Summary
RNA turnover is a primary source of gene expression variation, in turn promoting cellular adaptation. Mycobacteria leverage reversible mRNA stabilization to endure hostile conditions. Although RNase E is essential for RNA turnover in several species, its role in mycobacterial single-cell physiology and functional phenotypic diversification remains unexplored. Here, by integrating live-single-cell and quantitative-mass-spectrometry approaches, we show that RNase E forms dynamic foci, which are associated with cellular homeostasis and fate, and we discover a versatile molecular interactome. We show a likely interaction between RNase E and the nucleoid-associated protein HupB, which is particularly pronounced during drug treatment and infection, where phenotypic diversity increases. Disruption of RNase E expression affects HupB levels, impairing Mycobacterium tuberculosis growth homeostasis during treatment, intracellular replication, and host spread. Our work lays the foundation for targeting the RNase E and its partner HupB, aiming to undermine M. tuberculosis cellular balance, diversification capacity, and persistence.
Subject areas: Environmental science, Ecology, Biological sciences, Microbiology
Graphical abstract

Highlights
-
•
Single mycobacterial cells exhibit phenotypic variation in RNase E expression
-
•
RNase E is implicated in the maintenance of mycobacterial cell growth homeostasis
-
•
RNase E and HupB show a functional interplay in single mycobacterial cells
-
•
RNase E-HupB disruption impairs Mycobacterium tuberculosis fate under drug and in macrophages
Biological sciences; Microbiology; Tuberculosis; Single-cell biology; Proteomics; Post-transcriptional regulation; Bacterial persistence
Introduction
A long-standing question in bacterial cell biology is what governs microbial adaptation. Genetic mutations occur seldom in a population, providing slow but stable responses to environmental changes (Castro et al., 2020; Robert et al., 2018). In contrast, variation in gene expression virtually occurs in the whole population and is responsible for fast events of phenotypic diversification among genetically identical cells (Shis et al., 2018). The formation of phenotypic variants can provide a transient fitness advantage and has been associated with persistent infections (Dhar et al., 2016; Fisher et al., 2017; Schröter and Dersch, 2019; Bakkeren et al., 2020). Given the clonal population structure of Mycobacterium tuberculosis (Castro et al., 2020), both inherent and causal phenotypic variation offer a functional diversification potential, beneficial to cope with environmental onslaughts, but the mechanisms remain largely unclear (Dhar et al., 2016; Sakatos et al., 2018; Manina et al., 2019; Goossens et al., 2020).
Both intrinsic and extrinsic sources of noise influence the cell biochemical reactions, contributing to the dynamics and magnitude of gene-expression variation and to its attenuation (Shis et al., 2018; McAdams and Arkin, 1997; Mitchell and Hoffmann, 2018; Patange et al., 2018; Megaridis et al., 2018; Dame et al., 2020; Kim et al., 2020). Although all stages of gene expression are subject to variation, low transcript levels translated at a high rate are usually associated with enhanced variation, compared to high transcript levels translated at a low rate (McAdams and Arkin, 1997; Swain, 2004; Arbel-Goren et al., 2014; Hansen and Weinberger, 2019). In other words, the contribution of mRNA turnover to variation is considered to dominate that of proteins, primarily for a difference in their stability. Bacterial mRNA half-life is of the order of min at steady state, shorter than the growth-dependent dilution rate; conversely, proteins’ half-life is longer and dominated by dilution. Furthermore, the overall lack of correlation between mRNA and proteins points to the relevance of post-transcriptional regulation and RNA stability in gene expression variation (Swain, 2004; Hansen and Weinberger, 2019; Taniguchi et al., 2010; Arbel-Goren et al., 2016).
RNA decay represents a cost-effective and irreversible means for the cell to rapidly fine-tune gene expression (Hui et al., 2014; Van Assche et al., 2015). RNA stability is contingent on the formation of intramolecular stem loops, on the interaction with small RNAs, and on the activity of RNA degrading machineries, also acting in competition with ribosomes (Hui et al., 2014). Mycobacteria can reversibly stabilize their transcriptome during stresses (Vargas-Blanco et al., 2019), such as starvation and hypoxia, implying the importance of RNA degrading machineries in transcriptional remodeling and adaptive diversification. In the slow-growing pathogen M. tuberculosis the average mRNA half-life is longer than in fast-growing model microorganisms (Rustad et al., 2013), and this relative mRNA stability was attributed to the initial step of destabilization and to the energetic state of the cell (Rustad et al., 2013; Vargas-Blanco et al., 2019).
