Abstract
The gene regulatory network for segmentation in arthropods offers valuable insights into how networks evolve owing to the breadth of species examined and the extremely detailed knowledge gained in the model organism Drosophila melanogaster. These studies have shown that Drosophila’s network represents a derived state that acquired changes to accelerate segment patterning, whereas most insects specify segments gradually as the embryo elongates. Such heterochronic shifts in segmentation have potentially emerged multiple times within holometabolous insects, resulting in many mechanistic variants and difficulties in isolating underlying commonalities that permit such shifts. Recent studies identified regulatory genes that work as timing factors, coordinating gene expression transitions during segmentation. These studies predict that changes in timing factor deployment explain shifts in segment patterning relative to other developmental events. Here, we test this hypothesis by characterizing the temporal and spatial expression of the pair-rule patterning genes in the malaria vector mosquito, Anopheles stephensi. This insect is a Dipteran (fly), like Drosophila, but represents an ancient divergence within this clade, offering a useful counterpart for evo-devo studies. In mosquito embryos, we observe anterior to posterior sequential addition of stripes for many pair-rule genes and a wave of broad timer gene expression across this axis. Segment polarity gene stripes are added sequentially in the wake of the timer gene wave and the full pattern is not complete until the embryo is fully elongated. This ‘progressive segmentation’ mode in Anopheles displays commonalities with both Drosophila’s rapid segmentation mechanism and sequential modes used by more distantly related insects.
Keywords: Anopheles, mosquito, evo-devo, segmentation, pair-rule patterning, gene regulatory network
Graphical Abstract
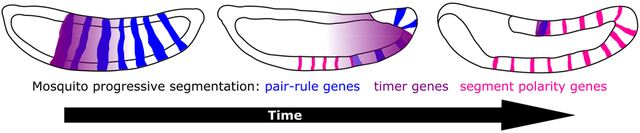
1. Introduction:
Arthropods share a segmented body plan, composed of repeating units, which is specified early in development. Maternal inputs initiate a gene regulatory network in which gap genes are expressed in broad regions along the anterior-posterior axis of the embryos. The gap genes work in combination to define the repeating patterns of the pair-rule genes in alternating segment primordia. Finally, the pair-rule genes direct the expression of segment polarity genes in each segmental unit. This gene network is best understood in the fruit fly, Drosophila melanogaster, in which the gap, pair-rule and segment polarity genes were first discovered (Nüsslein-Volhard & Wieschaus, 1980). Much of the body of work on segmentation over the last few decades has aimed to compare and contrast segmentation gene regulatory networks across the breadth of arthropod species, offering an unparalleled view into the mechanisms underlying the evolution of development (for review Chipman, 2020; Clark et al., 2019; Damen, 2007; Davis & Patel, 2002; Peel et al., 2005).
One hurdle to comparing different arthropod segmentation networks in a meaningful way is that Drosophila’s mode of segment patterning is not representative of the majority of arthropods, but instead is found in only holometabolous insects (Davis & Patel, 2002). Holometabolous insects, which undergo metamorphosis from a larvae to a distinct adult form, include Coleoptera (beetles), Hymenoptera (bees, wasps, and ants), Lepidoptera (moths and butterflies), and Diptera (flies). Within this clade, a wide range of segmentation modes including long-, intermediate-, short-germ, and even dual mechanisms, have been reported (Davis & Patel, 2002; Rosenberg et al., 2014). The current view of the differences between these modes focuses on how many segments are specified prior to gastrulation, but also takes into account traditional criteria such as whether or not the embryo elongates posteriorly as segments are added (Davis & Patel, 2002; Krause, 1939). Drosophila, with its long-germ mode, represents one extreme in which all segments are patterned more or less simultaneously prior to gastrulation. This mode is thus frequently referred to as “simultaneous” segmentation, which is contrasted to “sequential” segmentation in other species, where new segments are added from a posterior segment addition zone (SAZ) throughout the entirety of germband elongation. For simultaneous segmentation, segment patterning is completed during blastoderm stages before gastrulation and germband elongation begin and is thus very rapid. For sequential segmentation, segment patterning is gradual, occurring in a ‘one at a time’ fashion in an anterior to posterior order. Sequential segmentation is believed to be the ancestral mode for arthropods (Clark et al., 2019; Davis & Patel, 2002). Even among long-germ insects, Drosophila and other Brachyceran flies enjoy greater pre-patterning of the blastoderm by gap genes and more rapid segment generation than Nematoceran flies (a paraphyletic grouping of mosquitoes, midges, gnats), which delay posterior segment generation (Rohr et al., 1999). Although Drosophila’s segmentation gene network is intimately understood after decades of intense study, it is challenging to understand how and when its distinct mechanism emerged from an ancestral mode.
For many years, it has been hypothesized that the differences in segmentation modes are generated by heterochronic shifts in segment patterning (Davis & Patel, 2002). Recent work has led to several models for underlying genetic mechanisms (Clark et al., 2019; El-Sherif, Lynch, et al., 2012, p.; Zhu et al., 2017). One model proposes that a morphogen gradient of variable steepness explains different segment patterning speeds among insects (Zhu et al., 2017). A maternal gradient of hb is a good candidate for the morphogen executing this “speed regulation” model in Drosophila and Tribolium castaneum due to its effects on anterior-posterior patterning of the gap genes (Rudolf et al., 2020). Another model relies on the discovery that odd-paired (opa) works as a timing factor in Drosophila segmentation, promoting the transition from pair-rule to segment-polarity patterning (Clark & Akam, 2016; Koromila et al., 2020; Soluri et al., 2020). In Drosophila, this gene is expressed broadly across the whole trunk, overlaying completed pair-rule patterns and prompting a simultaneous transition to segment polarity stripe patterns. The different spatial-temporal deployment of opa and other timing factor genes in other insects led to the suggestion that opa serves as a “timing factor” throughout insects, potentially explaining heterochronic shifts in segmentation modes (Clark & Peel, 2018; Taylor & Dearden, 2021). Both the “Timing Factor” model and the Speed Regulation model also implicate posterior-anterior gradients of caudal (cad) and Wnt signaling in the evolution of segmentation modes (Clark et al., 2019; Zhu et al., 2017).
We previously published the expression patterns of the Anopheles stephensi orthologs of the Drosophila melanogaster pair-rule genes at the cellular blastoderm stage, at which time the patterns of seven broad stripes correspond extremely well between these two Dipteran species in spite of 272 million years of divergence (Cheatle Jarvela et al., 2020; Hedges et al., 2015). However, that work also demonstrated that one Drosophila pair-rule gene, paired (prd) has been lost from mosquito genomes entirely and its pair-rule function replaced by gooseberry (gsb), a segment polarity gene in Drosophila. In Anopheles, gsb is expressed in a pattern that combines features of a pair-rule and a segment polarity gene, starting out as wide stripes at blastoderm stages that resolve into narrow segmental stripes that persist through germband extension. Unlike Drosophila pair-rule genes, Aste-gsb stripes are generated gradually and sequentially in anterior-posterior order. We wondered whether this is a quirk of gsb, which has evolved to accommodate a novel function in the mosquito lineage, or indicative of an overall distinct developmental mechanism in Anopheles vs. Drosophila. Here, we investigated the dynamic temporal and spatial patterns of Drosophila pair-rule gene orthologs in Anopheles, uncovering differences in emergence and subsequent fading compared to Drosophila expression patterns. Interestingly, opa, and the nuclear receptor, ftz transcription factor-1 (ftz-f1), are expressed as waves that progress from anterior to posterior throughout Anopheles segmentation. Anopheles completes pair-rule patterning during blastoderm stages but adds stripes sequentially and converts pair-rule patterns to segmental expression over the whole course of germband elongation. Thus, our data indicate that Anopheles does not fit neatly into a sequential vs. simultaneous mechanism binary, but instead represents a progressive segmentation intermediate.
2. Methods:
All protocols and reagents have been previously described in detail (Cheatle Jarvela et al., 2020).
2.1. Mosquito Culture
Anopheles stephensi India strain was maintained at 29 °C with 80% humidity and a 12-h light/dark cycle. Larvae were fed a diet of pulverized fish food (Tetramin) and adults provided 10% (wt/vol) sucrose solution. Bovine blood (Lampire, Pipersville, PA) was provided by artificial membrane feeder 3–5 days prior to oviposition.
2.2. Gene Identification and Amplification
Anopheles stephensi orthologs of Drosophila segmentation gene network genes were identified by reciprocal BLAST (Altschul et al., 1990) as described previously (Cheatle Jarvela et al., 2020). Sequences of interest were amplified for in situ probe production using primer sets (Sup Table 1) with the T7 polymerase recognition sequence at the 5’ end of reverse primers.
2.3. Embryo Fixation and Gene Expression Analysis
Anopheles embryos were collected over the course of two hours on wet filter paper, then removed from the cage and aged at 29 °C to obtain three to six hour old embryos representing blastoderm, gastrulation and germband extension stages. Staged embryos were dechorionated in bleach, fixed in 4% paraformaldehyde, and peeled from their endochorions as described in previous work (Cheatle Jarvela et al., 2020; Goltsev et al., 2004; Juhn & James, 2012).
in situ hybridization was performed essentially as established for Drosophila embryos (Cheatle Jarvela et al., 2020; Goltsev et al., 2004; Tautz & Pfeifle, 1989). Antibody staining against En was performed prior to in situ hybridization when desired according to established protocols (Gutjahr et al., 1994). Briefly, fixed embryos were washed three times in PBTH (1 x PBS, 0.1% Tween-20, 50 μg/ml heparin, and 250 μg/ml tRNA) and incubated in 1:5 anti-En (catalog number 4D9, Developmental Studies Hybridoma Bank) diluted in PBTH with 1:1000 RNase inhibitor overnight at 4°C. The following day, the embryos were washed three times in PBTH and then incubated with 1:200 biotinylated goat anti-mouse secondary antibody (BA9200, Vector Labs) in PBTH plus 1:1000 RNase inhibitor for 1 hour. After three more washes in PBTH the embryos were incubated for 30 minutes in ABC reagent (Vectastain Elite ABC kit, Vector Labs), then washed another three times. Finally, expression was detected by DAB reaction (SigmaFast reagent, Sigma-Aldrich). After several washes in PBST to remove DAB reaction, embryos were processed for in situ hybridization.
All stained embryos were visualized using DIC on a Zeiss Axio Imager.M1 microscope. Using the 20x objective, we were able to capture entire whole-mount samples in a single image, yet also obtained sufficient resolution to view and count individual cells. Uniform brightness/contrast adjustments were applied to images using ImageJ (Schneider et al., 2012). Figures were arranged using Inkscape open-source software (https://inkscape.org/).
