Summary
Cell shape changes from locomotion to cytokinesis are, to a large extent, driven by myosin-driven remodeling of cortical actin patterns. Passive crosslinkers such as α-actinin and fascin as well as actin nucleator Arp2/3 complex largely determine actin network architecture and, consequently, membrane shape changes. Here we reconstitute actomyosin networks inside cell-sized lipid bilayer vesicles and show that depending on vesicle size and concentrations of α-actinin and fascin actomyosin networks assemble into ring and aster-like patterns. Anchoring actin to the membrane does not change actin network architecture yet exerts forces and deforms the membrane when assembled in the form of a contractile ring. In the presence of α-actinin and fascin, an Arp2/3 complex-mediated actomyosin cortex is shown to assemble a ring-like pattern at the equatorial cortex followed by myosin-driven clustering and consequently blebbing. An active gel theory unifies a model for the observed membrane shape changes induced by the contractile cortex.
Subject areas: Mechanobiology, Biological sciences, Cell biology
Graphical abstract

Highlights
-
•
Myosin, α-actinin, and fascin drive the formation of diverse actin patterns in GUVs
-
•
A self-assembling membrane-bound contractile ring pattern slightly deforms GUVs
-
•
Dendritic actomyosin requires actin crosslinkers for clustering and GUV blebbing
-
•
A physical model describes membrane deformation at the site of actomyosin cluster
Mechanobiology; Biological sciences; Cell biology
Introduction
Cell morphogenesis, shape change, and motility are highly dependent on actin networks acting in different regions of a cell (Blanchoin et al., 2014; Svitkina, 2018). In the thin actin cortex under the cell membrane, the pool of actin filaments is spatiotemporally regulated by actin regulators, resulting in different functionalities of the actin networks (Lappalainen, 2016; Pollard, 2016). The actin cortex supports the cell’s morphology and can rearrange into structures such as lamellipodia or lead to the formation of protrusions such as filopodia at the cell periphery (Mejillano et al., 2004). Actin networks at the cell periphery are organized by myosin motors, passive crosslinkers, and nucleation factors under a variety of conditions. Detachment of actin from the membrane and depletion of Arp2/3 complex both contribute to the initiation of cell protrusion at the cell periphery whereas short crosslinkers, such as fascin, participate in the assembly of actin bundles for cell protrusion (Blanchoin et al., 2014; Dimchev et al., 2021; Welf et al., 2020). In cytokinesis, another large-scale self-organization event, myosin and α-actinin intensely localize at the cell equatorial plane to assemble the contractile ring, the positioning of which is regulated by Arp2/3 complex-mediated actin cortex (Fujiwara et al., 1978; Pollard and O’Shaughnessy, 2019). Simulation of actomyosin networks in confinement also showed that myosin-driven contractility has a major role in the formation of ring-like cortex structures (Ni et al., 2021), which are found at the periphery of T cell immunological synapse, axons, and ring complex of podosomes (Collin et al., 2008; Hammer et al., 2019; Xu et al., 2013). It was shown that actin ring-like vortices and aster structures in the cortex upon adherence of HeLa cells to a substrate are highly regulated by the Arp2/3 complex whereas myosin motors were not involved in these transitions (Fritzsche et al., 2017).
The interplay between different actin-binding proteins in living cells makes the mechanistic study of actin network morphogenesis challenging. There has been a myriad of studies on in vitro reconstitution of active and passive actin networks that have unraveled the role of actin binding proteins in network self-organization. By selective activation of myosin in a pool of actin filaments on supported lipid bilayers, it has been shown that actomyosin networks contract radially toward the center of the activation region in a cooperative manner (Linsmeier et al., 2016). Myosin contractility toward the barbed end of crosslinked actin filaments can induce polarity sorting and formation of aster-like structures. However, the role of myosin as an active motor or passive bundler in the formation of actin patterns depends on myosin concentration as well as crosslinker type and concentration. Myosin at low concentrations acts as an actin crosslinker in the presence of a short crosslinker fascin whereas it actively contracts actin bundles to form asters at high concentrations (Backouche et al., 2006). Myosin was recently shown to have more motility and higher velocities on fixed actin filaments bundled by fascin whereas it becomes almost immobile and trapped in a mixed polar network crosslinked by alpha-actinin (Weirich et al., 2021). Considering the diverse set of actin binding proteins and different members of motor proteins in cells, how specific structures arise from the same pool of proteins remains unknown.
Addition of biomembranes in actin reconstitution platforms made it possible to reconstitute more sophisticated structures and study the physics and mechanism governing the interaction of actin networks with cellular membrane (Bashirzadeh et al., 2020, 2021a; Bashirzadeh and Liu, 2019; Groaz et al., 2021; Liu et al., 2008; Liu and Fletcher, 2006; Sharma et al., 2021). In the absence of molecular motors, branched actin networks reconstituted on lipid bilayer membranes demonstrated the physics and mechanism governing membrane protrusion by cortical actin (Cáceres et al., 2015). In the presence of myosin motors and depending on network tension, actin cortex polymerized on the outside of giant unilamellar vesicles (GUVs) could crush the vesicle or peel the membrane (Carvalho et al., 2013b). When encapsulated inside the GUVs, however, the actomyosin cortex was shown to cluster at the periphery of membrane or in the center of the vesicle depending on the strength of actin network-membrane attachment (Carvalho et al., 2013b). Of interest, this phenomenon has been investigated recently in Xenopus egg extract droplet in oil emulsions and is modeled as a tug-of-war between contraction waves and spontaneous bridge formation between the cluster and the membrane. It has been shown that the contraction wave period and bridge formation timescale are dependent upon the confinement size (Sakamoto et al., 2020). In such a confined environment and in the presence of actin crosslinkers and crowding agents, reconstituted actomyosin bundles formed rings whereas increasing their contractility led to cluster formation (Miyazaki et al., 2015). Contractile actomyosin rings linked to the membrane of GUVs and formed by crosslinking of actin by vinculin and talin have been reconstituted to mimic cell division (Litschel et al., 2021). These contractile rings induce transient shape changes in GUVs and contract to a final cluster form. However, the involvement of a branched actin cortex in contractile ring positioning and membrane constriction is missing in this work despite its important role in cytokinesis. Recently, it was shown that actin clustering could occur solely because of α-actinin crosslinking activity in GUVs and that α-actinin-actin bundles could form rings or peripheral asters depending on GUV size (Bashirzadeh et al., 2021a). When reconstituted together, α-actinin and fascin sorted in separate domains to form central aster patterns in GUVs (Bashirzadeh et al., 2021a). However, the physical conditions and the contribution of individual actin crosslinkers and nucleation factors which induce the dynamic evolution of contractile actomyosin networks into diverse patterns and actomyosin interaction with the membrane in confinement have not yet been studied.
Here, force generation and size-dependent pattern formation of luminal, membrane-bound, and branched actomyosin networks in confinement is investigated. By tuning the amount of active and passive actin crosslinkers, we determine the optimal conditions for pattern formation in GUVs in the form of luminal and peripheral clusters, central and peripheral asters, and contractile rings. By changing membrane surface properties, we demonstrate the formation of cortical rings and asters with inhibited membrane protrusion. Membrane-bound actomyosin rings exert contractile force and slightly deform GUV membrane. We then show that Arp2/3 complex-dendritic actomyosin networks in the presence of actin crosslinkers assemble contractile ring-like patterns at the equatorial plane of GUVs and condense into clusters to induce membrane deformation. An active gel model describes membrane deformation caused by dendritic cortex tension at the cluster site. Our results highlight the central role of closed boundary conditions in the formation of emergent actomyosin patterns and consequently biological membrane shape changes by these patterns.
