Abstract
We investigated the patterns of recurrence and primary endocrine resistance according to estrogen receptor (ER) alpha gene (ESR1) mutations, as assessed by digital droplet (dd) PCR, in patients with non-metastatic ER+ breast cancer. We collected 121 formalin-fixed paraffin-embedded (FFPE) surgical specimens from ER+ breast cancer patients who had relapsed after surgery. Genomic DNA was extracted from the FFPE samples and ESR1 mutations were evaluated using ddPCR. ESR1 mutations were detected in 9 (7.4%) of 121 primary breast cancer specimens. The median recurrence-free interval and overall survival were significantly lower in patients with ESR1 mutations than in those without. Of the patients treated with ET (N = 98), eight had ESR1 mutations. Of these, six (75.0%) had primary endocrine resistance and two (25.0%) had secondary endocrine resistance. By contrast, only 22 of 90 (24.4%) patients without ESR1 mutations had primary endocrine resistance. A multivariable model showed that an ESR1 mutation is a significant risk factor for primary endocrine resistance. Our findings provide clinical evidence that the presence of rare ESR1 mutant clones identified by ddPCR in primary tumors is associated with primary endocrine resistance in an adjuvant setting.
Subject terms: Breast cancer, Cancer genomics, Predictive markers
Introduction
Mutations in the estrogen receptor (ER) alpha gene, ESR1, were first described in cell lines, when mutations in the ligand-binding domain (LBD), including Y537S and E380Q, were shown to constitutively activate the receptor1. Experiments with breast cancer cells harboring mutations in the LBD-encoding region of the ESR1 gene have shown that mutant cells require a higher anti-estrogen drug concentration and that they proliferate in an estradiol-independent manner through constitutive activation of the ER pathway1. However, previous large-scale studies, such as The Cancer Genome Atlas project, have found that ESR1 mutations are rarely detected in primary breast tumors (0.5% in 962 samples)2.
With the introduction of next-generation sequencing (NGS) technology in genomic research, ESR1 mutations have been re-analyzed in samples from metastatic ER+ breast cancer. A series of studies has demonstrated that the incidence of ESR1 mutations is as high as 11–55% in metastatic tumors samples from patients who previously underwent aromatase-inhibitor (AI) treatment3–7. Furthermore, using a hybridization capture-based NGS assay, known as the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay8, ESR1 mutations have been detected in 3.5% (11 of 313) of primary breast cancer and 13.6% (84 of 616) of metastatic tumor samples9. These studies have collectively shown that ESR1 mutations present rarely in primary treatment-naive ER+ breast cancer, whereas they are highly prevalent in metastatic tumors, suggesting that these mutations may potentially arise from rare clones of primary tumors through clonal selection against endocrine therapy (ET)10–13.
Despite the scarcity of ESR1 mutations in primary ER+ breast cancer, several lines of evidence suggest that ESR1-mutated clones may be identified in primary tumors by droplet digital PCR (ddPCR)14–16. The rates of ESR1 mutation detection are 2.6% to 12.0% in primary cancer when using ddPCR14–16. In ddPCR, template DNA is partitioned into approximately 20,000 droplets in a single reaction well and is then amplified within individual droplets. Therefore, this highly sensitive method has the capacity of providing accurate quantification without external references and is considered to be a useful tool to detect rare mutant alleles17–19. However, the clinical outcomes of patients with ESR1-mutated primary breast cancer are not well understood.
In this study, we sought to detect ESR1 mutations using ddPCR in non-metastatic ER+ breast cancer. Moreover, we hypothesized that breast cancers harboring an ESR1 mutation may show a different recurrence pattern compared to those with wild-type ESR1. We further addressed the relationship between the presence of an ESR1 mutation and primary endocrine resistance in patients receiving adjuvant ET.
Results
Baseline characteristics and ESR1 mutations
A total of 121 patients with recurrences were included in the study (Fig. 1). The median age at surgery for all patients was 45 years (range, 23–77 years). Among the 121 patients, 36 (29.8%) had stage I, 53 (43.8%) had stage II, and 32 (26.4%) had stage III breast cancer. Adjuvant chemotherapy was administered to 97 (80.8%) patients. In all, 98 (81.0%) patients received adjuvant ET, including tamoxifen (N = 59) and aromatase inhibitors (AIs, N = 39), whereas upfront anti-estrogen therapy was not administered to 23 (19.0%) patients (Supplementary Table 1).
Fig. 1. Consort diagram.
