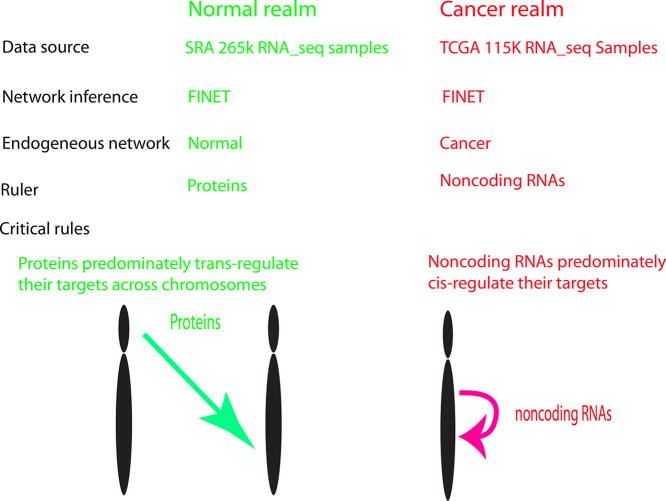
Graphical abstract
Keywords: Noncoding RNA, Cancer, Regulatory network, Systems, Endogenous, FINET, Big data, Pseudogene
Abstract
Cancers evolve from normal tissues and share an endogenous regulatory realm distinctive from that of normal human tissues. Unearthing such an endogenous realm faces challenges due to heterogeneous biology data. This study computes petabyte level data and reveals the endogenous regulatory networks of normal and cancers and then unearths the most important endogenous regulators for normal and cancerous realm. In normal, proteins dominate the entire realm and trans-regulate their targets across chromosomes and ribosomal proteins serve as the most important drivers. However, in cancerous realm, noncoding RNAs dominate the whole realm and pseudogenes work as the most important regulators that cis-regulate their neighbors, in which they primarily regulate their targets within 1 million base pairs but they rarely regulate their cognates with complementary sequences as thought. Therefore, two distinctive mechanisms rule the normal and cancerous realm separately, in which noncoding RNAs endogenously regulate cancers, instead of proteins as currently conceptualized. This establishes a fundamental avenue to understand the basis of cancerous and normal physiology.
1. Introduction
All cancers generally result from genome abnormality and they share an endogenous regulatory realm distinct from normal human tissues[1], [2], [3], [4]. Understanding the regulatory realm endogenous in all cancers and normal humans helps to advance our deep insights toward fundamental mechanisms of cancer and normal human physiology and to develop a general strategy to combat all cancers and to maintain physiological homeostasis.
Numerous gene regulations and networks have been identified for tumorigenesis, but most of these gene interactions are specific for a given cancer type, which refers to a specific organ and tissue [5], [6], [7], [8], [9]. Thus tumorigenesis mechanisms have been mostly marked as cancer type specific. However, certain regulations have been found endogenously in all cancer types. For example, TP53 has been typically characterized as an universal suppressor endogenous for all cancers [5]. More recently, a pseudogene PTENP1 has also been identified to be an endogenous regulator that regulates PTEN in examined cancer types [8], [10]. Given million gene regulations in the human genome, the magnitude of endogenous cancerous regulations for all cancer types should be very large. These gene regulations usually assemble a systems cancerous regulatory network distinct from that of normal humans, yet revealing such a network faces challenges due to two principal reasons.
First of all, human genome data is heterogeneous. Computationally searching a systems network from this type of data suffers high noise, with low accuracy < 50% [11], [12]. Moreover, the human regulatory network is complex, and emerging noncoding RNAs complicate this network [8], [9], [13]. This network complexity adds extra challenges in inferring a reliable network from heterogeneous data.
Secondly, there are not any appropriate biological approaches to reveal a real natural gene regulation. Current biological approaches like knockout suffer several limitations such as transcript compensation and genome alteration [14]. Knocking out a single gene normally results in alterations of thousand gene activation, leading to a biased picture of gene regulations.
To overcome these limitations above, we previously developed an algorithm called FINET to infer endogenous gene interactions from heterogeneous big data with high accuracy (>92% precision) [12]. In this present study, we utilized FINET [12] to infer a systems regulatory network endogenous in all cancers and human normal respectively from massive heterogeneous data, including all human RNAseq data available from Sequence Read Archive (SRA 265361 samples) and The Cancer Genome Atlas (TCGA 11574 samples). After inferring networks, we generated quantitative patterns from these networks to reveal endogenous rulers for all cancers and normal humans.
2. Materials and methods
2.1. Data resources
We downloaded human RNAseq data from two data resources, SRA (265361 samples) and TCGA (11,574 samples). Total 265361 SRA samples were searched by Homo sapiens and RNA_seq from the SRA database. Total 11,574 RNAseq samples from the TCGA portal website were downloaded for 36 cancer types. All detailed IDs and networks were available on our project website [15].
2.2. Download and alignment
The sra format files for total 265361 SRA samples were prefetched by their running ID (SRR#) via sratoolkit.2.8, and then were converted to fastq file via fastq-dump. The fastq files were aligned to GRCh38.p10.v27 containing 63,925 unique genes by using STAR-2.5 [16] with following settings, runThreadN 30 --genomeDir GRCh38.p10.v27 --outSAMtype BAM Unsorted SortedByCoordinate --outFilterMultimapNmax 20 --outFilterType BySJout --chimSegmentMin 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --quantMode TranscriptomeSAM GeneCounts --outFilterIntronMotifs RemoveNoncanonical --twopassMode Basic.
The files of total 11,574 cancer samples were directly downloaded from TCGA data portal website in.htseq.counts file format, which counted 60,483 genes annotated by GRCh38.p2.v22 done by TCGA.
2.3. Sample filtered
We focused on high quality samples with whole transcriptome RNAseq, and automatically filtered out any abnormal samples. All TCGA RNAseq data were sequenced for the whole transcriptome and we used the distribution of zero count in each TCGA sample as a normal reference of full transcriptome (Fig. S1A). The distribution of zero count in each sample was close to normal distribution, with the majority containing 30,000 zero count and few containing <10,000 or >40,000 zero (zero contained in each sample, X-axis). Rare are found samples carrying 50,000 zero count, which was used as a cutoff for SRA samples as described below. We treated all TCGA samples as high quality samples, but filtered out the control samples (no cancer samples) and got 8,972 samples for 36 cancer types.