Results
The formation of contractile actomyosin ring and aster patterns depend on GUV size and the relative concentration of myosin and passive crosslinkers
We wanted to first establish the contractility of actomyosin networks and transmission of the contractile forces in vitro. To do this, we used Fourier transform traction cytometry (FTTC) (Bashirzadeh et al., 2019; Butler et al., 2002; Schwarz et al., 2002) to measure the traction stresses exerted by actomyosin networks to a soft substrate (Young’s modulus ∼0.75 kPa) through biotinylated actin-streptavidin linkages (Figure S1A and see STAR Methods). Non-uniform distribution of NeutrAvidin beads bound to biotinylated actin resulted in the formation of an F-actin ‘template’ pattern upon polymerization (Figures S1B and S1C). Upon the addition of 150 nM myosin, F-actin immediately bundled, contracted (Video S1), and deformed the substrate (Videos S2 and S3), resulting in instant increase of traction stresses (Figures S1D, S1E, and Video S4) and strain energy the rate of which gradually decreased (Figures S1F). A second burst of contraction could be induced by adding more ATP (Figures S1D–S1F). An inverse contraction force-velocity relationship was observed in the system upon addition of myosin (Figure S1G) (Thoresen et al., 2011). Local divergence of bead displacement field upon the addition of myosin revealed a convergent contraction toward the template and actin enrichment in the template (Figures S1H–S1J). These results provide a quantitative means of traction stresses and work produced by actomyosin networks and show that active filaments of muscle myosin at 150 nM concentration deliver cellular-scale mechanical energy to an attached substrate at time scales on the order of seconds (Bashirzadeh et al., 2019). The results also show that non-covalent bonds of biotin-avidin efficiently transmit actomyosin contractile forces to induce deformation.
To reconstitute contractile actomyosin ring, we sought to find the molar concentrations of crosslinkers and myosin that can induce formation of rings in GUVs. Knowing that our system of actin filament and myosin creates robust contraction, we first explored how actomyosin networks self-organize in GUVs in the absence of membrane attachment and how actin crosslinkers fascin or α-actinin influence actomyosin network organization. In the absence of actin crosslinkers, encapsulation of actomyosin networks at myosin molar ratios ranging from 0.0125 to 0.05 (63 nM–250 nM) in DOPC/cholesterol GUVs all resulted in instant contraction and accumulation of actin filaments at either the center or periphery of GUVs (Figures 1A and S2), as expected. In small GUVs (diameter <15 μm), the majority of GUVs had accumulated actin and appear in the form of a short but thick bent bundle at the GUV periphery. We next encapsulated a mix of myosin, α-actinin, and fascin, each at different molar ratios, with actin inside GUVs. Irrespective of myosin concentration, addition of α-actinin did not change actomyosin condensation into a cluster (Figure 1B). Myosin-fascin-actin bundles, however, formed actin meshworks with filopodia-like membrane protrusions (Figure 1C), without actin condensation. In the presence of both α-actinin and fascin, when α-actinin concentration was lower than fascin concentration, stiff actin rings were formed in small GUVs (diameter <15 μm) (Figure 1D, green arrows, and Figure 1E, green arrows, Video S5). In large GUVs (diameter >20 μm), centripetal contraction condensed actin into clusters resulting in the formation of aster structures (Figure 1D, red arrow, and Figure 1E, left, Video S5) following elongation of actin bundle arms outside clusters (Figures S3A, S3B, and Video S6). α-Actinin localized in the cluster region of asters (Figures S3C and S3D). Stiffer actin bundle arms outside the cluster often formed membrane protrusions (Figure 1D, yellow arrow, and Figure S4). Any structure that possesses multiple actin bundle arms radiating outside a clustered actin region is referred to as an aster in this work.
Figure 1.
Diverse actomyosin network patterns emerge in confinement
(A-F) Representative z-projected confocal images of actin networks in the presence of myosin, α-actinin, and/or fascin at the indicated concentrations. Actin, 5 μM. Scale bars, 10 μm. n = 2 experiments per condition.
(D) Green arrows represent actin bundle rings in small GUVs. Red arrow points to a large GUV with encapsulated actin aster and ring.
(F) Yellow arrows show actin aster with long actin bundles bent at the GUV periphery. These actin bundles turned and elongated toward GUV lumen and may appear as axially symmetric structures in 2D (green arrow). Cyan arrow shows an actin aster pattern with bundle length shorter or equal to GUV radius. Pink arrow points to two peripheral actin asters formed at two poles of a GUV. Also see Figures S2–S4.
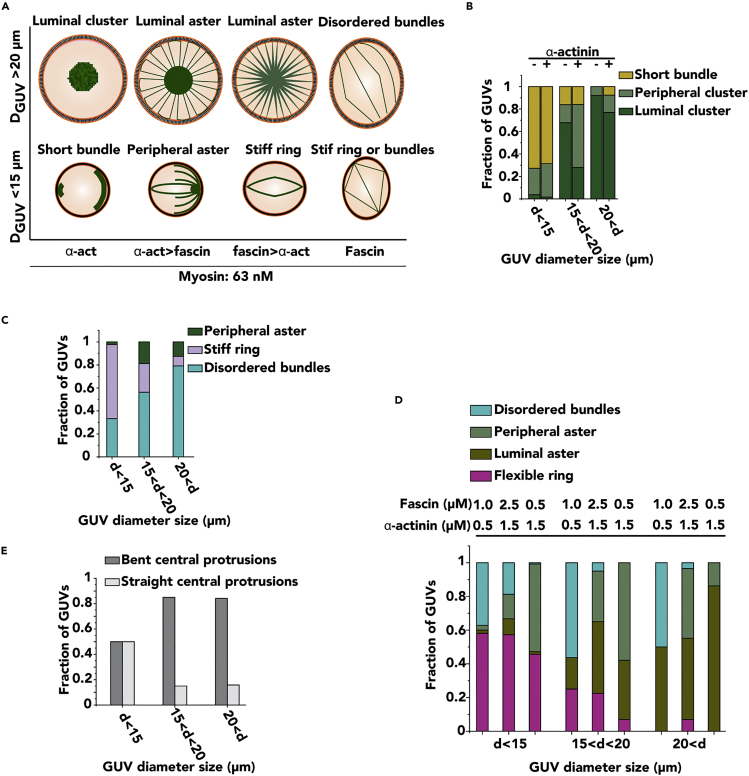
At high α-actinin/fascin ratios, most actin bundles formed aster-like patterns at the GUV center (Figure 1F). Large actin bundles were bent at the GUV periphery and looped backed into the GUV lumen (Figure 1F, yellow arrows). Almost axially symmetric structure with regular spacing between actin bundles was observed (Figure 1F, green arrow). However, actin bundle arms whose radius was less than GUV radius, stayed straight (Figure 1F, cyan arrow). Occasionally, peripheral actin bundle arms were joined at single or multiple clusters to form peripheral actomyosin asters (Figure 1F, magenta arrow). It should be noted that, in the absence of myosin, α-actinin-fascin-actin bundles form rings and asters at high α-actinin concentrations (Bashirzadeh et al., 2021a). Most actin bundle rings in this condition were positioned at the equatorial plane of GUVs and parallel to the substrate a few hours post-encapsulation (Video S7). Figure 2A summarizes the distinct actomyosin patterns that most GUVs formed by tuning the concentrations of fascin and α-actinin. The fraction of GUVs that exhibited these patterns is summarized in Figures 2B-2E. Myosin-driven buckling of actin resulted in the formation of a short filament bundle or cluster in all GUVs in the absence or presence of α-actinin (Figure 2B). These filaments formed a large cluster in more than 92% of large GUVs (diameter >20 μm). In the presence of myosin and fascin, disordered actin bundles or a stiff ring was formed in more than 81% of GUVs without any clustering activity (Figure 2C). The bundles formed disordered structures in 79% of large GUVs whereas they formed a stiff ring in 64% of small GUVs (diameter <15 μm). In the presence of α-actinin, fascin, and myosin, 57% of actin filaments formed a ring at high fascin/α-actinin ratios in small GUVs (d < 15 μm). In large GUVs, greater than 90% of GUVs formed aster structures when α-actinin concentration was 1.5 μM (Figure 2D). 85% of the actin bundle arms in aster structures bent and extended in to the GUV lumen when fascin concentration (0.5 μM) was below α-actinin concentration in medium- (15μm < diameter <20 μm) and large-sized (diameter >20 μm) GUVs (Figure 2E). Together these results showed that α-actinin-actin bundles with large filament spacing contribute to actomyosin clustering which, in turn, results in the formation of aster-like patterns in the presence of fascin. Fascin-parallel actin bundles do not cluster in the presence of low myosin concentrations, and together with a lower concentration of α-actinin, form a flexible contractile actomyosin ring in the periphery of the majority of small GUVs.