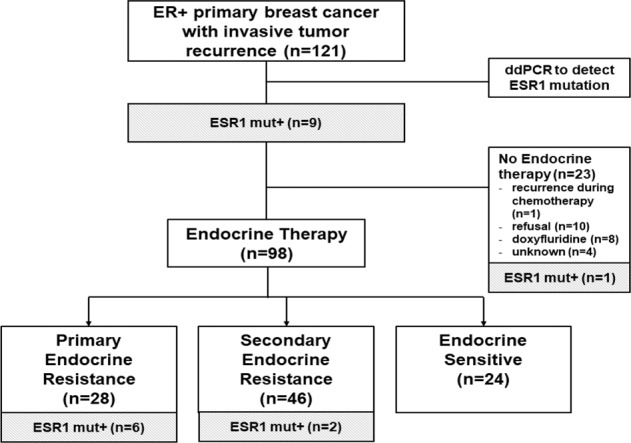
The definitions of primary endocrine resistance, secondary endocrine resistance, and endocrine sensitivity followed the 5th International Consensus Conference for Advanced Breast Cancer guidelines and are provided in the Patients and methods section.
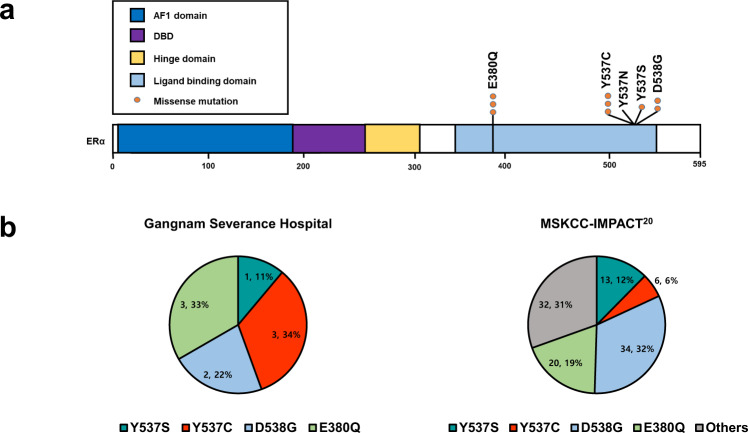
ESR1 mutations (E380Q, Y537C, Y537N, Y537S, and D538G) were detected by ddPCR. ESR1 mutations were detected in 9 (7.4%, 95% Wald asymptotic confidence interval (CI) 2.8–12.1%) out of 121 primary breast cancer specimens (Fig. 2A). Y537C and E380Q mutations were found in three patients (33%), D538G mutation was found in two patients (22%), and Y537S mutation was found in one patient (11%). No Y537N ESR1 mutations were detected. The median number of mutant allele copies was 2 (range, 2–587), and the median mutant allele fraction was 0.32% (0.01–8.37). The distribution of ESR1 mutations in our cohort compared to that in the MSKCC-IMPACT series20 is illustrated in Fig. 2B.
Fig. 2. ESR1 missense mutation in nine patients.
a Locations and frequencies. b Distributions of ESR1 missense mutation sites: comparison the Gangnam Severance Hospital with the MSKCC-IMPACT series20.
When clinical and pathologic characteristics were compared based on the presence of ESR1 mutations, the ESR1-mutation group had a higher T stage than the wild-type ESR1 group (Supplementary Table 1). Other factors did not differ between the two groups.
The sites of the first recurrence are summarized in Supplementary Table 2. The most common site of the first tumor relapse was the bone (33.9%), followed by the lungs (24.0%) and distant lymph nodes (17.4%). There were no differences in the metastatic sites based on the ESR1 mutations.
Survival according to ESR1 mutation occurrence
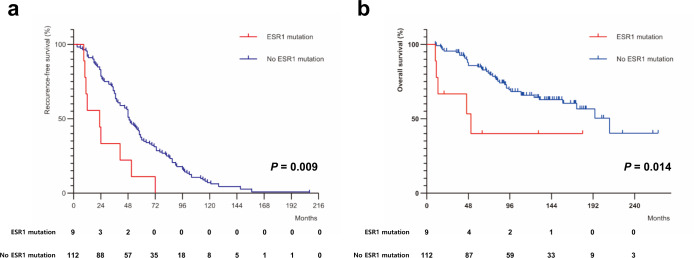
Recurrence-free interval (RFI) was defined as the time from the date of breast cancer surgery to the time of the first breast cancer recurrence, including locoregional and distant recurrences. Overall survival (OS) was defined as the time from the date of breast cancer surgery to the date of death from any cause or the last censored follow-up. The median follow-up time for the study population was 140 months (95% CI, 126–154 months). Since we selected patients with tumor recurrence, we compared the median RFI and OS using the Mann-Whitney U test. The median RFI was significantly lower in patients with an ESR1 mutation than in those without an ESR1 mutation (23.0 versus 49.0 months; p = 0.009). The median OS was 51 months in the ESR1-mutant group versus 211 months in the ESR1-wild-type group (p = 0.014). Survival plots for RFI and OS are presented in Fig. 3.