For SRA samples, we first filtered out any abnormal samples from downloaded and aligned steps, such as unauthorized, unpublic, undownloadable, unaligned to the whole genome, and uncountable for the whole genome. These filtered steps generated 65,314 samples from 265361 aligned samples. Based on zeros distribution (Fig. S1B), we further filtered samples with zero count >50,000 to get the overall zero distribution close to normal distribution, finally getting 26,896 high quality samples for the rest of the analysis.
2.4. Calculating TPM and filtering genes
To make gene expression comparable for each sample, we normalized RNAseq data by calculating TPM (Transcripts Per Kilobase Million) for each sample as following.
TPM = ratio / sum(ratio) * 1,000,000
ratio = read counts /gene lengths
After TPM, we filtered out genes with all zeros and kept genes with nonzero counts >3 samples, and finally got 58,871 genes in 26,896 SRA samples, and 58,517 genes in 8,972 TCGA samples.
2.5. Regulatory network construction
The normalized TPM data for SRA and TCGA were used respectively to build the normal and cancerous regulatory networks. The current software for this task fails to produce reliable results efficiently [11]. We employed our algorithm called FINET to infer regulatory networks, which produces a network with >92% precision [12]. Briefly, FINET treats each gene as a target (set as y) and searches its regulators from the rest of genes (set as X) in the TPM matrix, in which samples were arranged as rows and genes as columns. FINET randomly split total samples into m groups(m = 8 in this study) and select target-regulator interactions from each group via elastic net. If an interaction like A to B is consistently shown in each group, this interaction (A to B) could be true and it has a frequency of 8. This random sampling repeats n times (n = 50 in this study). A frequency score, which is equal to frequency/(m*n), is calculated for each interaction. If A to B still showed up in all iterations (m*n), this A to B had a frequency score of 1 and it was treated as true endogenous interaction, independent of any conditions. This study used diverse heterogeneous data and applied frequency score > 0.95 as a cutoff, meaning that an interaction showing in 380 out of 400 random trials was selected, in which the error is less than 5% in all conditions. This ensures our results are truly robust. These selected interactions were assembled into an endogenous regulatory network for all cancers and humans respectively.
We run FINET as: julia finet.jl -c 120 -k 5 -n 50 -m 8 -a 0.5 -p 0.95 -i mydata.txt -o mynetwork
The results of frequency score with 0.95 was reported here, but the network data deposited in our server [15] were permitted to search networks with p from 0.9 to 1.0 (frequency score cutoff 0.9 to 1), allowing more flexibility to users.
2.6. Survival analysis and hazard ratios estimation
The survival analysis and hazard ratios (HR) estimation was performed by ISURVIVAL [17] as we previously described [18]. The software implementation of ISURVIVAL was available [17]. Briefly, we inserted stability-selection into Cox Proportional-Hazards Model to run the modified survival analysis and to estimate HR. All RNAseq and clinical data available in TCGA were used to run this model. The top 480 deadliest inducers were extracted from top 525 deadliest regulators with cutoff abs(coef) > 1 & p-value < 1.0e-9, which included 480 inducers and 45 repressors [18]. The significance shown here was derived from the wald-test.
2.7. Network centrality
Network centrality was calculated by using NetworkX implemented in python [19]. To avoid biases, we calculated two types of centrality, degree and eigenvector, for normal and cancer networks respectively. Genes with degree and eigenvector centrality for each network were ranked separately on the basis of ranking score as approached for network node ranking [11]. The final ranking was made based on the sum of two ranking scores as practiced in gene ranking in a regulatory network [11].
The cancerous network was filtered by gene interactions significant in survival analysis (p-value < 0.01 an HR < 0.9 or HR > 1.1), which was performed as our paralleled clinical data study [18] as described above.
2.8. Module identification and category
Network modules were clustered by network topology via MCODE [20] as following settings, degree cutoff: 3, K-score: 3, node score cutoff:0.2, max.depth:100, finding: haircut.
A module was clustered into the protein or noncoding category depending on the proportion of proteins and non-codings in a module. If protein or noncoding members occupied more than 50% of total members in a module, this module was referred to as protein or noncoding module respectively. If protein and noncoding members were equal, 50% for each, this module was ignored and was not classified into any category.
2.9. Statistics
All statistics like chi-squared test and figure drawings were performed by the R3.6 library. Network was visualized by cytoscape3.8 [21].
3. Results
3.1. Systems regulatory networks endogenous in cancers and normal human
To assemble a systems regulatory network endogenous for all conditions in all cancers and normal humans respectively, we first need a complete set of data representing endogenous genomic activation for all conditions. SRA and TCGA provided such data. SRA RNAseq contained various heterogeneous data sets. Endogenous gene interactions in total SRA data sets reasonably represent interactions endogenous in normal humans (Fig. S1, materials and methods). Similarly, TGCA provided RNAseq data for 32 cancer types [18] and endogenous gene regulations in this data set represent gene interactions endogenous for common cancer types.
Secondly, we need software that can infer an accurate unbiased gene regulation. We previously developed algorithms and a software, FINET [12]. Compared to the current software with high noise during inference, FINET significantly improves the accuracy, and it can efficiently and accurately infer unbiased gene interactions with >94% precision, true positives/(true positives + false positives) (materials and methods). FINET filters out all condition-dependent interactions and only keeps the true endogenous ones independent from any conditions such as biological sample heterogeneity and sequencing technique variations.