Figure 2.
Phase diagram and analysis of actomyosin pattern formation as a function of GUV size and concentration of α-actinin and fascin
(A) Phase diagram of encapsulated actomyosin patterns that are likely to form as a function of GUV size and relative concentration of actin crosslinkers. (B-E) Bar graphs of the frequency of actomyosin phenotypes depicted in Figure 1. Actin, 5 μM. Myosin, 63 nM.
(B) Fraction of GUVs with size-dependent actomyosin network phenotypes in the presence (0.5 μM) or absence of α-actinin.
(C) Fraction of GUVs with size-dependent actomyosin network phenotypes in the presence of 1 μM fascin.
(D) Bar graphs demonstrating quantification of different GUV size-dependent phenotypes in the presence of α-actinin and fascin at the indicated concentrations.
(E) Fraction of GUVs of either kinked or straight actomyosin asters in the presence of α-actinin (1.5 μM) and fascin (0.5 μM). For bar graphs, n = 2 experiments per condition. Number of analyzed GUVs >42 per experiment per condition.
Anchoring actin to GUV membrane localizes actomyosin patterns at the GUV periphery but does not change the architecture of actomyosin ring and aster patterns
Because contractile actomyosin rings and asters assemble in the cell cortex and interact with the plasma membrane (Fritzsche et al., 2017; Pollard and O’Shaughnessy, 2019), we next sought to test the formation of cortical actomyosin patterns bound to GUV membrane. Membrane binding via biotinylated actin-streptavidin-biotinylated lipid linkages resulted in the formation of peripheral patterns of actin bundles. Membrane-bound α-actinin-actin bundles formed peripheral rings (Figure 3A). In the presence of α-actinin and myosin, actin filaments condensed into clusters to form peripheral aster-like patterns (Figure 3B). Increasing myosin concentration further enhanced clustering at the periphery (Figure S5). Membrane-bound actomyosin asters resembled aster-like cortical actin networks of cells during cell adhesion (Fritzsche et al., 2017). Replacing α-actinin with fascin induced the formation of peripheral actomyosin meshworks (Figure S6 and Video S8), in stark contrast to unanchored fascin-actomyosin system (Figure 1C). Complete suppression of fascin-induced membrane protrusions is again in line with recent observations that membrane attachment reduces actin network protrusions in cells (Welf et al., 2020). The observed fascin-actomyosin meshworks were also reported to form in bulk at low myosin and high fascin concentrations (Backouche et al., 2006). Like unanchored actomyosin networks, our observations indicate that fascin facilitates the formation of actomyosin bundles whereas α-actinin contributes to myosin-driven cluster formation in membrane-bound actomyosin networks. Like unanchored networks, reconstitution of membrane-bound actomyosin networks in the presence of both fascin and α-actinin resulted in the formation of stiff aster-like patterns. However, the clusters were located at the GUV periphery and packed bundles elongated into the GUV lumen with suppressed membrane protrusion (Figure 3C). The fraction of GUVs (Figure 3D) forming the phenotype of an aster-like pattern in the absence (Figure 3E) and presence (Figure 3F) of fascin is illustrated. It should be noted that, regardless of the crosslinker, we occasionally observed clustered actin bundles that were detached from the membrane and formed luminal networks (Figure S7). We suspect this could be because of either myosin-driven contraction of actin filaments detaching them from the membrane or weak binding of actin to the membrane via biotin actin-neutravidin-biotin lipid linkages (Carvalho et al., 2013b). Like unanchored actin, the growth of actin bundle arms formed an aster-like structure in these cases (Figure S7C).
Figure 3.
Enhanced actin-membrane interaction does not change the architecture of contractile actin patterns but suppresses actin bundle protrusion
(A) Representative z-projected confocal images of membrane-bound actin bundles in the presence of α-actinin at concentrations indicated. Actin, 5 μM. Scale bars, 10 μm n = 2 experiments.
(B) Representative z-projected confocal images of short peripheral actin asters formed in the presence of myosin and α-actinin at concentrations indicated. Actin, 5 μM. Scale bar, 10 μm. n = 2 experiments.
(C) Representative z-projected confocal images of membrane-bound actin in the presence of myosin, α-actinin, and fascin at concentrations indicated. White arrows show aggregated actin-streptavidin linkages at the site of actin cluster. Actin, 5 μM. Scale bar, 10 μm. n = 2 experiments.
(D) Schematic illustration of clustering of membrane-bound actin networks and the emergence of short actin asters (left) and normalized frequency of GUV size-dependent appearance of different actomyosin network phenotypes (right) in the presence of myosin and α-actinin under the conditions in (b).
(E) Schematic illustration of the effect of fascin on the architecture of cortical actomyosin patterns in large GUVs (diameter >20 μm). Although attachment to GUV membrane suppresses protrusion, α-actinin and fascin together form aster structures elongating into the lumen of GUVs (left). Normalized frequency of GUV size-dependent appearance of different actomyosin network phenotypes under this condition (c) is shown on the right. For bar graphs, n = 2 experiments per condition. Number of analyzed GUVs >33 per experiment per condition. Also see Figures S5 and S6.
Membrane-bound actomyosin ring patterns deform GUV membrane during self-assembly
Because our long-term goal is to generate a contractile ring in a GUV to mimic cytokinesis, we wanted to understand better how these actomyosin rings form. Using a high fascin molar ratio in the presence of α-actinin, a condition we found to form rings in small GUVs, we successfully captured time evolution of the assembly of a membrane-bound contractile actomyosin ring over ∼40 min. Like the formation of contractile bundles on soft substrates, we first noticed the formation of a thin template of an actin ring that evolved over time (Figures 4A, Video S9, and Figures S8). During contractile ring assembly we noticed that GUV membrane was constricted (Figure 4A, red arrows, and Video S10, dashed box) at the locus of accumulated actin in the ring. By measuring membrane angles with respect to the plane of actomyosin ring (Figure 4B), and assuming membrane tensions, σ1 and σ2 are equal to their average, σ (Stachowiak et al., 2014), we found the maximum contractile force of the ring (Fc), measured at time point 16 min to be 5.89 × 10−6σ [N]. Membrane tension for DOPC GUVs of similar size under a mild tension, which was induced by membrane extrusion, is estimated to be ∼3 × 10 −5 [N/m] under iso-osmotic conditions (Borghi et al., 2003). Using this value, contractile force of the ring would then be ∼176 [pN], which is comparable to 390 [pN] generated by sliding contractile ring in fission yeast protoplasts (Stachowiak et al., 2014). Concurrent to force generation, more actin accumulated over time to complete ring assembly (Figures 4C–4D).
Figure 4.
Membrane-bound contractile ring patterns deform GUV membrane
(A) Representative fluorescence confocal actin images showing a time-lapse of contractile ring assembly in the presence of myosin, fascin, and α-actinin as well as z-projected confocal images of α-actinin, myosin, and merged images of the ring at the last time point (38 min). At the first two time points, actin images were captured as single 2D images before GUV settlement. The red arrow shows the region where maximum ring constriction occurs. Dashed lines show the line along which actin, myosin, and α-actinin intensities were quantified. Actin 5 μM. Myosin, 63 nM α-Actinin, 1.5 μM. Fascin, 2.5 μM. Scale bar, 10 μm.
(B) Schematic illustration of parameters used to calculate contractile force (Fc) as a function of membrane tensions (σ1,σ2).
(C) Average values of maximum actin intensity along the three white dashed lines in (a). Intensity values were normalized to average intensity along each line.
(D) Schematic illustration of myosin-induced clustering and contractile ring assembly in a GUV.
Dendritic actomyosin cortex in the presence of actin crosslinkers assembles into a ring-like contractile pattern at the GUV equator and induces membrane deformation.