Fig. 3. Kaplan-Meier survival plots according to ESR1 mutation.
a Recurrence-free interval. b Overall survival.
We analyzed whether ET type affected RFI stratified by ESR1 mutation. The median RFI was not significantly different according to ET (tamoxifen vs. AIs), both in the ESR1 mutation group (17.0 vs. 18.0 months; p > 0.999) and non-ESR1 mutation group (48.0 vs. 58.0 months; p = 0.551). The survival plots are shown in Supplementary Fig. 1.
ESR1 mutation and primary endocrine resistance
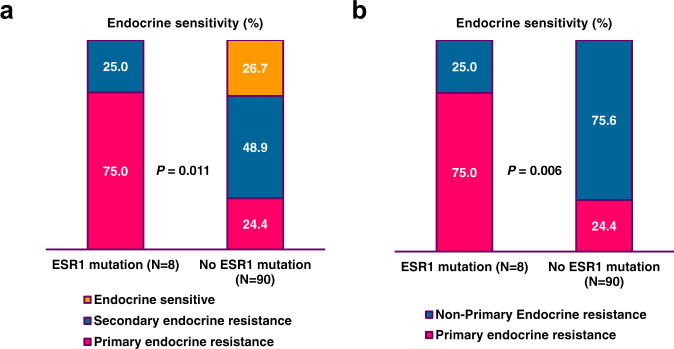
Next, to assess the influence of ESR1 mutations on primary endocrine resistance, we excluded patients without adjuvant ET (N = 23). The reasons for patients not undergoing ET are shown in Fig. 1. We classified the patients receiving adjuvant ET (N = 98) into three groups: (i) primary endocrine resistance (n = 28), defined as relapse during the first 2 years of adjuvant ET, (ii) secondary endocrine resistance (N = 46), defined as relapse during adjuvant ET, and (iii) endocrine sensitivity (N = 41), defined as not belonging to primary or secondary endocrine resistance21. The clinical and pathological characteristics of the three groups are presented in Table 1. Twenty-eight patients (28.6%) had primary endocrine resistance. Out of the eight patients with ESR1 mutation, six (75.0%) had primary endocrine resistance and two (25.0%) had secondary endocrine resistance (Fig. 4). None of the endocrine sensitivity group had ESR1 mutations detected. In contrast, only 22 out of 90 (24.4%) patients without an ESR1 mutation had primary endocrine resistance, whereas 52 (48.9%) and 16 (26.7%) patients had secondary endocrine resistance and endocrine sensitivity, respectively (Fig. 5A, B).
Table 1.
Baseline characteristics according to endocrine resistance in patients who received adjuvant endocrine therapy.
| Primary endocrine resistance (N = 28) |
Secondary endocrine resistance (N = 46) |
Endocrine Sensitive (N = 24) |
Total (N = 98) |
P value | |
|---|---|---|---|---|---|
| Age (median, range) | 45 (28–77) | 44 (30–74) | 48 (23–75) | 45 (23–77) | 0.973 |
| Histologic type | 0.789a | ||||
| IDC | 23 (82.1%) | 42 (91.3%) | 21 (87.5%) | 86 (87.8%) | |
| ILC | 2 (7.1%) | 2 (4.3%) | 1 (4.2%) | 5 (5.1%) | |
| Others | 3 (10.7%) | 2 (4.3%) | 2 (8.3%) | 7 (7.1%) | |
| HGb | 0.493a | ||||
| 1 or 2 | 21 (84.0%) | 37 (86.0%) | 21 (95.5%) | 79 (87.8%) | |
| 3 | 4 (16.0%) | 6 (14.0%) | 1 (4.5%) | 11 (12.2%) | |
| LVIb | 0.316 | ||||
| No | 14 (66.7%) | 27 (73.0%) | 12 (92.3%) | 53 (74.6%) | |
| Yes | 7 (33.3%) | 10 (27.0%) | 1 (7.7%) | 18 (25.4%) | |
| T stage | 0.011a | ||||
| 1 | 8 (28.6%) | 23 (50.0%) | 13 (54.2%) | 44 (44.9%) | |
