Abstract
microRNA-290 (miR-290) clusters are highly expressed in mouse preimplantation embryos, but their specific role and regulatory mechanisms in the development of mouse preimplantation embryos remain unclear. Here, we found that miR-291a-5p and miR-291b-5p, as mature microRNA molecules of miR-290 clusters, were dynamically expressed in mouse preimplantation embryos. The expression of miR-291a-5p and miR-291b-5p in mouse embryos increased during the 2–4-cell stages and was accompanied by the decreasing expression of the autophagy-related genes Atg5 and Becn1 in mRNA. Immunofluorescence studies showed that the formation of autophagosomes and autophagic lysosomes increased in the 1-cell stage, decreased in the 2-cell stage, and rapidly decreased during the 4–8-cell stage. Transmission electron microscopy (TEM) also demonstrated that there were autophagosomes in the cytoplasm of fertilized eggs with a double-layer membrane structure, whereas this structure was not observed in the unfertilized oocyte cytoplasm. Moreover, miR-291a/b-5p inhibited the protein and mRNA expression of Atg5 and Becn1 in NIH/3T3 cells. A dual-luciferase reporter assay confirmed that miR-291a/b-5p directly targeted the Atg5 and Becn1 genes. MiR-291a/b-5p repressed rapamycin-induced autophagy-related LC3-I to LC3-II conversion, ultimately inhibiting the formation of autophagosomes. Furthermore, the microinjection of mouse zygote cytoplasm with miR-291a-5p inhibitors increased the mRNA expression of Atg5 and Becn1 in mouse embryos and facilitated the first cleavage of mouse embryos and blastocyst formation. Our results suggest the important role of miR-291a/b-5p during mouse preimplantation embryo development.
MiR-291a-5p and MiR-291b-5p, was dynamically expressed and inhibited autophagy by targeting Atg5 and Becn 1 during mouse preimplantation embryo development.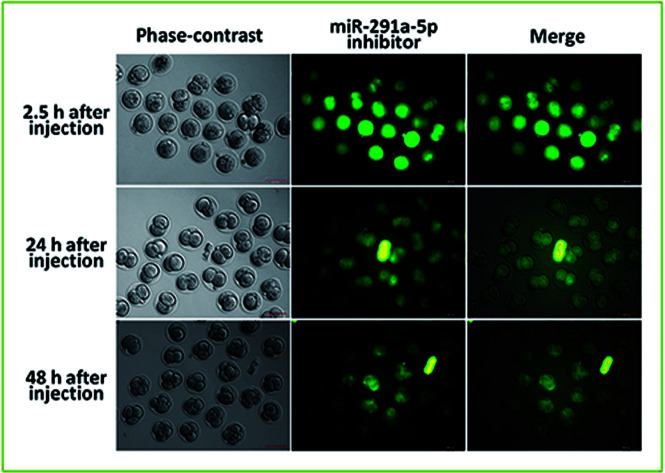
Introduction
The mammalian embryo development begins with the combination of sperm and ovum. When the sperm is combined with the ovum, the fertilized oocyte is activated and the early embryo development begins. After fertilization, the number of cells in the embryo increases rapidly, and the number of nuclei and amount of DNA increase exponentially, but the total amount of cytoplasm remains constant.1 In fact, mammals begin to prepare and accumulate a set of maternal mRNAs and proteins required for embryonic development as early as oocyte growth and maturation. After fertilization, the maternal mRNA and protein stored in the cytoplasm provide important nutritional support for embryonic development, which is also unique to preimplantation embryo development. After fertilization, these maternal stocks are rapidly degraded and replaced by new substances composed of fertilized ovum genes.2 In mice, fertilization activates the degradation process of the transcripts stored in the ovum, which is approximately 90% completed in the 2-cell phase.3
Autophagy is a highly conserved and critical metabolic degradation system in the cell. Through the autophagy system, cells can cope with hunger, hypoxia, immune response, etc., and gain survival advantage.4 As mentioned earlier, after fertilization, the cytoplasmic content of the embryo undergoes an “oocyte-to-embryo transition” process in which maternal RNA and proteins are degraded and used to provide amino acid energy.5 The embryonic genome performs new RNA and protein synthesis as well as organelle remodeling. Previous studies have shown that many of the maternal proteins in embryos are degraded via the ubiquitinated proteasome system.6 Recent new research suggests that autophagy, as another important degradation system, participates in the turnover of cytosolic proteins and plays an important role in this process.7–10 Atg5 and Ben1 genes are two critical regulatory molecules in the process of autophagy. Atg5-deficient oocytes, due to limited autophagy function, cannot develop further when the zygote develops to the 4–8-cell embryo phase.9 Becn1-deficient mutant mouse embryos have been shown to exhibit significant developmental delay and death at E7.5 days.11 The embryonic body composed of undifferentiated embryonic stem cells, after the deletion of Atg5 or Becn1, has defects in the function of recruiting and clearing dead cells, exhibits low intracellular ATP levels and is unable to develop cavities.12 These studies suggest that the autophagy-related genes Atg5 and Becn1 are important for early embryo development, energy supply, and maintenance of intracellular homeostasis.
MicroRNA (miRNA, miR) is a class of small single-stranded non-coding RNA molecules of approximately 22 nucleotides in length. The function of miRNA in inhibiting gene expression is based on its complementary to the 3′ untranslated region (3′UTR) of target mRNA through its seed sequence region. Binding to the target mRNA, miRNA acts to degrade the target mRNA or inhibit the translation of the target protein.13 The miR-290 gene cluster, cloned from mouse embryonic stem cells, ranges from miR-290 to miR-295 and is the most abundantly expressed miRNA in mouse embryonic stem cells.14 The miR-290 gene cluster is also abundantly expressed in mouse preimplantation embryos.15 After implantation in mouse embryos, the expression level of the miR-290 cluster rapidly declines to no expression.16 In various organs of adult rats, including heart, liver, spleen and lung, the miR-290 cluster is not expressed either.17 Recent studies have shown that the miR-290 cluster promotes mouse embryonic stem cell proliferation18 and also promotes mesoderm and endoderm differentiation and development by targeting Pax6.19 During mouse embryonic development, vitellicle and somite development defects occur in miR-290 cluster-deficient mice.20 These studies suggest that the miR-290 gene cluster may play an important role in regulating the development and differentiation of preimplantation embryos. However, the mechanism of miR-290 clusters on embryonic development through autophagy systems is not well understood.