Actomyosin ring contraction and membrane constriction in the cell is highly regulated by a branched actomyosin cortex under the membrane and adjacent to the ring. To test the role of branched actin cortex on the organization of actomyosin patterns and contractile ring assembly, we reconstituted an Arp2/3-branched dendritic actin cortex on the inner surface of GUVs in the presence and absence of myosin, α-actinin, and fascin. Arp2/3 complex was activated by membrane-bound constitutively active VCA domain of neural Wiskott Aldrich syndrome protein. The dendritic actin patterns formed an actin shell at the GUV periphery regardless of the presence of α-actinin and fascin (Figure S9). Inclusion of myosin, and together with the concentrations of α-actinin and fascin known to form rings (Figure 2A), in the dendritic networks caused symmetry breaking and actin clustering at the periphery and the emergence of small spherical membrane protrusions (Figures 5A–5C). A myosin molar ratio of 0.0125 was sufficient for symmetry breaking, and consequently clustering actin toward the membrane because of strong attachment of actin to the membrane (Carvalho et al., 2013a, 2013b). Interestingly, however, actin was intensely localized at the equatorial plane of GUVs in the form of a ring-like pattern (Figure 5C). 3D intensity analysis of the contractile cortex structures showed that the contractile cortex not only promoted the formation of ring-like patterns at the equatorial cortex (Figures 5D–5G) but tended to induce actin clustering, often in multiple locations (Figure 5F, red arrows). To rule out the existence of artifacts in the formation of ring-like patterns, we also analyzed 3D intensities of GUV membrane and dendritic actin cortex structures formed in the absence of myosin and actin crosslinkers and confirmed that actomyosin cortex structures indeed induced the formation of ring-like patterns (Figure S10). Membrane deformation was frequently observed at the location of actin clusters, which often appeared in the form of a ring-like pattern at the neck of the deformed membrane (Figure 5D, white arrow). The mode and timescale of membrane deformation resembled the constriction of membrane-bound actomyosin bundles formed in the presence of talin and vinculin (Litschel et al., 2021). We occasionally observed a large membrane bleb at the site of actomyosin cluster in the active networks (Figure 5H). The absence of actin cortex in the example shown suggested the possibility of bleb formation because of the generation of cytoplasmic pressure induced by cortex tension in the reconstituted active networks (Loiseau et al., 2016). Together these data demonstrated that myosin-driven contractility contributes to both the formation of ring-like patterns at the GUV equator and a build-up of cortex tension for membrane protrusion in confinement.
Figure 5.
Dendritic actomyosin networks in the presence of actin crosslinkers self-assemble into a ring-like equatorial cortex
(A) Representative fluorescence confocal actin images of a crosslinked actomyosin cortex 0 and 10 min after imaging. Representative 3D reconstructed fluorescence confocal images of a membrane-bound actomyosin cortex in the presence of VCA, Arp2/3 complex, α-actinin, and fascin at different time points.
(B) Color map of actin intensities in (A).
(C) Representative z-projected confocal actin images of a crosslinked actomyosin cortex 0 and 70 min after imaging.
(D) Another example of a 3D reconstructed fluorescence confocal actin images of a crosslinked actomyosin cortex 0 and 70 min after imaging. White arrow points at the ring-like cluster around the neck of the deformed membrane.
(E) Schematic representation of 2D top view (left) and 3D side view (right) of an encapsulated actin cortex with highly enriched actin at its mid plane. The red dashed lines represent radial lines along which maximum actin intensities are plotted in (F).
(F) Actin cortex intensity around equally spaced circular z-planes from the top plane to mid plane of the GUV in (D). Actin intensity (y axis) at each plane (color map) was measured by detecting maximum intensity along 30 radial lines (red dashed lines in (E) drawn with an equal angular interval (x-axis). Red arrows point at the intensity of actin clusters at the GUV equatorial plane.
(G) The mean value of normalized actin cortex intensity from top plane to mid plane of six GUVs. Normalized mean intensity (y axis) for each cortex was measured by taking the mean value of maximum intensities along 30 radial lines (red dashed lines in (E)) from top plane to mid plane of each GUV (x-axis) with z-interval of 2.4 μm averaged over the mean value at each z-plane.
(H) Representative fluorescence confocal images of actin, lipid, and merged lipid-actin for a GUV with a bleb-like protrusion. Actin: 5 μM, Arp2/3 complex: 1 μM; His6-tagged VCA: 0.5 μM, α-actinin: 0.5 μM, and fascin: 1 μM for all panels. All scale bars are 10 μm. n = 2 experiments. Also see Figures S7 and S8.
Discussion
We encapsulated membrane-bound and –unbound actin networks in the presence of myosin motors and passive crosslinkers, α-actinin and fascin, and showed that diverse actomyosin patterns in the form of clusters, rings, and asters emerged, the architecture of which depend on confinement size and the concentration of myosin and passive crosslinkers.
The active nature of the networks was demonstrated using traction force microscopy of muscle myosin filaments acting on substrate-anchored actin filaments. This platform provides a robust way to quantify traction stress field in any region of interest on the top surface of a soft substrate with no background noise from the substrate lower planes (Kollimada et al., 2021). However, it is limited to 2D actin networks crowded on the substrate and it does not allow actin networks to dynamically reorganize. Traction force resolution is another limitation of this platform as embedded fluorescent beads are not finely and uniformly distributed.
Although α-actinin forms flexible actin bundles in the form of rings in confinement, it promotes entrapment of myosin and clustering of actin bundles. α-actinin-actomyosin clustering and disassembly to monomers could thus facilitate the elongation of parallel fascin-bundles and consequently the formation of aster patterns (Reymann et al., 2012). Parallel fascin-actin bundles are less prone to motor-induced clustering which, at certain concentrations of α-actinin and myosin, gain enough flexibility to promote contractile ring assembly. Membrane-bound contractile ring could deform the lipid membrane during self-assembly, but it lacks a branched actomyosin cortex to position, stabilize, and build tension in the membrane. We were unable to induce large-scale ring contraction and GUV constriction as in the work of Litschel et al. (Litschel et al., 2021) using biotin actin-neutravidin-biotin lipid linkages. We could however reconstitute slight GUV constriction using this approach (Figure4). It should be noted that capturing ring constriction in the current study was challenging because of slow GUV settlement and rapid myosin activity in confinement. Capturing a z-projection of ring assembly and constriction in time was possible at slow myosin activity and low light exposure time (Video S9) to avoid photo-bleaching or photo-induced damage to actin bundles (Bashirzadeh et al., 2020).
A unique part of our study was that we demonstrated cortex assembly and GUV shape changes using Arp2/3 complex-mediated dendritic actomyosin networks. These branched networks in the presence of fascin have been shown to form clusters with short actin bundle arms in the form of aster-like structures but protrusions were inhibited when the networks were attached to GUV membrane in the form of a dendritic actin cortex (Haviv et al., 2006; Ideses et al., 2008; Wubshet et al., 2021). Similarly, here, a dendritic actomyosin cortex formed an actin shell at the periphery. In the presence of α-actinin and fascin, the dendritic actomyosin cortex intensely localized at the equatorial plane in a ring-like pattern. The simulation of α-actinin-actomyosin networks in confinement showed that high actin treadmilling rate shifts myosin-driven centripetal actin clustering toward the periphery which consequently results in the formation of a ring-like actomyosin cortex. It was further shown in simulations and cultured T cells that inhibition of treadmilling induces collapse of ring-patterns into clusters (Ni et al., 2021). Here, actomyosin clustering occurred more frequently in the plane of ring-like actomyosin patterns in the cortex. Peripheral collapse of the actomyosin networks into clusters rather than centripetal clustering was indicative of strong attachment of the cortex to the membrane. Actomyosin collapse and disruption of the cortex are expected to cause cytoplasmic pressure buildup and consequently bleb-like protrusion of the membrane as observed at the site of cluster (Charras and Paluch, 2008; Tinevez et al., 2009; Sheetz et al., 2006). The presence of a ring-like actin pattern at the neck of the deformed bilayer also suggests the presence of a constricting force which may contribute to bleb formation and equilibration. Thus, as there was no evidence of membrane detachment at the site of the cluster, it is expected that actomyosin networks contribute to membrane deformation in two ways, by cortex tension and generation of excess cytoplasmic pressure, and by exertion of force on the membrane.