| 2 | 15 (53.6%) | 23 (50.0%) | 11 (45.8%) | 49 (50.0%) | |
| 3 | 5 (17.9%) | 0 | 0 | 5 (5.1%) | |
| N stage | 0.336a | ||||
| 0 | 11 (39.3%) | 20 (43.5%) | 11 (45.8%) | 42 (42.9%) | |
| 1 | 8 (28.6%) | 16 (34.8%) | 9 (37.5%) | 33 (33.7%) | |
| 2 | 3 (10.7%) | 4 (8.7%) | 4 (16.7%) | 11 (11.2%) | |
| 3 | 6 (21.4%) | 6 (13.0%) | 0 | 12 (12.2%) | |
| Stage | 0.424 | ||||
| 1 | 6 (21.4%) | 14 (30.4%) | 10 (41.7%) | 30 (30.6%) | |
| 2 | 12 (42.9%) | 22 (47.8%) | 10 (41.7%) | 44 (44.9%) | |
| 3 | 10 (35.7%) | 10 (21.7%) | 4 (16.7%) | 24 (24.5%) | |
| Breast surgery | 0.083 | ||||
| BCS | 6 (21.4%) | 18 (39.1%) | 12 (50.0%) | 36 (36.7%) | |
| TM | 22 (78.6%) | 28 (60.9%) | 12 (50.0%) | 62 (63.3%) | |
| Axillary surgery | 0.142 | ||||
| SLNB | 9 (26.5%) | 8 (17.4%) | 9 (37.5%) | 26 (26.5%) | |
| ALND | 19 (67.9%) | 38 (82.6%) | 15 (62.5%) | 72 (73.5%) | |
| Adjuvant chemotherapy | 0.426 | ||||
| No | 4 (14.3%) | 10 (21.7%) | 7 (29.2%) | 21 (21.4%) | |
| Yes | 24 (85.7%) | 36 (78.3%) | 17 (70.8%) | 77 (78.6%) | |
| Adjuvant endocrine | 0.466 | ||||
| Tamoxifen | 17 (60.7%) | 30 (65.2%) | 12 (50.0%) | 59 (60.2%) | |
| AI | 11 (39.3%) | 16 (34.8%) | 12 (50.0%) | 39 (39.8%) | |
| Adjuvant radiotherapy | 0.402 | ||||
| No | 8 (28.6%) | 20 (43.5%) | 8 (33.3%) | 36 (36.7%) | |
| Yes | 20 (71.4%) | 26 (56.5%) | 16 (66.7%) | 62 (63.3%) |
IDC invasive ductal carcinoma, ILC invasive lobular carcinoma, HG histologic grade, LVI lymphovascular invasion, BCS breast-conserving surgery, TM total mastectomy, SLNB sentinel lymph node biopsy, ALND axillary lymph node dissection, AI aromatase inhibitor.
aP values were obtained using Fisher’s exact test.
bMissing values.
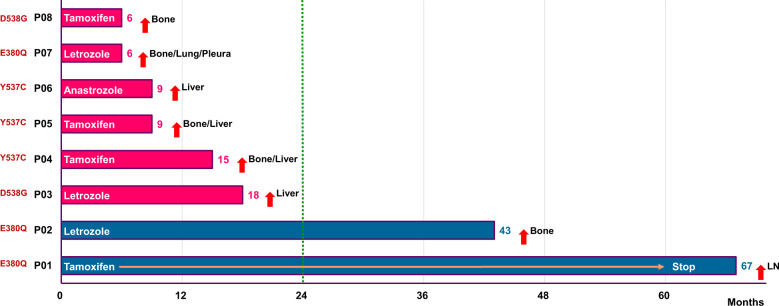
Fig. 4. Recurrence types according to responsiveness to adjuvant endocrine therapy in eight patients with ESR1 mutations who received adjuvant endocrine therapy.
Among eight patients, six patients belonged to primary endocrine resistance, and two patients belonged to secondary endocrine resistance.
Fig. 5. The relationship between ESR1 mutation and recurrence type according to responsiveness to adjuvant endocrine therapy.
a Primary endocrine resistance, secondary endocrine resistance, and endocrine sensitivity (p = 0.011; chi-square test) b Primary endocrine resistance versus non-primary endocrine resistance (p = 0.006; Fisher’s exact test).
In comparison with other clinical characteristics, T stage was higher in the group with primary endocrine resistance than in the groups with secondary endocrine resistance or endocrine sensitivity (Table 1). Accordingly, the group with primary resistance was more likely to receive a total mastectomy.