The ribonuclease E (RNase E) is a conserved and broad-spectrum single-stranded endonuclease, mainly responsible for the initial cleavage of A/U-rich and 5′-monophosphorylated RNA substrates (Mackie, 2013; Durand et al., 2015). In Escherichia coli, RNase E activity is complemented by helicases and exonucleases, forming a macromolecular complex, referred to as RNA degradosome (Mackie, 2013), which is partly understood in mycobacteria (Płociński et al., 2019). The mycobacterial RNase E is essential for ribosomal maturation (Zeller et al., 2007; Taverniti et al., 2011), and its depletion remodels M. tuberculosis transcriptome (Płociński et al., 2019). Importantly, the RNase E catalytic potential is controlled by binding metabolites, other proteins, and by cellular compartmentalization (Hui et al., 2014; Tejada-Arranz et al., 2020).
In this study, we probe the role of RNase E and its interactome in mycobacterial cell physiology and functional phenotypic diversification, by combining spatiotemporal analyses of single cells with quantitative mass spectrometry. We found that fluorescently tagged RNase E is unevenly expressed in isogenic mycobacterial cells during optimal growth and that one subpopulation forms transient localization foci, associated with RNA. Targeting RNase E caused foci abrogation and affected growth, and targeting essential cellular processes caused RNase E subcellular delocalization. Collectively, our results point to a strategic role of RNase E in mycobacterial cell homeostasis, consistent with the dynamics of the RNase E interactome. In particular, we found a possible interaction between RNase E and the nucleoid-associated protein HupB, both under homeostatic and stressful conditions. We found that the phenotypic variation of RNase E and HupB was not only associated with drug tolerance but also with long-term intracellular survival. Lastly, our findings offer mechanistic and functional insights into M. tuberculosis phenotypic diversification for the design of enhanced anti-persistence strategies.
Results
Mycobacterium smegmatis RNase E-mKate2 reporter exhibits dynamic localization events associated with single-cell homeostasis
To explore the dynamics of RNase E in single M. smegmatis cells, we constructed a fluorescent reporter of RNase E protein expression, referred to as RNase E-mKate2 (Figures S1A−S1C). As we were unable to fluorescently label the native rne locus, most likely due to its essentiality, we generated a stable merodiploid strain (Figure S1A). This strategy was preferable to the use of episomal plasmids, which typically amplify cell-to-cell variation, as the second chromosomal copy of rne fused to mKate2 was expected to robustly correlate with the native copy of rne (Manina et al., 2019). We confirmed that the levels of expression of the native and the fluorescently-tagged rne copy are similar (Figure S1C). Next, we carried out time-lapse microfluidic imaging (Manina et al., 2019) of exponentially growing RNase E-mKate2 cells (Figure 1A and Video S1), followed by single-cell multiparametric analysis. We found that RNase E fluorescence was heterogeneously expressed in clonal cells (Figures 1B and 1C), in that about one-third of the population exhibited low-intensity patchy fluorescence, and the remaining two-thirds exhibited dynamic localization events during the cell lifetime, also referred to as foci. We confirmed that the two subpopulations were significantly different from each other (Figure 1D). The merodiploid RNase E-mKate2 strain had a moderate variation of cell-growth associated parameters compared to control cells at the single-cell level, not detectable in bulk assay (Figures 1E, 1F, and S1B), implying a relationship between small variations in RNase E levels and single-cell growth heterogeneity. RNase E foci were mainly located toward the mid-cell position or the new cell pole and were positively correlated between two consecutive generations, and between siblings regardless of cell age (Figures 1G, 1H, S1D, and S1E).
Figure 1.
Dynamic RNase E-mKate2 foci are associated with single-cell homeostasis
(A) Representative time-lapse images of exponential-phase M. smegmatis RNase E-mKate2 fluorescent reporter. Phase-contrast (blue) and fluorescence (magenta) are merged. Scale bar, 10 μm. Numbers represent hours. Boxes zoom in on two first-generation (G1) cells, without (white arrow) or with (magenta arrow) RNase E fluorescent focus, which disappears in the second generation (G2). Scale bar, 5 μm. See also Video S1.
(B) Representative heatmaps of single-cell RNase E-mKate2 fluorescence as a function of generation time and percent of cell length.
(C and D) RNase E-mKate2 fluorescence frequency distribution (C) or averaged over the cell lifetime (D) in single cells segregated by the absence (purple, n = 180) or presence (magenta, n = 330) of RNase E foci. Black lines indicate mean ± SD. Significance by unpaired t-test. Data are from two independent experiments.
(E and F) Single-cell size (E) and growth rate (F) averaged over the cell lifetime in M. smegmatis wild type (CT) and in RNase E-mKate2 subpopulations identified in (C). Black lines indicate mean ± SD (100 ≤ n ≤ 330). Significance by one-way ANOVA followed by Tukey’s multiple comparison test: ∗p = 0.012 and 0.023; ∗∗∗p = 0.0003; ∗∗∗∗p < 0.0001.
(G) Distribution of RNase E foci as a function of cell length (n = 734). Black line is the fitting with a sum of two Gaussians. Old (O) and new (N) cell-pole are indicated. Data are from two independent experiments.