3. Results:
3.1. A subset of pair-rule gene seven-stripe expression patterns emerge in anterior to posterior order
The temporal dynamics of Aste-gsb, a gene with newly acquired pair-rule function in Anopheles, suggested a progressive mode of segment specification for this species. To assess whether this expression mode is representative of canonical pair-rule gene expression, we performed in situs for each Drosophila pair-rule gene ortholog to collect detailed time series encompassing early cellularization of the blastoderm through the end of germband elongation.
even-skipped (eve) was first detected as a broad domain of expression, as has been observed in another mosquito, Anopheles gambiae, as well as in Drosophila (Frasch et al., 1987; Goltsev et al., 2004). The first eve pair-rule stripe emerged at the anterior end of this broad domain (Figure 1a) and a second such stripe appeared in a similar fashion soon after (Figure 1b). Stripes 3–6 appeared next, the strength and definition of the stripes suggesting that 3 and 4 emerged prior to 5 and 6 (Figure 1d). Finally, seven stripes of roughly equal intensity were briefly present at the end of cellular blastoderm (Figure 1f). The stripes are dynamic over time, emerging as four-cells wide (Figure 1c), briefly reaching as large as six-cells wide (Figure 1e), and ultimately narrowing to two-cells wide (Figure 1g). From this point on, as gastrulation and germband extension proceeded, the stripes faded in anterior to posterior order starting with the one labeled “1” (Figure1a–k). No secondary stripes were ever observed. At the end of germband extension, the only remaining expression was in the proctodeum with pair-rule stripes no longer detectable, in agreement with observations from Anopheles gambiae (Goltsev et al., 2004). Previous analysis of eve expression in the species Anopheles gambiae indicated that it is much like Drosophila, with a broad region of expression resolving into seven stripes that fade all at once upon completion of germband extension (Frasch et al., 1987; Goltsev et al., 2004). While this may represent a difference between gambiae and stephensi, it is of note that the progressive pattern we observed was only clear when many embryos was examined, capturing intermediate timepoints.
Figure 1: Expression of Anopheles stephensi even-skipped and fushi-tarazu occur in an anterior to posterior progression.
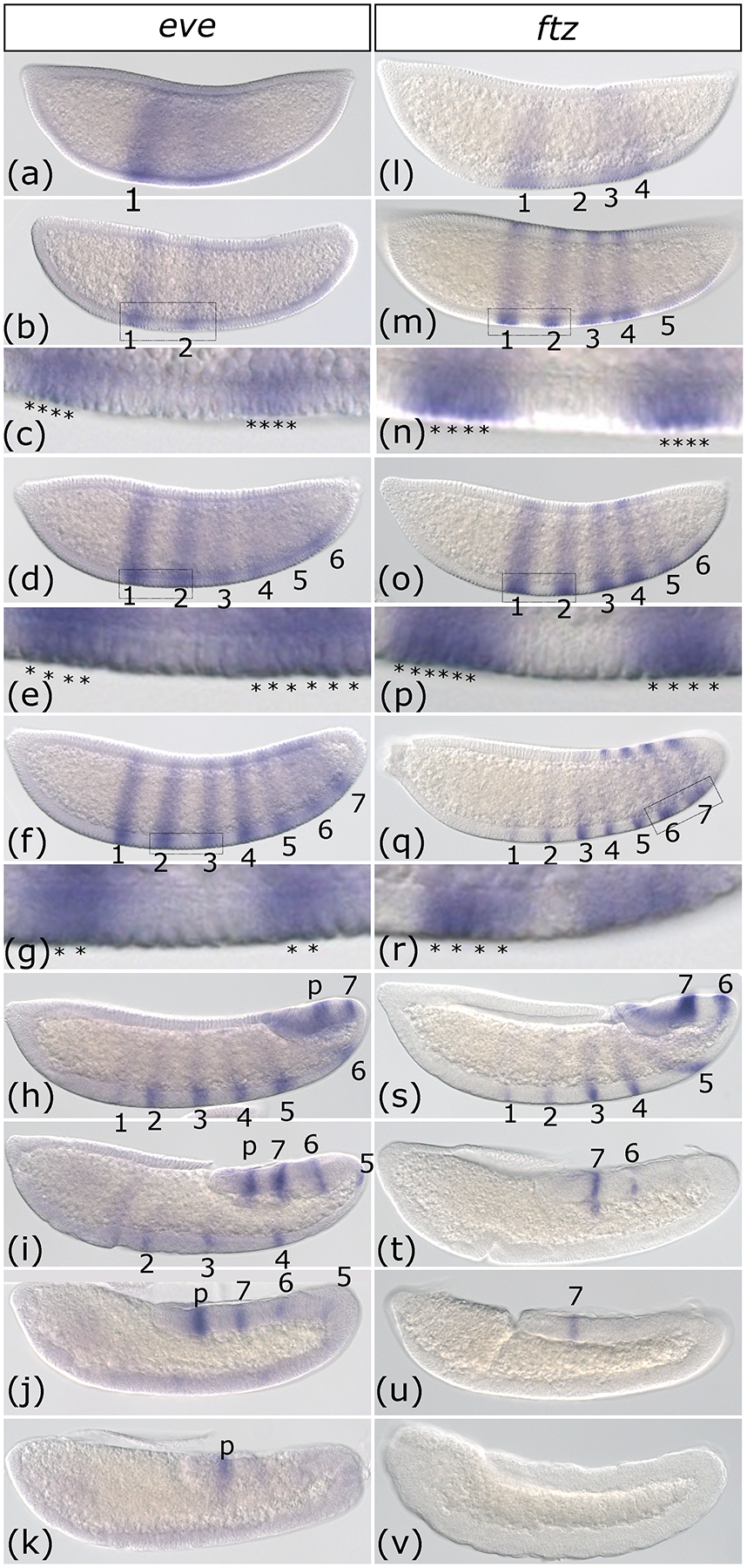
(a-k) Whole-mount in situ hybridization of even-skipped (eve) from early cellular blastoderm (a) to the end of germband extension (k). Expression begins as a broad domain (a) followed by just stripes 1 and 2 (b). (c) is an inset of the region indicated by a box in (b), with stained cells indicated by asterisks. Stripes continue to emerge in the posterior of the embryo (d) culminating in seven pair-rule stripes (f) plus an additional stripe of expression that lingers in the proctodeum (labeled “p”) (h-k). (e) and (g) are insets of the regions indicated by boxes in (d) and (f) respectively, with stained cells indicated by asterisks. Then the stripes fade in intensity and ultimately disappear, starting with stripe 1 (h), quickly followed by 2 and 3 (i), 4 and 5 (j) and finally 6 and 7 (k) leaving only the proctodeum stripe, which is not considered part of the pair-rule pattern. (l-v) Whole-mount in situ hybridization of fushi-tarazu (ftz) from early cellular blastoderm (l) to the end of germband extension (v). Expression begins with pair-rule stripes 1,2,3 and 4 (l), followed by addition of stripe 5 (m) and 6 (o) and 7 (q). ftz is briefly expressed in seven stripes at the end of the blastoderm stage (q). Panels n, p, and r are insets of the regions indicated by rectangles in m, o, and q respectively. As the germband elongates, stripes fade in anterior to posterior order starting with 1 and 2 (s), followed by 3, 4, and 5 (t) and finally 6 (u) and 7 (v). No ftz expression is detected in the fully elongated germband (v). Images are shown in order of increasing time. All embryos are depicted from a lateral view with the anterior on the left side.
Similarly, fushi tarazu (ftz) stripes emerged in the anterior half of the embryo prior to the posterior half (Figure1l), although these were not perfectly in order as stripe 2 lagged behind 1, 3, and 4. Yet, these first four stripes fully developed prior to the emergence of stripe 5 (Figure 1m) and likewise, stripe 6 emerged just after stripe 5 and before stripe 7 (Figure 1o). This is in contrast to Drosophila where ftz stripes emerge in a seemingly random order (Schroeder et al., 2011; Surkova et al., 2008; Yu & Pick, 1995). By the time stripe 7 appeared, stripe 1 had already become narrower and faded (Figure 1q). In general, the stripes are four-cells wide, however, stripe 1 appears to be wider and stripe spacing is not uniform (Figure 1n, p, and r). During early germband elongation, although all seven stripes were still visible, there was an anterior to posterior gradient of intensity and thickness with stripe 1 barely detectable and 7 still very strong (Figure 1s). As germband extension continued, the stripes faded out in order until no expression was detectable (Figure 1t–v). Surprisingly, at the end of germband extension, neurogenic expression was not observed, although this has been reported in several other arthropods, including Drosophila, and is thought to be well conserved (Heffer et al., 2013; Yu & Pick, 1995). This expression may emerge later during development or this difference could reflect the divergent nervous system patterning network described for mosquitoes (Suryamohan et al., 2016).
3.2. Aste-ftz-f1 and -opa are expressed in a wave that progresses from anterior to posterior during development.
As we previously described, Aste-ftz-f1 and -opa exhibit broad expression domains in Anopheles (Cheatle Jarvela et al., 2020). What was not evident from the single described stage in our previous work, but that can be seen here, is that expression of each of these genes moves across the anterior-posterior axis as a wave over time. During cellular blastoderm opa and ftz-f1 were each expressed in a band towards the anterior of the zone where pair-rule patterning will ultimately emerge (Figure 2a, i). Both patterns broadened to ultimately include the whole central region of the blastoderm, corresponding roughly to the outer limits of the seven-stripe pattern of genes like ftz and eve (Figure 2b,c, j, k). Initially, the anterior range of the pattern appeared stronger, likely because expression here was initiated earlier. Eventually, the anterior end of these broad domains faded as the posterior ends reached peak expression. This is much more evident for opa in the images shown (Figure 2d and l). By gastrulation, both patterns shifted to the posterior end of the embryo (Figure 2e and m). During germband extension, both ftz-f1 and opa were detected only in the posterior end as small diffuse bands (Figure 2f, g n, o). Upon completion of germband extension, opa developed a new, weak segmental stripe pattern, reminiscent of a segment polarity pattern (Figure 2h). ftz-f1 expression was no longer detectable by the end of germband extension (Figure 2p).
Figure 2: Expression of Anopheles stephensi odd-paired and ftz transcription factor-1 occur in an anterior to posterior wave.