To construct a multivariate model for primary endocrine resistance, univariate binary logistic regression analyses were first performed. ESR1 mutation and T stage were found to be significant in these analyses (Table 2). The binary multivariate model demonstrated that an ESR1 mutation was a significant factor for primary endocrine resistance, independent of T stage. The odds ratio of an ESR1 mutation was 8.334 (95% CI, 1.524–45.561; Table 2), and the area under the curve (AUC) of the model consisting of an ESR1 mutation and T stage was 0.698 (95% CI, 0.583–0.812; Fig. 6). The AUC of this model was numerically higher than that of the model with T stage alone (AUC, 0.658; 95% CI, 0.545–0.770). Within the subgroup (N = 74) nested by excluding endocrine-sensitive patients, an ESR1 mutation was the only significant risk factor for primary endocrine resistance (Supplementary Table 3).
Table 2.
Univariate and multivariate analysis of primary endocrine resistance in patients who received endocrine therapy.
| Univariable | Multivariable | |||
|---|---|---|---|---|
| Variable | OR (95% CI) | P value | OR (95% CI) | P value |
| Age | 0.999 (0.962–1.038) | 0.976 | ||
| Histologic type | ||||
| IDC | Ref. | |||
| Others | 1.479 (0.704–3.104) | 0.301 | ||
| HG | ||||
| 1 or 2 | Ref. | |||
| 3 | 1.578 (0.419–5.944) | 0.500 | ||
| T stage | ||||
| 1 | Ref. | Ref. | ||
| 2 or 3 | 2.647 (1.029–6.807) | 0.043 | 2.419 (0.907–6.453) | 0.078 |
| N stage | ||||
| Negative | Ref. | |||
| Positive | 1.228 (0.503–3.001) | 0.652 | ||
| Adjuvant chemotherapy | ||||
| No | Ref. | |||
| Yes | 1.925 (0.585–6.333) | 0.281 | ||
| Adjuvant radiotherapy | ||||
| No | Ref. | |||
| Yes | 1.667 (0.645–4.306) | 0.292 | ||
| ESR1 mutation | ||||
| No | Ref. | Ref. | ||
| Yes | 9.273 (1.744–49.305) | 0.009 | 8.334 (1.524–45.561) | 0.014 |
OR odds ratio, CI confidence interval, IDC invasive ductal carcinoma, HG histologic grade, LVI lymphovascular invasion.
Fig. 6. The areas under the curve (AUC) of two models.
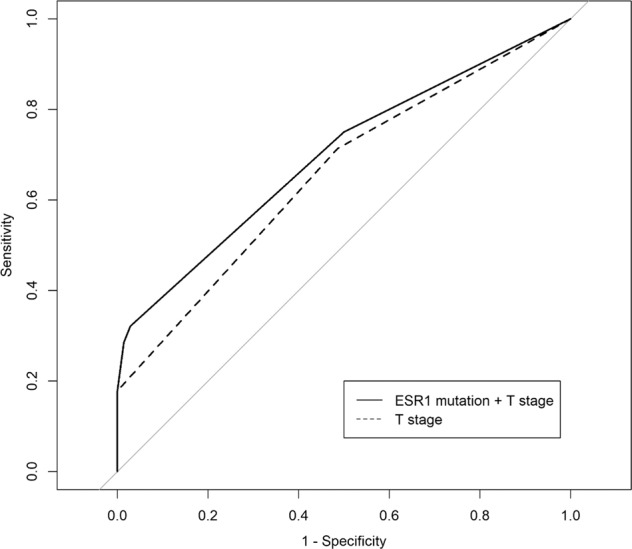
The AUC of the model with an ESR1 mutation and T stage was numerically higher than that of the model with T stage alone. The AUCs of the two models were 0.698 (95% CI, 0.583–0.812) and 0.658 (95% CI, 0.545–0.770), respectively. There was no statistical difference when using the DeLong method.
Discussion
Using ddPCR, we detected rare ESR1 mutant clones in 9 of 121 (7.4%) primary ER+ breast cancer patients with relapse after surgery. Furthermore, we correlated the presence of an ESR1 mutation with survival outcomes and found that the presence of ER+ treatment-naive tumors bearing an ESR1 mutation was associated with primary endocrine resistance, despite their clonal rarity. This is the first report to provide clinical evidence that an ESR1 mutation interrogated by ddPCR is linked with primary resistance to adjuvant ET in ER+ breast cancer.
Our ddPCR-based ESR1 mutation detection rate was consistent with the rates reported in previous studies. These earlier studies identified ESR1 mutant clones in 2.6% (7 of 270)16, 7.0% (3 of 43)14, and 12.0% (for Y537N)15 of primary cancers, respectively. As the study by Takeshita et al. included non-ER+ breast cancer16, it can be assumed that ESR1 mutant clones may be present in more than 5% of primary ER+ breast cancers.
