Abstract
N6-methyladenosine (m6A) is the most abundant posttranscriptional methylation modification that occurs in mRNA and modulates the fine-tuning of various biological processes in mammalian development and human diseases. In this study we investigated the role of m6A modification in the osteogenesis of mesenchymal stem cells (MSCs), and the possible mechanisms by which m6A modification regulated the processes of osteoporosis and bone necrosis. We performed systematic analysis of the differential gene signatures in patients with osteoporosis and bone necrosis and conducted m6A-RNA immunoprecipitation (m6A-RIP) sequencing to identify the potential regulatory genes involved in osteogenesis. We showed that fat mass and obesity (FTO), a primary m6A demethylase, was significantly downregulated in patients with osteoporosis and osteonecrosis. During the differentiation of human MSCs into osteoblasts, FTO was markedly upregulated. Both depletion of FTO and application of the FTO inhibitor FB23 or FB23-2 impaired osteogenic differentiation of human MSCs. Knockout of FTO in mice resulted in decreased bone mineral density and impaired bone formation. PPARG, a biomarker for osteoporosis, was identified as a critical downstream target of FTO. We further revealed that FTO mediated m6A demethylation in the 3’UTR of PPARG mRNA, and reduced PPARG mRNA stability in an YTHDF1-dependent manner. Overexpression of PPARG alleviated FTO-mediated osteogenic differentiation of MSCs, whereas knockdown of PPARG promoted FTO-induced expression of the osteoblast biomarkers ALPL and OPN during osteogenic differentiation. Taken together, this study demonstrates the functional significance of the FTO-PPARG axis in promoting the osteogenesis of human MSCs and sheds light on the role of m6A modification in mediating osteoporosis and osteonecrosis.
Keywords: FTO, N6-methyladenosine, mesenchymal stem cells, osteogenesis, PPARG, FB23
Introduction
Mesenchymal stem cells (MSCs) from bone marrow are common progenitor cells of osteoblasts and adipocytes. To maintain skeletal health, our bodies strictly modulate the fine balance between osteogenic and adipogenic differentiation of MSCs [1–3]. Osteoporosis is evaluated by bone mineral density (BMD) assessment and is characterized by excessive adipose tissue but low bone mass in bone marrow, while age-related osteonecrosis is characterized by disruption of osteoblast differentiation by gradual lipid accumulation within osteoblasts and osteocytes, increased cell death, and defective bone repair [4, 5]. In addition, osteoporotic bone is prone to fracture, resulting in blocked blood supply to the bone and causing bone necrosis, which is characterized by bone ischemia, joint pain, and bone destruction [6]. The prevalence of osteoporosis increases with age and varies by race. As evaluated by BMD, approximately 4.3% of men over the age of 50 have osteoporosis, while approximately 15.4% of women over the age of 50 have osteoporosis [7]. Bone necrosis frequently occurs in the jaw [8], femoral head [9], and hip [10] and in patients with systemic lupus erythematosus [11] and is linked to long-term steroid treatment and excessive alcohol consumption. Current treatments for osteoporosis target bone remodeling, including formation, reversal, resorption, and quiescence [12, 13], are represented by hormone replacement therapy (estrogen) and bisphosphonate therapy, both of which have been shown to inhibit bone resorption and prevent bone loss [14]. Anabolic drugs such as teriparatide promote bone formation. However, long-term use of estrogen can increase the risk of coronary heart disease, stroke, and diabetes, while long-term use of bisphosphonates may increase the risk of jaw osteonecrosis by inhibiting the normal repair of bone microdamage, and long-term use of teriparatide can increase the risk of osteosarcoma [15–17]. Therefore, it has been considered urgent to investigate the regulatory mechanisms that are involved in osteoporosis and bone necrosis and to seek a safe and effective strategy to restore bone formation for the treatment of osteoporosis and bone necrosis.
N6-methyladenosine (m6A) is the most abundant posttranscriptional methylation modification that occurs in mRNA and modulates the fine-tuning of various biological processes. m6A modification is primarily installed by methyltransferases, including METTL3, METTL14, and WTAP; removed by the demethylases fat mass and obesity (FTO) and ALKBH5; and recognized by readers that prioritize the identification of m6A-methylated RNA transcripts, including the YTH domain-containing family members, HNRNPA2B1, and IGF2BP [18–20]. At the molecular level, m6A marks are spatiotemporally installed and removed from their modulated RNA transcripts to mediate RNA stability, alternative splicing processes, and translation efficiency upon exposure to various stimuli during different physiological events or human diseases [21–25]. Subsequently, changes in the expression of RNAs with m6A modification affect cell homeostasis, metabolism, and stemness, which eventually control cell fate determination under exposure to various stimuli. Examining these dynamic changes may provide technical support for the diagnosis of human diseases and the development of targeted therapeutic drugs.
FTO was the first m6A demethylase identified in humans and is one of the members of the AlkB-related protein family. It is localized in the nucleus and mediates genome-wide m6A demethylation [26]. A previous study screened small molecule inhibitors of FTO to disrupt the process of mRNA methylation, and rhein was identified as a potential FTO inhibitor to increase the global level of m6A in mRNA [27]. Structurally, rhein interacts with the AlkB enzymatic site and causes inactivation of FTO or AlkB. Subsequently, another anti-inflammatory inhibitor, meclofenamic acid (MA), was reported to specifically interact with FTO but not ALKBH5; it blocked the activity of FTO and resulted in an elevated level of m6A in cells [28, 29]. Based on the structure of MA, the more potent FTO inhibitor fluorescein was developed to simultaneously block the activity of FTO and label FTO with a probe. Alternatively, other compounds harboring tethered nucleotides with structures similar to 2OG-binding sites were identified to inhibit the activity of FTO [30]. More recently, two potent FTO inhibitors named FB23 and FB23-2 were developed based on a structural design approach and exhibited the promising effect of mimicking FTO depletion [27]. Subsequently, nonconserved binding sites in FTO were used to screen potential inhibitors, and high-throughput screening assays were established based on fluorescence emission from RNA aptamers upon FTO-mediated demethylation. While emerging FTO inhibitors have been identified, validation of the effects of these FTO inhibitors in cell activity and their applications in human diseases are still limited; in particular, the in vivo effects of these inhibitors have yet to be determined.
In this study, we aimed to determine whether m6A modification plays a role in the osteogenesis of human MSCs and to explore the possible mechanism by which m6A modification regulates the processes of osteoporosis and bone necrosis. We first showed that FTO was downregulated in patients with osteoporosis and osteonecrosis. Silencing FTO or inhibiting FTO significantly inhibited the osteogenic differentiation of human MSCs. We further identified the functional significance of the FTO-PPARG axis in mediating osteogenic differentiation of human MSCs and found that blocking this signaling pathway may promote osteoporosis and osteonecrosis.
Materials and methods
Osteoporosis and bone necrosis samples and cell culture
Our clinical samples were obtained from the sample bank of femoral heads of patients with fracture and osteonecrosis at the First Clinical Medical College, Guangzhou University of Chinese Medicine. Clinical sample collection was approved by the Institutional Ethics Committee of the hospital (K[2020]131). hMSCs (HUXMA-01001, Cyagen Biosciences, Suzhou, China) were cultured in α-MEM (C12571500BT, Gibco, Amarillo, TX, USA) containing 10% fetal bovine serum (10270-106, Gibco) and 100 units/mL Pen-Strep (15140-122, Gibco) at 37 °C in an atmosphere of 5% CO2. The reagents used in this study included 50 μg/mL L-ascorbic acid (A92902, Sigma, Germany), β-glycerophosphate disodium salt hydrate (5 mM, G9422, Sigma), and 100 nM dexamethasone (D1756, Sigma).