In the present study, we investigated the dynamic expression profile of miR-291a/b-5p as well as Atg5 and Becn1 genes in preimplantation embryos. We found that miR-291a/b-5p inhibited autophagosome formation by targeting autophagy-related genes Atg5 and Becn1. We also found that inhibition of miR-291a-5p by embryonic cytoplasmic microinjection of miR-291a-5p inhibitors promoted the development of preimplantation embryos from the 1-cell phase to the 2-cell phase.
Experimental
Animals
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of The Fourth Military Medical University and approved by the Animal Ethics Committee of The Fourth Military Medical University (Animal Protocol no. 20151104). C57BL/6J mice were purchased from the experimental animal center of the Fourth Military Medical University. The male mice used for mating were 8 weeks old and replaced for more than 3 months. The female mice used for superovulation were 4 weeks old. The experimental mice were housed in the SPF (Specific Pathogen Free) animal room of the Fourth Military Medical University, with a constant 12 h light/dark cycle.
Cell culture and transfection
The mouse embryo-derived fibroblast cell line NIH/3T3 (gifted by Dr Shan Wang, Department of Biochemistry and Molecular Biology, Fourth Military Medical University) was cultured in DMEM medium supplemented with 10% fetal calf serum (FBS) and penicillin streptomycin mixture (penicillin 100 U ml−1, streptomycin 100 μg ml−1). The cells were cultured in a saturated humidity incubator at a temperature of 37 °C and 5% CO2. Cell transfections were performed with Lipofectamine® 2000 transfection reagent (Invitrogen) according to the reagent manual.
Induction of superovulation in mice and acquisition of MII oocytes
Female mice were intraperitoneally injected with pregnant mare's serum gonadotropin (PMSG, Ningbo Second Hormone Factory, China) at a dose of 8 IU per mouse. After 46–48 h, female mice were intraperitoneally injected with human chorionic gonadotropin (HCG, Ningbo Second Hormone Factory, China) at a dose of 8 IU. MII oocytes were collected 15 h after HCG injection. Specifically, female mice were sacrificed by cervical dislocation. The dissection of the abdominal cavity and separation of the bilateral oviducts of the mice were performed. The oviducts were placed in a droplet of M2 medium prepared in advance in a 35 mm culture dish pre-warmed at 37 °C. The ampullas of the fallopian tubes were torn under a stereo microscope, so that the cell-encapsulated oocyte mass was released. The periplasmic space was digested with hyaluronidase (working concentration 300 μg ml−1, Sigma-Aldrich) to remove cumulus granulosa cells; then intact and refractive MII oocytes were collected.
Collection of mouse embryos at different stages
In order to collect a large number of preimplantation mouse embryos at different developmental stages, the mice were divided into 4 groups, each of which contained 10 male mice and 10 superovulation female mice. Group I mice were treated with PMSG or HCG on day 1 or day 3, respectively; group II mice were treated on day 2 or day 4; group III mice were treated on day 3 or day 5; group IV mice were treated on day 4 or day 6. The drugs PMSG and HCG were intraperitoneally injected into the mice, and the female mice that had been successfully mated were fed according to the sub-component cages. On the 7th day, the pre-implantation embryos of the mice at different developmental stages were collected. To collect mouse embryos at the 1-cell stage, embryos were collected 22 h after HCG injection, using the same method as that for the MII oocytes. To collect mouse embryos between the 2-cell stage and the morula or blastocyst stage, each female mouse was sacrificed using the cervical vertebrae dislocation method, the abdominal cavity was dissected, and the fallopian tube and uterine horn were separated and placed in M2 medium pre-warmed at 37 °C. Finally, the syringe needle was inserted into the end of the fallopian tube, the fallopian tube was gently rinsed with M2, and the embryos or blastocysts were collected.
Construction of dual-luciferase reporter gene vector
To construct the Atg5 or Becn1 dual-luciferase reporter vector, target sequences of Atg5 or Becn1 were predicted using the online bioinformatics software TargetScan 6.2 (http://www.targetscan.org/). The target sequences were synthesized (Sangon Biotech, Shanghai) and inserted into the Sac1/Xba1 sites of the double luciferase reporter plasmid pmirGLO (Promega) to obtain the Atg5 or Becn1 reporter plasmids. The target sequences were as follows (lowercase letters indicate the sequence of the mutation sites): Atg5-WT-F: 5′-CTA GCG GCC GCT AGT TTG TAA CTT TGA TAA TGA ACT TGT AAC TTT GAT AAT GAA CT-3′; Atg5-WT-R: 5′-CTA GAG TTC ATT ATC AAA GTT ACA AGT TCA TTA TCA AAG TTA CAA ACT AGC GGC CGC TAG AGC T-3′; Atg5-mut-F: 5′-CTA GCG GCC GCT AGT TTG TAA CTT ATT AAT GAA CTT GTA ACT TAT TAA TGA ACT-3′; Atg5-mut-R: 5′-CTA GAG TTC ATT AAT AAG TTA CAA GTT CAT TAA TAA GTT ACA AAC TAG CGG CCG CTA GAG CT-3′; Becn1-WT-F: 5′-CTA GCG GCC GCT AGT ATG CTG TTT GAT ACT GTG TGA TGC TGT TTG ATA CTG TGT GT-3′; Becn1-WT-R: 5′-CTA GAC ACA CAG TAT CAA ACA GCA TCA CAC AGT ATC AAA CAG CAT ACT AGC GGC CGC TAG AGC T-3′; Becn1-mut-F: 5′-CTA GCG GCC GCT AGT ATG CTG TTA TTA CTG TGT GAT GCT GTT ATT ACT GTG TGT-3′; Becn1-mut-R: 5′-CTA GAC ACA CAG TAA TAA CAG CAT CAC ACA GTA ATA ACA GCA TAC TAG CGG CCG CTA GAG CT-3′.
Dual-luciferase activity assay
The cell culture medium was removed and washed with 1× PBS. 100 μl per well of 1× PLB passive lysate (Promega) was added to the cells, and the cells were lysed in a room temperature shaker for 15 min. Then the cell lysate of each well was transferred to a 1.5 ml tube and centrifuged at 12 000 rpm for 5 min. The supernatant was collected to detect the luciferase activity using a Progema GLOMAX 20/20 Luminometer (Promega).