(H) Pearson correlation of RNase E-mKate2 fluorescence averaged over the cell lifetime between mother cells and their old- (black) and new-pole (white) daughters (left panel), and between siblings (right panel). Data are from two independent experiments (n = 170, per category).
(I) Representative snapshot images (left) of RNase E-mKate2 reporter stained with RNAselect dye. Phase-contrast (gray), RNase E-mKate2 (magenta), and RNAselect (green) fluorescence. Scale bar, 5 μm. Single-cell Pearson correlation of fluorescence (right panel) in exponential-phase cells without (gray) or with (magenta) RNase E foci. Data are from two independent experiments (n = 145).
(J) Representative time-lapse image series of exponentially growing RNase E-mKate2 reporter treated with 2.5 mM of the putative M5 RNase E inhibitor (Kime et al., 2015). Phase-contrast (blue) and fluorescence (magenta) are merged. Numbers represent hours. Scale bar, 10 μm. See also Video S2.
(K and L) Single-cell RNase E-mKate2 fluorescence (K) and size (L) from time-lapse microscopy (J). M5 inhibitor (gray shading). Black lines indicate mean ± SD (n = 142). Data are from two independent experiments. Significance by one-way ANOVA followed by Tukey’s multiple comparison test: ns, not significant; ∗∗∗∗p < 0.0001.
Representative video of exponential-phase bacteria seeded into our customized microfluidic device (Manina et al., 2019), and grown under constant flow of fresh 7H9 medium. Images were recorded at 30-minute intervals (15 fps). RNase E-mKate2 fluorescence (magenta) and phase-contrast (blue) channels are merged. Time is indicated in hours. Scale bar, 10 μm.
To examine the functional significance of the RNase E foci, we stained bacteria with an RNA-selective dye. RNAselect positively correlated with RNase E fluorescence mainly in cells forming foci and in cells treated with rifampicin (RIF) (Figures 1I and S1F−S1H), which causes transcription arrest and nucleoid compaction (Cabrera et al., 2009). Lastly, we carried out a two-phase time-lapse microscopy experiment (Figure 1J and Video S2), by exposing exponentially growing RNase E-mKate2 bacteria to a compound that inhibits RNase E in vitro, but whose mode of action in cells was not reported (Kime et al., 2015). RNase E foci disappeared about 6 h after treatment (Figure 1K), and bacteria experienced growth rate slowdown and reductive division (Figure 1L), which might also be due to some off-target effect. Overall, these results proved the cell-to-cell variation of RNase E expression and its functional role in the maintenance of mycobacterial cell growth homeostasis.
Representative video of exponential-phase bacteria, grown in fresh medium for 18 hours, and stressed with the putative M5 RNase E inhibitor for 14 hours. Images were recorded at 30-minute intervals (15 fps). RNase E-mKate2 fluorescence (magenta) and phase-contrast (blue) channels are merged. Time in hours and the presence of the inhibitor are indicated. Scale bar, 10 μm.
Deregulation of RNase E levels reshapes M. smegmatis whole-cell proteome
We found that overexpression and repression of RNase E encoding gene in M. smegmatis caused a moderate and marked reduction in growth rate, respectively (Figures S2A−S2D). Similar to other microorganisms, we also observed aberrant migration of M. smegmatis RNase E (Figure S2B), likely due to its particular amino acid composition (Casarégola et al., 1992; Cormack et al., 1993; Kovacs et al., 2005). Our results further pointed to the relevance of RNase E in mycobacterial cell physiology. Thus, we carried out mass spectrometry analysis of M. smegmatis following RNase E overexpression (oe_rne) and silencing (si_rne), compared to wild type (WT) M. smegmatis cells (Figure S2E). Comparative proteome profiling revealed significant alterations in M. smegmatis proteome, by 11.5% after RNase E overexpression, and by 23.6% after silencing (Figures 2A, 2B, S2F−S2H and Table S1). Consistent with the implication of RNase E in RNA metabolism (Hui et al., 2014; Płociński et al., 2019), the oe_rne strain was enriched in proteins responsible for RNA turnover and transcription. Transcription factors and alternate sigma factors involved in stress responses, such as WhiB2, SigB, and SigG were upregulated (Figure 2A and Table S1). Several ribosomal proteins were significantly downregulated. Proteins involved in environmental sensing and signal transduction (Prisic et al., 2014), including DevS and SenX3, redox regulation, different stress responses, iron scavenging, protein turnover, and cell wall biosynthesis and remodeling were upregulated (Figure 2A and Table S1). Collectively, these results are suggestive of a metabolic slowdown and cell alert state. Conversely, the si_rne strain was mainly enriched in proteins involved in primary and secondary metabolism, mainly nitrogen metabolism, which is associated with cellular detoxification (Amon et al., 2010), amino acids biosynthesis, and oxidation-reduction processes (Figure 2B and Table S1). In conclusion, exogenously induced alterations in RNase E levels caused considerable proteome remodeling, entailing a pleiotropic effect of RNase E on mycobacterial cell physiology.