Bioinformatic analysis
The gene expression profiles of patients with bone diseases were downloaded from the GSE35955 and GSE123568 datasets: the GSE35955 dataset contains data for MSCs isolated from the femoral heads of elderly individuals with osteoporosis symptoms, and the GSE123568 dataset contains data for MSCs isolated from patients with femoral head necrosis. Analysis of the m6A-related differentially expressed genes was performed with the edgeR package of R from the GEO database. Gene expression was quantified as fold change (FC) values using the threshold criteria of a FC ≥ 1.5 and a P value <0.05. In addition, the relative expression levels of FTO in IHC samples from the osteonecrosis population were calculated by comparison with the fracture population with ImageJ (v.1.8.0), and statistical significance was analyzed with GraphPad Prism 7.0. The values indicate the standard deviation of the average expression level between the fracture and osteonecrosis populations.
Immunohistochemistry
The femoral heads were excised from patients with fracture and osteonecrosis and were fixed with 4% polyoxymethylene for 48 h and subsequently embedded and decalcified in 10% ethylenediaminetetraacetic acid (EDTA) for 2 weeks before sectioning (5 μm). The sections were placed in sodium citrate buffer at 100 °C for 15 min for antigen retrieval and were then incubated with 3% H2O2 for 10 min at room temperature and blocked with 5% BSA in PBST for 1 h. The sections were incubated with a rabbit anti-FTO primary antibody (ab124892, 1:200, Abcam, UK) overnight at 4 °C. The sections were then incubated with a goat anti-rabbit IgG-HRP secondary antibody (1:200; Beyotime, A0208, Shanghai, China) for 1 h at room temperature. Images were acquired by microscopy (Leica-DM6000B, Germany).
RNA m6A dot blot assay
TRIzol reagent (10606ES60, YEASEN, China) was used to extract total RNA from femoral heads of patients with fracture or osteonecrosis. The concentration and purity of the RNA were determined with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). Total RNA (400/200/100 ng) was dropped onto a nylon membrane (FFN11, Beyotime) prior to UV crosslinking (254 nm) for 1 min and the membrane was blocked with 5% skim milk in PBST. The membrane was incubated with an anti-m6A primary antibody (ABE572, 1:1000, Merck Millipore, USA) overnight at 4 °C. The membrane was then incubated with a goat anti-mouse IgG-HRP secondary antibody (1:5000; Beyotime, A0216) at room temperature for 1 h. The nylon membrane with the m6A dots was visualized using a gel image analyzer (SYSTEM GelDoc XR + IMAGE LAB, Bio-Rad, USA). The same total RNA was dropped onto a nylon membrane and stained with methylene blue (D024, JianCheng, Nanjing, China) for 1 h prior to washing with ddH2O for 30 min. Finally, the nylon membrane with the m6A dots was visualized using a gel image analyzer (SYSTEM GelDoc XR + IMAGE LAB, Bio-Rad).
Alkaline phosphatase (ALP) staining and alizarin red S staining (ARS)
ALP staining was performed with a BCIP/NBT Alkaline Phosphatase Color Development Kit (C3206, Beyotime). In brief, hMSCs were inoculated in a six-well plate and cultured in osteogenic induction medium (OIM). BCIP/NBT working solution (500 µL) was placed in each well and incubated in the dark for ALP staining for 25 min; the staining signals were then recorded accordingly. Similarly, based on the manufacturer’s instructions for ARS, hMSCs were inoculated in a six-well plate and cultured in OIM. The cells were fixed with 4% polyoxymethylene for 15 min prior to staining with Alizarin red S (C0138, Beyotime) for 15 min. The cells were washed three times with double-distilled water and incubated for another 15 min before the staining signals were evaluated.
Quantitative real-time PCR
TRIzol reagent (10606ES60, YEASEN) was used to extract total RNA. The concentration and purity of the RNA were determined with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). One microgram of total RNA from each sample was reverse transcribed into cDNA with a Revert Aid First Strand cDNA Synthesis Kit (Takara, Japan). qRT-PCR was conducted according to the manufacturer’s instructions for SYBR Green (11195ES, YEASEN). Reactions were performed and data were analyzed by using a Bio-Rad CFX instrument. The relative FC in RNA expression was determined by the 2−ΔΔCT method with GAPDH used for normalization.
Western blot analysis
hMSCs were inoculated in six-well plates and treated with the FTO inhibitor FB23-2. Lysis was performed using RIPA lysis buffer at 4 °C. A PierceTM BCA Protein Assay Kit (23225, Thermo Fisher Scientific) was used to quantify the protein concentration. Samples (40 µg) were separated by SDS-PAGE for 1.5 h at 75 V. Proteins were transferred onto a nitrocellulose membrane (IPVH00010, Millipore) for 2 h at 200 mA, and the membrane was then blocked with 5% skim milk in TBST and incubated with a rabbit anti-FTO primary antibody (ab124892, 1:1000, Abcam) and a rabbit anti-GAPDH primary antibody (AF0006, 1:1000; Beyotime) at 4 °C overnight. After washing, the membrane was incubated with a goat anti-rabbit IgG-HRP secondary antibody (1:5000; Beyotime, A0208) for 1 h at room temperature. The bands were imaged with a gel image analyzer (SYSTEM GelDoc XR + IMAGE LAB, Bio-Rad).
RNA-seq and data analysis
Based on the manufacturer’s instructions, TRIzol reagent (10606ES60, YEASEN) was used to extract total RNA from osteogenic hMSCs treated with FB23-2 for 7 days. The cDNA library was constructed by Annoroad Gene Technology and subsequently sequenced on the Illumina HiSeq 2500 platform. Analysis of differential gene expression was performed in accordance with the standard Illumina sequence analysis process. The criteria for significant differential gene expression were set as a FC ≥ 1.5 and a P value <0.05. The differentially expressed genes were identified using Origin 2019 (9.0).
Mice and treatment protocols
B6.129-Ftotm1.1Pzg/YxtJ mice (027830) (16 weeks old) were obtained from The Jackson Laboratory, USA. Twist2-Cre mice (NM-KI-200138) were obtained from GemPharmatech, China. Conditional knockout of Fto in bone in mice was generated as previously described [31]. Throughout the study, mice were maintained on a 12 h:12 h light:dark cycle in a specific pathogen-free facility. All animal care and experiments were approved by the Institutional Animal Care and Use Committee of Guangdong Pharmaceutical University (Approval No. gdpulacSPF2017254). All mice were sacrificed, and the femurs were harvested for micro-CT analysis, histological staining, and immunohistochemical analysis.
Micro-CT scanning
The excised femurs of mice were fixed with 10% formalin and scanned by using a SkyScan 1176 micro-CT instrument (SkyScan, Bruker, Kontich, Belgium). The region of interest was defined as a volume of interest (1 mm) located 2 mm from the distal femur growth plate. The parameters of trabecular bone, including the cancellous bone density (Tb.BMD), bone volume fraction (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular spacing (Tb.Sp), and cortical bone density (Cort.BMD), were analyzed with the CT Analyzer program (Bruker micro-CT).
Histological examination and immunohistochemical staining
Excised femur samples were decalcified in 14% EDTA at 37 °C for 7 days. Femur sections (5 µm) were evaluated by hematoxylin and eosin (H&E) staining after being embedded into paraffin cassettes. For immunohistochemical staining, tissue was incubated with pepsin antigen repair solution at 37 °C for 20 min. After blocked in 3% BSA for 30 min, tissue sections were incubated with a goat anti-ALPL primary antibody (1:250; AF2910, R&D Systems, Minneapolis, MN, USA) overnight at 4 °C followed by a donkey anti-goat IgG-HRP secondary antibody (1:100; A0181, Beyotime) for 1 h at RT. Images were acquired by microscopy (DM6000B, Leica, Germany).