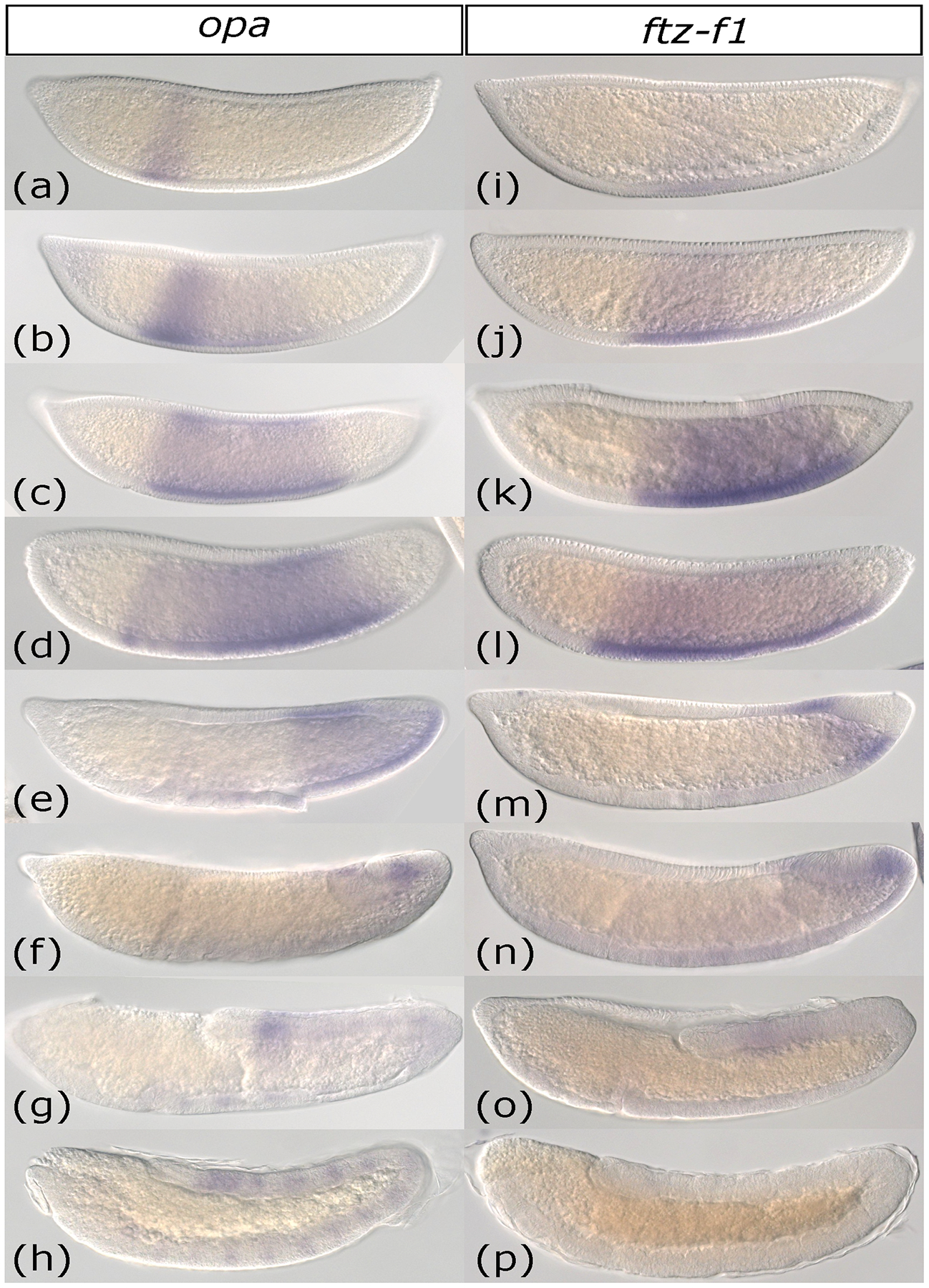
(a-h) Whole-mount in situ hybridization of odd-paired (opa) from early cellular blastoderm (a) to the end of germband extension (h). Expression is first detected as an anterior domain (a) which gradually expands posteriorly (b-c). Next, the anterior-most portion of the opa expression domain fades (d). By gastrulation, opa expression has retracted to the posterior half of the embryo (e). opa persists as a small expression domain near the posterior end of the elongating germband (f-h). Upon completion of germband extension, segmental stripes of opa appear (h). (i-p) Whole-mount in situ hybridization of ftz transcription factor-1 (ftz-f1) from early cellular blastoderm (i) to the end of germband extension (p). ftz-f1 expression begins as a faint band near the anterior of the embryo’s trunk (i). This band darkens (j) and then expands towards the posterior (k-l). ftz-f1 expression retracts posteriorly, leaving a narrow band at the posterior end of the embryo at gastrulation (m), which persists as expression near the end of the elongating germband (n-o). ftz-f1 expression completely fades by the end of germband extension (p).
Images are shown in order of increasing time. All embryos are depicted from a lateral view with the anterior on the left side.
3.3. Aste-slp, wg, and en stripes are activated in anterior to posterior order but persist through later stages
sloppy-paired (slp) expression was first observed as a wide anterior stripe (Figure 3a). Next, pair-rule stripes arose in a strict anterior to posterior order over the course of the blastoderm stage and into gastrulation (Figure 3b–g). Secondary stripes intercalated between the original pair-rule stripes during germband elongation, resulting in a segmental pattern (Figure 3f–g). This final pattern was similar to Aste-gsb stripes (Cheatle Jarvela et al., 2020) differing in that gsb stripes are initially four-cells wide, but later narrow to two-cells and split into separate stripes during cellular blastoderm stage, while slp stripes are always two-cells wide (Figure 3c, Supplemental Figure 1). Another difference is that while gsb stripes split soon after their initial generation, slp does not achieve a fourteen-stripe pattern until the end of germband elongation when another set of seven stripes arise in between the existing set all at once (Figure 3g, Supplemental Figure 1). This is in contrast to Drosophila, which also patterns one set of slp1 seven stripes prior to intercalation of a secondary set of seven, but all fourteen are complete prior to gastrulation (Cadigan et al., 1994; Clark & Akam, 2016; Grossniklaus et al., 1992). Drosophila slp2 is expressed with a slight temporal delay with respect to slp1 but is thought to be more closely related to Aste-slp, which could explain the slightly later expression pattern (Fujioka & Jaynes, 2012; Grossniklaus et al., 1992). The most intriguing difference is that in Anopheles, slp was expressed at the posterior end of the blastoderm as pair-rule stripe emergence began (arrow head, Figure 3b). This expression gradually disappeared as additional slp stripes were added (Figure 3d and e). This is not observed in Drosophila, even when embryos are meticulously staged and expression is quantified (Surkova et al., 2008).
Figure 3: Expression of Anopheles stephensi sloppy-paired, wingless, and engrailed occur in an anterior to posterior progression that is completed during germband elongation.
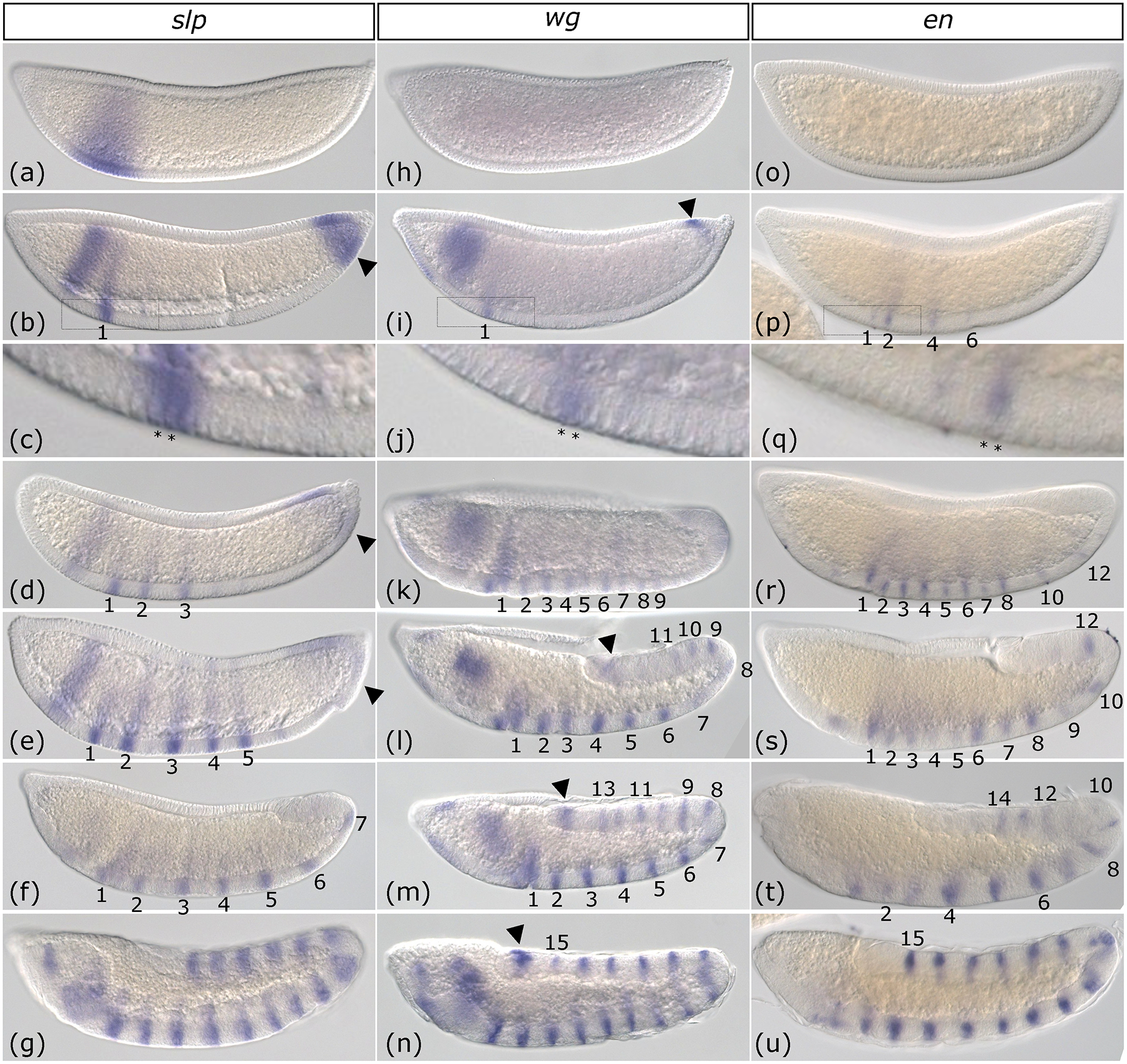
Whole-mount in situ hybridization of sloppy-paired (slp) (a-g), wingless (wg) (h-n), and engrailed (en) (o-u) from early cellular blastoderm to the end of germband extension. slp is first expressed as a broad anterior domain (a). Pair-rule stripe 1 emerges from this domain and a new domain of slp expression develops at the posterior terminus of the embryo (arrowhead) (b). Newly generated stripes are two-cells wide (c, inset of indicated region of b). Pair-rule stripes 2 and 3 (d) and 4 and 5 (e) are added sequentially and weak slp expression continues at the posterior terminus (arrowheads). At the beginning of germband extension, all seven pair-rule stripes are finally detectable, but secondary stripes are not (f). slp is expressed in a segmental pattern by the end of germband elongation (g). wg is not detected during early blastoderm (h). wg stripes (numbered) begin to develop later in blastoderm (i). wg stripes are two-cells wide (j). (j) is an inset of the indicated region in (i). Nine wg stripes are present at gastrulation (k). Stripes 10 through 15 develop sequentially as germband extension proceeds (l-n). A small domain of expression is also present at the posterior tip of the embryo throughout wg stripe addition (arrowhead i-n). en is not detected during early blastoderm (o). As the blastoderm stage progresses, even en stripes 2, 4, and 6 are visible and so is odd en stripe 1 (p). Stripes are two-cells wide once they have fully developed (q, an inset of the boxed region in p). At gastrulation, even en stripes 2–12 and odd en stripes 1–7 have developed (r). Both even and odd en stripes continue sequential addition in anterior-posterior order during germband extension (s-u). The full complement of fifteen en stripes is complete at the end of germband extension (u). Images are shown in order of increasing time. All embryos are depicted from a lateral view with the anterior on the left side.