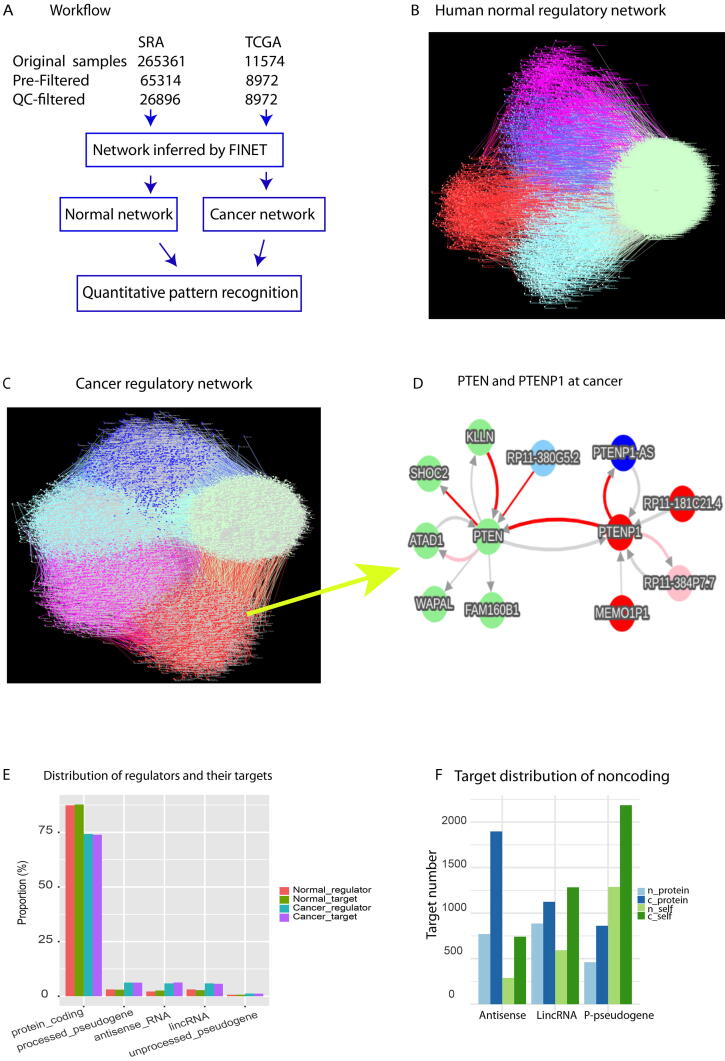
We employed FINET to search all possible gene regulations in the human genome by systematically treating each gene as a target and selecting its regulators from the rest of all annotated genes(Fig. 1A, materials and methods). In this way, each gene has an equal chance to be a target or a regulator without any presumption, regardless of its gene category in protein_coding or noncoding RNA. This search was separately performed for SRA and TCGA data, and the result eventually was assembled into a network for normal human and cancer respectively. The normal network contained 19,721 nodes (genes) and 63,878 edges (interactions), and the cancerous one included 25,402 nodes and 61,772 edges (Fig. 1B-1C).
Fig. 1.
Gene regulatory networks endogenous in cancers and normal humans. A, the workflow of this study. B-C, Completed gene regulatory network endogenous in normal (B) and cancer(C). The nodes (genes) and edges (interactions) were grouped into 5 gene category sets, including protein (light green), antisense(blue), lincRNA (pink), p_pseudogene (red), and the rest (other, lightblue). Interaction domains shift from protein interactions at normal(B) to noncoding interactions at cancer(C). D, an example of sub_network, PTEN interacting with PTENP1 in the cancer network directly extracted from our network database. Network annotation follows these 4 points: 1) Node color denotes gene category, lightGreen, blue, pink, red, lightSkyBlue respectively denote protein_coding, antisenseRNA, lincRNA, processed-pseudogene, other. 2) Edge color represents regulation strength: red, pink, blue, lightSkyBlue, and lightGray respectively represent strong positive, middle positive, strong negative, middle negative and weak regulation(positive or negative). 3) Edge thickness denotes confidence, thicker, more confident. 4) Edge arrow denotes the regulatory direction, from a regulator to a target. E, overall distribution of regulators and their targets at normal and cancer. The top 5 most abundant categories were shown. F, The target distribution of three categorized noncoding RNAs, antisense, lincRNA, and p-pseudogene, at normal (n_) and cancer (c_). Targets were counted separately when the three individual noncoding RNAs were regulators. Self denotes the targets as self-categorized genes. For example, c_self antisense represents cancerous antisense RNAs that were targeted by cancerous antisense RNAs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As expected, our networks were much less complex than those of current reports because we only collected the reliable interactions endogenous in all conditions, yet these two networks actually represented two distinct regulatory realms endogenous for normal human and cancer at systems level, in which all endogenous layered crosstalk of any pair of genes were included.
As a validation, our cancer network contained an interaction between PTEN (protein_coding) and PTENP1 (a pseudogene of PTEN) (Fig. 1D), which was validated by plotting Loess regression of PTENP1 and PTEN (Fig. S2). This PTENP1-PTEN interaction only existed in the cancer network but did not exist in our normal network [15], consistent with experimental reports showing it only in cancers [8], [10]. This indicated our network with high reliability and specificity. Furthermore, in contrast to conventional approaches showing PTENP1 as a regulator for PTEN only, our systems network expands this PTENP1-PTEN interaction to a cancerous PTENP1 regulatory network including several novel PTENP1 interactions (Fig. 1D). PTENP1 also interacted with its-own antisense_RNA (PTENP1_AS), two pseudogenes (RP11-181C21.4, MEMO1P1), and a lincRNA (RP11-384P7.7). These natural endogenous interactions provide a complete systems regulatory picture for PTENP1. Moreover, our network is universally true for all types of cancers as expected by our algorithm [12] (materials and methods), which was also validated by plots of 26 individual cancer types with sample size > 100 [15]. These indicated that our results are highly reproducible and suggested PTENP1 regulating PTEN as a universal endogenous regulation in all cancers.
Similarly, a complete systems regulatory picture of any universal endogenous regulation can be easily extracted from our network online [15]. Strikingly, an antisense RNA RP11-335k5.2, which was recently uncovered by our clinical data analysis as the most strongest inducer for all cancers [18], was consistently found here in our cancer network [15], but not in the normal network. This indicated RP11-335k5.2 indeed as an endogenous cancer driver for all types of cancers.
Overall, our networks provide a reliable and comprehensive resource for understanding the complete systems pictures of endogenous regulations in the cancer and normal genome.