Tartrate-resistant acid phosphate (TRAP) staining
TRAP staining was performed according to the instructions of a commercial TRAP staining kit (AAPR379, Pythonbio, China). In brief, TRAP staining solution mix was prewarmed to 37 °C and incubated with slides that were previously deparaffinized and rehydrated through a series of graded ethanol to distilled water. After rinsed with distilled water, the slides were counterstained with 0.08% Fast Green for 1.5 min and rinsed with water. The slides were air dried completely before being dipped into xylene and mounted. The number of TRAP-positive multinuclear cells in each well was determined.
MeRIP-seq and MeRIP-qPCR
TRIzol reagent (10606ES60, YEASEN) was used to extract total RNA from osteogenic hMSCs treated with FB23-2 for 7 days. The mRNA was enriched by using a Poly(A) mRNA Isolation System IV (Z5310, Promega, USA). Two micrograms of purified mRNA was fragmented in ZnCl2 buffer for 5 min at 94 °C to produce fragments with a length of approximately 100 nt. The mRNA fragments were isolated with Oligo Clean & ConcentratorTM (D4061, Zymo Research, USA) before immunoprecipitation with the anti-m6A rabbit antibody (ABE572, 1:100, Merck Millipore). After extensive washing, free m6A 5’-monophosphate sodium salt (sc-215524, Santa Cruz, USA) was used to remove methylated fragments, and a library was constructed using TruSeq Stranded mRNA Sample Prep Kits (RS-122-2101, Illumina, USA). The input and m6A MeRIP libraries were sequenced on the Illumina NextSeq 500 platform with a high output kit prior to MeRIP-seq or MeRIP-qPCR.
Plasmid constructions, cell transfection, and infection
Targeted genes were stably knocked down via lentivirus-delivered short-hairpin RNA (shRNA). By using the primers, PLKO.1 vectors with hygromycin and puromycin resistance cassettes were constructed. For shRNA knockdown and overexpression, the pCDH and pLKO.1 plasmids were transfected with the packaging and envelope plasmids pMD2.G and psPAX2 into 293T cells with a Calcium Phosphate Transfection Kit (CAPHOS-1KT, Sigma). The virus was collected and used to transduce hMSCs in the presence of 10 mg/mL polybrene (TR-1003-G, Sigma).
Statistical analysis
Data in the bar graphs are listed as mean ± SD. A value of P < 0.05 (versus the indicated group) was considered to indicate a statistically significant difference by one-way analysis of variance or Student’s t-test (GraphPad Prism 7.0).
Results
Downregulation of FTO in patients with osteoporosis and bone necrosis
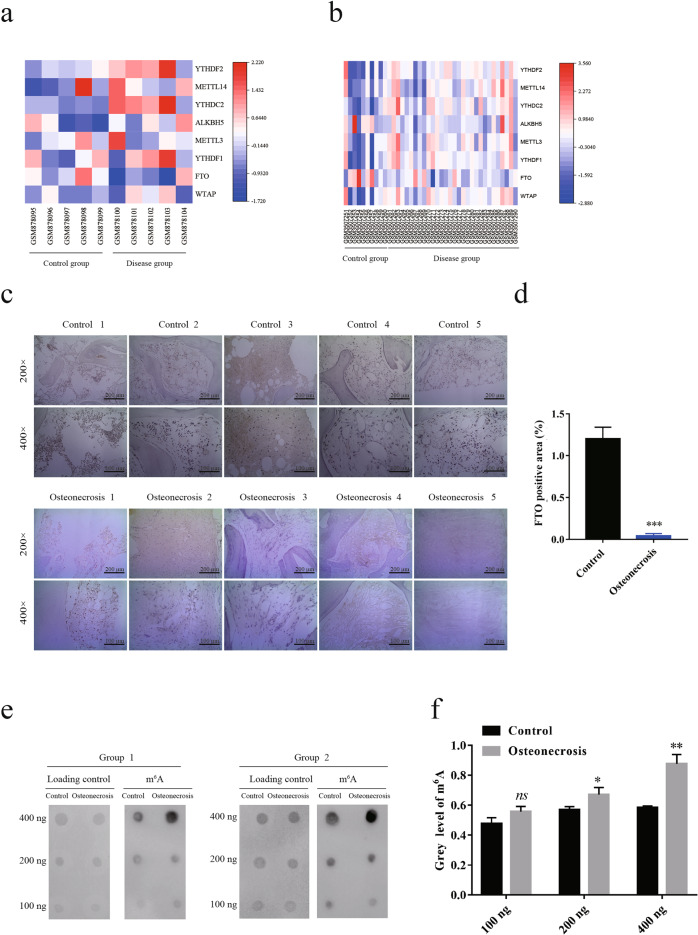
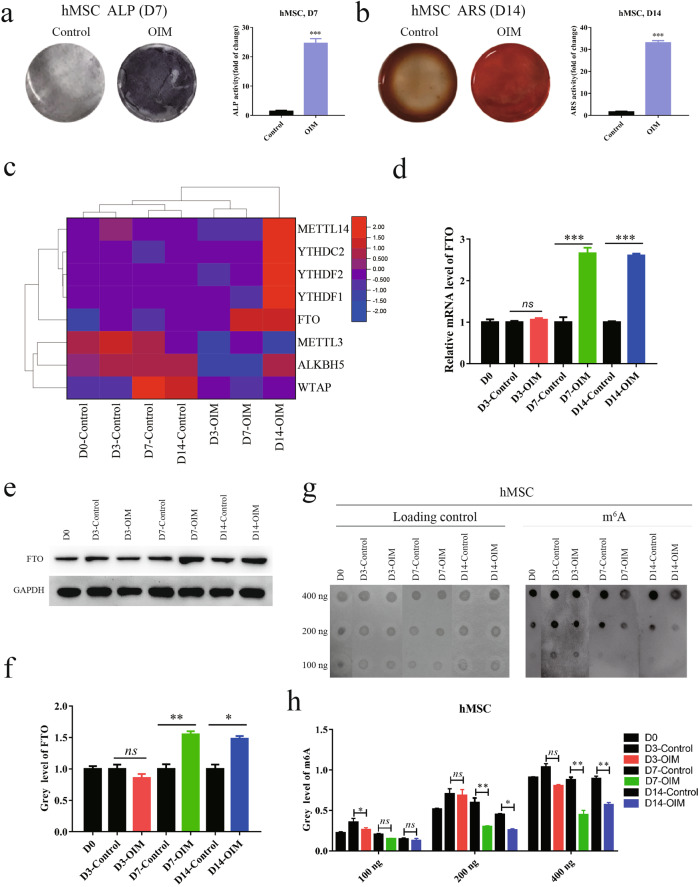
To investigate the potential molecular mechanisms involved in bone diseases, we systematically analyzed differential gene expression in patients with osteoporosis (GSE35955) and bone necrosis (GSE123568) [32]. FTO, a primary m6A demethylase, was significantly downregulated in tissues of osteoporosis and bone necrosis (Fig. 1a, b). We further validated the downregulation of FTO in our clinical samples of femoral heads from patients with fracture or osteonecrosis by immunohistochemistry (Fig. 1c, d). As expected, the global RNA m6A levels were increased in patients with osteoporosis compared with healthy people (Fig. 1e, f), indicating a potential role of FTO in mediating osteonecrosis by m6A modifications. To determine the role of FTO in osteoporosis and bone necrosis, we established an osteogenesis model by culturing human MSCs in OIM and confirmed the differentiation of human MSCs into osteoblasts by ALP staining and ARS (Fig. 2a, b). We further examined differential gene expression during osteoblast differentiation by RNA sequencing and found that FTO expression was dramatically induced on day 7 and day 14 after stimulation with OIM (Fig. 2c). We confirmed the upregulation of FTO during osteoblast differentiation at both the mRNA and protein levels (Fig. 2d–f). The global RNA m6A level was decreased during osteoblast differentiation induced with OIM, as indicated by the m6A dot blot assay, consistent with the upregulation of FTO expression (Fig. 2g, h). These results suggested that downregulation of FTO may be involved in mediating osteogenic differentiation during the processes of osteoporosis and bone necrosis.