Real-time PCR
Total RNA was extracted using a PicoPure® RNA Isolation Kit (Invitrogen) according to the manufacturer's instructions. Reverse transcriptase and real-time PCR were performed using the SYBR® PrimeScript miRNA RT-PCR Kit (TakaRa) according to the manufacturer's instructions. The fold change of expression was analyzed using the 2−ΔΔCt method. U6 or actin was used as an internal control for quantification. The primers used for PCR were as follows: miR-291a-5p F: 5′-CAT CAA AGT GGA GGC CCT CTC T-3′; miR-291b-5p F: 5′-GAT CAA AGT GGA GGC CCT CTC C-3′; Atg5 F: 5′-TGT GCT TCG AGA TGT GTG GTT-3′; Atg5 R: 5′-GTC AAA TAG CTG ACT CTT GGC AA-3′; Becn1 F: 5′-GGC CAA TAA GAT GGG TCT GA-3′; Becn1 R: 5′-CAC TGC CTC CAG TGT CTT CA-3′; Actin F: 5′-CAT CCG TAA AGA CCT CTA TGC CAA C-3′; Actin R: 5′-ATG GAG CCA CCG ATC CAC A-3′.
Western blot
The cell lysates were centrifuged at 12 000 rpm for 10 min at 4 °C. The supernatant was collected and the protein concentration of each sample was adjusted to 5 μg μl−1. The same amount of protein was separated on 10% SDS-PAGE gel and transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk at room temperature for 1 h, and then incubated with the indicated primary antibodies including rabbit anti-Atg5 (A0856, Sigma), rabbit anti-Becn1 (HPA028949, Sigma) or rabbit anti-LC3-I/II (ABC929, Sigma) overnight at 4 °C. After washing the membrane 3 times with each wash taking 5 min, the secondary antibodies (anti-actin, A4700, Sigma) were added for incubating for 1 h at room temperature. The bands on the membranes were visualized using an Immobilon Western Chemiluminescent HRP Substrate Chemiluminescence Kit (Millipore). The images were quantified using Image J 1.47 software.
Immunofluorescence
For fixation and infiltration, the collected oocytes and pre-implantation embryos at each stage were placed in a dark wet box with 1% paraformaldehyde (PFA, Sigma) and 0.2% Triton X-100 (Sigma) for 1 h at room temperature. The washing solution (3% BSA/PBS) was prepared, the infiltrated specimens were then washed sequentially in droplets 5 times with each wash taking 5 min. After washing, the samples were placed in the blocking solution (3% BSA, 10% FBS/PBS) at 4 °C overnight. Then the samples were incubated with primary antibody (1 : 500 dilution of rabbit anti-LC3, L8918, Sigma; 1 : 500 dilution of mouse anti-LAMP2, SAB1402250, Sigma) at 4 °C overnight. After incubation of the primary antibody, the samples were washed 3 times and then incubated with secondary antibodies (1 : 400 dilution of anti-rabbit, 1 : 400 dilution of anti-mouse, Sigma) at 4 °C overnight. Finally, the samples were observed under a fluorescence microscope (Olympus) in a dark room and photographed.
Transmission electron microscopy (TEM)
The mouse oviduct of each embedded sample was prepared, and the oviduct was transferred into pre-cooled 3% glutaraldehyde and fixed at 4 °C overnight. Then the samples were treated using the following steps: 1% osmic acid staining and fixation, ethanol gradient dehydration, acetone penetration and epoxy resin embedding and trimming. The semi-thin sections were prepared under light microscopy, and then the ultrathin sections were observed and captured using a JEM-2000EX transmission electron microscope.
Cytoplasmic microinjection
To prepare the microinjection plate, 37 °C preheated M2 medium was added to the bottom of the 35 mm plate and placed on the 37 °C Thermo Plate thermostat (Tokai Hit, Japan). A microloader capillary needle (Eppendorf) was used to absorb 3 μl microinjection reagent, and the injection needle was then added. Then the needle was installed on the FemtoJet 4i microinjection operator (Eppendorf) and the “Clean” button was pressed to unblock the needle. About 30 mouse embryos were transferred into the injection plate and microinjection was performed in batches. Microinjection of mouse embryonic cytoplasm was performed in a high-power field of the injection system by selecting a zygote that had a second polar body, a female pronucleus and a male pronucleus. After the injection was completed, the injured embryos were removed. The morphologically intact embryos were selected for further in vitro culture.
Statistical analysis
Data was expressed as mean ± standard deviation. The statistical analysis was performed with the student's t test, and p < 0.05 was considered statistically significant. GraphPad Prism 5 was used for statistical analysis, and Image-Pro Plus 6.0 was used for graphical analysis.
Results
Dynamic expression of miR-291a/b-5p, Atg5 and Becn1 in mouse MII oocytes and preimplantation embryos
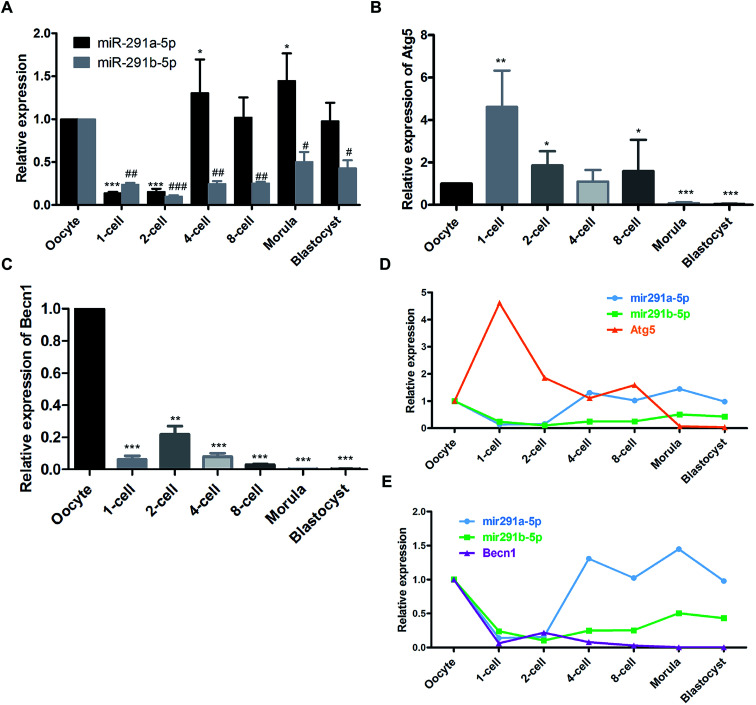
Firstly, we detected the dynamic expression of miR-291a-5p and miR-291b-5p at different developmental stages of mouse preimplantation embryos using real-time PCR assay. As shown in Fig. 1A, after fertilization, the expression of miR-291a-5p and miR-291b-5p in the 1-cell phase embryos decreased compared with that in oocytes. Subsequently, the expression of miR-291a/b-5p increased significantly from the 4-cell phase to the blastocyst phase. Then we measured the expression of the autophagy-related genes, Atg5 and Becn1, at different developmental stages of the mouse preimplantation embryos. The results demonstrate that after fertilization, the expression level of Atg5 mRNA in the 1-cell phase of the fertilized ova was significantly higher than that in the oocytes. The expression abundance decreased significantly at the 2-cell phase until the blastocyst phase (Fig. 1B). In contrast, the overall expression abundance of Becn1 mRNA was much lower at the 1-cell phase than that in the oocytes. During mouse preimplantation embryonic development, the overall expression of Becn1 increased at the 2-cell phase, then significantly declined at the 4-cell phase and gradually decreased until the blastocyst phase (Fig. 1C).