3.2. Overall noncoding RNA crosstalk are unexpectedly activated at cancers
Multilayered crosstalk among proteins and various types of noncoding RNAs play key roles in physiologic states but the complete picture of crosstalk endogenous in cancers and normal human tissues remains elusive [8], [9]. Here we first examined the picture by grouping activated genes into gene sets via set algorithms [22], which clusters a network into sub-network sets on the basis of node and edge properties. By using gene annotated categories as node attributes, we separated the entire network into 5 gene category sets, including protein_coding (referred as protein hereafter), lincRNA, processed-pseudogene (p-pseudogene), antisense RNA (antisense), and others that pooled the rest of gene categories (Fig. 1B-1C).
In normal tissue, the majority of proteins and p-pseudogenes were mostly either separated or self-targeted, in which targets and their regulators at the same gene category (referred as self-regulation thereafter), but most of antisense RNAs and lincRNAs were highly cross-talked to proteins (Fig. 1B). However, in cancer these 5 sets were overall separated, and the density of the protein set became less than normal (Fig. 1B_1C), indicating that cancerous protein–protein crosstalk declined but crosstalk within noncoding RNAs increased. Statistically, we counted the regulators and their targets in each gene category in both normal and cancer (Table S1, Table S2). Overall, at normal the total crosstalk around proteins occupied 87.7% (56039/63878), and the rest 12.3% was around noncoding RNAs (Table S1). However, for cancer the overall crosstalk around proteins significantly declined to 73.9% (45660/61774), and crosstalk around noncoding RNAs increased to 26.1% (Table S2) (p-value = 0.02157, Pearson's Chi-squared test with Yates' continuity correction, referred as chisq-test thereafter). We next counted the specific gene category interactions. The interactions from proteins to proteins at normal counted for 82.5% (52692/63878, Table S1), but declined to 64.8% at cancer (40053/61774, Table S2). The protein regulators and protein targets also decreased from normal to cancer, but noncoding RNA regulators and targets dramatically increased in cancer (Fig. 1E, p-value < 2.2e-16, Chisq-test). These indicated the primary regulatory crosstalk shifted from normal protein domination to cancerous noncoding RNAs.
To further explore the detailed targets of noncoding RNAs, we plotted the primary targets of three abundantly categorized noncoding RNAs, including antisense, lincRNA, and p-pseudogene. Targets of noncoding RNAs primarily contained not only proteins but also self-regulated genes such as p-pseudogenes primarily regulate p-pseudogenes (Fig. 1F, Table S1, Table S2). These self-targets of all three categorized noncoding RNAs were significantly induced by cancer (p-value = 1.46e-08, Fig. 1F). For example, p-pseudogenes targeted self-targets, p-pseudogenes, with significantly increasing from 1288 at normal to 2185 at cancer (Fig. 1F). This indicated that noncoding RNAs, especially p-pseudogenes, increase self-regulation in cancer.
Together, protein crosstalks dominate the normal network, but noncoding RNA crosstalks become unexpectedly activated in cancers. Noncoding RNAs significantly turn to self-regulation in cancers.
3.3. Network module composition shifts from normal proteins to cancerous noncoding RNAs
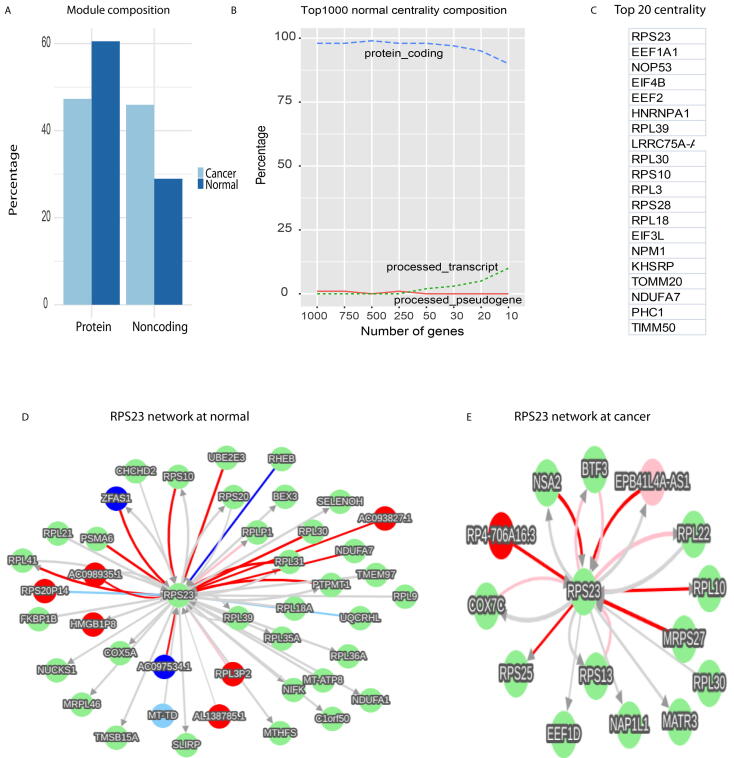
To understand the module differences between normal and cancer networks, we examined module member compositions. We identified modules by network topology [20] and then clustered modules into either protein modules (proteins occupied > 50% of members in a module) or noncoding modules (noncoding RNAs > 50% of members in a module, materials and methods). Modules with 50% of proteins or noncoding RNAs were ignored. At normal, protein modules occupied 60.52% out of total 38 modules and noncoding modules only took 28.94% (Fig. 2A, table S3), while cancerous modules significantly changed their compositions, in which protein modules reduced to 47.29% and noncoding modules increased to 45.94% of total 74 cancer modules (p-value = 0.02963, chisq-test) (Fig. 2A, table S4). Theoretically the network modules execute the primary functions for a network. This module pattern shifting from proteins to noncoding RNAs suggested noncoding RNAs as the key rulers in the cancer regulatory realm.
Fig. 2.
Network module compositions and normal network centrality. A, the module composition differences between normal and cancer network. B, Compositions of top 1000 normal network centrality. C, Top 20 normal centrality. D-E, RPS23 first neighbors in normal (D) and cancer(E).