Fig. 1. FTO was downregulated in patients with osteoporosis and bone necrosis.
a, b Heatmap of differential gene expression in tissue chips from patients with osteoporosis (a) and bone necrosis (b). c Expression of FTO in clinical samples of femoral heads from patients with fracture or osteonecrosis, as assessed by immunohistochemistry. d Quantification of FTO expression levels from the immunohistochemical staining results. e, f The global RNA m6A levels in clinical patients with osteonecrosis (e); quantification of m6A levels was performed with ImageJ (f). *P < 0.05; **P < 0.01; ***P < 0.001; n = 3.
Fig. 2. FTO was upregulated during the differentiation of human BMSCs into osteoblasts.
a, b Differentiation of human BMSCs into osteoblasts by osteogenic induction medium (OIM) and verification by alkaline phosphatase staining (a) and alizarin Red S staining (b). c Heatmap of differentially expressed m6A regulator genes during osteoblast differentiation. d Relative mRNA levels of FTO during osteoblast differentiation on day 3, day 7, and day 14, as assessed by quantitative PCR. e, f Relative protein levels of FTO during osteoblast differentiation on day 3, day 7, and day 14, as assessed by Western blot (e); quantification of FTO protein levels was performed with ImageJ (f). g, h The global RNA m6A levels during osteoblast differentiation (g) and quantification of m6A levels was performed with ImageJ (h). *P < 0.05; **P < 0.01; ***P < 0.001; n = 3.
Inhibition of FTO impaired osteogenic differentiation of human MSCs
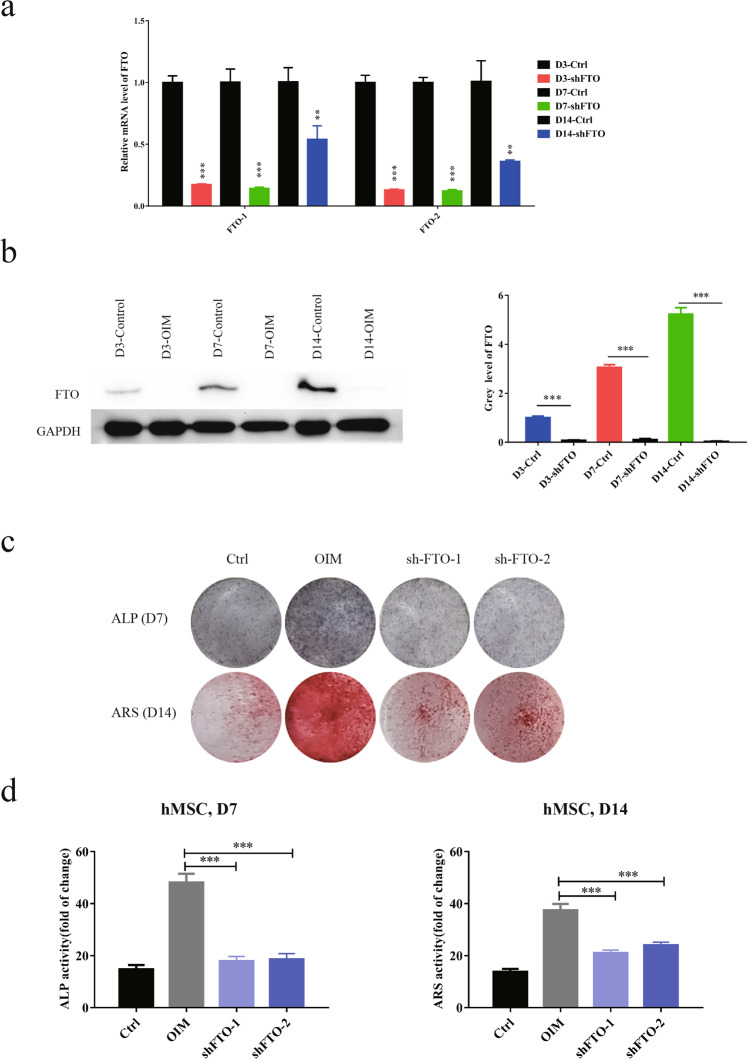
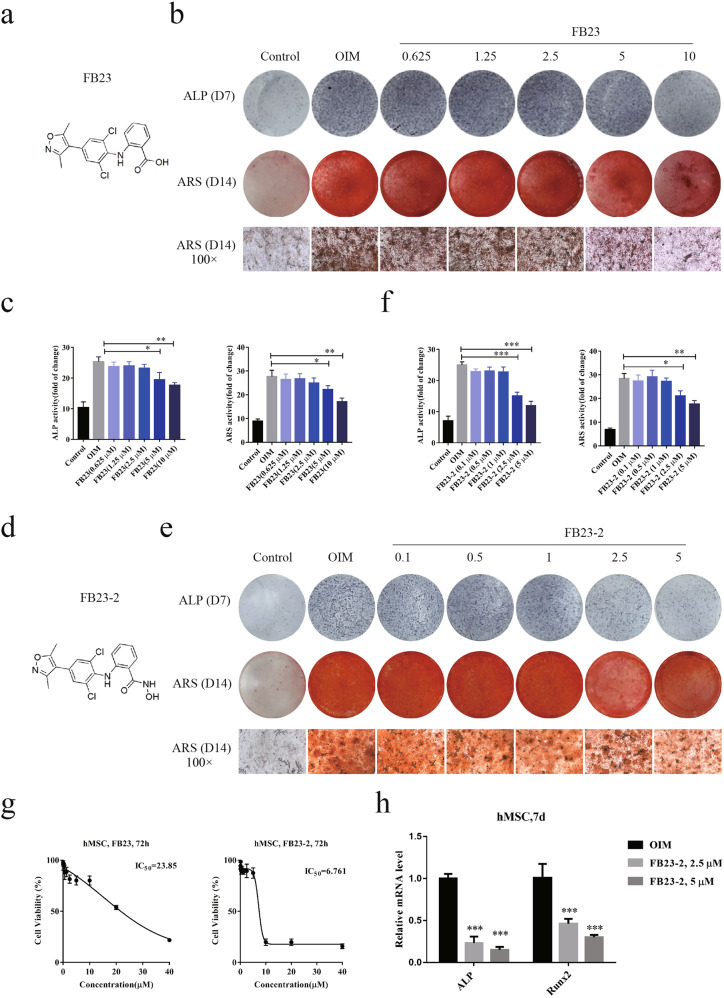
To validate the role of FTO in mediating osteogenic differentiation, we generated human MSCs with stable knockdown of FTO and validated the knockdown efficiency at both the mRNA and protein levels. We observed the long-term effects of FTO knockdown on day 3, day 7, and day 14 after treatment with OIM (Fig. 3a, b). Depletion of FTO diminished osteogenic differentiation of human MSCs, as indicated by the decreases in ALP activity and ARS staining (Fig. 3c, d). To determine the role of FTO activity in osteogenesis independent of its expression level, we utilized two specific potent inhibitors of FTO, FB23, and FB23-2 (Fig. 4a–f). Inhibition of FTO by FB23 and FB23-2 diminished the osteogenesis of human MSCs and impaired osteoblast formation in a dose-dependent manner. Treatment with 10 µM FB23 or 2.5 µM FB23-2 effectively blocked osteogenic differentiation upon culture with OIM (Fig. 4b, e). To avoid the impact of FTO inhibitors on the growth of human MSCs, we measured the growth inhibitory effects of these two potent FTO inhibitors with a CCK8 assay. FB23-2 treatment showed significant inhibitory effects on the growth of human MSCs at low concentrations, while FB23 treatment was relatively nontoxic in human MSCs (Fig. 4g). To validate the role of FTO in osteogenesis, we measured the mRNA expression levels of the osteogenic factors ALP and Runx2 in human MSCs treated with FTO inhibitors. Our results further confirmed that inhibition of FTO significantly reduced the expression of these osteogenic factors, indicating the crucial role of FTO in bone formation (Fig. 4h).