Fig. 1. Dynamic expression of (A) miR-291a/b-5p, (B) Atg5 and (C) Becn1 mRNA at different developmental stages of mouse preimplantation embryos measured using real-time PCR assay. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. miR-291b-5p expression in oocytes; *P < 0.05, **P < 0.01, ***P < 0.001 vs. miR-291a-5p, Atg5 or Becn1 expression in oocytes. Expression trends between miR-291a/b-5p and (D) Atg5 or (E) Becn1 in mouse preimplantation embryos.
Comprehensive analysis was performed to compare the expression of miR-291a-5p, miR-291b-5p, Atg5 and Becn1 at the different developmental stages of the mouse preimplantation embryos. The results showed that after fertilization, the expression of miR-291a-5p and miR-291b-5p increased gradually with the development of the mouse preimplantation embryos, and increased significantly at the 4-cell phase. The expression level of Atg5 mRNA increased significantly in the 1-cell phase of the fertilized ovum, and then gradually decreased from the 2-cell phase to the blastocyst phase (Fig. 1D). In contrast, the expression level of Becn1 mRNA was lower than that in the oocytes after fertilization. The Becn1 expression increased at the 2-cell phase, and then gradually decreased until the blastocyst phase (Fig. 1E). These results suggest that the expression trend of miR-291a/b-5p was reversed to the expression of Atg5 or Becn1 during the development of the preimplantation embryos.
Dynamic changes of autophagy in mouse MII oocytes and preimplantation embryos
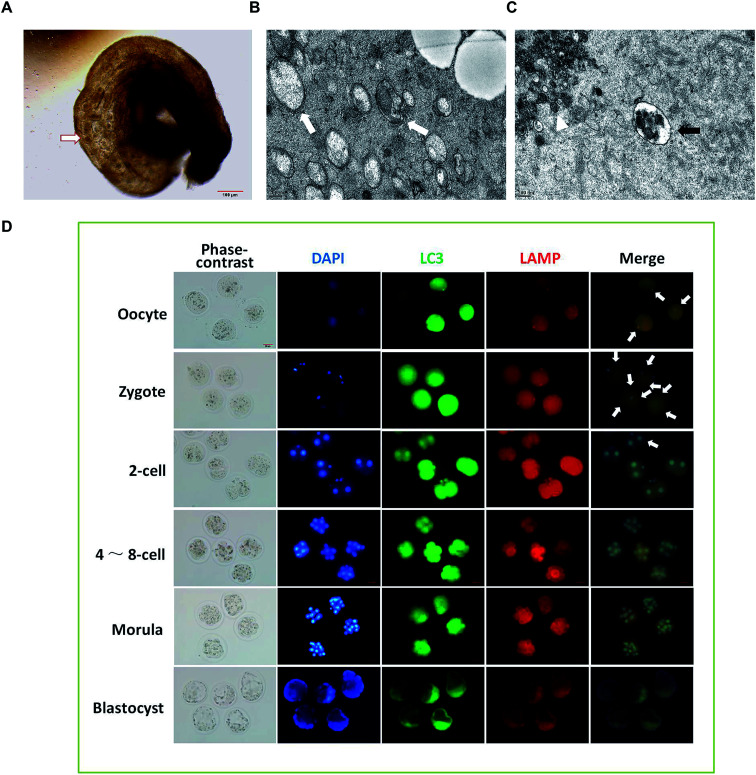
To observe the formation of autophagosomes in mice preimplantation embryos, the collected mouse preimplantation embryos were transferred into the swollen oviduct ampullas of the mice, and the dispersed embryos were embedded and fixed for TEM analysis (Fig. 2A). The ultrastructure of the mouse preimplantation embryonic cells was detected using TEM. The results show that the autophagy double-layer membrane structure appeared in the cytoplasm of the fertilized ovum at the 1-cell phase after fertilization, indicating the formation of initial autophagosomes (Fig. 2B). In the cytoplasm of the oocytes, the lysosomal structure was observed, but the double-layer membrane structure of autophagic vacuoles could not be detected (Fig. 2C).
Fig. 2. The dynamic changes of autophagy at different stages of embryo development. (A) Under microscope observation, the collected mouse preimplantation embryos were transferred into the swollen oviduct ampulla of the mice, and the dispersed embryos were embedded and fixed. (B) After fertilization, the autophagic vacuole bilayer membrane structure (white arrow) appeared in the 1-cell phase fertilized egg cytoplasm as revealed by TEM. (C) In the cytoplasm of the oocytes, TEM pictures show the lysosomal structure (black arrow), and cumulus granule cells (white Δ) were also observed. (D) Immunofluorescence was performed to detect the autophagy in preimplantation embryos of mice at different developmental stages. White arrows indicate the autophagic lysosomes.