3.4. Noncoding RNAs serve as the centrality in cancerous network but ribosomal proteins dominate in the normal
To understand the core controllers of the normal and cancerous networks, we investigated the centrality of normal and cancer networks (materials and methods). At the normal network proteins worked as the primary centrality (top 1000, Fig. 2B) and ribosomal proteins dominated the top 20 centrality in normal (Fig. 2C). The top 1 centrality, RPS23, abundantly interacted with proteins and noncoding RNAs (Fig. 2D), but at cancer the interactions of RPS23 declined dramatically (Fig. 2E).
To validate this network, we randomly selected a node, UBE2E3, which was presenting in normal (Fig. 2D) but absent in cancer (Fig. 2E), and plotted the LOESS regression of RPS23 and UBE2E3 with all normal and cancer data (Fig. S3). Consistent with interactions in the normal and cancer networks, there was strong correlation between RPS23 and UBE2E3 in normal but not in cancer (Fig. S3). Based on gene ontology (GO) [23], these top 20 centralities in normal networks performed crucial functions in translation (RPL18, RPL3, RPL30, RPL39, RPS10, RPS23, RPS28). Consistently, the functions for the whole normal network and modules (table S3) were also relevant to translation and negative regulations, suggesting ribosomal proteins as the delicately regulatory core of the normal human genome.
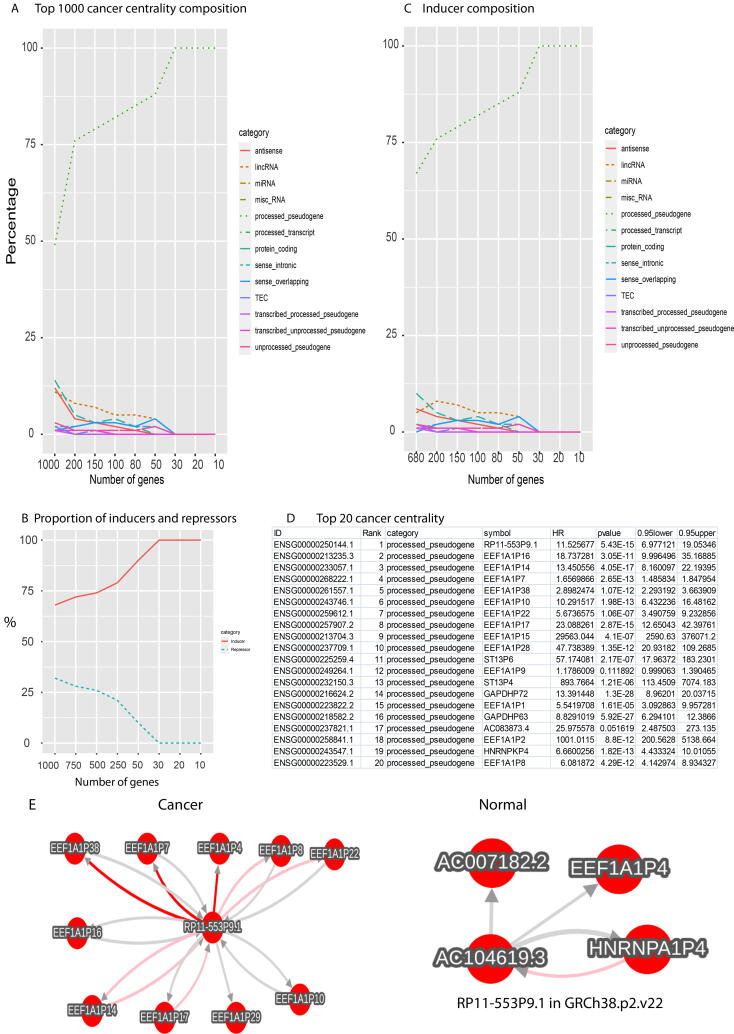
In contrast, p-pseudogenes dominated the cancerous centrality (Fig. 3A, materials and methods) and most of these centrality worked as cancer inducers (regulators with coefficient > 0 and < 0 were respectively referred as inducers and repressors during FINET inferences, materials and methods, Fig. 3B). Most of these inducers were p-pseudogenes (Fig. 3C), and all top 20 centralities were p-pseudogenes (Fig. 3D). This indicated p-pseudogenes as the primary rulers for cancers. Literature mining [24] showed no functions associated with these top 20 p-pseudogenes. Their functions remain to be further investigated.
Fig. 3.
Cancer network centrality. A, Compositions of top 1000 cancer network centrality. B, proportion of inducers and repressors in top 1000 cancer network centrality. C, compositions of inducers in top cancer centrality(B). D, top 20 cancer centrality. E, ENSG00000250144.1 has more interactions in cancers than normal. Please note that the gene symbol was different in the two annotation versions as labeled in the figure and our database.
These pseudo-gene interactions escalated in cancers have been shown in our database. For example, cancers activated much more interactions in the top listed pseudo-gene (Fig. 3D), ENSG00000250144.1, than normal (Fig. 3E).
These data suggest that ribosomal proteins serve as the most important regulatory core for the protein-dominated normal network, but noncoding RNAs, especially p-pseudogenes, primarily control the center of the cancerous realm.