Fig. 3. Knockdown of FTO diminished osteogenic differentiation of BMSCs.
a Stable knockdown of FTO in human BMSCs by lentiviral shRNA sequences (shFTO-1 and shFTO-2). The knockdown efficiency was verified by quantitative PCR after incubation in OIM for 3, 7, and 14 days. b The knockdown efficiency was verified by Western blot after incubation in OIM for 3, 7, and 14 days. Quantification of FTO protein levels was performed with ImageJ. c Knockdown of FTO diminished the differentiation of human BMSCs into osteoblasts, as determined by the ALP activity assay and ARS staining. d Quantification of ALP activity and the ARS staining was performed with ImageJ. **P < 0.01; ***P < 0.001; n = 3.
Fig. 4. Inhibition of FTO impaired osteogenic differentiation of BMSCs.
a Synthesis of the FTO inhibitor FB23 and validation of its effects on osteogenic differentiation of BMSCs. b An ALP activity assay and ARS staining were performed on BMSCs treated with various concentrations of FB23 for 7 and 14 days, respectively. c Quantification of ALP activity and ARS staining in BMSCs treated with FB23 was performed with ImageJ. d Synthesis of the FTO inhibitor FB23-2 and validation of its effects on osteogenic differentiation of BMSCs. e The ALP activity assay and ARS staining were performed on BMSCs treated with various concentrations of FB23-2 for 7 and 14 days. f Quantification of ALP activity and ARS staining in BMSCs treated with FB23-2 was performed with ImageJ. g The IC50 of FB23 and FB23-2 in hMSCs after treatment for 72 h. h The mRNA expression levels of ALP and Runx2 on day 7 in hMSCs treated with FB23 and FB23-2. *P < 0.05; **P < 0.01; ***P < 0.001; n = 3.
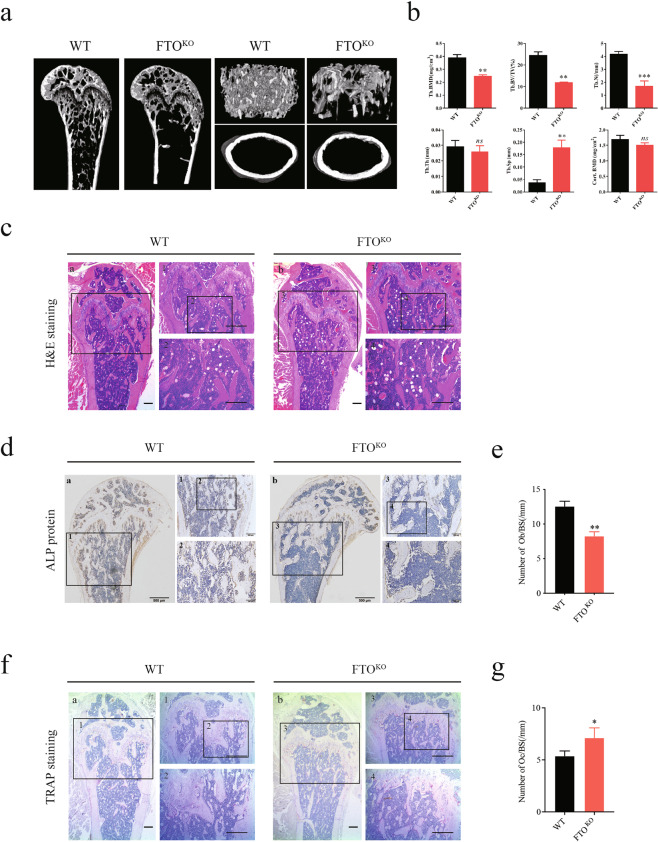
To further examine the in vivo effects of FTO depletion on bone formation, we generated mice with conditional knockout of Fto in bone by crossing Fto-floxed mice with Twist-Cre mice. Depletion of Fto in 16-week-old mice was accompanied by a moderate decrease in the femur length compared with that in control mice, as indicated by the 3D reconstruction results; in addition, lower Tb.BMD, Tb.N, and Tb.BV/TV were observed in Fto-knockout mice (Fig. 5a, b). By microcomputed tomography (μCT) analysis, we quantified bone histomorphometric parameters of femurs from 16-week-old Fto-knockout mice. Prominent decreases in the trabecular BMD, trabecular bone volume fraction, and trabecular number were observed in Fto-knockout mice (Fig. 5b). However, the trabecular thickness and cortical area BMD exhibited no significant differences based on the primary cortical parameters (Fig. 5b). H&E staining of tibia sections further demonstrated that depletion of Fto caused a decrease in the trabecular density, as indicated by the μCT results (Fig. 5c). The cortical bone and primary trabecular spongiosa were lined with alkaline phosphatase (ALP)-positive cells, showing that impaired bone formation in Fto-knockout mice resulted from decreased activity in osteoblasts (Fig. 5d, e) and the increase in the osteoclast number, as shown by TRAP staining (Fig. 5f, g).
Fig. 5. Knockout of FTO impaired osteogenic development in mice.
a Sagittal femur μCT, 3D trabecular reconstructions, and transverse scans at the mid-diaphysis in 16-week-old Fto-knockout mice. b Quantification of bone histomorphometric parameters. c H&E staining in paraffin sections of decalcified tibias from 16-week-old Fto-knockout mice. d, e Osteoblast localization and activity in decalcified tibias were shown by staining with an anti-ALP antibody (d); quantification was performed with ImageJ (e). f, g Osteoclast localization and activity in decalcified tibias were shown by TRAP staining (f), and quantification was performed with ImageJ (g). **P < 0.01; ***P < 0.001; n = 3.