Similar to slp, wingless (wg) was initially expressed in a broad anterior domain (Figure 3h–i). Stripes then appeared sequentially throughout blastoderm, gastrulation and germband extension stages (Figure 3i–n), which were two cells wide (Fig. 3j). All fifteen of wg’s segmental stripes arose in a strict anterior to posterior order (Figure 3i–n). wg was also expressed in the posterior-most region of the blastoderm and at the tip of the elongating germband (arrow heads Figure 3i, l-n). By the onset of germband extension, only nine of the segment polarity stripes had developed (Figure 3k). The full complement of wg segment polarity stripes was not observed until the end of germband extension (Figure 3n). This represents a temporal shift relative to Drosophila, which develops a full pattern of wg segment polarity stripes by gastrulation.
engrailed (en) stripes also emerged as two-cell wide stripes in an anterior to posterior progression (Figure 3o–u). However, the even-numbered en stripes emerged slightly before the odd-numbered stripes (Figure 3p–t), as they do in Drosophila, with each “cohort” emerging in an anterior-to posterior order when considered separately. Unlike in Drosophila, stripe addition was not complete at gastrulation, but instead continued throughout germband elongation (Figure 3r–u). Together, patterns of slp, wg, and en reveal differences in posterior patterning and a prolonged developmental progression that continues until germband extension is complete, in contrast to Drosophila, which develops fourteen-stripe patterns of each of these genes by gastrulation.
3.4. Aste-hairy, odd and runt do not have a strong anterior to posterior progression.
Three Drosophila pair-rule gene orthologs were not expressed progressively. The majority of hairy (h) stripes arose simultaneously (Figure 4a–b) and were six-cells wide at peak expression (Figure 4c). Only stripes 6–8 emerged in anterior to posterior order (Figure 4 b–e). h stripe development was roughly similar to Drosophila where the first six stripes develop before the seventh and the first three appear slightly ahead of the next three (Surkova et al., 2008). The three posterior-most stripes lingered longer into germband extension than the previous five (Figure 4f). In the fully elongated germband, faint and poorly defined stripes were visible along with stronger expression in the posterior tip of the germband and in parts of the head (Figure 4g).
Figure 4: Anopheles stephensi hairy, odd-skipped and runt are not expressed progressively.
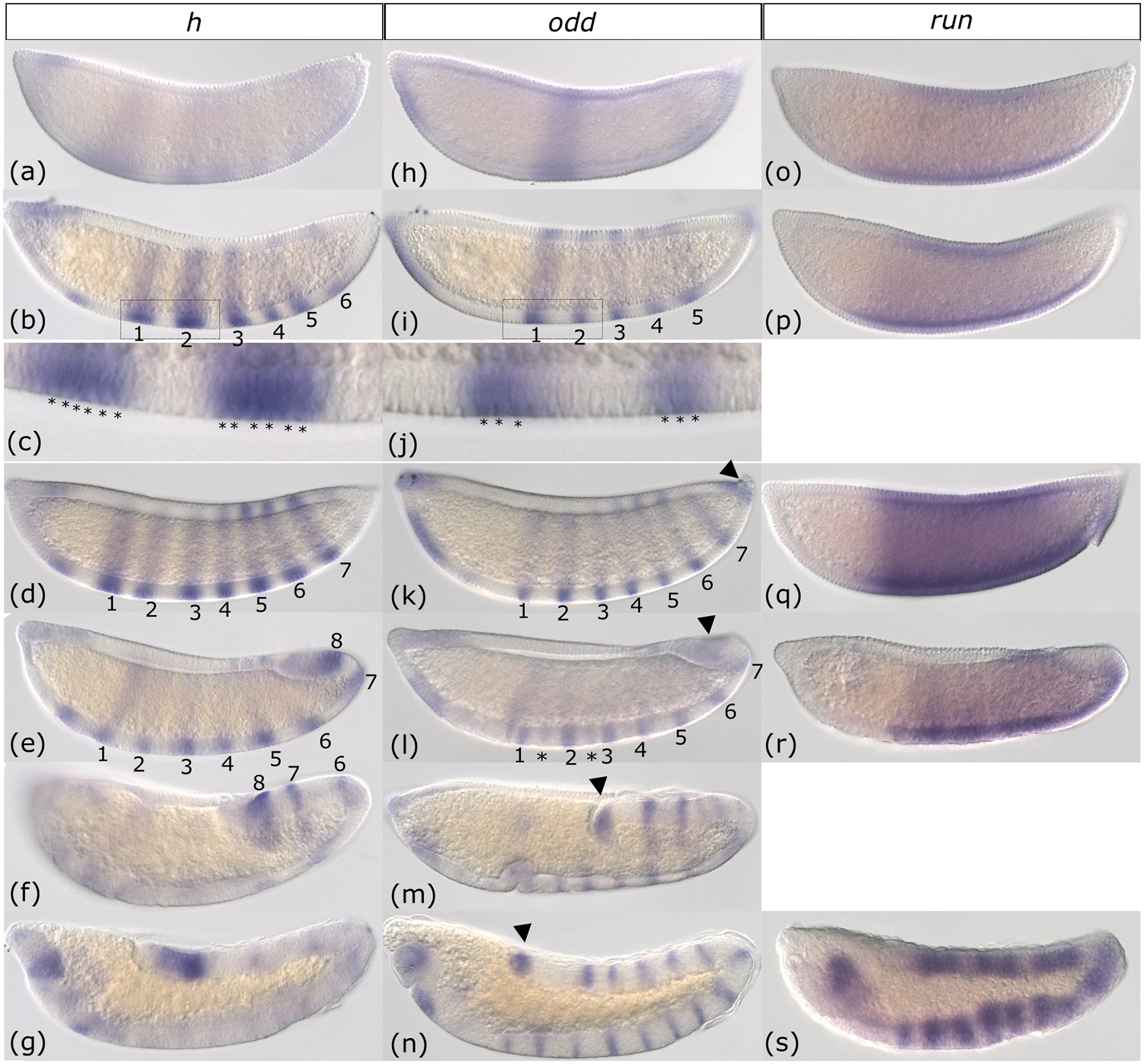
Whole-mount in situ hybridization of hairy (h) (a-g), odd-skipped (odd) (h-n), and runt (run) (o-s) from early cellular blastoderm to the end of germband extension. During early blastoderm, h stripes begin as faint stripes (a) and quickly resolve into six pair-rule stripes (b) that are six-cells wide (c). (c) represents an inset of the region indicated in (b). Seven h stripes are complete at the end of the blastoderm stage (d). An eight stripe is added as germband elongation begins (e). Partway through germband elongation, the anterior stripes fade but stripes 6, 7, and 8 remain (f). At the end of germband elongation, h is expressed in the head and at the posterior end of the germband (g). odd expression begins in a broad central domain (h) but five pair-rule stripes rapidly appear (i). These stripes are three-cells wide at their emergence (j). (j) represents an inset of the region indicated in (i). odd stripes 6 and 7 are detectable by the end of blastoderm stage, as well as a small domain at the posterior tip (k and arrowhead). At gastrulation, secondary stripes (asterisks) begin to intercalate between the pair-rule stripes (l). Both segmental and posterior tip odd expression patterns continue through germband elongation (m-n). run is initially expressed as a broad domain that spans the trunk of the blastoderm (o). This pattern continues through blastoderm (p-q) and gastrulation (r). By the end of germband elongation run transitions to a messy segmental pattern (s). Images are shown in order of increasing time. All embryos are depicted from a lateral view with the anterior on the left side.
For odd-skipped (odd), much like h, the first five pair rule stripes arose all at once from more indistinct patterns across the blastoderm (Figure 4 h–i). odd stripes were already narrow when they developed, with a width of 2–3 cells (Figure 4j). During the late blastoderm stage, all seven stripes were fully developed, and there was also a small region of expression at the very tip of the posterior, reminiscent of wg’s posterior expression (Figure 4k–n, arrowhead). Overall, the roughly simultaneous emergence of the first several stripes in early blastoderm and the later addition of stripe seven once the other stripes were established was much like Drosophila (Surkova et al., 2008). However, the non-conserved expression at the posterior end lingered throughout germband extension as secondary stripes of odd appeared (Figure 4l–n). Secondary stripe addition may occur progressively as only the first few were visible at the beginning of germband extension (Figure 4l).
Expression of runt (run) was highly unusual in that it appeared as a strong broad domain rather than as pair-rule stripes (Figure 4o–r). By the end of germband extension run developed segmental stripes, but they were broader and poorly defined when compared to stripes of odd, wg, and en (Figure 4s). This is distinct from run expression in Drosophila, which is expressed as a broad domain very early on, but quickly resolves into seven stripes (Gergen & Butler, 1988; Surkova et al., 2008). Unlike the broad expression domains of opa and ftz-f1 reported in Figure 2, run expression did not shift over time from the anterior to the posterior.
3.5. Gene expression order suggests conserved regulatory hierarchy but with different spatio-temporal dynamics.
We next sought to determine the relative timing of these different patterns by comparing the expression of progressively expressed pair-rule genes to a presumed segment polarity target, En. gsb, which is known to activate En, is expressed first as pair-rule stripes which progressively transition to segment polarity stripes (Cheatle Jarvela et al., 2020). When co-expression is examined in this known case of positive regulation, it is clear that gsb pair-rule stripes occur prior to En expression because they are visible more posteriorly (Supplemental Figure 1). The transition from pair-rule to segment polarity pattern is likely coordinated as we observe En stripes throughout the region where gsb stripes have resolved into their segment polarity phase.
At the blastoderm stage when all eight eve stripes were expressed, the first even-numbered En appeared in between eve stripes 1 and 2 (Figure 5a). As gastrulation and germband extension proceeded, En stripes appeared alternatively, with even stripes preceding odd stripes, and the latter overlapping with the eve stripes (Figure 5b–c). During germband elongation, En stripes retain a segmental pattern after eve has faded from the anterior-most stripes (Figure 5d). Overall, examining eve and En simultaneously demonstrates that eve’s anterior to posterior wave occurs earlier than En’s: within single embryos, eve stripes were observed at the posterior where En stripes were not yet detectable (Figure 5a–b) and En stripes were clear towards the anterior where eve had already faded (Figure 5d). When they coincide, eve stripes overlapped in expression with odd-numbered En stripes (Figure 5c), as in Drosophila, (Lawrence & Johnston, 1989) suggesting conserved regulation.
Figure 5: Engrailed stripes follow behind waves of pair-rule gene expression.