To investigate the dynamic changes of autophagy in preimplantation embryos, immunofluorescence was performed using Alexa Fluor® 488 green fluorescent secondary antibody labeled LC3B primary antibody and Alexa Fluor® 594 red fluorescent labeled LAMP2 primary antibody. As shown in Fig. 2D, the green spot shows the LC3 aggregate, which represents the formation of autophagic vacuoles. The red spot is located at a site of accumulated lysosomal-associated membrane protein (LAMP), representing the location of the lysosome. The position where the green spot coincides with the red spot represents the formation of autophagic lysosomes, which result from the fusion of autophagic vacuoles and lysosomes. The results show that the production of autophagic lysosomes increased after fertilization, and then decreased significantly after the 2-cell phase. When developing at the 4–8-cell phase, the autophagic lysosomes reduced continuously and were difficult to detect at the blastocyst phase.
MiR-291a/b-5p inhibits the formation of autophagosomes by targeting Atg5 and Becn1
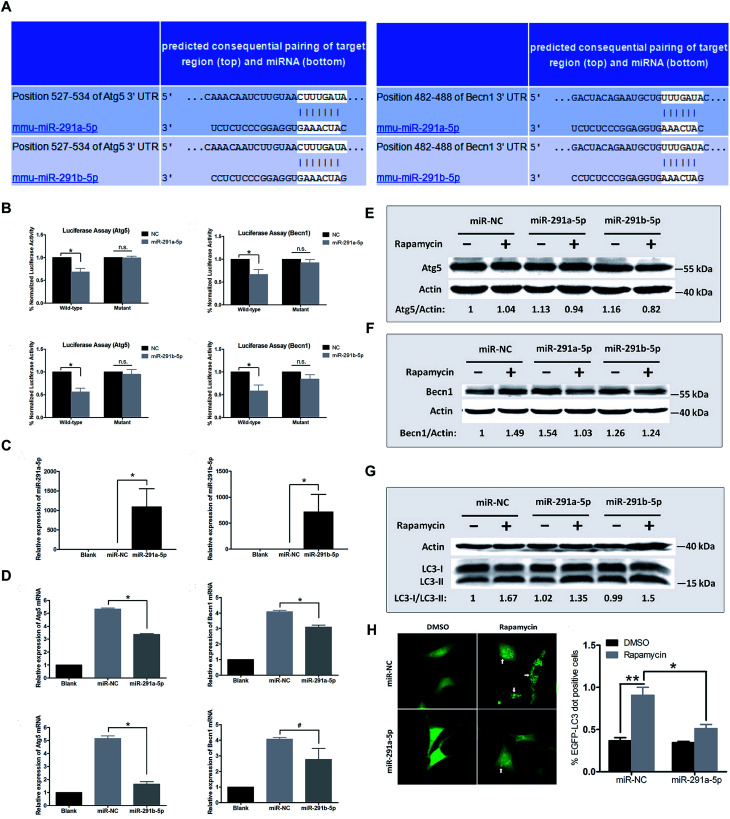
To further explore the relationship between miR-291a/b-5p and Atg5 or Becn1 genes, we employed the online bioinformatics software TargetScan6.2 for microRNA target prediction to analyze the potential miRNA binding sites in the 3′ untranslated region (3′UTR) of Atg5 and Becn1. As shown in Fig. 3A, the seed regions of miR-291a-5p and miR-291b-5p were predicted to bind the position 527–534 in the 3′UTR of Atg5 or position 482–488 in the 3′UTR of Becn1 mRNA. To verify the inhibitory effect of miR-291a/b-5p on the target genes, wildtype/mutant luciferase reporters containing the target/mutant region of Atg5 or Becn1 were constructed and a dual-luciferase activity assay was performed with NIH/3T3 cells. The results showed that miR-291a-5p and miR-291b-5p significantly inhibited the luciferase activity of wildtype Atg5 and Becn1 reporters but not the mutated reporters (Fig. 3B). To determine the effect of miR-291a/b-5p on the mRNA expression of Atg5 and Becn1 genes, miR-291a-5p or miR-291b-5p mimics were transfected into NIH/3T3 cells and the mRNA levels of Atg5 and Becn1 were examined using real-time PCR. After transfection, the expression levels of miR-291a-5p or miR-291b-5p were obviously upregulated compared with the miRNA negative control (miR-NC) transfected group (Fig. 3C). By overexpressing miR-291a/b-5p in NIH/3T3 cells, the expression levels of Atg5 or Becn1 mRNA were significantly inhibited in the miR-291a/b-5p mimics transfected group compared to the NC group (Fig. 3D). To further determine the effect of miR-291a/b-5p on the protein expression of Atg5 and Becn1, NIH/3T3 cells were transfected with miR-NC, miR-291a-5p or miR-291b-5p mimics. Then the transfected cells were treated with rapamycin for 24 h and the extracted proteins were collected for western blotting assay. The results showed that miR-291a-5p and miR-291b-5p slightly downregulated the Atg5 protein expression in cells after rapamycin-induced autophagy (Fig. 3E). In contrast, both miR-291a-5p and miR-291b-5p inhibited the expression of Becn1 protein. Among them, miR-291a-5p exhibited a more significant inhibitory effect than miR-291b-5p (Fig. 3F). Taken together, these results suggest that Agt5 and Becn1 are potential targets of miR-291a/b-5p.
Fig. 3. (A) Bioinformatics software TargetScan 6.2 was used to predict the binding region of miR-291a/b-5p in the 3′UTR sequence of Atg5 or Becn1. (B) Dual-luciferase reporter assay was performed in NIH/3T3 cells to verify the inhibition effect of miR-291a/b-5p targeting Atg5 or Becn1. (C) Real-time PCR was performed to obtain the expression levels of miR-291a-5p or miR-291b-5p in cells after transfection of miR-291a-5p or miR-291b-5p mimics in NIH/3T3 cells. (D) mRNA expression levels of Atg5 or Becn1 were detected in the NIH/3T3 cells transfected with miR-291a-5p, miR-291b-5p or miRNA negative control (miR-NC) mimics. After transfecting miR-NC, miR-291a-5p or miR-291b-5p mimics in NIH/3T3 cells, cells were treated with rapamycin for 24 h. Western blot was performed to detect the protein expression of (E) Atg5 or (F) Becn1. Actin was used as an internal control, which was set to 1. (G) After co-transfecting miR-NC, miR-291a-5p or miR-291b-5p as well as EGFP-LC3 plasmid in NIH/3T3 cells, cells were treated with rapamycin for 24 h, and the changes of LC3-I/II in cells were detected by western blot. Actin was used as an internal control, which was set to 1. (H) After co-transfecting miR-NC, miR-291a-5p and EGFP-LC3 plasmid in NIH/3T3 cells, cells were treated with rapamycin for 24 h. The LC3 aggregated particles (white arrow) were observed under a fluorescence microscope. Three fields were randomly selected to calculate the proportion of EGFP-LC3 aggregated particles (n = 3, *P < 0.05, **P < 0.01).