An elastic model of bleb formation has successfully described the effect of actomyosin cortex tension on Laplace pressure and consequently bleb expansion (Tinevez et al., 2009). To account for membrane constriction at the site of protrusion, here, we include a force, Fc, acting in the plane of constriction and opposing GUV total tension (cortex plus membrane tension), σg, and protruded membrane tension, σb (Figure 6). Given the balance of forces in the cortex and fluid pressure balance between the GUV and bleb at equilibrium, one would obtain σg = 1.41 σb + c, where c is the term corresponding to the elastic resistance of the cortex to its contraction and is negligible at equilibrium in the absence of cytoplasmic elasticity (Tinevez et al., 2009). Because GUV cytosol is mostly made up of water, and lacks crowded intracellular structures as opposed to a cell, it lacks elastic resistance thereby causing the term elastic resistance to vanish (i.e. c→0), which corresponds to the convergence of GUV cortex tension toward equilibrium (Tinevez et al., 2009). Balance of forces at the plane of constriction results in Fc = 2.75 × 10−6 (0.917 σg + 0.819 σb) [N]. Strikingly, at σg = 1.44 σb (compare with σg = 1.41 σb + c), Fc equals the maximum contractile force (Fc = 5.88 σb× 10−6) generated by the actomyosin ring at the same myosin concentration (Figure 4), and also holds the assumption of negligible cortex elastic resistance at equilibrium, i.e. c ∼ 1.44 σb - 1.41 σb = 0.03 σb. The ratio (σg/σb = 1.44) between membrane tension of branched actomyosin-bound lipids (σg) and bare lipids (σb) is very close to the ratio of tension (∼1.3) between a GUV doublet covered with a branched actomyosin cortex and its initial tension before myosin addition and continuation of actin polymerization (V. Caorsi et al., 2016), although, the cortex tension at the site of constriction can differ from the tension of floppy membrane domains caused by the presence of a corral domain and Marangoni effect (Guevorkian et al., 2015). The presence of dendritic actin cortex formed by Arp2/3 complex is the major difference between constriction force at the site of the bleb and the force generated by actomyosin ring in cortex-less GUVs. Because myosin drives constriction, and crosslinking forces have negligible effect on the contractile force (Stachowiak et al., 2014), our results suggest that Arp2/3 complex participates in bleb formation yet has minimal effect on constriction at the bleb site.
Figure 6.
A physical model explains how cortex tension and clustering induce GUV deformation
The schematic illustrates parameters used for modeling membrane deformation and bleb formation induced by actin cortex contraction and clustering.
From our findings, it is evident that the mechanics of actin binding proteins and their interaction in confinement allows the spontaneous formation of actomyosin patterns that provide insights in the formation of asters and rings in the cell cortex (Colin-York et al., 2019; Fritzsche et al., 2017; Ni et al., 2021). Branched actomyosin cortex is thought to be necessary for positioning of the contractile ring and regulation of the division of cells whose elasticity is key in membrane cleaving in the absence a cell wall (O’Shaughnessy and Thiyagarajan, 2018; Pollard and O’Shaughnessy, 2019). Our results suggested that the contractile ring assembly could be a result of the self-positioning of the cortex itself, and that actin crosslinkers are necessary for such positioning. The absence of a mechanism for actin turnover and treadmilling could be a drawback of the minimal mimics presented here. Sarcomere-like and disordered contractions, which have both been proposed for animal cell cytokinesis, has recently been reconstituted in vitro (Ennomani et al., 2016). The former perhaps does not require actin filament assembly in the ring upon network disassembly during ring contraction (Pollard and O’Shaughnessy, 2019). Different strategies could be devised to reconstitute and test these long-debated contractile mechanisms in GUVs as cell mimics. Another improvement for bottom-up reconstitution of cytokinesis could be testing actin filament barbed ends or their capping proteins for membrane interaction. Barbed end attachment to the membrane has been suggested to be necessary for in-plane generation of tension and membrane constriction during cytokinesis (Wang and O’Shaughnessy, 2019). Our model indicates that the biological consequence of global cortex tension and local force generation by accumulated actomyosin cortex is bleb formation in GUVs. A similar mechanism could occur in budding yeasts where budding and cytoplasmic flow from mother cell to the bud occur during the assembly of septin and contractile rings (Wloka and Bi, 2012). Measuring cytoplasmic dynamics during cortex contraction and membrane shape changes could be an interesting future direction for the bottom-up reconstitution of contractile actin networks (Malik-Garbi et al., 2019).
Limitations of study
In this study, actin binding proteins were encapsulated in GUVs in a single compartmentalization step. Simultaneous binding, nucleation, bundling, and contraction of actin filaments make it difficult to study the contribution of individual proteins in spatiotemporal organization of confined networks and membrane deformation. Studies can be designed in which the activation of encapsulated components is triggered for understanding the temporal evolution of the dynamics of the synthetic system. For example, photoinactivation of pharmacological inhibitor of myosin activity, blebbistatin (Lee et al., 2021; Linsmeier et al., 2016), can be used for tracing actomyosin clustering and membrane blebbing in time, which can provide valuable insights in the mechanism of blebbing in synthetic and living systems. Another caveat is possible heterogeneities in the synthetic cell population in terms of the encapsulated content as well as the transfer of ions and molecules from the membrane of GUVs that contain oil residues. Together, these can cause the formation of more than a single actomyosin phenotype across GUVs with similar sizes. The design of solvent-free microfluidics devices for GUV generation can provide a possible solution toward high-throughput assembly of macromolecules in GUVs (Deshpande et al., 2016).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21 (DE3) | NEB | Cat# C2527I |
| E. coli BL21 (DE3) RIPL | Agilent | Cat# 230280 |
| Chemicals, peptides, and recombinant proteins | ||
| Actin | Cytoskeleton | Cat# AKL99-A |
| α-actinin | Cytoskeleton | Cat# AT01-A |
| Myosin II | Cytoskeleton | Cat# MY02-A |
| Arp2/3 | Cytoskeleton | Cat# RP01P-A |
| Actin-biotin | Cytoskeleton | Cat# AB07-A |
| Adenosine triphosphate (ATP) | Cytoskeleton | Cat# BSA04-001 |
| Alexa Fluor 568 actin | Thermo Fisher Scientific | Cat# A12374 |
| ATTO488-Actin | Hypermol | Cat# 8153-02 |
| Neutravidin beads | Thermo Fisher Scientific | Cat# F8774 |
| TMR-alpha-actinin | David Kovar (Winkelman et al., 2016) | |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) | Avanti Polar Lipids | Cat# 850375C |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt) (biotinyl cap PE) | Avanti Polar Lipids | Cat# 870273C |
| 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS-NTA(Ni)) | Avanti Polar Lipids | Cat# 790404C |
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhod-PE) | Avanti Polar Lipids | Cat# 810150C |
| Cholesterol | Avanti Polar Lipids | Cat# 700100P |
| Silicone oil | Sigma-Aldrich | Cat# 317667 |
| Mineral oil | Acros organics | Cat# 8042-47-5 |
| Optiprep | Sigma-Aldrich | Cat# D1556 |
| Alexa Fluor™ 647 microscale protein labeling kit | Thermo Fisher Scientific | Cat# A30009 |
| Recombinant DNA | ||
| pGEX-6P-1 fascin | This lab | n/a |
| His6-VCA | Jack Taunton | n/a |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html |
| Origin | Headquartered in Northampton, Massachusetts, USA | http://www.originlab.com/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab |
| Deposted data | ||
| Data for Encapsulated actomyosin patterns drive cell-like membrane shape changes | This paper | Mendeley Data: https://dx.doi.org/10.17632/mmg7jz5587.2 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Allen Liu (allenliu@umich.edu).
Materials availability
All materials generated in this study are available from the lead contact without restriction.