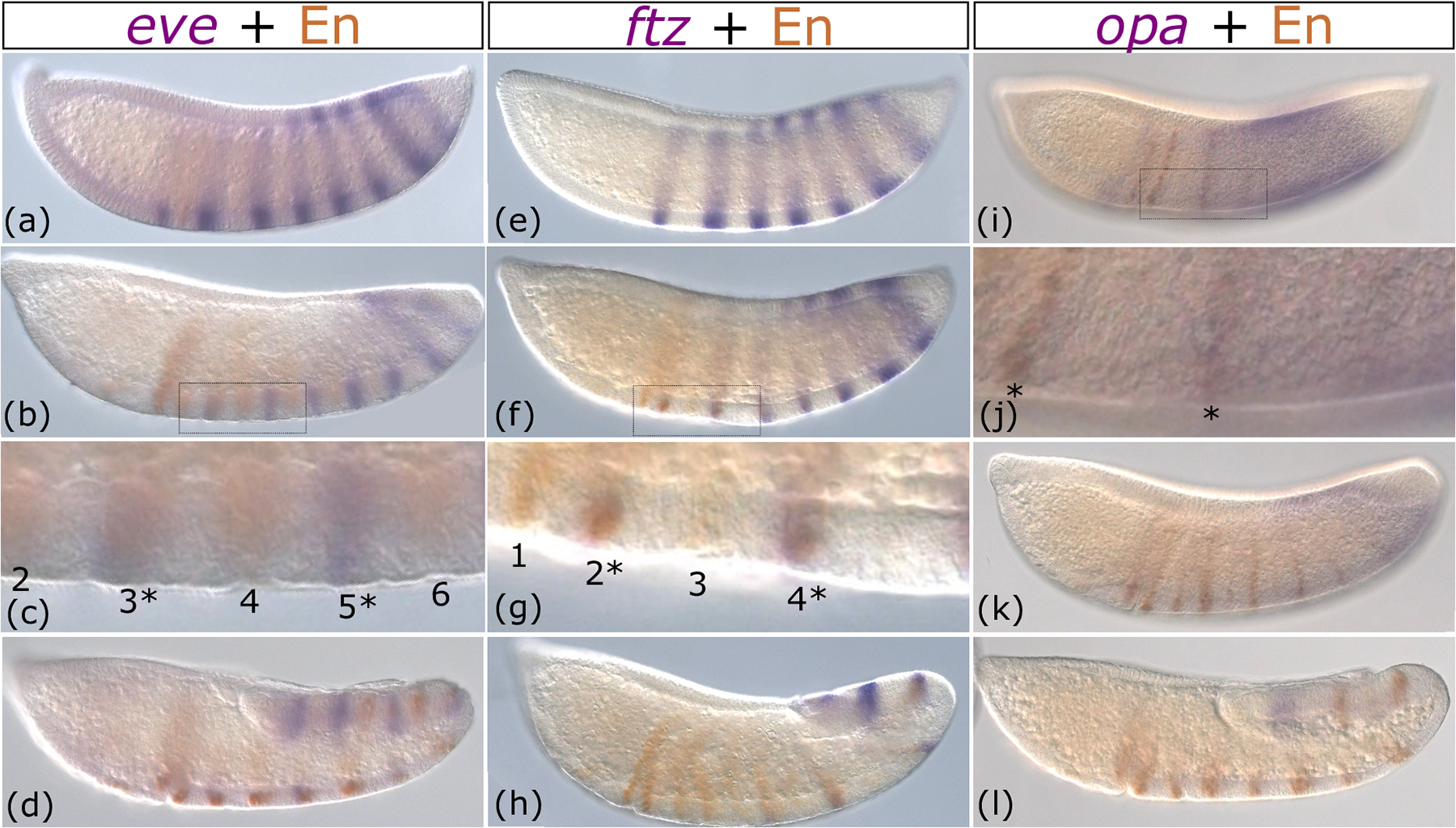
Immunohistochemistry of En (brown) combined with whole-mount in situ hybridization (purple) of eve (a-d) ftz (e-h) and opa (i-l) from late blastoderm through germband elongation. Images are shown in order of increasing time. All embryos are depicted from a lateral view with the anterior on the left side. In each column, pair-rule gene expression (purple) is first shown as a completed pattern that fades or retracts posteriorly in each successive row. En expression is first shown as just beginning as anterior stripes (a, e, i). In each successive row, more En stripes are added in anterior to posterior order in the wake of fading pair-rule gene patterns (b-d, f-h, and j-l). Panels c, g, and j are insets of the boxed regions shown in b, f, and I respectively.
Similarly, ftz expression preceded En expression, completing its seven-stripe pattern as the first, anterior-most En stripe was developing (Figure 5e). Over time, even-numbered En stripes briefly overlapped with fading ftz stripes (Figure 5f–h). By the time En stripe 12 developed, only three ftz stripes were observed (Figure 5h). Although we did not perform double in situs of ftz and eve, we can infer from their overlap with even and odd En stripes respectively that they lie in alternate stripe registers, as is true in Drosophila (Figure 5c vs. g). We can also predict that Ftz regulation of even-numbered En stripes (DiNardo & O’Farrell, 1987; Florence et al., 1997; Yu et al., 1997) is a conserved feature of Diptera.
Finally, En stripes developed progressively as opa’s broad expression domain retracted posteriorly (Figure 5i–l). That is, opa was expressed earlier in the cells that later expressed En. There were no detectable En stripes when opa was initially expressed across the trunk of the embryo (Figure 2j vs. 3o). In late blastoderm, once opa had cleared from the anterior and was expressed strongly at the posterior, En stripes 1, 2, 4, and 6 were observed (Figure 5i and j). In later germband extension stages, opa was expressed as a band in the posterior end where new En stripes had not yet emerged (Figure 5k). The posterior opa domain lingered in germband stages until the final En stripe developed (Figure 5l). These results indicate that the progression of opa occurs prior to the progressive addition of En stripes, suggesting conserved activation of en by Opa (Benedyk et al., 1994; Clark & Akam, 2016).
4. Discussion:
4.1. Anopheles deploys its segmentation network in an anterior-posterior progression.
Our previous work on Anopheles segmentation pointed to pair-rule patterning developing sequentially along the anterior-posterior axis (Cheatle Jarvela et al., 2020). This was surprising as previous work on another Anopheles species suggested pair-rule stripe development occurs as in Drosophila (Goltsev et al., 2004). However, our earlier work analyzed the temporal dynamics of only one gene - gsb, a gene with newly acquired pair-rule function in Anopheles, which would not necessarily be representative of canonical Anopheles pair-rule gene expression. Here, we analyzed the detailed spatio-temporal expression of all the Anopheles orthologs of Drosophila pair-rule genes, as well as the segment polarity genes en and wg. We found that that in Anopheles, the expression patterns of orthologs of Drosophila’s pair rule genes, eve, ftz, ftz-f1, opa, and slp arise in an anterior-posterior progression (Figures 1, 2, and 3). Non-progressive patterning of other pair-rule genes (run, h, odd, Figure 4) indicates that some steps towards the transition to simultaneous segmentation may have occurred in the Dipteran ancestor. We also found that segment polarity patterns of en and wg are not fully developed until the end of germband extension even though pair-rule stripe patterns are complete by the end of the blastoderm stage (Figure 3). This represents a temporal shift when compared to expression of the segment polarity genes in Drosophila. Together, these results indicate that segmentation in Anopheles does not occur as characterized in Drosophila and another mode must be employed.
4.2. Comparison to other insect species supports use of a progressive segmentation mode in Anopheles.
Simultaneous segmentation is highly correlated with long-germ embryos and sequential with short- and intermediate-germ embryos, but this is a generalization not a hard rule (Davis & Patel, 2002). These segmentation modes have been best studied in the model organisms, Drosophila melanogaster and Tribolium castaneum, respectively (Figure 6a). In sequential segmentation, gap and pair rule genes emanate from the posterior SAZ as waves (El-Sherif, Averof, et al., 2012; El-Sherif et al., 2014; Sarrazin et al., 2012; Zhu et al., 2017). In contrast, gap and pair-rule genes arise as mature domains in simultaneous segmentation thanks to highly pre-patterned eggs, with later posterior-anterior shifts in domain boundaries as the only remainder of dynamic ancestral patterning (El-Sherif, Lynch, et al., 2012; El-Sherif & Levine, 2016; Lim et al., 2018). Another distinction between these modes is that segment patterning is complete prior to gastrulation for simultaneous segmentation but is ongoing throughout germband elongation for sequentially-segmenting species. In contrast, progressive segmentation involves anterior-posterior addition of gene expression stripes that do not originate in a SAZ and so far has only been described in long-germ insects (Taylor & Dearden, 2021). This term was recently introduced to describe segmentation in the anterior of the jewel wasp (Nasonia vitripennis), which utilizes a dual mode in which distinct segmentation modes operate in different body regions (Rosenberg et al., 2014). Progressive segmentation is an intermediate between simultaneous and sequential segmentation (Figure 6a).
Figure 6: Evolutionary relationships among discussed insect species and their segmentation modes.
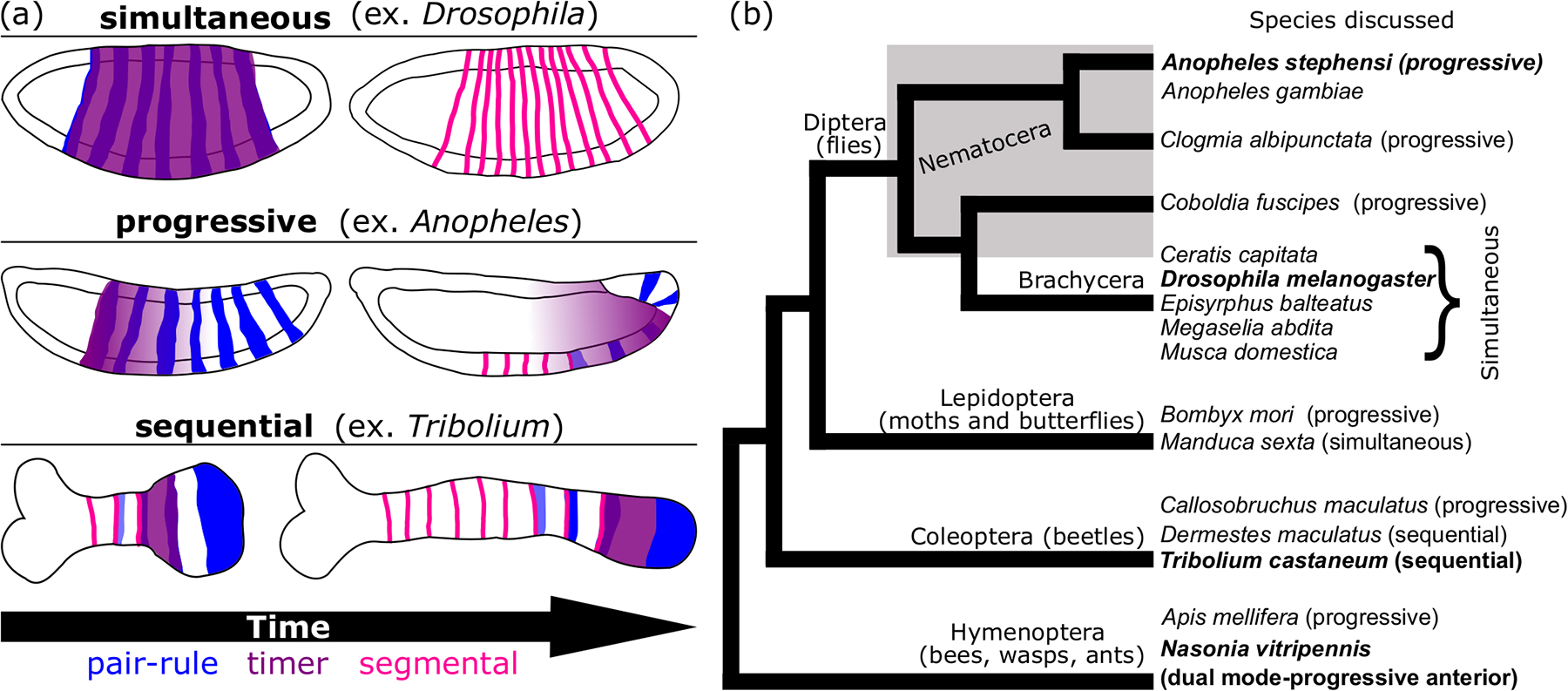
a) A schematic depicting each of the segmentation modes discussed in this work. Simultaneous (top), progressive (middle) and sequential (bottom) modes are depicted as characterized in the listed insect model (parentheses). General co-expression patterns of pair-rule (blue), timer (purple) and segmental (pink) genes are shown in representative locations on each embryo cartoon. For each mode, two embryo stages are shown, but note that the amount of time between the first and second stage is not the same for each mode. Each embryo is drawn with the anterior side on the left and the posterior on the right. (b) A cladogram depicting evolutionary relationships among holometabolous insects with major clades denoted. The paraphyletic grouping “Nematocera” is indicated by a gray box around included taxa. Species mentioned in the main text are listed and species discussed with great frequency are written in bold. Next to each species name, the presumptive segmentation mode is listed in parentheses. Modes are designated based on our assessment of the available literature (see Discussion and references therein).