During the formation of autophagosomes, free LC3-I in the cytosol is modified and converted to LC3-II and finally aggregates on the membrane of autophagosomes. Therefore, LC3 is an important protein in the process of autophagosome formation. To investigate the effect of miR-291a/b-5p on autophagosomes in NIH/3T3 cells, we transfected NIH/3T3 cells with miR-291a/b-5p mimics and then induced autophagy using rapamycin. The ratio of LC3-I to LC3-II protein was detected by western blot assay to monitor the transformation of LC3-I to LC3-II, which reflected the changes of autophagosome formation. As shown in Fig. 3G, rapamycin induced the conversion of LC3-I to LC3-II in cells, while miR-291a-5p and miR-291b-5p downregulated the conversion of LC3-I to LC3-II. The downregulation effect of miR-291a-5p was more obvious than that of miR-291b-5p. To further monitor the extent of autophagosome formation in cells, we observed the LC3 aggregated particles using fluorescence microscopy. EGFP-LC3, the green fluorescent protein-labeled LC3B eukaryotic expression vector, was co-transfected into NIH/3T3 cells with miR-NC or miR-291a-5p mimics. After treatment with rapamycin in cells for 24 h, the number of LC3 aggregated cells was observed under a fluorescence microscope. The results showed that the numbers of LC3 aggregated cells increased significantly after induction of autophagy by rapamycin, while the transfection of miR-291a-5p significantly inhibited the numbers of LC3 aggregated positive cells (Fig. 3H). Collectively, these results suggest that miR-291a/b-5p inhibited the conversion of LC3-I to LC3-II by downregulating Atg5 and Becn1 expression, ultimately inhibiting the formation of autophagosomes.
MiR-291a-5p inhibitors promote the development of preimplantation embryos
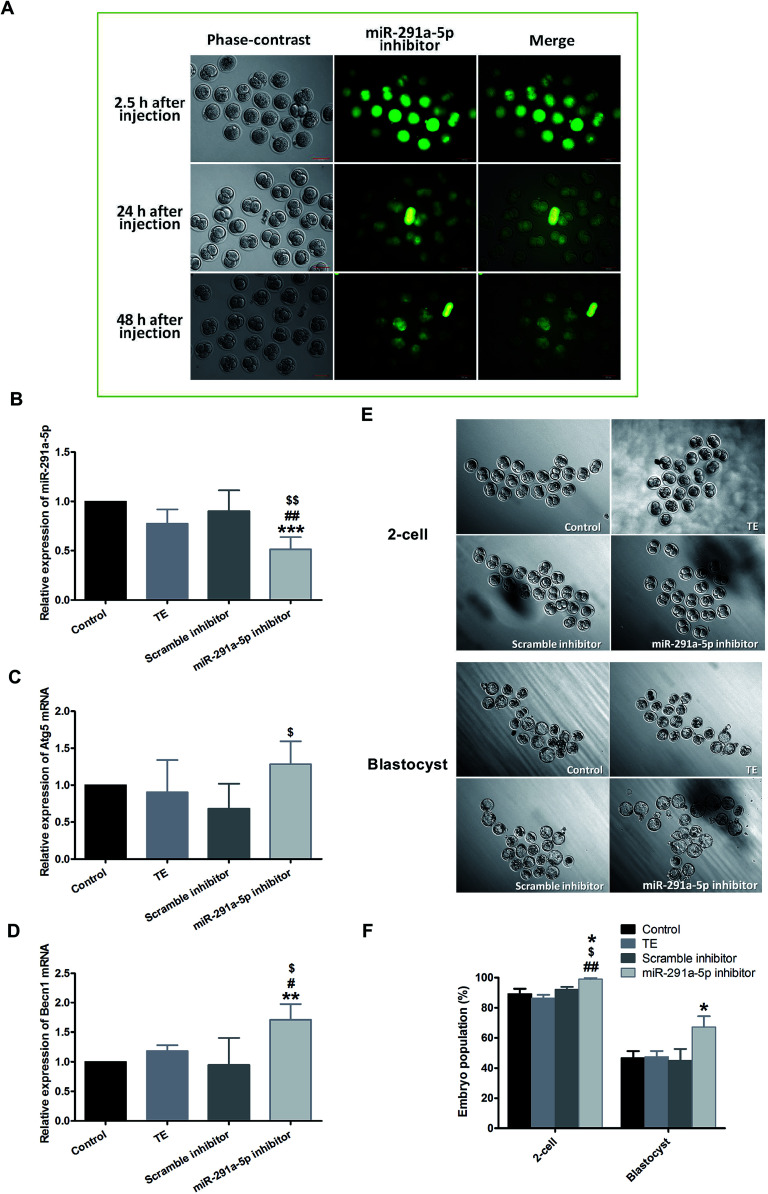
To explore the role of miR-291a-5p in the development of preimplantation embryos, the microinjection of mouse zygote cytoplasm with 5′ FAM-modified miR-291a-5p inhibitors was performed at the 0.5 days post-coitum (dpc) stage. After injection, the embryo was cultured in vitro, and the distribution of miR-291a-5p inhibitors was observed using a fluorescence microscope (Fig. 4A). The images show that miR-291a-5p inhibitors were evenly distributed in the embryonic cytoplasm 2.5 h after microinjection, and were further distributed in each blastomere following the fission of the embryo. Green fluorescence can be observed at 24 h and 48 h after injection, but the fluorescence intensity was weaker than that at 2.5 h after injection.