FTO regulated osteogenic differentiation of human MSCs by mediating PPARG signaling
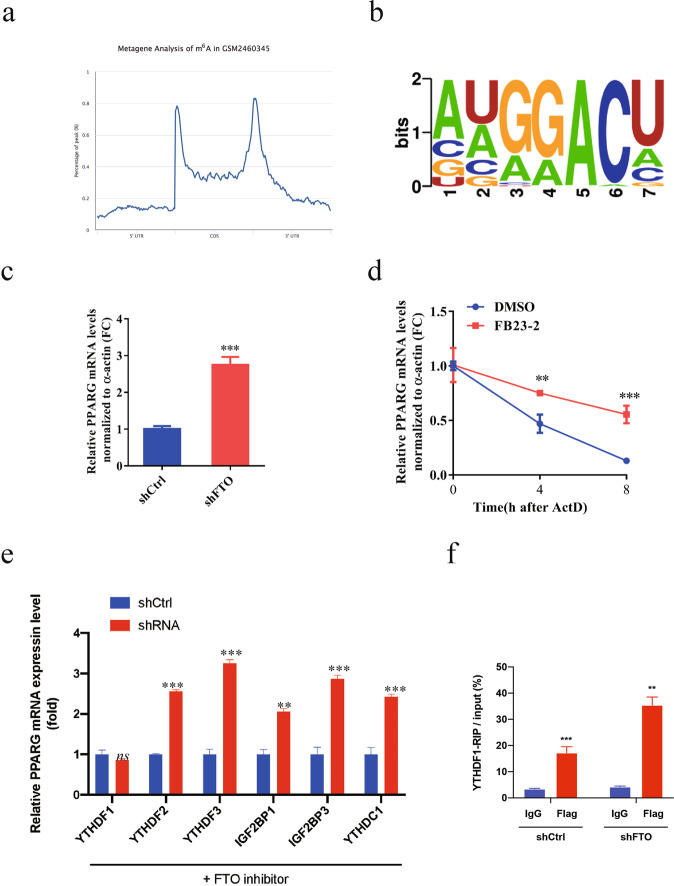
To determine the underlying mechanism by which FTO regulates the osteogenic differentiation of human MSCs, we performed m6A-Seq analysis in human MSCs treated with OIM and FB23-2 (2.5 and 5 µM) (Fig. 6a). Of note, the m6A level in PPARG, a biomarker for osteoporosis, was markedly increased in cells with FTO inhibition (Fig. 6b, c). Via m6A-RNA immunoprecipitation (MeRIP), we confirmed the increased m6A level in PPARG mRNA in MSCs treated with FB23-2 compared to that in cells treated with OIM (Fig. 6d). Consistent with these findings, m6A was enriched at the 3’-UTR of PPARG mRNA, as determined by metagene analysis (Fig. 7a). The motif of m6A in PPARG identified by sequencing contains a typical RRACH consensus motif (Fig. 7b). Knockdown of FTO significantly increased the mRNA expression level of PPARG (Fig. 7c). The half-life of PPARG mRNA was markedly increased in FTO knockdown cells after actinomycin D treatment (Fig. 7d), indicating that the induction of PPARG expression upon FTO depletion may be mediated through posttranscriptional regulation. To identify the bona fide reader for the m6A site on PPARG mRNA, we generated several stable cell lines with knockdown of the representative m6A readers, including YTHDF1, YTHDF2, YTHDF3, IGF2BP1, IGF2BP3, and YTHDC1, and confirmed their knockdown efficiency by RT-PCR (Fig. 7e). Knockdown of YTHDF1 diminished the increase in the mRNA level of PPARG in human MSCs treated with FTO inhibitors (Fig. 7e), while knockdown of FTO strikingly induced m6A modification of PPARG mRNA, as determined by YTHDF1-RIP analysis (Fig. 7f). Taken together, our results suggested that FTO-mediated m6A modification decreased PPARG expression in a YTHDF1-dependent manner.
Fig. 6. m6A-Seq identified PPARG as a downstream target of FTO-mediated m6A modification during osteogenic differentiation of BMSCs.
a Venn diagrams illustrating the overlap of differentially expressed genes in BMSCs treated with OIM and FB23-2. b m6A-Seq identified the m6A site in the 3’-UTR of PPARG mRNA. c Inhibition of FTO increased the m6A level of PPARG mRNA, as determined by m6A-Seq. d Inhibition of FTO promoted m6A methylation of PPARG mRNA, as determined by m6A MeRIP analysis. ***P < 0.001; n = 3.
Fig. 7. FTO-mediated m6A modification decreased the PPARG mRNA level in a YTHDF2-dependent manner.
a Metagene analysis of the m6A status of PPARG. b The m6A motif in PPARG. c Knockdown of FTO increased the mRNA expression level of PPARG. d Half-life of PPARG mRNA in BMSCs treated with an FTO inhibitor. e Knockdown of YTHDF1 diminished the induction of PPARG expression in BMSCs treated with an FTO inhibitor. f Knockdown of FTO induced m6A methylation of PPARG mRNA, as determined by YTHDF1-RIP. **P < 0.01; ***P < 0.001; n = 3.
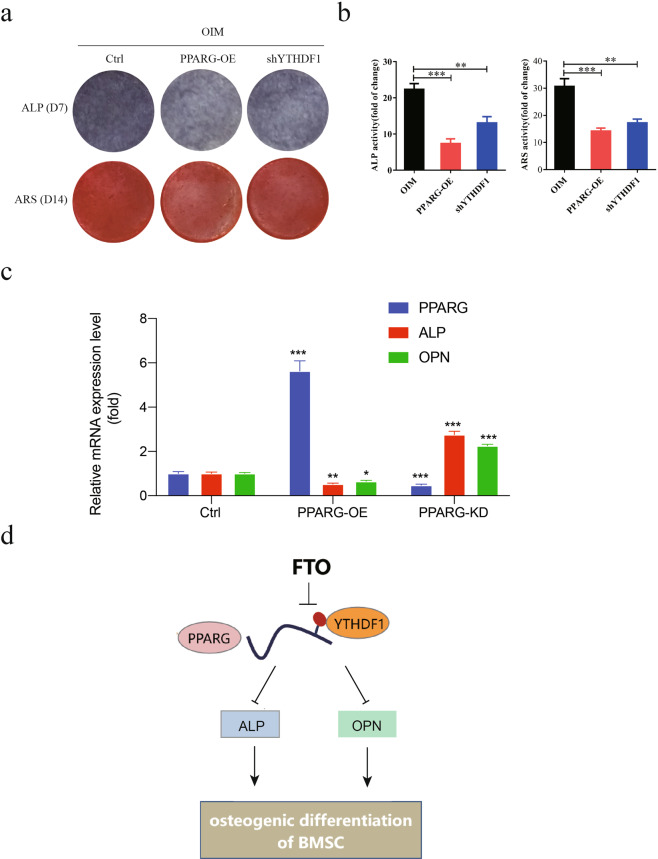
Overexpression of PPARG alleviated FTO-mediated osteogenic differentiation of human MSCs
To investigate whether PPARG plays a critical role in FTO-dependent osteogenic differentiation of human MSCs, we generated human MSCs with PPARG overexpression by lentiviral transduction. Overexpression of PPARG significantly reduced osteoblastic differentiation of human MSCs treated with OIM compared with control hMSCs, as indicated by the reductions in ALP activity and ARS staining (Fig. 8a). Interestingly, knockdown of YTHDF1 partially rescued the impairment of osteogenic differentiation, indicating that the FTO-PPARG-YTHDF1 axis plays an effective functional role in mediating osteogenic differentiation (Fig. 8b). Overexpression of PPARG caused decreased expression of ALP and osteopontin (OPN), two markers of osteoblasts, in human MSCs; conversely, knockdown of PPARG induced their expression (Fig. 8c). Taken together, these results demonstrated the functional significance of the FTO-PPARG axis in promoting the osteogenesis of human MSCs.
Fig. 8. Overexpression of PPARG significantly alleviated FTO-mediated osteogenic differentiation of BMSCs.
a Overexpression of PPARG or knockdown of YTHDF1 impaired osteogenic differentiation of BMSCs treated with OIM, as assessed with an ALP activity assay and ARS staining. b Quantification of ALP activity and ARS staining was performed with ImageJ. c PPARG negatively regulated the expression of the osteoblast marker genes ALP and OPN in BMSCs. d Working model. *P < 0.05; **P < 0.01; ***P < 0.001; n = 3.