Even though the term “progressive segmentation” is newly adopted, an analysis of older studies indicates that is likely applies to many of the holometabolous long-germ insects (Figure 6b). Anterior-posterior addition of pair-rule stripes has been observed in the Lepidopteran Bombyx mori (Nakao, 2010, 2015; Xu et al., 1997), the Hymenoptera Apis mellifera (Binner & Sander, 1997; Wilson & Dearden, 2012) and Nasonia vitripennis (Taylor & Dearden, 2021), the Coleopteran Callosobruchus maculatus (Patel et al., 1994), and Nematocera (Diptera) Clogmia albipunctata and Coboldia fuscipes (Bullock et al., 2004; Rohr et al., 1999) (Figure 6b). In many of these species, the eve pair-rule pattern is not complete before gastrulation. In Bombyx, progressive fading of eve stripes was also recorded, but it is uncertain how generally conserved the progressive pattern of fading may be due to lack of data from later stages in many of the other listed species (Xu et al., 1997). In contrast, the style of stripe development described in Drosophila, in which eve stripes arise out of order and are complete prior to gastrulation has also been observed in a variety of Brachyceran flies such as the scuttle fly Megaselia abdita (Bullock et al., 2004), the teprhitid fruit fly Ceratis capitata (Peterson et al., 2009), and the hoverfly Episyrphus balteatus (Lemke & Schmidt-Ott, 2009). Previous work on Anopheles eve lead to the hypothesis that the mosquitoes independently evolved an “extreme long-germ” mode of pair-rule patterning that is very similar to that of Drosophila (Jaeger, 2011). Our analysis confirms the finding that led to this hypothesis; like Drosophila, and unlike many of the other long-germ holometabola species discussed, all seven eve pair-rule stripes are formed by gastrulation in Anopheles. However, the progressive nature of the stripe development in Anopheles, described for the first time here, suggests a genetic mechanism that is actually more closely aligned with other progressively segmenting insects than with the Brachyceran flies. This idea is bolstered by the fact that slp’s pair-rule pattern (Figure 3) is not complete until germband extension begins. Similarly, patterns of en, and wg are not complete until germband extension is nearly complete while in Drosophila and other Brachyceran flies, en’s pattern is complete by gastrulation (Ingham, 1988; Sommer & Tautz, 1991). Together, our data indicate that Anopheles shares a progressive segmentation mode in common with a wide variety of holometabolous long-germ insects, and within Diptera, Drosophila’s extreme simultaneous mode likely evolved once at the base of Brachycera (Figure 6b). We hypothesize below, in agreement with others, that shifts in the expression of upstream gap patterning and downstream timer genes can lead to fluidity in how the segmentation gene network is deployed in developmental time and space (Clark & Peel, 2018; Schroeder et al., 2011; Zhu et al., 2017). This tunability could easily explain convergent emergence of a progressive segmentation mode in multiple holometabolous lineages (Figure 6b).
4.3. Differences in gap gene expression are correlated with segmentation mode
One possible explanation for the evolution of simultaneous segmentation within Diptera is differences in posterior gap gene expression (Jaeger, 2011). In Drosophila, a largely symmetrical arrangement of gap gene expression domains allows for the majority of pair-rule stripes to be specified using dual-stripe cis-regulatory elements (i.e. one element directs the expression of both eve stripe 3 and stripe 7, another ftz stripes 1 and 5) (Schroeder et al., 2011). Thus, the posterior stripes are specified at the same time as the anterior stripes, promoting simultaneous patterning. In other Brachyceran flies, posterior gap gene domains are expressed as in Drosophila, and these flies also undergo simultaneous segmentation (Bullock et al., 2004; Lemke et al., 2010; Lemke & Schmidt-Ott, 2009; Peterson et al., 2009; Rohr et al., 1999; Sommer & Tautz, 1991). In the mosquito Anopheles gambiae, anterior blastoderm expression of gap genes hunchback and giant are similar to Drosophila, but both lack the more posterior domain at this stage and only exhibit very weak posterior expression at gastrulation (Goltsev et al., 2004). In another progressively-segmenting nematoceran fly, the moth midge Clogmia albipunctata, differences in posterior gap gene expression are also observed, with no hunchback expression prior to gastrulation, and very weak and transient posterior expression of giant (García-Solache et al., 2010; Janssens et al., 2014). In contrast, the lepidopteran Manduca sexta, develops through a simultaneous mode and also exhibits posterior blastoderm expression of hb (Kraft & Jäckle, 1994). Together, these patterns of hb expression support the notion that the timing and strength of this gap gene’s expression has important implications for evolution of simultaneous segmentation mechanisms. Based on this, we hypothesize that without posterior domains of hb and gt, dual-stripe elements would not be functional in Nematoceran flies and instead individual stripe elements capable of responding to a dynamic anterior-posterior cue govern progressive stripe development.
Interestingly, overexpression of run in Drosophila alters gap gene expression patterns (Tsai & Gergen, 1994). Recent work demonstrated that when overexpressed, run specifically silenced expression from hb’s posterior stripe-generating element, but not the earlier-acting anterior stripe element (Koromila & Stathopoulos, 2017). Aste-run is unusual in that it is expressed ubiquitously through the trunk rather than as a striped pattern. One possible outcome of Aste-run’s comparatively expanded expression domain is prevention of posterior hb expression. Yet, predicted pair-rule and segment-polarity targets of Run, such as slp and en, appear to maintain conserved segmental expression patterns in Anopheles. Dmel-run’s repressor vs activator effects on these genes are determined by combinatorial interactions with Ftz and Opa (Hang & Gergen, 2017; Swantek & Gergen, 2004). Perhaps the dynamic expression of both ftz and opa in Anopheles limit Aste-run’s effects on segmental expression patterns to appropriate times and positions. Although Aste-run is one of the exceptions to the progressive expression trend recorded for other segmentation genes in Anopheles, it could nevertheless promote progressive vs. simultaneous patterning through potential regulation of gap genes like hb.
4.4. Differences in the spatio-temporal dynamics of timer gene expression are correlated with segmentation mode
Other differences in gene expression also offer clues about the transition from progressive to true simultaneous segmentation modes. In Drosophila, opa is expressed ubiquitously throughout the trunk during segmentation and provides a temporal cue necessary to transition pair-rule gene expression from seven-stripe/double segment patterns to fourteen- stripe/single segment patterns in concert with the onset of segment polarity gene expression (Benedyk et al., 1994; Clark & Akam, 2016). opa accomplishes this by acting as a pioneer factor, changing chromatin accessibility of late segmentation network cis-regulatory modules (Koromila et al., 2020; Soluri et al., 2020). In sequentially-segmenting beetles, Tribolium and Dermestes, a broad band of opa is seen in the anterior of the SAZ (Choe et al., 2017; Xiang et al., 2017). It has been hypothesized that this phase of opa expression provides a similar temporal input during segment addition as characterized in Drosophila, reconciling simultaneous and sequential mechanisms (Clark et al., 2019; Clark & Peel, 2018). In Anopheles, opa is expressed throughout the trunk (Figure 2) as in Drosophila, however, the dynamics of the Anopheles opa pattern is distinct from Drosophila’s. Aste-opa begins as a narrower, anteriorly positioned band that gradually expands, works its way towards the posterior of the blastoderm by retracting from the anterior, and lingers in the end of the elongating germband over time. Dmel-opa lacks these dynamics and is instead expressed uniformly across the trunk of the blastoderm stage (Benedyk et al., 1994; Clark & Akam, 2016). We find that expression of Aste-opa precedes En expression in each position along the AP axis, suggesting a conserved role in promoting the segment polarity phase of the segmentation gene network (Figure 5). This wavelike pattern of opa can explain the gradual transition from pair-rule to segment polarity type patterns. A similarly shifting opa expression pattern has recently been described in Nasonia, which undergoes progressive segmentation in the anterior portion of its trunk (Taylor & Dearden, 2021). This is suggestive of a common timing mechanism used in long germ insects to achieve progressive segmentation which was modified in the lineage leading to Drosophila to promote simultaneous segmentation.
We also describe new differences among Diptera in the expression of wg in the posterior blastoderm, and the elongating germband as well (Figure 3). This variation in wg expression likely represents a subtle shift in usage of a highly conserved regulator of segment phasing, considered to be among the suite of timer genes (Clark et al., 2019; Clark & Peel, 2018). In many insects, wg (or another wnt ligand) is used in the posterior-most SAZ and coordinates elongation and segment patterning (Bolognesi et al., 2008; Chesebro et al., 2013; Miyawaki et al., 2004; Oberhofer et al., 2014). In Drosophila, wg is expressed in the posterior at the cellular blastoderm stage, but as a stripe at the end of the segmenting trunk region, rather than as a persistent domain at the posterior-most cells of the embryo (Baker, 1987; Vorwald-Denholtz & De Robertis, 2011). It has been suggested that posterior wg expression in the Drosophila blastoderm is a vestige of this ancestral patterning mechanism, but it has long been difficult to test this function specifically (Vorwald-Denholtz & De Robertis, 2011). Future work on wg function in Anopheles could yield insights into the transition to simultaneous segmentation in Diptera.