3.5. Noncoding RNAs and proteins respectively serve as the strongest regulators in the cancerous and normal network
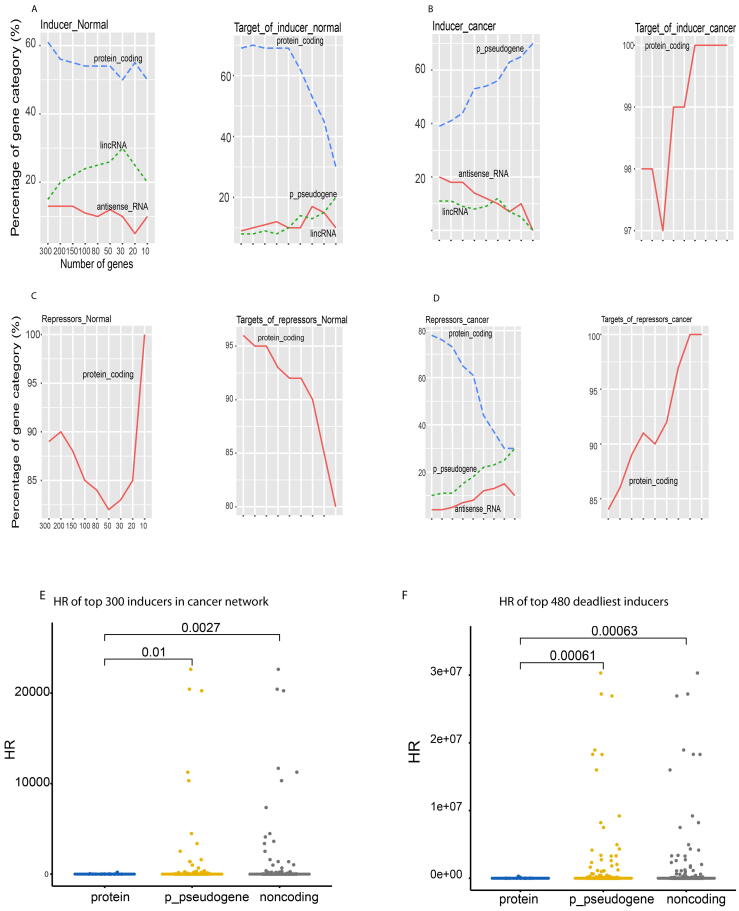
To understand the strongest regulators governing normal and cancer genomes and to make our pattern robust, we examined the composition of the top 300 regulators and their corresponding targets based on their absolute coefficient rankings. For pattern recognition and clear illustration, we only presented any gene category with abundance > 10%. At normal, proteins worked as the strongest inducers. From the top 300 to top 10 inducers, proteins occupied 60% to 50% respectively (Fig. 4A left). LincRNAs came next and occupied ∼20%. These inducers mostly targeted proteins and p-pseudogenes (Fig. 4A right). Yet in cancer, proteins even did not show up (<10%), instead, noncoding RNAs dominated the top inducers, including p-pseudogene, antisense RNA and lincRNA (Fig. 4B left). For example, p-pseudogenes counted 70% out of the top 10, suggesting p-pseudogenes as the primary strongest drivers in the cancer genome, instead of proteins as conventionally thought. Interestingly, these cancerous inducers mostly targeted proteins (Fig. 4B right). This suggested that proteins work as targets at cancer instead of as cancerous drivers. The conventional practice treating protein-coding genes as cancerous drivers is very misleading. Consistently, our results from big clinical data also found p-pseudogenes as the primary drivers universal for all types of cancers [18].
Fig. 4.
Top 300 strongest inducers and repressors at the normal and cancer network. The composition of strongest inducers and repressors at normal and cancer network (A-D). A, The top 300 strongest inducers (left) and their targets (right) of normal network. B, the top 300 strongest cancerous inducers and their targets. C, the top 300 strongest repressors and their targets at normal. D, The top 300 strongest cancerous repressors and their targets. Clinic data of top cancerous inducers(E-F). E, comparison of hazard ratio (HR) between protein, p-pseudogenes and noncoding RNAs in top 300 strongest inducers in the cancer network built by this present study. F, HR profiling of top 480 deadliest inducers directly extracted from Cox proportional-hazards model analysis of all TCGA RNAseq data [18]. P-values (above line) were calculated by t-test.
As for the strongest repressors, almost all repressors and their targets were proteins at normal (Fig. 4C). However, cancerous repressors contained proteins, p-pseudogenes, and antisense RNAs, with at least 10% at each (Fig. 4D). Surprisingly, regardless of normal and cancerous repressors, almost all their targets were proteins > 85%, and noncoding RNA targets in any categories were too low to show (<10%). This pattern revealing proteins as targets for both inducers and repressors in cancer interprets why the current observations have focused on proteins, yet treating protein-targets as cancerous drivers is fundamentally misleading. Noncoding RNAs, especially p-pseudogenes, serve as the primary universal drivers for all types of cancers.
3.6. Noncoding RNAs serve as the deadliest inducers in cancers
To understand the cancerous association of noncoding RNAs, we calculated the HR (hazard ratios) of top 300 inducers in the cancer network as described above (Materials and methods). Among these inducers, p-pseudogenes and all noncoding RNAs (including p-pseudogenes) had significant higher HR than proteins, with p-value = 0.01 between p-pseudogenes and proteins and p-value = 0.0027 between noncoding RNAs and proteins (Fig. 4E). To further confirm this result, we examined these HR differences between proteins, p-pseudogenes and noncoding RNAs in top deadliest inducers derived from unbiased survival analysis of all cancer type data from TCGA [18](Materials and methods). Similarly, both p-pseudogenes and noncoding RNAs had significantly higher HR than proteins, with p-value = 0.00061 and 0.00063 respectively (Fig. 4F). These results consistently indicated that noncoding RNAs, especially p-pseudogenes, play more important roles in causing cancer death than proteins. This and our previous results [18] provide strong systems evidence to validate our systems network results showing noncoding RNAs as the most important drivers for tumorigenesis, instead of proteins.
