates of adenylation (Step 2) and the strand connection (Step 3) in RNA ligation are significantly reduced, when mismatches are introduced into the 3′-OH terminal region.30,31 Apparently, strict freezing of molecular dynamics in the stems is important for efficient catalysis. Thirdly, the 3′-OH at the joining site must derive from ssRNA, although the 5′-phosphate can come from either ssRNA or ssDNA.10,16 The ssRNA in the 3′-OH side is indispensable to connect two single strands by Rnl2, since a stem must be formed in this side and it allows larger flexibility of the strand at 5′-side (Fig. S4†). In addition to this factor, 2′-OH (as well as 2′-OMe) is favourable to form 3′-endo pucker (RNA-like pucker), which is required for the ligation.16,21 Furthermore, the increased nucleophilicity of 3′-OH by the adjacent 2′-OH in the ssRNA may also promote the ligation (its pKa is lowered to around 12).21,32 Munafo et al. also reported that 2′-O-methyl modification of the ribose at the 3′-end decreased the ligation efficiency to some extent, which is consistent with the above discussion that nucleophilicity of 3′-OH is increased by 2′-OH.33 For mutated T4 Rnl2, the ligation of ssRNA was also observed.9,34 For example, T4 Rnl2tr, a truncated form of T4 Rnl2, was reported to be ten times more active than the full length T4 Rnl2 in 5′-adenylated RNA ligation.16 We believe that nick-like structure was also formed for these ssRNA ligations, although further detailed analysis is required.
Conclusion
The features of Rnl2 catalysis in two-ssRNA connection, found in this paper, are summarized as follows. (1) Only a substrate involving a nick or nick-like structure is satisfactorily ligated. (2) The ligation is very efficient, even when the duplex parts on both sides of the nick-like structure are only 4-bp length. (3) The ligation is successful, even when some mismatches are present around the nick-like structure. These results indicate that a “nick-like intermediate” is formed, at least temporarily, when this enzyme connects two ssRNA strands. Larger flexibility of RNA strands than DNA strands is an important factor to allow this catalysis. The mechanisms of Rnl2 for two-ssRNA connection and dsRNA-nick seal have been comprehensively understood.
Based on these findings, efficient systems of two-ssRNA connection can be straightforwardly designed in terms of secondary structures in the reaction mixtures. For example, various ssRNA rings can be prepared by Rnl2 ligation in high yields (without changing the sequence). There, desired ssRNA ring is hypothetically cut at appropriate position (on paper) so that the ligation position is located in “nick-like structure”. The resultant ssRNA should be used as the substrate for the ligation by Rnl2. This strategy can be easily extended to high-yield preparation of long circular RNA with hundreds of nucleotides, which are otherwise hardly producible. These studies are currently underway in our laboratory.
Conflicts of interest
None declared.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China [31571937 to X. L.]; China Postdoctoral Science Foundation [2018M632718 to R. A.]; the Fundamental Research Funds for the Central Universities [201713050 to R. A.]. Funding for open access charge: National Natural Science Foundation of China.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra01513b
References
- Chakraborty C. Sharma A. R. Sharma G. Doss C. G. P. Lee S. S. Mol. Ther.--Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. P. Sharei A. Ding X. Sahay G. Langer R. Jensen K. F. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- Zuckerman J. E. Davis M. E. Nat. Rev. Drug Discovery. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- Lachelt U. Wagner E. Chem. Rev. 2015;115:11043–11078. doi: 10.1021/cr5006793. [DOI] [PubMed] [Google Scholar]
- Teller C. Willner I. Curr. Opin. Biotechnol. 2010;21:376–391. doi: 10.1016/j.copbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Komiyama M. Yoshimoto K. Sisido M. Ariga K. Bull. Chem. Soc. Jpn. 2017;90:967–1004. doi: 10.1246/bcsj.20170156. [DOI] [Google Scholar]
- He Y. Ye T. Su M. Zhang C. Ribbe A. E. Jiang W. Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- Komiyama M. Mori T. Ariga K. Bull. Chem. Soc. Jpn. 2018;91:1075–1111. doi: 10.1246/bcsj.20180084. [DOI] [Google Scholar]
- Ho C. K. Wang L. K. Lima C. D. Shuman S. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Nandakumar J. Ho C. K. Lima C. D. Shuman S. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- Tessier D. C. Brousseau R. Vernet T. Anal. Biochem. 1986;158:171–178. doi: 10.1016/0003-2697(86)90606-8. [DOI] [PubMed] [Google Scholar]
- Bullard D. R. Bowater R. P. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S. Kirino Y. Nucleic Acids Res. 2017;43:e77. doi: 10.1093/nar/gkv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S. Shigematsu M. Morichika K. Telonis A. G. Kirino Y. RNA Biol. 2015;12:501–508. doi: 10.1080/15476286.2015.1031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N. Matsumoto K. Nishihara M. Nakano Y. Shibata A. Maruyama H. Shuto S. Matsuda A. Yoshida M. Ito Y. Abe H. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J. Shuman S. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Hinton D. M. Brennan C. A. Gumport R. I. Nucleic Acids Res. 1982;10:1877–1894. doi: 10.1093/nar/10.6.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. K. Shuman S. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S. Appel B. RNA Biol. 2017;14:1018–1027. doi: 10.1080/15476286.2016.1239009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Liang D. Chen C. Wang Y. Amu G. Yang J. Yu L. Dmochowski I. Tang X. Mol. Ther.--Nucleic Acids. 2018;10:237–244. doi: 10.1016/j.omtn.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J. Shuman S. Lima C. D. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Pascal J. M. Curr. Opin. Struct. Biol. 2008;18:96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Petkovic S. Muller S. FEBS Lett. 2013;587:2435–2740. doi: 10.1016/j.febslet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Yin S. Ho C. K. Miller E. S. Shuman S. Virology. 2004;319:141–151. doi: 10.1016/j.virol.2003.10.037. [DOI] [PubMed] [Google Scholar]
- Wang J. Pan X. Liang X. J. Anal. Methods Chem. 2016;2016:5318935. doi: 10.1155/2016/5318935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard J. M. Zuker M. Gulari E. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi G. Zhou Y. Zhong Z. Toh D. F. K. Chen G. Wiley Interdiscip. Rev.: RNA. 2015;6:111–128. doi: 10.1002/wrna.1261. [DOI] [PubMed] [Google Scholar]
- Makimura H. Mizuno T. M. Mastaitis J. W. Agami R. Mobbs C. V. BMC Neurosci. 2002;3:18. doi: 10.1186/1471-2202-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y. Han X. An R. Zhang Y. Cheng K. Liang X. Komiyama M. Nucleic Acids Res. 2018;46:e132. doi: 10.1093/nar/gky365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J. Shuman S. J. Biol. Chem. 2005;280:23484–23489. doi: 10.1074/jbc.M500831200. [DOI] [PubMed] [Google Scholar]
- Chauleau M. Shuman S. RNA. 2013;19:1840–1847. doi: 10.1261/rna.041731.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama M. Takeda N. Shigekawa H. Chem. Commun. 1999;16:1443–1451. doi: 10.1039/A901621J. [DOI] [Google Scholar]
- Munafo D. B. Robb G. B. RNA. 2010;16:2537–2552. doi: 10.1261/rna.2242610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet S. Fuchs R. T. Munafo D. B. Zhuang F. Robb G. B. BMC Biotechnol. 2011;11:72. doi: 10.1186/1472-6750-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.