Figure 2.
Whole-cell mass spectrometry in M. smegmatis following deregulation of RNase E
(A and B) STRING protein-protein interaction analyses of the whole-cell proteome after rne overexpression (A) and silencing (B). Proteins were grouped according to their functional category using Mycobrowser and color-coded. Symbols represent exclusive, upregulated and downregulated proteins compared to control bacteria. The PANTHER classification system was used for gene ontology analysis, to identify additional clusters. Data are from five independent replicates. See also Table S1.
The mycobacterial RNase E pull-down includes proteins involved in RNA turnover, cell division, stress response, and virulence
To probe the RNase E protein interactors, we carried out pull-down assays against the whole-cell extracts of M. smegmatis oe_rne compared to control (CT) strain, followed by label-free quantitative mass spectrometry (Figures 3A, S1A, S3A, and S3B). Differential analysis revealed that 224 proteins were specific to the RNase E pull-down and 380 to the CT. Moreover, 301 proteins were enriched and 36 lowered in the RNase E pull-down (Figure 3A and Table S2).
Figure 3.
Mass spectrometry-based proteomics of M. smegmatis RNase E pull-down and single-cell analysis of putative interactors
(A) Differential analysis of proteomics data. Proteins are ranked in a volcano plot according to their statistical p value derived from a LIMMA t-test (y-axis), and their relative abundance ratio between the RNase E overexpressing and CT strains (x-axis). Off-centered spots are those that vary the most between groups. Dotted lines indicate a doubling or halving of the ratio. The dashed line indicates a 1% false discovery rate (FDR). Proteins with FDR <0.01 are considered significantly more abundant in one condition than the other. Names or gene identifiers of some relevant proteins are indicated, and functional categories are color-coded. Data are from seven independent replicates. See also Table S2.
(B) Number of proteins (x-axis) attributed to different functional categories (y-axis). Gray bars represent proteins lowered (n = 36), and magenta bars indicate proteins enriched (n = 301) or exclusively present (n = 224) in the RNase E pull-down compared to the CT.
(C−J) Single-cell Pearson correlation of fluorescence in M. smegmatis dual fluorescent reporters, between RNase E (x-axes) and possible protein interactors (y-axes). DeaD (C, MSMEG_5042); RraA (D, MSMEG_6439); InfA (E, MSMEG_1519); UvrA (F, MSMEG_3808); RelA (G, MSMEG_2965); Ppk (H, MSMEG_2391); Obg (I, MSMEG_4623) and Lsr2 (J, MSMEG_1060). Exponential phase cells (white circles), and cells starved for 6 h (gray circles) are shown. Nonlinear regression analyses of the data fitted with a log-log function [Y = 10ˆ(slope∗log(X)) + Yintercept] are shown for exponential (solid black lines) and starved (dashed lines) cells. Significant positive correlations suggest an interaction between protein pairs. Data are from two independent experiments (n = 150, per strain).
Next, we carried out the functional analysis of proteins that were significantly enriched in the RNase E pull-down (Figures 3A and 3B). We identified 14 proteins implicated in RNA turnover, including a ribonucleoprotein (MSMEG_1193), a ribokinase (MSMEG_4661), and MutT2, belonging to the nudix family and responsible for generating 5′-monophosphorylated transcripts, which are more susceptible to RNase E (Deana et al., 2008). As expected, we measured enrichment of GpsI (Płociński et al., 2019) and of Rph, both having 3′-5′-exoribonuclease activity that typically follows RNase E endonucleolytic cleavage (Mackie, 2013). We also detected several ATP-dependent helicases, including DEAD/DEAH box helicases, HelY, DeaD, and HrpA, as well as RraA, which was shown to negatively regulate RNase E in E. coli (Lee et al., 2003). Proteins involved in transcription, metabolism, and signal transduction were also highly represented (Figures 3A and 3B). Consistent with the role of RNase E in rRNA processing and with the protective role of translation on transcripts degradation (Taverniti et al., 2011; Hui et al., 2014), some ribosomal proteins and the translation initiation factor InfA were not only enriched but also ranked among the proteins with the highest relative stoichiometry (Figure 3A and Table S2). Consistent with former studies (Kovacs et al., 2005; Płociński et al., 2019), we found significant enrichment of chaperones and proteases, such as GrpE, DnaJ, ClpB, ClpX, and HtpX. Remarkably, proteins implicated in DNA damage (Warner et al., 2013) and stringent response (Prusa et al., 2018), and in cell-wall remodeling and division (Hett and Rubin, 2008), including FtsZ, PonA1, Wag31, RipA, and MSMEG_5691, were also enriched (Figures 3A, 3B, and Table S2). DNA-binding and nucleoid-associated proteins (NAPs) involved in nucleoid coating and remodeling, cell cycle, and global gene regulation, such as Lsr2 and HupB (Kołodziej et al., 2021; Pandey et al., 2014; Hołówka and Zakrzewska-Czerwińska, 2020), were significantly enriched too.