4.5. Other differences in pair-rule gene deployment among Dipterans may affect segmentation mode
To our knowledge, orthologs of slp genes are not expressed in the posterior-most blastoderm of other insects. In Drosophila, a related forkhead domain transcription factor, forkhead, is expressed in this region and is important for repressing segment development in this region (Jürgens & Weigel, 1988; Weigel et al., 1989). FoxG (Slp) and FoxA (Fkh) recognize highly similar binding sites, suggesting that Slp might be able to repress the segmentation network in the posterior blastoderm in place of, or redundantly with, Fkh (UniPROBE database http://thebrain.bwh.harvard.edu/uniprobe/index.php) (Hume et al., 2015; Mariani et al., 2017; Shokri et al., 2019). We recently demonstrated that transcription factor paralogs can substitute for each other in gene networks and speculate that this could be a widespread phenomenon (Cheatle Jarvela et al., 2020). Perhaps slp plays a role in Anopheles’s alternate posterior patterning mechanism in the absence of posterior gap gene expression.
ftz-f1 encodes a nuclear receptor, which in addition to its pair-rule patterning role, and in common with nuclear receptors in general, mediates a wide variety of important developmental transitions in insects (for review Cheatle Jarvela & Pick, 2017). In Drosophila, ftz-f1 is expressed in blastoderm embryos but it’s pair-rule function is dependent upon co-expression with Ftz, limiting its activity in spite of its broad expression (Guichet et al., 1997; Yu et al., 1997; Yussa et al., 2001). It is notable that unlike in Anopheles, the entire anterior-posterior axis of Drosophila, and thus all seven Ftz stripes, have simultaneous access to Ftz-F1 at the blastoderm stage. This likely influences the simultaneous development of segmental gene expression patterns, such as en, which is a known target of the Ftz/Ftz-F1 complex (Florence et al., 1997; Guichet et al., 1997; Yu et al., 1997). In Anopheles, ftz-f1 is also expressed broadly, but the broad domain moves in a wave across the embryo, very similar to that of opa (Figure 2). If Ftz and Ftz-F1 interact to regulate en in Anopheles as they do in Drosophila, the wave of ftz-f1 expression would explain the anterior-posterior progression of Aste-en stripes. We note that in Tribolium, activation of en by Ftz-F1 is conserved, but ftz-f1 is expressed in pair-rule stripes rather than ubiquitously (Heffer et al., 2013). ftz-f1 is also expressed as stripes in other sequentially-segmenting insects (Reding et al., 2019; Xiang et al., 2017). Together, shifts from striped to progressive to ubiquitous expression during insect evolution, plus ftz-f1’s overall function in promoting developmental transitions, suggest a potential role for ftz-f1 in determining segmentation mode.
Data availability statement:
No large datasets were generated for this study. Information about experimental procedures, including primer sequences, is included in the manuscript.
Supplementary Material
Supplemental Figure 1: Co-expression of gsb and En demonstrate coordinated anterior to posterior progression from pair-rule to segment polarity pattern. Immunohistochemistry of En (brown) combined with whole-mount in situ hybridization of gsb (purple). A late blastoderm stage embryo is shown in both a lateral (a) and dorsal (b) view. A dotted line indicates the interface between pair-rule and segment polarity patterns. Pair-rule pattern occurs further towards the posterior, indicating that it occurs prior to segment polarity gene expression.
Acknowledgements:
We thank members of the Pick (UMD-College Park) and Chipman (HUJI) labs for thoughtful feedback on this work. We thank Channa Aluvihare (UMD-IBBR) for the kind gift of our Anopheles starter colony and for invaluable advice and protocols on mosquito rearing. Funding for this work was provided by the NIH (R01GM113230) to L.P.
Footnotes
Conflict of Interest: The authors declare no conflict of interest
References:
- Altschul SF, Gish W, Miller W, Myers EW, & Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Baker NE (1987). Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: The spatial distribution of a transcript in embryos. The EMBO Journal, 6(6), 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, & DiNardo S (1994). odd-paired: A zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes & Development, 8(1), 105–117. [DOI] [PubMed] [Google Scholar]
- Binner P, & Sander K (1997). Pair-rule patterning in the honeybee Apis mellifera: Expression of even-skipped combines traits known from beetles and fruitfly. Development Genes and Evolution, 206(7), 447–454. 10.1007/s004270050074 [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Farzana L, Fischer TD, & Brown SJ (2008). Multiple Wnt Genes Are Required for Segmentation in the Short-Germ Embryo of Tribolium castaneum. Current Biology, 18(20), 1624–1629. 10.1016/j.cub.2008.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Stauber M, Prell A, Hughes JR, Ish-Horowicz D, & Schmidt-Ott U (2004). Differential cytoplasmic mRNA localisation adjusts pair-rule transcription factor activity to cytoarchitecture in dipteran evolution. Development, 131(17), 4251–4261. 10.1242/dev.01289 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, & Gehring WJ (1994). Functional redundancy: The respective roles of the two sloppy paired genes in Drosophila segmentation. Proceedings of the National Academy of Sciences of the United States of America, 91(14), 6324–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, & Pick L (2017). The Function and Evolution of Nuclear Receptors in Insect Embryonic Development. Current Topics in Developmental Biology, 125, 39–70. 10.1016/bs.ctdb.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Trelstad CS, & Pick L (2020). Regulatory gene function handoff allows essential gene loss in mosquitoes. Communications Biology, 3(1), 1–10. 10.1038/s42003-020-01203-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro JE, Pueyo JI, & Couso JP (2013). Interplay between a Wnt-dependent organiser and the Notch segmentation clock regulates posterior development in Periplaneta americana. Biology Open, 2(2), 227–237. 10.1242/bio.20123699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD (2020). Chapter Ten—The evolution of the gene regulatory networks patterning the Drosophila Blastoderm. In Peter IS (Ed.), Current Topics in Developmental Biology (Vol. 139, pp. 297–324). Academic Press. 10.1016/bs.ctdb.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Choe CP, Stellabotte F, & Brown SJ (2017). Regulation and function of odd-paired in Tribolium segmentation. Development Genes and Evolution, 1–9. 10.1007/s00427-017-0590-7 [DOI] [PubMed] [Google Scholar]
- Clark E, & Akam M (2016). Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. ELife, 5, e18215. 10.7554/eLife.18215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E, & Peel AD (2018). Evidence for the temporal regulation of insect segmentation by a conserved sequence of transcription factors. Development, 145(dev155580). 10.1242/dev.155580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E, Peel AD, & Akam M (2019). Arthropod segmentation. Development, 146(dev170480). 10.1242/dev.170480 [DOI] [PubMed] [Google Scholar]
- Damen WGM (2007). Evolutionary conservation and divergence of the segmentation process in arthropods. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 236(6), 1379–1391. 10.1002/dvdy.21157 [DOI] [PubMed] [Google Scholar]
- Davis GK, & Patel NH (2002). SHORT, LONG, AND BEYOND: Molecular and Embryological Approaches to Insect Segmentation. Annual Review of Entomology, 47(1), 669–699. 10.1146/annurev.ento.47.091201.145251 [DOI] [PubMed] [Google Scholar]
- DiNardo S, & O’Farrell PH (1987). Establishment and refinement of segmental pattern in the Drosophila embryo: Spatial control of engrailed expression by pair-rule genes. Genes & Development, 1(10), 1212–1225. 10.1101/gad.1.10.1212 [DOI] [PubMed] [Google Scholar]
- El-Sherif E, Averof M, & Brown SJ (2012). A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development (Cambridge, England), 139(23), 4341–4346. 10.1242/dev.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E, & Levine M (2016). Shadow Enhancers Mediate Dynamic Shifts of Gap Gene Expression in the Drosophila Embryo. Current Biology, 26(9), 1164–1169. 10.1016/j.cub.2016.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E, Lynch JA, & Brown SJ (2012). Comparisons of embryonic development in Drosophila, Nasonia, and Tribolium. Wiley Interdisciplinary Reviews. Developmental Biology, 1(1), 16–39. 10.1002/wdev.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E, Zhu X, Fu J, & Brown SJ (2014). Caudal regulates the spatiotemporal dynamics of pair-rule waves in Tribolium. PLoS Genetics, 10(10), e1004677. 10.1371/journal.pgen.1004677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B, Guichet A, Ephrussi A, & Laughon A (1997). Ftz-F1 is a cofactor in Ftz activation of the Drosophila engrailed gene. Development (Cambridge, England), 124(4), 839–847. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, & Levine M (1987). Characterization and localization of the even-skipped protein of Drosophila. The EMBO Journal, 6(3), 749–759. 10.1002/j.1460-2075.1987.tb04817.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, & Jaynes JB (2012). Regulation of a Duplicated Locus: Drosophila sloppy paired is Replete with Functionally Overlapping Enhancers. Developmental Biology, 362(2), 309–319. 10.1016/j.ydbio.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Solache M, Jaeger J, & Akam M (2010). A systematic analysis of the gap gene system in the moth midge Clogmia albipunctata. Developmental Biology, 344(1), 306–318. 10.1016/j.ydbio.2010.04.019 [DOI] [PubMed] [Google Scholar]
- Gergen JP, & Butler BA (1988). Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes & Development, 2(9), 1179–1193. 10.1101/gad.2.9.1179 [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Hsiong W, Lanzaro G, & Levine M (2004). Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Developmental Biology, 275(2), 435–446. 10.1016/j.ydbio.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, & Gehring WJ (1992). The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes & Development, 6(6), 1030–1051. 10.1101/gad.6.6.1030 [DOI] [PubMed] [Google Scholar]
- Guichet A, Copeland JW, Erdélyi M, Hlousek D, Závorszky P, Ho J, Brown S, Percival-Smith A, Krause HM, & Ephrussi A (1997). The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature, 385(6616), 548–552. 10.1038/385548a0 [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Vanario-Alonso CE, Pick L, & Noll M (1994). Multiple regulatory elements direct the complex expression pattern of the Drosophila segmentation gene paired. Mechanisms of Development, 48(2), 119–128. [DOI] [PubMed] [Google Scholar]
- Hang S, & Gergen JP (2017). Different modes of enhancer-specific regulation by Runt and Even-skipped during Drosophila segmentation. Molecular Biology of the Cell, 28(5), 681–691. 10.1091/mbc.e16-09-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, & Kumar S (2015). Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution, 32(4), 835–845. 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffer A, Xiang J, & Pick L (2013). Variation and constraint in Hox gene evolution. Proceedings of the National Academy of Sciences of the United States of America, 110(6), 2211–2216. 10.1073/pnas.1210847110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, & Bulyk ML (2015). UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Research, 43(Database issue), D117–122. 10.1093/nar/gku1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW (1988). The molecular genetics of embryonic pattern formation in Drosophila. Nature, 335(6185), 25–34. 10.1038/335025a0 [DOI] [PubMed] [Google Scholar]