We detected a low ESR1 mutation allele frequency, ranging from 0.01 to 8.37%, in primary ER+ breast cancer. This is similar to the findings of Wang et al., who reported ESR1 mutant allele frequencies of 0.07 to 0.2% in ER+ primary breast cancer14. Due to the small number of cases with an ESR1 mutation, we could not determine whether the mutant allele fraction was correlated with type of endocrine resistance. More data with a larger sample size are required to address this issue.
When we compared the distribution of ESR1 LBD mutation sites between our cohort and the MSKCC-IMPACT series mainly consisting of ER+ metastatic breast cancer (Fig. 2B), Y537S (33%) and Y537C (33%) were observed most frequently in our cohort, while E380Q was observed at the highest frequency (32%) in the external cohort. Because the MSKCC-IMPACT used the NGS technique, they found additional ESR1 mutations outside the LBD, including frame shifts or indels outside ESR1 LBD, with low frequency (N ≤ 2)20. To determine whether there is a difference in the ESR1 mutation site between primary and metastatic breast cancer, further research is required.
Robust preclinical and clinical data suggest that ESR1 mutations are associated with resistance to ET5–7,10,13,15,22,23. Mutations in the LBD-encoding region of the ESR1 gene alter the structure of the ER protein, leading to ligand-independent activity7,23 and the recruitment of coactivators, such as SRC-1 and SRC-35,22, which confer endocrine resistance. We investigated whether the presence of mutant ESR1 in primary tumors affects endocrine resistance. Based on recent guidelines for the classification of endocrine resistance, we found that ER+ breast cancer patients bearing an ESR1 mutation at surgery may have primary endocrine resistance. None of the eight subjects with an ESR1 mutation showed a relapse pattern consistent with endocrine sensitivity.
If novel therapeutics that effectively eradicate mutant ESR1 are employed clinically, the upfront use of the agents in an adjuvant setting has the potential to reduce failure of endocrine treatment. In the SoFEA (Study of Faslodex Versus Exemestane with or without Arimidex) trial, which was conducted in ER+ metastatic breast cancer, fulvestrant was shown to improve progression-free survival compared with exemestane in patients with an ESR1 mutation, as detected in plasma samples by ddPCR13. Furthermore, novel selective estrogen receptor degraders (SERDs), which potentiate the degradation of mutant ER, have been under development and evaluated in clinical trials by several pharmaceutical companies24. For instance, an in vitro study showed that cancer cells with Y537S mutant ESR1 are resistant to fulvestrant but sensitive to potent SERDs, such as AZD949620.
Because we retrospectively identified and included only recurring patients with available primary surgical samples, our study has an inherent limitation of selection bias. Considering breast cancer recurrences continued to occur steadily after the end of ET25, the proportion of primary or secondary endocrine resistance was relatively high, at approximately 75%. This bias may have affected the ESR1 mutation rate and its subsequent prognostic impact. Therefore, our findings should be interpreted with caution. In addition, the lack of inclusion of non-recurring patients prevents formal assessment of primary ESR1 mutations as biomarkers to guide ET. Further studies in a large prospective cohort with a sufficient follow-up period, including patients without relapse, are required to verify the findings.
Another limitation of our study was that we did not investigate ESR1 mutations in serial tissue and blood samples from index patients. Analyses of matched primary and metastatic samples or serial plasma samples may elucidate how rare ESR1-mutant clones arise in primary tumors and become metastatic through dissemination in the bloodstream, in accordance with clinical tumor progression. However, metastatic tissues or blood samples were not available for these analyses. In future studies, the assessment of ESR1 mutations in primary tumors and serial plasma samples will be essential for establishing therapies targeting mutant ESR1.
Furthermore, we could not assess the analytical sensitivity of this assay using synthetic templet or tumor genomic DNA which represented different levels of mutation abundance (%) for each mutation-specific probe in this study. However, according to Chu et al.26, the analytical sensitivity of ESR1 mutation assay was reported at approximately 0.007%, and it was validated with clinical samples with 0.01% mutant alleles. Therefore, we determined 0.01% mutant alleles as a cut-off threshold and considered clinical samples with less than 0.01% of mutation fraction as an “ESR1 mutation-negative”.
Lastly, other ESR1 mutations, such as K303R, L536P, and S463P14,27–29, were not included in our ddPCR panel. ddPCR assays targeting a larger number of mutations may affect the detection rate of ESR1 mutations in primary cancer. In addition, our ESR1 mutant detection rate should be interpreted with caution, considering that we only used tumor samples from subjects with relapse.