Experimental model and subject details
Microbe strains
BL21(DE3) and BL21(DE3) RIPL were purchased from NEB and Agilent, respectively, and cultured in LB broth with appropriate antibiotics at 37 °C after transformation with the recombinant plasmid.
Method details
Materials
Actin, α-actinin, Arp2/3 complex, and myosin II were purchased from Cytoskeleton Inc, USA. ATTO 488 actin was purchased from Hypermol Inc, Germany. Alexa Fluor 568 actin was purchased from Thermo Fisher Scientific. TMR-α-actinin was gifted by David Kovar (University of Chicago). Myosin was labeled using Alexa Fluor 647 microscale protein labeling kit (Thermo Fisher Scientific). Fascin was purified from E. coli as Glutathione-S-Transferase (GST) fusion protein (Bashirzadeh et al., 2021a). Briefly, BL21(DE3) E. coli cells were transformed with pGEX-6P-1 fascin. Cells were grown at 37°C while shaking at 220 rpm until the OD600 reached 0.5–0.6. Protein expression was induced with 0.1 mM IPTG and cell cultures were incubated at 24°C for 8 h. Cells were harvested and resuspended in lysis buffer (20 mM K-HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF) and ruptured by sonication. Cell lysates were centrifuged at 45,000 × g for 25 min and supernatants were loaded on a GSTrap FF 1 mL column (GE Healthcare) using an AKTA Start purification system (GE Healthcare). The column was next washed with 15 mL cleavage buffer (20 mM K-HEPES pH 7.5, 500 mM NaCl). Next, 2 mL cleavage buffer containing 160 μg Pre-Scission protease (Tri-Altus Bioscience) was loaded on the column and incubated overnight at 4°C for cleavage. The proteins were then eluted with 5 mL elution buffer (20 mM K-HEPES pH 7.5, 100 mM NaCl). Purified products were dialyzed against 1 L elution buffer twice for 3 h and once overnight at 4°C. We similarly purified hexa-histidine-tagged VCA (His6-tagged VCA) domain from neural Wiskott Aldrich syndrome protein following the procedures described previously (Wubshet et al., 2021). Briefly, BL21(DE3) RIPL cells transformed with His6-tagged VCA plasmid were grown and induced with 0.5 mM IPTG when O.D. reached 0.4-0.5. The culture was then incubated at 37°C for 3 h. Cells were harvested and resuspended in lysis buffer containing 20 mM HEPES pH 7.5, 200 mM NaCl, 10 mM imidazole, 1 mg/mL lysozyme, and 1 mM PMSF. Cells were then flash frozen with liquid nitrogen, thawed, and then lysed by sonication. The lysate was incubated with Ni-NTA resin (600 μL resin for 2 mL of lysates) for 2.5 h, washed several times with 20 mM HEPES pH 7.5 containing 200 mM NaCl, and eluted with an elution buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 250 mM imidazole) from a column into aliquots. Fractions containing purified His6-VCA were dialyzed in 20 mM HEPES pH 7.5, 200 mM NaCl, and 1 mM TCEP overnight.
Measurement of contractile stresses via traction force microscopy
Traction stresses exerted by actomyosin networks were quantified by using Fourier transform traction cytometry (FTTC) (Bashirzadeh et al., 2019; Butler et al., 2002; Schwarz et al., 2002). Actin filaments were coupled to a soft Matrigel (Young’s modulus ∼0.75 kPa and Poisson’s ratio ∼0.5) allowing the measurement of contractile stresses exerted as traction forces on the gel. For this purpose, 200 nm yellow-green neutravidin-coated beads (Invitrogen) were embedded on the top surface of a soft gel following previous protocols (Bashirzadeh et al., 2018a, 2018b). The substrate was prepared in an open chamber, allowing the addition of actin and other components (i.e., myosin and extra ATP) for time-lapse measurements. 3 μM monomeric actin (including 10% Alexa Fluor 568 actin) and 10 mol % biotinylated actin in polymerization buffer (2 mM MgCl2, 4.2 mM ATP, 0.2 mM CaCl2, and 50 mM KCl, in 15 mM Tris, pH 7.5) was added to the chamber. Monomeric actin (including 10% Alexa Fluor 568 actin) with 10 mol % biotinylated actin in polymerization buffer were added to the chamber. Coupling of biotinylated actin to the fluorescent neutravidin beads enabled the transmission of contraction forces to the substrate. The displacement field of the embedded fluorescent beads enabled the extraction of exerted traction forces using FTTC. Strain energy was measured using custom-written MATLAB routines (Sabass et al., 2008).
GUV generation
Lipids containing 70% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 30% cholesterol in a 1:4 mixture of mineral oil and silicone oil was first made in a glass tube. The total concentration of lipids was 0.4 mM in the oil mixture. For membrane-bound actomyosin networks (without VCA and Arp2/3 complex), lipid mixture was composed of 69% DOPC, 30% cholesterol, and 1% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt) (biotinyl cap PE). For dendritic actomyosin networks (with VCA and Arp2/3 complex), lipid mixture was composed of 70% DOPC, 25% cholesterol, and 5% 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (DGS-NTA(Ni)). For labeling GUVs, 0.1% of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rhod-PE) was added to the lipid mixture. All lipids were purchased form Avanti Polar Lipids. Mineral oil and silicone oil were purchased from Sigma-Aldrich.
Then, 5 μM actin, including 10% ATTO 488 actin and 4% biotinylated actin (in the case of membrane-bound networks without VCA and Arp2/3 complex), in polymerization buffer and 7.5% OptiPrep was prepared and kept in ice for 15 min. Myosin (0.06-0.25 μM), α-actinin (0.5-1.5 μM), fascin (0.5-1.5 μM), VCA (1 μM), Arp2/3 complex (1 μM), or their combinations was then added to the sample. Our previous observations found that, in contrast to reconstituted dendritic networks outside GUVs, high molar ratios of Arp2/3 complex with respect to actin (∼0.2) were needed for the formation of cortical dendritic patterns inside GUVs (Wubshet et al., 2021).
GUVs were produced by a modification of the cDICE method (Bashirzadeh et al., 2021a,2021b; Abkarian et al., 2011). A rotor chamber was 3D-printed with clear resin and mounted on the motor of a stir plate and rotated at around 1,200 rpm. 0.7 mL outer solution (200 mM glucose matching the osmolarity of inner solution) and 5 mL of lipid-in-oil dispersion were sequentially transferred into the rotating chamber. The difference in density between the two solutions results in the formation of two distinct layers with a water/oil interface. Then, a water-in-oil emulsion was created by adding 0.7 mL of lipid/oil mixture to 20 μL of protein mixture and rigorously pipetting up and down 6–7 times. The emulsion was then pipetted into the rotating chamber. It should be noted that the prepared mixture of actin binding proteins was added to the actin solution 2–3 s before encapsulation in droplets, and droplets traveled through the lipid dispersion in the rotating chamber. As the droplets cross the water-oil interface in the rotating chamber, they form a bilayer and are released in the outer solution as GUVs.
Microscopy and image analysis
GUVs were transferred to a 96 well plate for microscopy. Other than time-lapse images, all other images were captured at least 40 min after GUV generation at the steady state of actin patterns. Images were taken using an oil immersion Plan-Apochromat 60 ×/1.4 NA objective on an inverted microscope (Olympus IX-81) equipped with an iXON3 EMCCD camera (Andor Technology), AOTF-controlled lasers (Andor Technology), and a Yokogawa CSU-X1 spinning disk confocal. Acquisition of images was controlled by MetaMorph (Molecular Devices). Single and z stack images of lipid and actin were captured with 561 nm excitation at exposure time of 20–25 ms and 488 nm excitation at exposure time of 350–500 ms, respectively. 3D z-projected images were produced by brightest point projection of z stack image sequences in Fiji/ImageJ. Z-projected processed images in Figures 5C and 5D were generated using Fiji/ImageJ complemented with the plugin Squassh (Rizk et al., 2014).