Fig. 4. (A) After microinjection of miR-291a-5p inhibitors, the expression of miR-291a-5p inhibitors in mouse embryos was observed using fluorescence microscopy. The mouse zygote was injected at 0.5 dpc with 5′ FAM-modified miR-291a-5p inhibitor molecules, which showed green fluorescence. 2.5 h, 24 h and 48 h after injection, green fluorescence was observed. Scale bar = 100 μm. After microinjection of the control, TE buffer, scramble inhibitor or miR-291a-5p inhibitor for 24 h, real-time quantitative PCR was performed to detect the expression of (B) miR-291a-5p, (C) Atg5 mRNA and (D) Becn1 mRNA in mouse embryos. $P < 0.05, $$P < 0.01 vs. scramble inhibitor group; #P < 0.05, ##P < 0.01 vs. TE group; **P < 0.01, ***P < 0.001 vs. control group. (E) After microinjection of control, TE buffer, scramble inhibitor or miR-291a-5p inhibitor for 24 h, the cleavage rates of mouse embryos at the 2-cell and blastocyst phases were observed under a microscope. (F) Analysis of mouse embryo development rate at 1.5 dpc (2-cell) and 4.5 dpc (blastocyst) after microinjection of various reagents, as above (n = 3, *P < 0.05 vs. control group, $P < 0.05 vs. scramble inhibitor group, ##P < 0.01 vs. TE group).
We subsequently examined the effect of miR-291a-5p inhibitors on miR-291a-5p expression in mouse embryos as well as the expression of Atg5 and Becn1 mRNA in embryonic cytosol. Real-time PCR results demonstrated that the expression of miR-291a-5p in the inhibitor group was significantly lower than that in the other three control groups (Fig. 4B, p < 0.05 compared with the TE buffer group or the scramble inhibitor group, p < 0.01 compared with the normal culture control group). After injection with miR-291a-5p inhibitors, the expression of Atg5 mRNA in the embryo cytoplasm was higher than that in the other control groups, but was only statistically significant compared with the scramble inhibitor group (Fig. 4C, p < 0.05). In contrast, the expression of Becn1 mRNA was significantly higher than that in the other control groups (Fig. 4D, p < 0.05 compared with the TE group or the scramble inhibitor group, p < 0.01 compared with the normal culture control group).
To observe the effect of miR-291a-5p inhibitors on mouse preimplantation embryo development, we finally collected mouse zygotes and performed cytoplasmic microinjection of 1-cell phase mouse embryos at 0.5 dpc stage. After the injection, the developmental rates of embryo development to the 2-cell phase and blastocyst phase were observed at 1.5 dpc and 4.5 dpc, respectively. The results demonstrate that when the mouse embryos developed from the 1-cell to the 2-cell phase, the embryo cleavage rate in the miR-291a-5p inhibitor group was higher than that in the other control groups (p < 0.05). At 4.5 dpc, the blastocyst formation rate in the miR-291a-5p inhibitor group was statistically higher than that in the other control groups (p < 0.05) (Fig. 4E and F). These results indicate that microinjection of miR-291a-5p inhibitors significantly inhibited the expression of miR-291a-5p in embryonic cytoplasm, increased the development rate of mouse embryos from the 1-cell stage to the 2-cell stage, and promoted blastocyst formation.
Discussion
During ovum development, a large number of maternal substances, including mRNA and protein, are stored in the oocytes. After fertilization, these substances are rapidly degraded, and the genes of the fertilized ovum encodes lots of new intracytoplasmic substances. This process by which this cytoplasmic content in the ovum is transformed into the cytoplasmic content in the fertilized ovum is called the oocyte-to-embryo transition or the maternal-to-zygotic transition.21 This transition mode is conserved across multiple species and is critical for embryonic development. If the cytoplasmic material cannot be degraded, it will impair the further development of the embryo. The known ubiquitin/proteasome-mediated pathways for protein degradation are not sufficient to clean up the maternal products during the transformation process.22 Recent studies have suggested that autophagy degradation systems play an important role in this cleanup process.23,24 In the present study, we demonstrated that the inhibition of early embryonic miR-291a-5p promotes the expression of the autophagy-related genes Atg5 and Becn1, which may play a role in promoting autophagy, accelerating the degradation of maternal substances and the production of amino acid energy, thereby advancing the early development of mouse embryos.
Becn1 is one of the earliest autophagy-related genes in mammals. Becn1 is a coiled-coil protein of approximately 60 kDa in size and contains a binding domain that interacts with Bcl-2 protein.25 Becn1 forms a complex by binding to phosphatidylinositol-3-kinase (PI3K) vacuolar sorting protein 34 (VPS34),26 which is a key protein in the initial stage of autophagosome formation and promotes the formation of the bilayer membrane structure of autophagosomes. Yue et al. constructed a mutant mouse with Becn1 deletion, and found that these mutant mouse embryos showed significant developmental delay, and death occurred on E7.5 days. Moreover, the visceral endoderm size and cell structure exhibited abnormal defects.11 Embryoid bodies (EBs) are composed of undifferentiated embryonic stem cells, which can further develop cavities.27 Studies have confirmed that EBs composed of Becn1-deficient cells are unable to develop normally due to defects in the function of recruiting and clearing dead cells, and exhibit low intracellular ATP levels.12 These studies suggest that Becn1 is important for the early development of embryos, energy supply, and maintenance of homeostasis in the intracellular environment.
The Atg5–Atg12 covalent ubiquitination system is important for the formation of autophagosomes. Studies in mouse embryonic stem cells have shown that Atg5–Atg12 is covalently bound to the surface of the autophagosome bilayer membrane structure at the early stage of autophagy.28 Atg5 is localized on this bilayer membrane structure with the growth of the membrane structure. When Atg5 is deficient, the development of the bilayer membrane structure of autophagosomes in mouse embryonic stem cells is defective. Moreover, Atg5–Atg12 is also involved in assisting LC3 in binding to the developing autophagosome bilayer membrane structure. Thus, the Atg5–Atg12 covalent system plays a crucial role in the growth and closure of the bilayer membrane structure in the early stage of autophagosomes. Tsukamoto S. et al.9 fertilized Atg5-deficient oocytes with Atg5-deficient sperm and found that fertilized ovum developed to the 4–8-cell stage without further development afterwards. However, if the defective oocytes were fertilized with normal sperm, they could develop normally further, and the level of protein synthesis was also downregulated in the autophagy-deficient embryo. Therefore, during the early development of the embryo, the autophagy degradation system and the autophagy-related gene Atg5 are critical for mammalian preimplantation embryo development.