Discussion
Osteoporosis is an age-related metabolic skeletal disorder characterized by low bone mass and accumulating deposition of adipose tissue in bone marrow and is a major public health issue with increasing prevalence. Osteoporosis and obesity are closely genetically associated, which means that the regulation of bone marrow stem cell (BMSC) differentiation plays a crucial role in determining the fate of BMSCs as osteoblasts or adipocytes. Previous attempts used BMSCs to treat bone necrosis of the femoral head by cell transplantation and revealed the involvement of microRNA modulation [33, 34] and long noncoding RNA regulation [35, 36]. Osteoblasts are characterized as polarized cuboidal cells that are activated to secrete extracellular matrix (ECM) during the differentiation of BMSCs. The ECM is rich in type I collagen, which is required for the accumulation of calcium phosphate on hydroxyapatite and results in the formation of hard bone [37]. Osteoblasts transform into osteocytes that are located in the mineralized bone matrix and do not contain most cytoplasmic organelles; alternatively, they die via apoptosis or become inactive (quiescent). However, the molecular mechanisms underlying the switch in BMSC differentiation into osteoblasts or adipocytes, particularly during the process of osteoporosis and bone necrosis, remain largely unknown. Recently, a functional link between dysregulation of m6A modification and bone pathological disorders was identified [38]. Dysregulation of the PTH/PTH1R signaling pathway showed the critical effect of m6A modification on determining the fate of BMSCs [38], suggesting the importance of the involvement of epigenetic regulation in pathological development of bone disorders.
m6A is a widespread and abundant methylation modification that occurs at the N6 position of adenosine in coding and noncoding RNAs and is the most common internal mRNA methylation modification in most eukaryotes [18, 39]. The m6A demethylase FTO, which belongs to the AlkB-related protein family, was the first m6A demethylase found to have a demethylation function in humans [40, 41]. FTO is localized in the nucleus and mediates the removal of m6A modifications, and dysregulation of FTO expression is associated with various human diseases, including cancers and cardiovascular diseases [42]. Genome-wide association studies have revealed that single-nucleotide polymorphisms at the FTO gene locus are closely related to obesity and energy homeostasis, suggesting the role of FTO in various biological processes [43–45]. In this study, we found that FTO was significantly downregulated in osteoporotic and osteonecrotic tissue compared to normal bone tissue, suggesting its potential role in mediating the pathological development of bone diseases. We further found the essential role of FTO in regulating osteogenic differentiation in our established model. Knockdown of FTO failed to initiate the process of osteogenesis in BMSCs. To assess the role of FTO in mediating osteogenesis, we further used two specific and potent inhibitors of FTO, FB23, and FB23-2, in a BMSC model and found that treatment with these FTO inhibitors suppressed osteogenic differentiation of BMSCs. To validate the role of FTO in mediating bone formation in vivo, we utilized mice with conditional knockout of Fto in bone. Our results demonstrated that all Fto-knockout mice displayed a dramatically decreased BMD, indicating a positive correlation between Fto and bone formation in vivo. Previously, Mi et al. showed that m6A modification was decreased during fracture healing and that downregulation of METTL3 promoted osteogenesis [46]. In line with our result that knockout of Fto in mice led to an increase in osteoclasts, Li et al. reported that during osteoclast differentiation, the expression of METTL3 and the m6A level increased, by which METTL3 regulated the mRNA degradation of ATP6V0D2 in a YTHDF2-dependent manner and the nuclear export of Traf6 mRNA [47]. Collectively, these results suggest a direct role of FTO in mediating osteogenic differentiation of BMSCs and indicate that dysregulation of FTO may be involved in aging-induced osteoporosis and osteonecrosis.
To decipher the underlying molecular mechanisms by which FTO mediates the differentiation of BMSCs into osteocytes, we performed MeRIP sequencing and RNA sequencing to analyze the direct downstream targets of FTO by means of m6A demethylation. We found that PPARG is the direct target of FTO and plays a crucial role in osteogenic differentiation. Previously, PPARG was shown to serve as a critical regulator of adipogenesis, and the Wnt signaling pathway was shown to promote the osteogenesis of mesenchymal precursors through transrepression of PPARG by the histone methyltransferase complex NLK-SETDB1 [48]. Activation of PPARG by the thiazolidinedione class of insulin sensitizers showed adverse effects on bone by promoting adipogenesis [49], while inhibition of PPARG with its antagonist SR1664 promoted the induction of osteogenic differentiation in isolated BMSCs [50]. In this study, we revealed a posttranscriptional mechanism by which PPARG is regulated by FTO-mediated m6A demethylation. Inhibition of FTO increased the RNA expression level of PPARG via m6A modification, consistent with a previous study showing that PPARG was associated with FTO [51, 52]. Mechanistically, FTO mediated m6A demethylation in the coding region of PPARG and promoted the decay of PPARG RNA in a YTHDF1-dependent manner. YTHDF1 is the classical m6A reader that promotes efficient mRNA translation [53], while YTHDF2 acts as a negative regulator to influence mRNA stability [54]. Knockdown of YTHDF1 failed to induce the expression of PPARG in FTO-silenced cells, indicating that YTHDF1 mediates the translation efficiency of PPARG. Previously, PPARG was found to inhibit osteogenesis by downregulating the COX-2-iNOS axis [55]. Here, we further showed that PPARG suppressed the differentiation of BMSCs through the activation of two representative markers of osteoblasts, ALPL and OPN. Overexpression of PPARG alleviated FTO-mediated osteogenic differentiation of BMSCs, suggesting the importance of the FTO-PPARG-ALPL/OPN axis in promoting the osteogenesis of BMSCs.
In summary, we report that the m6A demethylase FTO is downregulated in osteoporosis and osteonecrosis and that FTO functions as an inducer to promote osteogenic differentiation of BMSCs. Knockdown or inhibition of FTO disrupted the differentiation of BMSCs into osteoblasts. Mechanistically, FTO mediated m6A methylation of PPARG RNA by promoting PPARG RNA decay via YTHDF1, thus enhancing the expression of ALPL and OPN and promoting the osteogenesis of BMSCs. Our findings on the implication of FTO-mediated m6A modification in epigenetic silencing of PPARG in BMSCs exemplify the critical role of dynamic changes in m6A in the processes of osteoporosis and osteonecrosis.
Supplementary information
Acknowledgements
This work was supported by the Guangdong Provincial Science and Technology Plan Project [No. 2017A030303083], Guangzhou Science and Technology Plan Project [No. 201804010387], National Natural Science Foundation of China (NSFC) [No. 82074462, 81774339, 81872860], National Major Special Projects for the Creation and Manufacture of New Drugs [No. 2019ZX09301104], Guangdong Provincial Science and Technology Plan Project [No. 2017A030303083], Guangzhou Science and Technology Plan Project [No. 201804010387], and Natural Science Foundation of Guangdong Province, China [No. 2021A1515011003].
Author contributions
JS, JJW, and PQL designed the experiments; LSC and MZ performed the experiments; JS, JJW, XFX, LSC, PC, and HBW analyzed the data; JS and LSC drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Jun-jian Wang, Email: wangjj87@mail.sysu.edu.cn.
Juan Shen, Email: shenjuan0412@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00756-8.