In conclusion, we showed that ddPCR detected rare clones with ESR1 mutations in primary ER-positive cancer and we provided clinical evidence that the presence of rare ESR1 mutant clones is associated with primary endocrine resistance in the adjuvant setting. We suggest that the detection of ESR1 mutations in primary cancer by ddPCR may help predict failure during the early period of ET and help guide the early use of novel ESR1-mutant-targeting therapy.
Methods
Study population
Our study was approved by the Institutional Review Board of Gangnam Severance Hospital, Yonsei University, Seoul, Republic of Korea (IRB no. 3-2017-0349) and followed the Good Clinical Practice guidelines and the principles of the Declaration of Helsinki. The requirement for informed consent was waived due to the retrospective study design.
The medical records of 1667 patients with breast cancer who underwent breast surgery followed by adjuvant treatment at Gangnam Severance Hospital between January 1997 and December 2015 were reviewed. We identified 225 patients with primary non-metastatic ER+ breast cancer who experienced invasive tumor relapse after surgery. Formalin-fixed paraffin-embedded (FFPE) samples of primary tumors were available for ddPCR from 121 patients.
None of the patients had distant metastasis at the time of surgery. The available clinicopathologic data, including age; type of surgery; adjuvant treatment, including chemotherapy and ET; survival; ER status; HER2 status; histological type; histological grade; lymphovascular invasion status; and pathological stage. The consort diagram for the study population is displayed in Fig. 1. In the study population, 23 patients did not receive adjuvant ET due to the patients’ refusal (Fig. 1).
Patient’s classification according to endocrine resistance
According to the 5th International Consensus Conference for Advanced Breast Cancer guidelines, we classified the 98 patients treated with adjuvant ET into the following three groups: (i) primary endocrine resistance, (ii) secondary endocrine resistance, and (iii) endocrine sensitivity21. Primary endocrine resistance was defined as relapse during the first two years of adjuvant ET. Secondary endocrine resistance was defined as relapse while on adjuvant ET, but after the first two years, or relapse within 12 months of completing adjuvant ET. The other patients were classified as endocrine sensitive.
Droplet digital PCR
We collected 121 FFPE surgical specimens from patients with ER+/HER2− non-metastatic breast cancer. Representative tumor areas were identified, out of which at least three 10-μm-thick sections from the same FFPE samples were obtained, deparaffinized, and macrodissected. Genomic DNA was extracted using the QIAamp FFPE Tissue Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocol. Digital PCR reactions were performed using a QX200 Droplet Digital PCR System and custom ddPCR assays (Bio-Rad Laboratories, Hercules, CA, USA). We detected E380Q, Y537C, Y537N, Y537S, and D538G mutations in the ESR1 gene using probes targeting mutant and wild-type sequences, as previously described by Chu et al. and Jeselsohn et al.26,30. The primer and probe sequences are shown in Supplementary Table 4. The 20 μL PCR mix was composed of 10 μL of Bio-Rad ddPCR Supermix, 2 μL of each amplification primer/probe mix, and 8 μL of template DNA. Droplets then underwent the following thermal cycling protocol: one cycle of 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s and 46 °C (for E380Q) or 65 °C (for Y537 and D538G) for 1 min, followed by one cycle of 98 °C for 10 min. Results were analyzed using QuantaSoft v.1.7.2 software (Bio-Rad) and expressed as a percentage or fractional abundance of mutant DNA alleles compared to total DNA alleles.
When 20 non-tumorous samples were tested, 1 droplet/reaction was detected in 3 samples using E380Q mutation probe, and in 1 sample each using D538G and Y537N mutation probes. No positive droplets were detected in the results of the remaining mutation probes. Based on this, we set the limit of blank (LOB) at 0.857 copies/reaction (the highest value among LOB of mutation probes), and ddPCR result with a value of less than 2 positive droplets was reported as “not detected”.
Statistical analysis
Categorical values were compared by chi-square or Fisher’s exact tests. The RFI was measured as the period from the date of breast cancer surgery to the first breast cancer recurrence, including locoregional and distant recurrences. OS was defined as the period from the date of breast cancer surgery to death from any cause or the last censored day. The medians of survival outcomes were compared using a Mann–Whitney U test because we only included patients with tumor recurrence. Binary logistic regression analysis was performed to identify independent factors associated with primary endocrine resistance. We determined the AUC of the multivariable model using a receiver operating characteristic curve with the DeLong method31. Variables with p < 0.05, in univariate analysis, were included in the multivariate analysis. All analyses were performed using IBM SPSS Statistics for Windows 23.0 (IBM Corp., Armonk, NY, USA) and SAS (version 9.3, SAS Inc., Cary, NC, USA). Statistical significance was defined as p < 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was supported by the Hao Lin Chu Research Grant of Gangnam Severance Cancer Hospital. This study was supported by Alvogen Korea.