In recent years, a large number of studies have shown that post-transcriptional and translational regulation mediated by non-coding miRNA molecules are involved in autophagy in tumors.29 The modulation mediated by miRNAs gives cells a survival advantage in response to hunger, genotoxic stress, and hypoxia. Chen et al.30 found that the miR-290 gene cluster was upregulated in the B16F1 progeny cell line by comparing the mouse melanoma cell line B16 with its passaged progeny cell line. This upregulation had no significant change in cell proliferation, migration or anchor-independent growth, but had an effect against glucose starvation. The miR-290 gene cluster inhibited autophagic death of mouse melanoma cells in a glucose starvation environment through the downregulation of various autophagy genes including Atg7 and ULK1. Based on the above studies, we hypothesized that the miR-290 gene cluster may be involved in the regulation of preimplantation embryo development by targeting autophagy-related genes. Therefore, we examined the dynamic expression of miR-291a/b-5p in different stages of development. We showed that miR-291a/b-5p had a low expression abundance after fertilization, and a significant increase from the 4-cell phase to the blastocyst stage. In contrast, the expression trend of Atg5 and Becn1 mRNA showed an inverse relationship with miR-291a/b-5p expression. Furthermore, we confirmed that Atg5 and Becn1 were the direct targets of miR-291a/b-5p using a dual-luciferase reporter assay.
In conclusion, as mature molecules of the miR-290 cluster, miR-291a/b-5p was dynamically expressed and the expression trend of miR-291a/b-5p showed an inverse relationship with the expression of the autophagy-related genes Atg5 or Becn1 during mouse preimplantation embryo development. MiR-291a/b-5p inhibited the formation of autophagosomes and exhibited targeted inhibition effects on Atg5 and Becn1 in NIH/3T3 cells. Repression of miR-291a-5p with miRNA inhibitors in fertilized ova upregulated the mRNA levels Atg5 and Becn1, promoting the first cleavage and blastocyst formation in mouse embryos. Our study suggests the crucial role of miR-291a/b-5p during mouse preimplantation embryo development.
Conflicts of interest
There are no conflicts to declare.
Abbreviations
- TEM
Transmission electron microscopy
- MiRNA
MicroRNA
- 3′UTR
3′ Untranslated region
- SPF
Specific pathogen free
- FBS
Fetal calf serum
- PMSG
Pregnant mare's serum gonadotropin
- HCG
Human chorionic gonadotropin
- PFA
Paraformaldehyde
- Dpc
Day post-coitum
Supplementary Material
Acknowledgments
This study was supported by National Natural Science Foundation of China (Grant No. 81501272, No. 81070496).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra00017h
References
- Arnold S. J. Robertson E. J. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Schier A. F. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- Nothias J. Y. Majumder S. Kaneko K. J. DePamphilis M. L. J. Biol. Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- Wada Y. Sun-Wada G. H. Kawamura N. Aoyama M. Curr. Opin. Genet. Dev. 2014;27:60–66. doi: 10.1016/j.gde.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Stitzel M. L. Seydoux G. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- DeRenzo C. Seydoux G. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Aburto M. R. Hurle J. M. Varela-Nieto I. Magarinos M. Cells. 2012;1:428–448. doi: 10.3390/cells1030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adastra K. L. Chi M. M. Riley J. K. Moley K. H. Reproduction. 2011;141:607–615. doi: 10.1530/REP-10-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S. Kuma A. Murakami M. Kishi C. Yamamoto A. Mizushima N. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Kuma A. Hatano M. Matsui M. Yamamoto A. Nakaya H. Yoshimori T. Ohsumi Y. Tokuhisa T. Mizushima N. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Yue Z. Jin S. Yang C. Levine A. J. Heintz N. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X. Zou Z. Sun Q. Luby-Phelps K. Cheng P. Hogan R. N. Gilpin C. Levine B. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- He L. Hannon G. J. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Houbaviy H. B. Murray M. F. Sharp P. A. Dev. Cell. 2003;5:351–358. doi: 10.1016/S1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Yang Y. Bai W. Zhang L. Yin G. Wang X. Wang J. Zhao H. Han Y. Yao Y.-Q. Dev. Dyn. 2008;237:2315–2327. doi: 10.1002/dvdy.21666. [DOI] [PubMed] [Google Scholar]
- Mineno J. Okamoto S. Ando T. Sato M. Chono H. Izu H. Takayama M. Asada K. Mirochnitchenko O. Inouye M. Kato I. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S. Berezikov E. Yamashita Y. Lagos-Quintana M. Kloosterman W. P. Enomoto M. Hatanaka H. Fujiwara S. Watanabe H. Soda M. Choi Y. L. Plasterk R. H. Cuppen E. Mano H. Nucleic Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X. Ravi A. Calabrese J. M. Medeiros L. A. Kirak O. Dennis L. M. Jaenisch R. Burge C. B. Sharp P. A. PLoS Genet. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspi H. Chapnik E. Levy M. Beck G. Hornstein E. Soen Y. Stem Cells. 2013;31:2266–2272. doi: 10.1002/stem.1465. [DOI] [PubMed] [Google Scholar]
- Medeiros L. A. Dennis L. M. Gill M. E. Houbaviy H. Markoulaki S. Fu D. White A. C. Kirak O. Sharp P. A. Page D. C. Jaenisch R. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D. Lee L. A. Curr. Biol. 2006;16:R93–R95. doi: 10.1016/j.cub.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Bowerman B. Kurz T. Development. 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Levine B. Cuervo A. M. Klionsky D. J. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. Klionsky D. J. Dev. Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liang X. H. Kleeman L. K. Jiang H. H. Gordon G. Goldman J. E. Berry G. Herman B. Levine B. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A. Kabeya Y. Ohsumi Y. Yoshimori T. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E. Martin G. R. Cell. 1995;83:279–287. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Yamamoto A. Hatano M. Kobayashi Y. Kabeya Y. Suzuki K. Tokuhisa T. Ohsumi Y. Yoshimori T. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai H. Fesler A. Ju J. Cell Cycle. 2013;12:246–250. doi: 10.4161/cc.23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Liersch R. Detmar M. Sci. Rep. 2012;2:808. doi: 10.1038/srep00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.