Lastly, we examined a shortlist of alleged RNase E protein interactors, belonging to diverse functional categories, and constructed a panel of dual-fluorescent reporters in the RNase E-mKate2 background (Figures S3C and S3D). By quantitative snapshot microscopy of individual bacteria (Figure S3E), we measured low but significant positive correlations between all protein pairs in the exponential phase that increased during starvation (Figures 3C−3J). The strongest correlations were found between RNase E and constituents of the RNA degradosome (Figures 3C and 3D), with the translation initiation factor InfA (Figure 3E), with the DNA repair enzyme UvrA (Figure 3F), and with the stress-response proteins Obg (Kumar et al., 2017) and Lsr2 (Figures 3I and 3J). In summary, the combination of proteomic and single-cell analyses pointed to possible interactions of RNase E with known and unknown members of the RNA degradosome (Bandyra et al., 2018; Płociński et al., 2019), and with proteins involved in critical aspects of mycobacterial cell physiology. Further analysis will be required to determine whether these potential interactions are direct or indirect, and whether they have a functional role in cellular adaptation to changing environmental conditions.
RNase E and HupB interact and jointly respond to the inhibition of key cellular processes
Among the proteins identified in the RNase E pull-down, the NAP HupB was enriched more than 3-fold and had the highest relative stoichiometry (Figure 3A and Table S2), prompting us to further examine their relationship. HupB is a conserved small histone-like protein, with a critical structural role in nucleoid topology. It can bind both double- and single-stranded DNA and has higher propensity for curved and AT-rich DNA, typically present at the origins of replication and in regulatory regions (Pandey et al., 2014; Kumar et al., 2010; Hołówka et al., 2018).
To characterize the molecular association between these two possible interactors, we generated a HupB fluorescent reporter in the RNase E-mKate2 background, and in the oe_rne and si_rne strains (Figures S4A and S4B). We confirmed the co-immunoprecipitation of RNase E with HupB-mCitrine, implying their likely interaction (Figure 4A). In contrast, RNase E did not co-immunoprecipitate with Lsr2-mCitrine (Figure S4C), suggesting that the interaction of RNase E with HupB is not merely due to the presence of DNA substrate. However, owing to the high instability of RNase E during treatment with nucleases at 37°C, we were unable to directly assess whether the interaction between RNase E and HupB is mediated by their respective RNA or DNA substrates. Next, we analyzed HupB-mCitrine by snapshot fluorescence microscopy, after the alteration of RNase E cellular levels (Figures 4B and 4C). In particular, single-cell length decreased following RNase E deregulation (Figures 4D and 4E), and HupB fluorescence significantly increased after RNase E induction, and decreased after RNase E depletion (Figures 4F and 4G), entailing their co-regulation.
Figure 4.
Characterization of the relationship between RNase E and HupB
(A) Representative Coomassie-stained SDS-PAGE and Western blot of soluble protein fractions from M. smegmatis HupB-mCitrine reporter, co-transformed either with empty control (CT) or with RNase E-His expressing plasmid (oe_rne). RNase E-His was co-immunoprecipitated with HupB-mCitrine, using an anti-GFP nanobody covalently bound to agarose beads. The presence of RNase E-His (113 kDa migrating at ∼180 kDa, black arrowheads) was confirmed using an antibody against the His tag, and the presence of HupB-mCitrine (50 kDa, yellow arrowheads) was detected using an antibody against GFP in cell extract (CE), flow through (FT) and eluate (EL) samples.
(B and C) Representative snapshot images of M. smegmatis HupB-mCitrine carrying either the inducible RNase E-His overexpressing plasmid (B, oe_rne), or the inducible sgRNA-dCas9 repression system (C, si_rne). Strains were imaged in the absence (left panels) and presence (right panels) of inducer, in exponential (6 h) and stationary phase (24 h). Phase-contrast (blue) and fluorescence (yellow) are merged. Fluorescence images are scaled to the brightest frame. Scale bars, 5 μm.
(D−G) Snapshot analysis of single-cell length (D and E) and fluorescence (F and G) of M. smegmatis HupB-mCitrine_oe_rne (D and F), and of HupB-mCitrine_si_rne (E and G) strains. The absence (−) or presence (+) of inducer and time of induction are indicated. Experiments were repeated twice (n = 150 per condition). Black lines indicate median and quartiles. Significance by one-way ANOVA, followed by Sidak’s multiple comparisons test.
(H) Representative heat maps of fluorescence as a function of cell length of the dual RNase E-mKate2 and HupB-mCitrine reporter in exponential growth-phase (EXP), and following 6 h of exposure to MMC (200 ng/mL), FtsZ inhibitor (C109, 16 μg/mL), RIF (20 μg/mL), CAP (16 μg/mL), and INH (20 μg/mL). New (0%) and old cell pole (100%).