3.7. Noncoding RNAs primarily turn to regulate their local targets at cancer
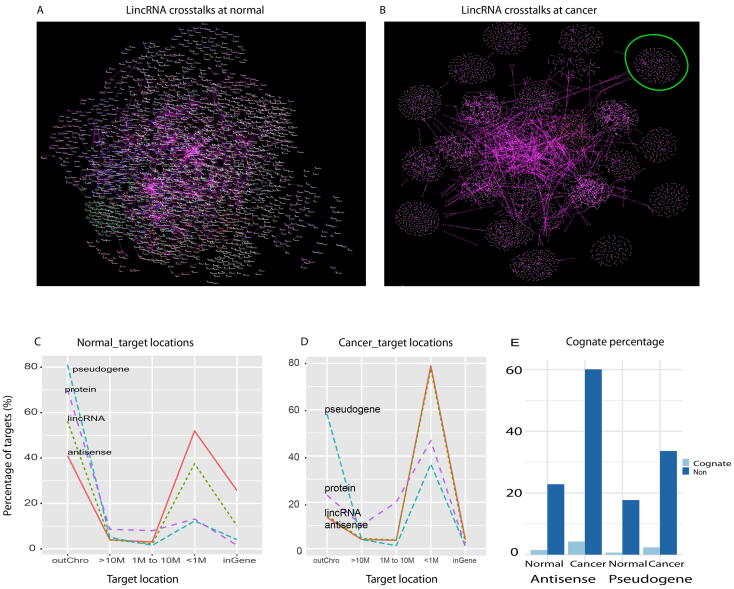
Understanding the systems distribution of distances between regulators and their targets helps to understand the functional framework of genome regulations but it remains debated [8], [9], [25], [26], [27]. To capture the systems profiling of target distances altered by cancer, we compared it to that of normal in top four gene categories, including protein, p-pseudogene, lincRNA, and antisense. To overlook the profiling, we clustered the targets by chromosomes via using set-algorithm as done in Fig. 1 above. At normal, all chromosome sets were mixed up but these sets were clearly separated at cancer (Fig. 5A-5B, Fig. S4), indicating that noncoding RNAs increasingly regulated their self-chromosome targets at cancer compared to normal. Statistically, most normal genes worked as trans-regulators regulating their targets outside their chromosomes (Fig. 5C). Especially, more than 80% p-pseudogene targets located outside the chromosome, and 70% protein and 55% lincRNA targets were also located outside the chromosome. Furthermore, p-pseudogenes and proteins rarely regulated their targets with overlapped sequences (inside genes, Fig. 5C). However, in cancer, most regulators of all categories turn to regulate their local targets (<1M bp Fig. 5D). Specifically, more than 80% of lincRNAs and antisense RNAs worked locally.
Fig. 5.
Target distance distribution. A-B, lincRNA regulatory network in normal (A) and cancer(B). These two networks were grouped by chromosome to show crosstalk between chromosomes. The chromosome 14 section (circle in B) was detailed in Fig. S4. LincRNAs trans-regulate their targets at normal(A) but cis-regulate their targets at cancer(B). C-D, target location distribution of top abundant gene categories (>10%) in normal (C) and cancer (D). OutChro represents targets located outside the chromosome, and M denotes million bp inside the chromosome. E, the percentage of cognates and non-cognates targeted by antisense RNAs and pseudogenes at normal and cancer. Non denotes non-cognate.
However, these antisense RNAs and pseudogenes rarely regulated their cognates at both normal and cancer conditions (Fig. 5E). Furthermore, cancer stimulated the non-cognate proportion. The non-cognate rate for antisense increased from normal 22.8% to cancerous 60%, and this for p-pseudogene also increased from 17.6% (normal) to 33.5% (cancer). In contrast, the cognate proportion shifted slightly from 85 (1.4% normal) to 241 (4.2% cancer) for antisense, and from 73 (0.6% normal) to 254 (2.3% cancer) for pseudo-gene (Fig. 5E). This suggested primary noncoding RNAs as cis-regulators in cancers, but not as cognate regulators as recently proposed [28].
This together suggests that regulations switch from normal trans-regulations to cancerous cis-regulations, yet noncoding RNAs do not serve as cognate-regulators.
4. Discussion
This study revealed a complete systems picture of endogenous regulatory mechanisms regulating the cancer and normal realm, in which noncoding RNAs endogenously rule the cancerous regulatory realm while proteins govern the normal. Numerous regulatory mechanisms have been uncovered for regulating cancers and normal physiology, but they are biased to a given biological experiment and are condition-dependent and thus are not universally endogenous for all conditions. The systems mechanism endogenous across all conditions remains unknown. Here, we revealed that proteins control the normal human regulatory realm at systems level and ribosomal proteins endogenously govern the core of the normal realm. Ribosomal proteins have been known as important factors in controlling cell type specific physiology and pathology [29], but we found more important role for them in which they actually work as an universal endogenous center to regulate whole human normal realm via interacting with other proteins and noncoding RNAs. This realm is dominated by proteins working as trans-regulators to regulate proteins as their primary targets, consistent with current practices in biology in which proteins are treated as both key regulators and targets. However, this normal protein-dominant realm cannot be applied to cancers. Cancers are endogenously regulated by noncoding RNAs. Noncoding RNAs, especially p-pseudogenes, serve as the primary centrality and the strongest inducers, and they also control the cancerous modules functioning for the entire systems realm. This parallels our recent observation from clinical data showing noncoding RNAs as the universal deadliest drivers for all types of cancers [18]. Our finding conceptually refreshes cancer systems mechanisms in which noncoding RNAs drive cancers, instead of proteins as conventionally thought [30], [31], [32]. This presents a novel basis for understanding the cancerous fundamental.
Pseudogenes were once thought as junk DNAs but recently they have been reported as regulators for cognate genes, in which they might regulate their corresponding protein-coding genes [8]. For example, pseudo-gene PTENP1 regulates PTEN in cancer. However, the number of known functional pseudogenes are very limited and the functions of these pseudo-genes have been thought as secondary. Here, we systematically revealed that the abundant pseudogenes were activated in cancer and these pseudogenes functionally worked as the most important cancer drivers instead of secondary regulators as thought. This was validated by clinical data in our paralleled study [18]. In contrast to the conventional validation via biochemistry in vitro in which results might not be applied to in vivo regulations, we systematically validated these noncoding RNA regulations by clinical data as in vivo evidence [18], which ensures our results are more reliable than in vitro results.
Pseudogenes rarely target their cognate genes, but they mostly regulate their remote targets outside the chromosomes. Pseudogenes should execute their functions in a way similar to proteins as trans regulators and drivers. This further suggested that pseudogenes might act as flexible and energy-saving activators for various physiologic conditions. This opens the block around pseudo-genes to explore their functions in other physiologic conditions like stress stimulation.
Understanding the majority of noncoding RNAs working as cis- versus trans-regulators provides the first step to understand their functions and mechanisms, but it remains controversial due to lack of knowing the complete crosstalk involved in all noncoding RNAs [8], [9], [13]. Here, we revealed that different types of noncoding RNAs have their own target-distance patterns varying with physiologic states, but universally, the majority of noncoding RNAs works as trans in normal, even antisense RNAs have only ∼ 50% working in local (<1M). This parallels the recent observation showing trans-regulation patterns in noncoding RNAs [33]. However, in cancer the majority of noncoding RNAs such as antisense RNAs and lincRNAs turns to target the local genes (<1Mb) as cis-regulators but not their cognates. Only a very limited number of noncoding RNAs target their cognates. Therefore, the hypothesized mechanism of noncoding RNAs executing their functions via bindings to complementary sequences of their cognates is misleading. In general, normal noncoding and coding genes primarily work as trans-regulators, but cancerous noncoding RNAs primarily serve as cis-regulators but not cognate-regulators.