References
- 1.Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772–82. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai M, Devlin MJ, Rosen CJ. Fat targets for skeletal health. Nat Rev Rheumatol. 2009;5:365–72. doi: 10.1038/nrrheum.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devlin MJ, Rosen CJ. The bone-fat interface: basic and clinical implications of marrow adiposity. Lancet Diabetes Endocrinol. 2015;3:141–7. doi: 10.1016/S2213-8587(14)70007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheller EL, Rosen CJ. What’s the matter with MAT? Marrow adipose tissue, metabolism, and skeletal health. Ann N Y Acad Sci. 2014;1311:14–30. doi: 10.1111/nyas.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurra S, Fink DA, Siris ES. Osteoporosis-associated fracture and diabetes. Endocrin Metab Clin. 2014;43:233–43. doi: 10.1016/j.ecl.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Pijnenburg L, Felten R, Javier RM. [A review of avascular necrosis, of the hip and beyond] Rev Med Interne. 2020;41:27–36. doi: 10.1016/j.revmed.2019.10.332. [DOI] [PubMed] [Google Scholar]
- 7.Ensrud KE, Crandall CJ. Osteoporosis. Ann Intern Med. 2017;167:Itc17–Itc31. doi: 10.7326/AITC201708010. [DOI] [PubMed] [Google Scholar]
- 8.Eguia A, Bagan-Debon L, Cardona F. Review and update on drugs related to the development of osteonecrosis of the jaw. Med Oral Patol Oral Cir Bucal. 2020;25:e71–e83. doi: 10.4317/medoral.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalavras CG, Lieberman JR. Osteonecrosis of the femoral head: evaluation and treatment. J Am Acad Orthop Surg. 2014;22:455–64. doi: 10.5435/JAAOS-22-07-455. [DOI] [PubMed] [Google Scholar]
- 10.Lespasio MJ, Sodhi N, Mont MA. Osteonecrosis of the hip: a primer. Perm J. 2019;23:18–100. doi: 10.7812/TPP/18-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin W, Yang X, Lu M. Juvenile-onset multifocal osteonecrosis in systemic lupus erythematosus: a case report. Medicine (Baltimore) 2021;100:e24031. doi: 10.1097/MD.0000000000024031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen GS, Zhou HB, Zhang H, Chen B, Liu ZP, Yuan Y, et al. The GDF11-FTO-PPAR gamma axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Bba-Mol Basis Dis. 2018;1864:3644–54. doi: 10.1016/j.bbadis.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 13.USPST Force, Curry SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, et al. Screening for osteoporosis to prevent fractures: US Preventive Services Task Force recommendation statement. JAMA. 2018;319:2521–31. doi: 10.1001/jama.2018.7498. [DOI] [PubMed] [Google Scholar]
- 14.Qaseem A, Forciea MA, McLean RM, Denberg TD, Clinical Guidelines Committee of the American College of Physicians. Barry MJ, et al. Treatment of low bone density or osteoporosis to prevent fractures in men and women: a clinical practice guideline update from the American College of Physicians. Ann Intern Med. 2017;166:818–39. doi: 10.7326/M15-1361. [DOI] [PubMed] [Google Scholar]
- 15.Fink HA, MacDonald R, Forte ML, Rosebush CE, Ensrud KE, Schousboe JT, et al. Long-term drug therapy and drug discontinuations and holidays for osteoporosis fracture prevention: a systematic review. Ann Intern Med. 2019;171:37–50. doi: 10.7326/M19-0533. [DOI] [PubMed] [Google Scholar]
- 16.Adler RA, El-Hajj Fuleihan G, Bauer DC, Camacho PM, Clarke BL, Clines GA, et al. Managing osteoporosis in patients on long-term bisphosphonate treatment: report of a Task Force of the American Society for Bone and Mineral Research. J Bone Min Res. 2016;31:16–35. doi: 10.1002/jbmr.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellmeyer DE. Atypical fractures as a potential complication of long-term bisphosphonate therapy. JAMA. 2010;304:1480–4. doi: 10.1001/jama.2010.1360. [DOI] [PubMed] [Google Scholar]
- 18.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–55. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell. 2020;181:1582–95. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540:242–7. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Wan A, Zhou Z, Chen D, Liang H, Liu C, et al. RNA-binding protein RALY reprogrammes mitochondrial metabolism via mediating miRNA processing in colorectal cancer. Gut. 2020;0:1–15. doi: 10.1136/gutjnl-2020-320652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–85. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–91. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toh JDW, Sun L, Lau LZM, Tan J, Low JJA, Tang CWQ, et al. A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor of N(6)-methyladenosine demethylase FTO. Chem Sci. 2015;6:112–22. doi: 10.1039/C4SC02554G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benisch P, Schilling T, Klein-Hitpass L, Frey SP, Seefried L, Raaijmakers N, et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One. 2012;7:e45142. doi: 10.1371/journal.pone.0045142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao W, Ning Y, Xu HJ, Zou WZ, Hu J, Liu XZ, et al. BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin Sci (Lond) 2019;133:1955–75. doi: 10.1042/CS20181064. [DOI] [PubMed] [Google Scholar]
- 34.Xu HJ, Liao W, Liu XZ, Hu J, Zou WZ, Ning Y, et al. Down-regulation of exosomal microRNA-224-3p derived from bone marrow-derived mesenchymal stem cells potentiates angiogenesis in traumatic osteonecrosis of the femoral head. FASEB J. 2019;33:8055–68. doi: 10.1096/fj.201801618RRR. [DOI] [PubMed] [Google Scholar]
- 35.Xiang S, Li Z, Weng X. The role of lncRNA RP11-154D6 in steroid-induced osteonecrosis of the femoral head through BMSC regulation. J Cell Biochem. 2019;120:18435–45. doi: 10.1002/jcb.29161. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY) 2019;11:8777–91. doi: 10.18632/aging.102264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granero Molto F, Weis JA, Miga MI, Landis B, Myers TJ, O’Rear L, et al. Regenerative effects of transplanted mesenchymal stem cells in fracture healing. Stem Cells. 2009;27:1887–98. doi: 10.1002/stem.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newrick PG, Langton-Hewer R. Motor neurone disease: can we do better? A study of 42 patients. Br Med J (Clin Res Ed) 1984;289:539–42. doi: 10.1136/bmj.289.6444.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S, Wei JB, Cui YH, Parka G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782–95. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao YR, Thakkar KN, Zhao HJ, Broughton J, Li Y, Seoane JA, et al. The m6A RNA demethylase FTO is a HIF-independent synthetic lethal partner with the VHL tumor suppressor. Proc Natl Acad Sci USA. 2020;117:21441–9. doi: 10.1073/pnas.2000516117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu Y, Wan A, Lin Z, Lu X, Wan GN. (6)-Methyladenosine modification: a novel pharmacological target for anti-cancer drug development. Acta Pharm Sin B. 2018;8:833–43. doi: 10.1016/j.apsb.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speakman JR. The ‘Fat Mass and Obesity Related’ (FTO) gene: mechanisms of impact on obesity and energy balance. Curr Obes Rep. 2015;4:73–91. doi: 10.1007/s13679-015-0143-1. [DOI] [PubMed] [Google Scholar]
- 44.Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–74. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang QY, Xiao TC, Guo J, Su ZQ. Complex relationship between obesity and the fat mass and obesity locus. Int J Biol Sci. 2017;13:615–29. doi: 10.7150/ijbs.17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi B, Xiong Y, Yan C, Chen L, Xue H, Panayi AC, et al. Methyltransferase-like 3-mediated N6-methyladenosine modification of miR-7212-5p drives osteoblast differentiation and fracture healing. J Cell Mol Med. 2020;24:6385–96. doi: 10.1111/jcmm.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D, Cai L, Meng R, Feng Z, Xu Q. METTL3 modulates osteoclast differentiation and function by controlling RNA stability and nuclear export. Int J Mol Sci. 2020;21:1660–77. doi: 10.3390/ijms21051660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–24. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 49.Viccica G, Francucci CM, Marcocci C. The role of PPARgamma for the osteoblastic differentiation. J Endocrinol Invest. 2010;33:9–12. [PubMed] [Google Scholar]
- 50.Marciano DP, Kuruvilla DS, Boregowda SV, Asteian A, Hughes TS, Garcia-Ordonez R, et al. Pharmacological repression of PPARgamma promotes osteogenesis. Nat Commun. 2015;6:7443–58. doi: 10.1038/ncomms8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marvelle AF, Lange LA, Qin L, Adair LS, Mohlke KL. Association of FTO with obesity-related traits in the Cebu Longitudinal Health and Nutrition Survey (CLHNS) cohort. Diabetes. 2008;57:1987–91. doi: 10.2337/db07-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–35. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m6A promotes Cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N(6)-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin TH, Yang RS, Tang CH, Lin CP, Fu WM. PPARgamma inhibits osteogenesis via the down-regulation of the expression of COX-2 and iNOS in rats. Bone. 2007;41:562–74. doi: 10.1016/j.bone.2007.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.