(I−K) Single-cell snapshot analysis in the conditions indicated in (H). Coefficient of correlation of fluorescence between the two reporters over the cell length (I) and mean fluorescence (J and K). Data are from two independent experiments (150 ≤ n ≤ 299). Black lines indicate median and quartiles. Significance by one-way ANOVA followed by Dunn’s multiple comparisons test: ∗p = 0.01; ∗∗p = 0.002; ∗∗∗∗p < 0.0001. Gray shading (I) indicates non-significant correlations.
To further examine the functional association between RNase E and HupB, we simultaneously monitored their expression in the exponential growth phase and after short treatment with molecules that impair essential cellular processes (Figure S4D). We generated submicrometric maps of both RNase E-mKate2 and HupB-mCitrine fluorescence as a function of single-cell length (Figures 4H and S4E). Exponential-phase cells exhibited a longitudinal distribution of HupB along the nucleoid region resembling a string of pearls (Figure S4D), consistent with other HupB reporters (Hołówka et al., 2018). Moreover, RNase E and HupB fluorescence were positively correlated at the single-cell level (Figure 4I). Treatment with the DNA crosslinker mitomycin C (MMC) caused significant cell elongation, similar to FtsZ inhibition by C109 compound (Chiarelli et al., 2020), and fluorescence decrease of both proteins with no apparent delocalization (Figures S4F, 4J, and 4K). In contrast, treatment with different macromolecular synthesis inhibitors caused substantial delocalization of both proteins. In particular, RIF exposure resulted in tight compaction of both RNase E and HupB fluorescence toward the center of the cell (Figures 4H and S4D), suggestive of transcription arrest and chromosomal condensation (Cabrera et al., 2009). RIF also caused marked fluorescence increase, confirmed by a strong positive correlation between the two markers at the single-cell level (Figures 4I−4K). Inhibition of translation with chloramphenicol (CAP) caused a pattern similar to RIF but less pronounced (Figures 4I−4K). Lastly, inhibition of cell wall biosynthesis with the frontline tuberculosis drug isoniazid (INH) caused typical cell size decrease (Figure S4F), massive induction of fluorescence and delocalization of both RNase E and HupB, which were strongly correlated (Figures 4I−4K and S4D). Collectively, these observations confirmed the co-regulation of RNase E and HupB, and their possible interplay at the nucleoid level mainly during stressful conditions, consistent with the nexus between nucleoid architecture and post-transcriptional regulation.
Concerted expression of RNase E and HupB in M. tuberculosis predicts single-cell responses to isoniazid
We hypothesized that the interaction between RNase E and HupB may occur in the pathogenic strain, promoting stress adaptation and survival. Thus, we investigated the single-cell dynamics of RNase E and HupB in M. tuberculosis, by multi-phasic time-lapse fluorescence microscopy (Figure S5A and Video S3). During optimal growth M. tuberculosis formed two subpopulations, characterized by either low levels of RNase E-mCherry fluorescence or brighter localization foci (Figures 5A, S5B and S5C). These two subpopulations had significantly different RNase E levels, relatively more homogeneous HupB levels, and grew at a similar rate (Figures 5B and 5C). Consistent with M. smegmatis, RNase E and HupB fluorescence were positively correlated, even more robustly in cells forming RNase E foci (Figure 5D). Additionally, both RNase E and HupB foci were mostly found between the mid-cell position and the new pole (Figures 5E and 1G). Although the strong but not absolute correlation between RNase E and HupB suggests that approximately 60% of RNase E foci may be contingent on HupB (Figure 5D), the moderate correlation between the ribosomal protein RpsJ (Figure S5D) suggests that about 25% of RNase E localization in M. tuberculosis may be related to the ribosomal activity.
Figure 5.
M. tuberculosis RNase E-mCherry and HupB-sfGFP are positively correlated and predictive of single-cell fate under isoniazid
(A) Representative heatmaps of RNase E and HupB fluorescence as a function of single-cell generation time and length. See also Video S3.
(B and C) Single-cell RNase E and HupB fluorescence (B) and growth rate (C) over the lifetime of the cell in subpopulations segregated by the absence (0) or presence (1) of RNase E foci. Black lines indicate mean ± SD (100 ≤ n ≤ 179). Significance by unpaired t-test (B), or one-way ANOVA followed by Tukey’s multiple comparisons test (C). Data are from two independent experiments.
(D) Single-cell Pearson correlation of fluorescence in cells having none (gray) or one (white) RNase E focus. Data are from two independent experiments (120 ≤ n ≤ 179).
(E) Histograms showing the distribution of RNase E-mCherry and HupB-sfGFP foci, as in (B), expressed as a function of cell length. Black lines indicate fitting of the data with a Lorentzian function. New (0%) and old cell poles (100%) are relative to cell length.