Gene regulatory networks have been widely studied, but most of them have been derived from gene pair studies and condition-dependent experiments [8], [13]. In addition, the current network inference approaches have suffered high noises and recently increasing noncoding RNA species have complicated the network inferences [8], [9], [11], [12], [13], resulting in seriously biased observations and leaving an actual blackbox of gene crosstalk. Here, we revealed the all endogenous crosstalk as systems networks hidden in massive data. Without any presumption, we generated unbiased quantitative patterns from systems networks and revealed the systems mechanisms from the data patterns, which made our results reliable. To ensure our networks were robust, we only included interactions with high precision. High precision selections dramatically reduced the false positives and all interactions in our networks do not depend on any conditions. Obviously, some conventional interactions might not be found in our network due to they are conditional-dependent, not endogenous. Indeed we intentionally missed numerous interactions that were conditionally dependent because including those condition-dependent interactions could dramatically introduce noise [12]. This practice to filter out noise to ensure reproducibility is also of first concern in experimental biology, in which biologists normally conducted many experiments to prove true gene regulation. Here our computational algorithm has systematically revealed thousands of reliable regulations in two systems networks. These networks are invaluable and provide a novel foundation to advance our insights into cancer and human normal physiology.
The limitations of this study reside in the lack of biological validations and functional data, but data derived from biological experiments are avoidably biased to experimental conditions like tissue types, leading to biased results. For example, only 22% of lncRNAs annotated by the GENCODE project are endogenous and 88% of them are condition-dependent [34]. Yet we filtered out the interactions with frequency score > 0.95 to ensure our final network reproducible as evidenced by overall data plots (Figs. S2-S4) and individual cancer type plots [15]. In contrast, conventional software without stability-selection and frequency score could lead to 90% false positives [12]. The functions of pseudo-genes generated by this study remain elusive, but our recent study promised their application in cancer discrimination [35]. Future research on these noncoding RNAs helps to understand the big picture of cancer mechanisms.
Acknowledgments
Acknowledgments
The data were downloaded from TCGA and SRA database
Funding
No funding associated with this project.
Availability of data and materials
All data resources and detailed network results were available on our project website (https://combai.org/network/)
Competing interests
No conflict of interests
Authors' contributions
A.W. designed project, developed algorithm, coded software, and wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.04.015.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howard T.P., Vazquez F., Tsherniak A., Hong A.L., Rinne M., Aguirre A.J., et al. Functional genomic characterization of cancer genomes. Cold Spring Harb Symp Quant Biol. 2016;81:237–246. doi: 10.1101/sqb.2016.81.031070. [DOI] [PubMed] [Google Scholar]
- 3.Calabrese C., Davidson N.R., Demircioğlu D., Fonseca N.A., He Y., Kahles A., et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Haar J., Canisius S., Yu M.K., Voest E.E., Wessels L.F.A., Ideker T. Identifying epistasis in cancer genomes: a delicate affair. Cell. 2019;177:1375–1383. doi: 10.1016/j.cell.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine A.J., Momand J., Finlay C.A. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 6.Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 7.Moya I.M., Castaldo S.A., den Mooter L.V., Soheily S., Sansores-Garcia L., Jacobs J., et al. Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science. 2019;366:1029–1034. doi: 10.1126/science.aaw9886. [DOI] [PubMed] [Google Scholar]
- 8.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang A., Hai R. FINET: Fast Inferring NETwork. BMC Res Notes. 2020;13:521. doi: 10.1186/s13104-020-05371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 14.El-Brolosy M.A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang A. human regulatory network http://combai.org/network/. 2019;
- 16.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang A. ISURVIVAL. combai.org/software/survival/. 2019;1.
- 18.Wang A., Hai R. Noncoding RNAs serve as the deadliest universal regulators of all cancers. Cancer Genomics Proteomics. International Institute of Anticancer Res. 2021;18:43–52. doi: 10.21873/cgp.20240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centrality — NetworkX 1.10 documentation [Internet]. [cited 2019 Sep 23]. Available from: https://networkx.github.io/documentation/networkx-1.10/reference/algorithms.centrality.html.
- 20.Bader G.D., Hogue C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris JH, Lotia S, Wu A, Doncheva NT, Albrecht M, Pico AR, et al. setsApp for Cytoscape: Set operations for Cytoscape Nodes and Edges. F1000Research. 2014;3:149. [DOI] [PMC free article] [PubMed]
- 23.The Gene Ontology Consortium The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cytoscape App Store - AgilentLiteratureSearch [Internet]. [cited 2022 Mar 2]. Available from: https://apps.cytoscape.org/apps/agilentliteraturesearch.
- 25.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarropoulos I., Marin R., Cardoso-Moreira M., Kaessmann H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature. 2019;571:510. doi: 10.1038/s41586-019-1341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarver A.L., Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731–733. doi: 10.6026/97320630008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills E.W., Green R. Ribosomopathies: There’s strength in numbers. Science. 2017;358 doi: 10.1126/science.aan2755. [DOI] [PubMed] [Google Scholar]
- 30.Blum A., Wang P., Zenklusen J.C. SnapShot: TCGA-Analyzed Tumors. Cell. 2018;173:530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- 31.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol Poznan Pol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., et al. Extensive and coordinated transcription of noncoding RNAs within cell cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A. Distinctive functional regime of endogenous lncRNAs in dark regions of human genome. bioRxiv. Cold Spring Harbor Laboratory; 2020;2020.12.06.413880. [DOI] [PMC free article] [PubMed]
- 35.Wang A., Hai R., Rider P.J., He Q. Noncoding RNAs and deep learning neural network discriminate multi-cancer types. Cancers. Multidisciplinary Digital Publishing Institute. 2022;14:352. doi: 10.3390/cancers14020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data resources and detailed network results were available on our project website (https://combai.org/network/)