- Jaeger J (2011). The gap gene network. Cellular and Molecular Life Sciences: CMLS, 68(2), 243–274. 10.1007/s00018-010-0536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens H, Siggens K, Cicin-Sain D, Jiménez-Guri E, Musy M, Akam M, & Jaeger J (2014). A quantitative atlas of Even-skipped and Hunchback expression in Clogmia albipunctata (Diptera: Psychodidae) blastoderm embryos. EvoDevo, 5(1), 1. 10.1186/2041-9139-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhn J, & James AA (2012). Hybridization in situ of salivary glands, ovaries, and embryos of vector mosquitoes. Journal of Visualized Experiments: JoVE, 64. 10.3791/3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, & Weigel D (1988). Terminal versus segmental development in the Drosophila embryo: The role of the homeotic gene fork head. Roux’s Archives of Developmental Biology: The Official Organ of the EDBO, 197(6), 345–354. 10.1007/BF00375954 [DOI] [PubMed] [Google Scholar]
- Koromila T, Gao F, Iwasaki Y, He P, Pachter L, Gergen JP, & Stathopoulos A (2020). Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. ELife, 9, e59610. 10.7554/eLife.59610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromila T, & Stathopoulos A (2017). Broadly expressed repressors integrate patterning across orthogonal axes in embryos. Proceedings of the National Academy of Sciences, 114(31), 8295–8300. 10.1073/pnas.1703001114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R, & Jäckle H (1994). Drosophila mode of metamerization in the embryogenesis of the lepidopteran insect Manduca sexta. Proceedings of the National Academy of Sciences of the United States of America, 91(14), 6634–6638. 10.1073/pnas.91.14.6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G (1939). Die Eitypen der Insekten. Thieme. [Google Scholar]
- Lawrence PA, & Johnston P (1989). Pattern formation in the Drosophila embryo: Allocation of cells to parasegments by even-skipped and fushi tarazu. Development (Cambridge, England), 105(4), 761–767. [DOI] [PubMed] [Google Scholar]
- Lemke S, Busch SE, Antonopoulos DA, Meyer F, Domanus MH, & Schmidt-Ott U (2010). Maternal activation of gap genes in the hover fly Episyrphus. Development (Cambridge, England), 137(10), 1709–1719. 10.1242/dev.046649 [DOI] [PubMed] [Google Scholar]
- Lemke S, & Schmidt-Ott U (2009). Evidence for a composite anterior determinant in the hover fly Episyrphus balteatus (Syrphidae), a cyclorrhaphan fly with an anterodorsal serosa anlage. Development, 136(1), 117–127. 10.1242/dev.030270 [DOI] [PubMed] [Google Scholar]
- Lim B, Fukaya T, Heist T, & Levine M (2018). Temporal dynamics of pair-rule stripes in living Drosophila embryos. Proceedings of the National Academy of Sciences, 115(33), 8376–8381. 10.1073/pnas.1810430115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L, Weinand K, Vedenko A, Barrera LA, & Bulyk ML (2017). Identification of Human Lineage-Specific Transcriptional Coregulators Enabled by a Glossary of Binding Modules and Tunable Genomic Backgrounds. Cell Systems, 5(3), 187–201.e7. 10.1016/j.cels.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, Ohuchi H, & Noji S (2004). Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mechanisms of Development, 121(2), 119–130. 10.1016/j.mod.2004.01.002 [DOI] [PubMed] [Google Scholar]
- Nakao H (2010). Characterization of Bombyx embryo segmentation process: Expression profiles of engrailed, even-skipped, caudal, and wnt1/wingless homologues. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 314(3), 224–231. 10.1002/jez.b.21328 [DOI] [PubMed] [Google Scholar]
- Nakao H (2015). Analyses of interactions among pair-rule genes and the gap gene Krüppel in Bombyx segmentation. Developmental Biology, 405(1), 149–157. 10.1016/j.ydbio.2015.06.012 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, & Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature, 287(5785), 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oberhofer G, Grossmann D, Siemanowski JL, Beissbarth T, & Bucher G (2014). Wnt/β-catenin signaling integrates patterning and metabolism of the insect growth zone. Development, 141(24), 4740–4750. 10.1242/dev.112797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH, Condron BG, & Zinn K (1994). Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature, 367(6462), 429–434. 10.1038/367429a0 [DOI] [PubMed] [Google Scholar]
- Peel AD, Chipman AD, & Akam M (2005). Arthropod Segmentation: Beyond the Drosophila paradigm. Nature Reviews Genetics, 6(12), 905–916. 10.1038/nrg1724 [DOI] [PubMed] [Google Scholar]
- Peterson BK, Hare EE, Iyer VN, Storage S, Conner L, Papaj DR, Kurashima R, Jang E, & Eisen MB (2009). Big Genomes Facilitate the Comparative Identification of Regulatory Elements. PLOS ONE, 4(3), e4688. 10.1371/journal.pone.0004688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reding K, Chen M, Lu Y, Cheatle Jarvela AM, & Pick L (2019). Shifting roles of Drosophila pair-rule gene orthologs: Segmental expression and function in the milkweed bug Oncopeltus fasciatus. Development (Cambridge, England), 146(17). 10.1242/dev.181453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr KB, Tautz D, & Sander K (1999). Segmentation gene expression in the mothmidge Clogmia albipunctata (Diptera, Psychodidae) and other primitive dipterans. Development Genes and Evolution, 209(3), 145–154. 10.1007/s004270050238 [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Brent AE, Payre F, & Desplan C (2014). Dual mode of embryonic development is highlighted by expression and function of Nasonia pair-rule genes. ELife, 3, e01440. 10.7554/eLife.01440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf H, Zellner C, & El-Sherif E (2020). Speeding up anterior-posterior patterning of insects by differential initialization of the gap gene cascade. Developmental Biology, 460(1), 20–31. 10.1016/j.ydbio.2019.04.015 [DOI] [PubMed] [Google Scholar]
- Sarrazin AF, Peel AD, & Averof M (2012). A Segmentation Clock with Two-Segment Periodicity in Insects. Science, 336(6079), 338–341. 10.1126/science.1218256 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, & Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Greer C, & Gaul U (2011). How to make stripes: Deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development (Cambridge, England), 138(14), 3067–3078. 10.1242/dev.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri L, Inukai S, Hafner A, Weinand K, Hens K, Vedenko A, Gisselbrecht SS, Dainese R, Bischof J, Furger E, Feuz J-D, Basler K, Deplancke B, & Bulyk ML (2019). A Comprehensive Drosophila melanogaster Transcription Factor Interactome. Cell Reports, 27(3), 955–970.e7. 10.1016/j.celrep.2019.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soluri IV, Zumerling LM, Payan Parra OA, Clark EG, & Blythe SA (2020). Zygotic pioneer factor activity of Odd-paired/Zic is necessary for late function of the Drosophila segmentation network. ELife, 9, e53916. 10.7554/eLife.53916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R, & Tautz D (1991). Segmentation gene expression in the housefly Musca domestica. Development, 113(2), 419–430. 10.1242/dev.113.2.419 [DOI] [PubMed] [Google Scholar]
- Surkova S, Kosman D, Kozlov K, Manu, null, Myasnikova E, Samsonova AA, Spirov A, Vanario-Alonso CE, Samsonova M, & Reinitz J (2008). Characterization of the Drosophila segment determination morphome. Developmental Biology, 313(2), 844–862. 10.1016/j.ydbio.2007.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan K, Hanson C, Andrews E, Sinha S, Scheel MD, & Halfon MS (2016). Redeployment of a conserved gene regulatory network during Aedes aegypti development. Developmental Biology, 416(2), 402–413. 10.1016/j.ydbio.2016.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swantek D, & Gergen JP (2004). Ftz modulates Runt-dependent activation and repression of segment-polarity gene transcription. Development (Cambridge, England), 131(10), 2281–2290. 10.1242/dev.01109 [DOI] [PubMed] [Google Scholar]
- Tautz D, & Pfeifle C (1989). A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma, 98(2), 81–85. 10.1007/BF00291041 [DOI] [PubMed] [Google Scholar]
- Taylor SE, & Dearden PK (2021). Nasonia segmentation is regulated by an ancestral insect segmentation regulatory network also present in flies. BioRxiv, 2021.03.23.436706. 10.1101/2021.03.23.436706 [DOI] [Google Scholar]
- Tsai C, & Gergen JP (1994). Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development (Cambridge, England), 120(6), 1671–1683. [DOI] [PubMed] [Google Scholar]
- Vorwald-Denholtz PP, & De Robertis EM (2011). Temporal pattern of the posterior expression of Wingless in Drosophila blastoderm. Gene Expression Patterns : GEP, 11(7), 456–463. 10.1016/j.gep.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jürgens G, Küttner F, Seifert E, & Jäckle H (1989). The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell, 57(4), 645–658. 10.1016/0092-8674(89)90133-5 [DOI] [PubMed] [Google Scholar]
- Wilson MJ, & Dearden PK (2012). Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera). PloS One, 7(9), e46490. 10.1371/journal.pone.0046490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Reding K, Heffer A, & Pick L (2017). Conservation and variation in pair-rule gene expression and function in the intermediate-germ beetle Dermestes maculatus. Development, 144(24), 4625–4636. 10.1242/dev.154039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu P-X, Amanai K, & Suzuki Y (1997). Double-segment defining role of even-skipped homologs along the evolution of insect pattern formation. Development, Growth & Differentiation, 39(4), 515–522. 10.1046/j.1440-169X.1997.t01-3-00013.x [DOI] [PubMed] [Google Scholar]
- Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, & Pick L (1997). The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature, 385(6616), 552–555. 10.1038/385552a0 [DOI] [PubMed] [Google Scholar]
- Yu Y, & Pick L (1995). Non-periodic cues generate seven ftz stripes in the Drosophila embryo. Mechanisms of Development, 50(2–3), 163–175. [DOI] [PubMed] [Google Scholar]
- Yussa M, Löhr U, Su K, & Pick L (2001). The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mechanisms of Development, 107(1–2), 39–53. [DOI] [PubMed] [Google Scholar]
- Zhu X, Rudolf H, Healey L, François P, Brown SJ, Klingler M, & El-Sherif E (2017). Speed regulation of genetic cascades allows for evolvability in the body plan specification of insects. Proceedings of the National Academy of Sciences, 114(41), E8646–E8655. 10.1073/pnas.1702478114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Co-expression of gsb and En demonstrate coordinated anterior to posterior progression from pair-rule to segment polarity pattern. Immunohistochemistry of En (brown) combined with whole-mount in situ hybridization of gsb (purple). A late blastoderm stage embryo is shown in both a lateral (a) and dorsal (b) view. A dotted line indicates the interface between pair-rule and segment polarity patterns. Pair-rule pattern occurs further towards the posterior, indicating that it occurs prior to segment polarity gene expression.
Data Availability Statement
No large datasets were generated for this study. Information about experimental procedures, including primer sequences, is included in the manuscript.