(F) Representative time-lapse image series of M. tuberculosis RNase E and HupB dual reporter stressed with INH (250 ng/mL, 10X-MIC). Phase-contrast (gray), RNase E -mCherry (magenta), and HupB-sfGFP (green) fluorescence are merged and scaled to the respective brightest frame. Numbers represent hours. Arrows track a regrowing cell. Scale bar, 5 μm. See also Video S3.
(G) Microcolony fluorescence (mean ± SD). INH (gray shadings). Data are from two independent experiments (n = 26).
(H) Representative single-cell heatmaps of fluorescence. Cells with three different fates are shown. Division events (dotted lines); INH (gray shadings).
(I and J) Single-cell RNase E (I) and HupB (J) fluorescence before (Pre-INH), during the first (INH-1st) and the second (INH-2nd) drug exposure period, and during the recovery phase between the two INH exposures (Washout). Cells that lysed (L), remained intact (I), and regrew (RG) after treatment are shown. Black lines indicate mean ± SD. Data are from two independent experiments (34 ≤ n ≤ 100). Significance by one-way ANOVA followed by Tukey’s multiple comparisons test: ns = not significant; ∗p < 0.05; ∗∗p = 0.0024; ∗∗∗p ≤ 0.0009; ∗∗∗∗p < 0.0001.
(K) Single-cell fate as a function of the absence (0) or presence (1) of RNase E foci before INH treatment. Significance by Chi-square test of independence for cells that died (n = 180) and regrew (n = 24).
Representative video of exponential-phase bacilli seeded into our customized microfluidic device (Manina et al., 2019), grown in fresh 7H9 medium for 4 days, exposed to INH (10X-MIC) for 2 days, allowed to recover in fresh 7H9 medium for 10 days, and exposed a second time to INH (10X-MIC) for 7 days. Images were recorded at 3-hour intervals (15 fps). RNase E-mCherry (magenta) and HupB-sfGFP (green) fluorescence and phase-contrast (gray) channels are merged. Time in hours and INH are indicated. Scale bar, 5 μm.
Next, we probed the implication of RNase E and HupB in the single-cell responses to INH treatment (Figure 5F and Video S3). After 4 days of growth in optimal conditions, we stressed M. tuberculosis with high concentration of INH for two days, followed by 10-day recovery in the absence of drug. Lastly, we exposed the regrown bacilli to INH a second time, to assess whether they had acquired genetic resistance, or they were still sensitive to the drug and thus derived from INH-tolerant cells. Consistent with M. smegmatis (Figure 4), we measured marked fluorescence induction of RNase E and HupB upon INH exposure, and strong positive correlation at both microcolony and single-cell level (Figures 5G, S5E, and S5F). In the recovery phase, we could distinguish three main cell fates, namely, cells that lysed, remained intact, or regrew (Figure 5H). Surprisingly, bacilli experiencing lysis and regrowing had significantly higher RNase E levels compared to those that remained intact (Figure 5I), which were most likely dead, and a similar pattern held true for HupB (Figure 5J). However, only lysing cells maintained very high induction of both RNase E and HupB during INH washout, unlike cells that survived, whose expression reverted to basal levels during the recovery phase (Figures 5I and 5J). We also found that regrowers were more likely to have formed foci prior to INH exposure (Figure 5K), entailing a protective role of RNase E foci relative to INH treatment.
In summary, these results not only proved the interplay between RNase E and HupB in M. tuberculosis during both normal growth and drug treatment, but also implied that their faulty expression is detrimental in a subpopulation of bacilli subject to INH.
RNase E and HupB jointly assist M. tuberculosis to endure drug and host stress
To further clarify the association between RNase E and HupB in the pathogenic strain, and the consequences relative to its fitness for the environment, we silenced rne expression in the HupB-sfGFP background strain, referred to as si_rne, using the anhydrotetracycline (ATC)-inducible CRISPRi repression system (Rock et al., 2017). In M. tuberculosis, similar to M. smegmatis (Figure S2C), rne repression caused not only a growth rate defect compared to the control strain (ct_rne) but also significant decrease of rne, hupB, infA transcripts and 16S rRNA, accounting for metabolic slowdown (Figures S6A and S6B). Consistent with our proteomic data (Figures 2 and 3), we also measured an increase of furA and katG transcripts (Figure S6B), which are co-transcribed and cleaved by the RNase E (Taverniti et al., 2011).