Author Contributions
S.G.A., S.J.B., and Y.K. are co-first authors. K.-A.L. and J.J. are considered as corresponding authors. Study concept and design: S.G.A., S.J.B., Y.K., K.-A.L., and J.J. Acquisition, analysis, or interpretation of data: S.G.A., S.J.B., Y.K., J.H.J., C.C., D.K., J.L., Y.J.C., K.-A.L., and J.J. Statistical analysis: S.G.A., S.J.B., Y.K., J.H.J., C.C., D.K., J.L., and Y.J.C. Drafting of the manuscript: S.G.A., S.J.B., Y.K., K.A.L., and J.J. The final draft of the manuscript was reviewed and approved by all authors.
Data availability
We identified the distribution of ESR1 mutation in next-generation sequencing MSK-IMPACT data from the previous study20. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sung Gwe Ahn, Soong June Bae, Yoonjung Kim.
Change history
5/18/2022
A Correction to this paper has been published: 10.1038/s41523-022-00436-8
Contributor Information
Kyung-A Lee, Email: kal1119@yuhs.ac.
Joon Jeong, Email: gsjjoon@yuhs.ac.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-022-00424-y.
References
- 1.Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol. Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 2.Comprehensive molecular portraits of human breast tumours. Nature490, 61–70, (2012). [DOI] [PMC free article] [PubMed]
- 3.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeselsohn R, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin. Cancer Res. 2014;20:1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merenbakh-Lamin K, et al. D538G mutation in estrogen receptor-α: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73:6856–6864. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 6.Robinson DR, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng DT, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razavi P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438.e426. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeselsohn R, et al. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell. 2018;33:173–186.e175. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yates LR, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32:169–184.e167. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinert T, Saad ED, Barrios CH, Bines J. Clinical implications of ESR1 mutations in hormone receptor-positive advanced breast cancer. Front. Oncol. 2017;7:26. doi: 10.3389/fonc.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fribbens C, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 2016;34:2961–2968. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 14.Wang P, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin. Cancer Res. 2016;22:1130–1137. doi: 10.1158/1078-0432.CCR-15-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelsomino L, et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res. Treat. 2016;157:253–265. doi: 10.1007/s10549-016-3829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeshita T, et al. Droplet digital polymerase chain reaction assay for screening of ESR1 mutations in 325 breast cancer specimens. Transl. Res. 2015;166:540–553.e542. doi: 10.1016/j.trsl.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl Acad. Sci. USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo YM, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl Acad. Sci. USA. 2007;104:13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toy W, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7:277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardoso F, et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5) Ann. Oncol. 2020;31:1623–1649. doi: 10.1016/j.annonc.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gates LA, et al. Proteomic profiling identifies key coactivators utilized by mutant ERα proteins as potential new therapeutic targets. Oncogene. 2018;37:4581–4598. doi: 10.1038/s41388-018-0284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlin M, et al. A computational assay of estrogen receptor α antagonists reveals the key common structural traits of drugs effectively fighting refractory breast cancers. Sci. Rep. 2018;8:649. doi: 10.1038/s41598-017-17364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shagufta, Ahmad I, Mathew S, Rahman S. Recent progress in selective estrogen receptor downregulators (SERDs) for the treatment of breast cancer. RSC Med. Chem. 2020;11:438–454. doi: 10.1039/C9MD00570F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan H, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 2017;377:1836–1846. doi: 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu D, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin. Cancer Res. 2016;22:993–999. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway K, et al. The estrogen receptor-alpha A908G (K303R) mutation occurs at a low frequency in invasive breast tumors: results from a population-based study. Breast Cancer Res. 2005;7:R871–R880. doi: 10.1186/bcr1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies MP, O’Neill PA, Innes H, Sibson DR. Hypersensitive K303R oestrogen receptor-alpha variant not found in invasive carcinomas. Breast Cancer Res. 2005;7:R113–R118. doi: 10.1186/bcr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghimenti C, Mello-Grand M, Regolo L, Zambelli A, Chiorino G. Absence of the K303R estrogen receptor α mutation in breast cancer patients exhibiting different responses to aromatase inhibitor anastrozole neoadjuvant treatment. Exp. Ther. Med. 2010;1:939–942. doi: 10.3892/etm.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeselsohn R, et al. Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc. Natl Acad. Sci. USA. 2017;114:E4482–e4491. doi: 10.1073/pnas.1620993114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We identified the distribution of ESR1 mutation in next-generation sequencing MSK-IMPACT data from the previous study20. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.