Quantification and statistical analysis
For quantification of actin phenotypes, the number of GUVs encapsulating an actomyosin pattern with defined phenotype was normalized to the total number of GUVs that formed actin structures. Actin phenotypes were classified manually. At least 78 GUVs were analyzed per condition. We observed a small number of GUVs without any actin bundling or clustering activity. These GUVs were not counted for statistical analysis. Origin software was used to compile data and plotting bar graphs. No statistical analysis was performed.
Intensities in Figure 5F was obtained using the Fiji/ImageJ plugin ‘Oval Profile’. Using this plugin, maximum intensities (cortical actin intensity in actin images and membrane intensity in lipid images) were detected along 30 radii from the center to the points of a circle surrounding the GUV of interest from the top plane to mid plane (equatorial plane) of the GUV (Figure 5E).
Acknowledgments
We thank David Kovar (University of Chicago) for TMR-α-actinin. We also thank Ulrich Schwarz (Heidelberg University), Benedikt Sabass (Ludwig-Maximilian University), and Venkat Maruthamuthu (Old Dominion University) for providing Matlab codes for FTTC. This work is supported by the National Science Foundation (1844132, 099332, and 2201236) and National Institutes of Health (NIBIBEB030031) to A.P.L.
Author contributions
Y.B. and A.P.L. designed the experiments. Y.B. and H.M. conducted the experiments. Y.B. analyzed data. Y.B. and H.M. prepared purified proteins. Y.B. and A.P.L. wrote the paper. All authors discussed the results and commented on the paper.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104236.
Supplemental information
Data and code availability
-
•
Data reported in this paper are available at Mendeley Data: https://dx.doi.org/10.17632/mmg7jz5587.2.
-
•
A custom-written MATLAB routine originally developed by Ulrich Schwartz, Benedikt Sabass, and Venkat Maruthamuthu was used with permission for image processing and calculation of displacements and traction forces by FTTC and measuring strain energies.
-
•
MATLAB routines were also used to plot intensity profiles which are available from the Lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Backouche F., Haviv L., Groswasser D., Bernheim-Groswasser A. Active gels: dynamics of patterning and self-organization. Phys. Biol. 2006;3:264. doi: 10.1088/1478-3975/3/4/004. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y., Chatterji S., Palmer D., Dumbali S., Qian S., Maruthamuthu V. Stiffness measurement of soft silicone substrates for mechanobiology studies using a widefield fluorescence microscope. J. Visualized Experiments. 2018:57797. doi: 10.3791/57797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y., Dumbali S., Qian S., Maruthamuthu V. Mechanical response of an epithelial island subject to uniaxial stretch on a hybrid silicone substrate. Cell Mol. Bioeng. 2019;12:33–40. doi: 10.1007/s12195-018-00560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y., Liu A.P. Encapsulation of the cytoskeleton: towards mimicking the mechanics of a cell. Soft Matter. 2019;15:8425–8436. doi: 10.1039/c9sm01669d. [DOI] [PubMed] [Google Scholar]
- Bashirzadeh Y., Qian S., Maruthamuthu V. Non-intrusive measurement of wall shear stress in flow channels. Sensors Actuators A: Phys. 2018;271:118–123. doi: 10.1016/J.SNA.2018.01.012. [DOI] [Google Scholar]
- Bashirzadeh Y., Redford S.A., Lorpaiboon C., Groaz A., Moghimianavval H., Litschel T., Schwille P., Hocky G.M., Dinner A.R., Liu A.P. Actin crosslinker competition and sorting drive emergent GUV size-dependent actin network architecture. Commun. Biol. 2021;4:1136. doi: 10.1038/s42003-021-02653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y., Wubshet N., Litschel T., Schwille P., Liu A.P. Rapid encapsulation of reconstituted cytoskeleton inside giant unilamellar vesicles. J.Vis. Exp. 2021:e63332. doi: 10.3791/63332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirzadeh Y., Wubshet N.H., Liu A.P. Confinement geometry tunes fascin-actin bundle structures and consequently the shape of a lipid bilayer vesicle. Front. Mol. Biosciences. 2020;7:610277. doi: 10.3389/FMOLB.2020.610277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Borghi N., Rossier O., Brochard-Wyart F. Hydrodynamic extrusion of tubes from giant vesicles. EPL (Europhysics Letters) 2003;64:837. [Google Scholar]
- Butler J.P., Toli-Nørrelykke I.M., Fabry B., Fredberg J.J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. - Cell Physiol. 2002;282:C595–C605. doi: 10.1152/AJPCELL.00270.2001. [DOI] [PubMed] [Google Scholar]
- Cáceres R., Abou-Ghali M., Plastino J. Reconstituting the actin cytoskeleton at or near surfaces in vitro. Biochim. Biophys. Acta. 2015;1853:3006–3014. doi: 10.1016/j.bbamcr.2015.07.021. [DOI] [PubMed] [Google Scholar]
- Carvalho K., Lemière J., Faqir F., Manzi J., Blanchoin L., Plastino J., Betz T., Sykes C. Actin polymerization or myosin contraction: two ways to build up cortical tension for symmetry breaking. Philosophical Trans. R. Soc. B: Biol. Sci. 2013;368:20130005. doi: 10.1098/RSTB.2013.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho K., Tsai F.C., Lees E., Voituriez R., Koenderink G.H., Sykes C. Cell-sized liposomes reveal how actomyosin cortical tension drives shape change. Proc. Natl. Acad. Sci. U S A. 2013;110:16456–16461. doi: 10.1073/pnas.1221524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G., Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- Colin-York H., Li D., Korobchevskaya K., Chang V.T., Betzig E., Eggeling C., Fritzsche M. Cytoskeletal actin patterns shape mast cell activation. Commun. Biol. 2019;2:93. doi: 10.1038/s42003-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin O., Na S., Chowdhury F., Hong M., Shin M.E., Wang F., Wang N. Self-organized podosomes are dynamic mechanosensors. Curr. Biol. 2008;18:1288–1294. doi: 10.1016/J.CUB.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S., Caspi Y., Meijering A.E.C., Dekker C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016;7:10447. doi: 10.1038/ncomms10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimchev V., Lahmann I., Koestler S.A., Kage F., Dimchev G., Steffen A., Stradal T.E.B., Vauti F., Arnold H.-H., Rottner K. Induced Arp2/3 complex depletion increases FMNL2/3 formin expression and filopodia formation. Front. Cell. Dev. Biol. 2021;9:65. doi: 10.3389/fcell.2021.634708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennomani H., Lle Letort G., De E.M., Cruz L., Thé M., Correspondence L.B. Architecture and connectivity govern actin network contractility. Curr. Biol. 2016;26:616–626. doi: 10.1016/j.cub.2015.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M., Li D., Colin-York H., Chang V.T., Moeendarbary E., Felce J.H., Sezgin E., Charras G., Betzig E., Eggeling C. Self-organizing actin patterns shape membrane architecture but not cell mechanics. Nat. Commun. 2017;8:14347. doi: 10.1038/ncomms14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K., Porter M.E., Pollard T.D. Alpha-actinin localization in the cleavage furrow during cytokinesis. J. Cell Biol. 1978;79:268–275. doi: 10.1083/JCB.79.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groaz A., Moghimianavval H., Tavella F., Giessen T.W., Vecchiarelli A.G., Yang Q., Liu A.P. Engineering spatiotemporal organization and dynamics in synthetic cells. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology. 2021;13:e1685. doi: 10.1002/wnan.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevorkian K., Manzi J., Pontani L.L., Brochard-Wyart F., Sykes C. Mechanics of biomimetic liposomes encapsulating an actin shell. Biophysical J. 2015;109:2471–2479. doi: 10.1016/J.BPJ.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J.A., Wang J.C., Saeed M., Pedrosa A.T. Origin, organization, dynamics, and function of actin and actomyosin networks at the T cell immunological synapse. Annu. Rev. Immunol. 2019;37:201–224. doi: 10.1146/annurev-immunol-042718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv L., Brill-Karniely Y., Mahaffy R., Backouche F., Ben-Shaul A., Pollard T.D., Bernheim-Groswasser A. Reconstitution of the transition from lamellipodium to filopodium in a membrane-free system. Proc. Natl. Acad. Sci.U S A. 2006;103:4906–4911. doi: 10.1073/PNAS.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideses Y., Brill-Karniely Y., Haviv L., Ben-Shaul A., Bernheim-Groswasser A. Arp2/3 branched actin network mediates filopodia-like bundles formation in vitro. PLoS One. 2008;3:e3297. doi: 10.1371/JOURNAL.PONE.0003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollimada S., Senger F., Vignaud T., Théry M., Blanchoin L., Kurzawa L. The biochemical composition of the actomyosin network sets the magnitude of cellular traction forces. Mol. Biol. Cell. 2021;32:1737–1748. doi: 10.1091/mbc.E21-03-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P. Actin-binding proteins: the long road to understanding the dynamic landscape of cellular actin networks. Mol. Biol. Cell. 2016;27:2519–2522. doi: 10.1091/mbc.e15-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Leech G., Lwin P., Michel J., Currie C., Rust M.J., Ross J.L., McGorty R.J., Das M., Robertson-Anderson R.M. Active cytoskeletal composites display emergent tunable contractility and restructuring. Soft Matter. 2021;17:10765–10776. doi: 10.1039/d1sm01083b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmeier I., Banerjee S., Oakes P.W., Jung W., Kim T., Murrell M.P. Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nat. Commun. 2016;7:12615. doi: 10.1038/ncomms12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschel T., Kelley C.F., Holz D., Adeli Koudehi M., Vogel S.K., Burbaum L., Mizuno N., Vavylonis D., Schwille P. Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 2021;12:2254. doi: 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.P., Fletcher D.A. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophysical J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.P., Richmond D.L., Maibaum L., Pronk S., Geissler P.L., Fletcher D.A. Membrane-induced bundling of actinfilaments. Nat. Phys. 2008;4:789–793. doi: 10.1038/nphys1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau E., Schneider J.A.M., Keber F.C., Pelzl C., Massiera G., Salbreux G., Bausch A.R. Shape remodeling and blebbing of active cytoskeletal vesicles. Sci. Adv. 2016;2:e1500465. doi: 10.1126/sciadv.1500465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Garbi M., Ierushalmi N., Jansen S., Abu-Shah E., Goode B.L., Mogilner A., Keren K. Scaling behaviour in steady-state contracting actomyosin networks. Nat. Phys. 2019;15:509–516. doi: 10.1038/s41567-018-0413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abkarian Manouk, Loiseau Etienne, Massiera Gladys. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter. 2011;7:4610–4614. doi: 10.1039/C1SM05239J. [DOI] [Google Scholar]
- Mejillano M.R., Kojima S.I., Applewhite D.A., Gertler F.B., Svitkina T.M., Borisy G.G. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Chiba M., Eguchi H., Ohki T., Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nat. Cell. Biol. 2015;17:480–489. doi: 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- Ni Q., Wagh K., Upadhyaya A., Papoian G.A. A tug of war between filament treadmilling and myosin induced contractility generates actin cortex. bioRxiv. 2021 doi: 10.1101/2021.06.06.447254. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy B., Thiyagarajan S. Mechanisms of contractile ring tension production and constriction. Biophysical Rev. 2018;10:1667–1681. doi: 10.1007/S12551-018-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. Actin and actin-binding proteins. Cold Spring Harbor Perspect. Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., O’Shaughnessy B. Molecular mechanism of cytokinesis. Annu. Rev. Biochem. 2019;88:661–689. doi: 10.1146/annurev-biochem-062917-01253088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A.C., Boujemaa-Paterski R., Martiel J.L., Guérin C., Cao W., Chin H.F., De La Cruz E.M., Théry M., Blanchoin L. Actin network architecture can determine myosin motor activity. Science. 2012;336:1310–1314. doi: 10.1126/SCIENCE.1221708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk A., Paul G., Incardona P., Bugarski M., Mansouri M., Niemann A., Ziegler U., Berger P., Sbalzarini I.F. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 2014;9:586–596. doi: 10.1038/nprot.2014.037. [DOI] [PubMed] [Google Scholar]
- Sabass B., Gardel M.L., Waterman C.M., Schwarz U.S. High resolution traction force microscopy based on experimental and computational advances. Biophysical J. 2008;94:207–220. doi: 10.1529/BIOPHYSJ.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto R., Tanabe M., Hiraiwa T., Suzuki K., Ishiwata S., Maeda Y.T., Miyazaki M. Tug-of-war between actomyosin-driven antagonistic forces determines the positioning symmetry in cell-sized confinement. Nat. Commun. 2020;11:30–63. doi: 10.1038/s41467-020-16677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz U.S., Balaban N.Q., Riveline D., Bershadsky A., Geiger B., Safran S.A. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophysical J. 2002;83:1380–1394. doi: 10.1016/S0006-3495(02)73909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B., Moghimianavval H., Hwang S.W., Liu A.P. Synthetic cell as a platform for understanding membrane-membrane interactions. Membranes. 2021;11:912. doi: 10.3390/membranes11120912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M.P., Sable J.E., Döbereiner H.G. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 2006;35:417–434. doi: 10.1146/ANNUREV.BIOPHYS.35.040405.102017. [DOI] [PubMed] [Google Scholar]
- Stachowiak M.R., Laplante C., Chin H.F., Guirao B., Karatekin E., Pollard T.D., O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev. Cell. 2014;29:547. doi: 10.1016/J.DEVCEL.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. The actin cytoskeleton and actin-based motility. Cold Spring Harbor Perspect. Biol. 2018;10:a018267. doi: 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen T., Lenz M., Gardel M.L. Reconstitution of contractile actomyosin bundles. Biophysical J. 2011;100:2698–2705. doi: 10.1016/j.bpj.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.-Y., Schulze U., Salbreux G., Roensch J., Joanny J.-F., Paluch E. Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. U S A. 2009;106:18581–18586. doi: 10.1073/PNAS.0903353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caorsi V., Lemière J., Campillo C., Bussonnier M., Manzi J., Betz T., Plastino J., Carvalho K., Sykes C. Cell-sized liposome doublets reveal active tension build-up driven by acto-myosin dynamics. Soft Matter. 2016;12:6223–6231. doi: 10.1039/C6SM00856A. [DOI] [PubMed] [Google Scholar]
- Wang S., O’Shaughnessy B. Anchoring of actin to the plasma membrane enables tension production in the fission yeast cytokinetic ring. Mol. Biol. Cell. 2019;30:2053–2064. doi: 10.1091/mbc.E19-03-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich K.L., Stam S., Munro E., Gardel M.L. Actin bundle architecture and mechanics regulate myosin II force generation. Biophysical J. 2021;120:1957–1970. doi: 10.1016/j.bpj.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welf E.S., Miles C.E., Huh J., Sapoznik E., Chi J., Driscoll M.K., Isogai T., Noh J., Weems A.D., Pohlkamp T., et al. Actin-membrane release initiates cell protrusions. Dev.Cell. 2020;55:723–736.e8. doi: 10.1016/J.DEVCEL.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman J.D., Suarez C., Hocky G.M., Harker A.J., Morganthaler A.N., Christensen J.R., Voth G.A., Bartles J.R., Kovar D.R. Fascin- and α-actinin-bundled networks contain intrinsic structural features that drive protein sorting. Curr. Biol. 2016;26:2697–2706. doi: 10.1016/J.CUB.2016.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C., Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton. 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Wubshet N.H., Bashirzadeh Y., Liu A.P. Fascin-induced actin protrusions are suppressed by dendritic networks in GUVs. Mol. Biol. Cell. 2021;32:1634–1640. doi: 10.1091/mbc.E21-02-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Zhong G., Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/SCIENCE.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in this paper are available at Mendeley Data: https://dx.doi.org/10.17632/mmg7jz5587.2.
-
•
A custom-written MATLAB routine originally developed by Ulrich Schwartz, Benedikt Sabass, and Venkat Maruthamuthu was used with permission for image processing and calculation of displacements and traction forces by FTTC and measuring strain energies.
-
•
MATLAB routines were also used to plot intensity profiles which are available from the Lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.