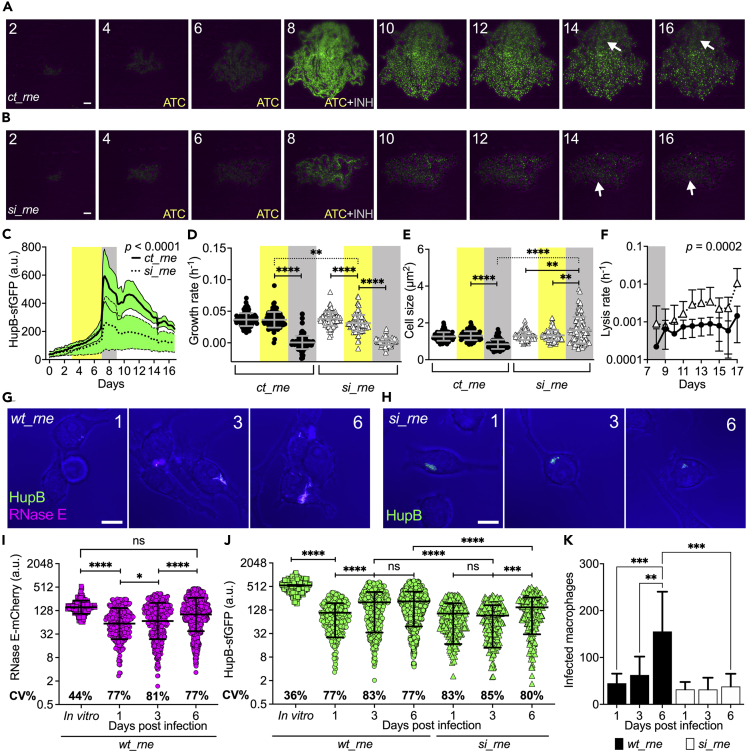
Given the role of the catalase-peroxidase KatG in both INH resistance and tolerance (Ando et al., 2011; Niki et al., 2012; Wakamoto et al., 2013), we investigated the effect of rne silencing on the single-cell responses of M. tuberculosis to INH treatment, by time-lapse microscopy (Figures 6A, 6B, S6C, S6D, and Video S4). In particular, we grew the ct_rne and si_rne strains under optimal conditions for three days, followed by ATC induction for five days, and by INH exposure in the presence of inducer for two days. Lastly, we monitored the recovery in fresh medium for a week. As expected, ct_rne bacilli responded to INH with a robust induction of HupB fluorescence, which was about 3-fold lower in the si_rne strain, confirming that HupB expression is largely RNase E-dependent during INH stress (Figures 6C, 5G and S6E). Silencing of rne also affected the single-cell growth rate, which further decreased in the presence of INH (Figure 6D). Surprisingly, only the control strain experienced single-cell shrinkage, typical of INH exposure (Wakamoto et al., 2013; Manina et al., 2015), whereas bacilli devoid of rne markedly elongated during INH treatment, and lysed at faster rate (Figures 6E and 6F). Collectively, these results imply that RNase E depletion impairs cell growth and division homeostasis, with detrimental effects during treatment with a cell-wall targeting drug.
Figure 6.
Silencing rne decreases HupB levels and impairs M. tuberculosis growth homeostasis during drug and host stress
(A and B) Representative time-lapse images of HupB-sfGFP reporter carrying the pLJR965 empty vector (ct_rne, A), or the ATC-inducible sgRNA-dCas9 repression system against rv2444c (si_rne, B). See also Video S4. ATC, INH, and days are indicated. Phase-contrast (magenta) and sfGFP fluorescence (green) are merged and scaled to the brightest frame. Arrows point to bacilli that resumed growth. Scale bars, 5 μm.
(C) Microcolony fluorescence. Lines and green shadings represent the mean ± SD (10 ≤ n ≤ 12 microcolonies). Data are from two independent replicates. Significance by unpaired t-test.
(D and E) Single-cell growth rate (D) and size (E) averaged over the cell lifetime during 7H9 growth (unlabeled), ATC induction (yellow shadings), and INH exposure (gray shadings). Gray lines indicate mean ± SD (n = 100), from two independent replicates. Significance by one-way ANOVA followed by Tukey’s multiple comparisons tests: ∗∗p < 0.005; ∗∗∗∗p < 0.0001.
(F) Single-cell lysis rate following INH exposure (gray shading) in ct_rne (black circles) and si_rne (white triangles) strains. Data are mean ± SD (10 ≤ n ≤ 12 microcolonies), from two independent replicates. Significance by two-way ANOVA.
(G and H) Representative snapshot images of RAW 264.7 macrophages infected with HupB-sfGFP_RNase E-mCherry strain (wt_rne, G), or with HupB-sfGFP_si_rne strain (si_rne, H) in the presence of ATC inducer from day 0. Bright field (blue) and color-coded fluorescence channels are merged. Numbers indicate days post-infection. Scale bars, 10 μm.
(I and J) Single-cell fluorescence of in vitro-grown (squares) and intracellular bacilli (circles). Black lines indicate mean ± SD (163 ≤ n ≤ 1259). Significance by one-way ANOVA followed by Tukey’s multiple comparisons test: ns = not significant; ∗p = 0.014; ∗∗∗p = 0.0017; ∗∗∗∗p < 0.0001. Data are from six independent experiments.
(K) Number of infected macrophages. Significance by two-way ANOVA, followed by Tukey’s multiple comparisons test. Data indicate mean ± SD and are from six independent experiments (102 ≤ n ≤ 263 macrophages).
Representative videos of exponential-phase bacilli grown in fresh 7H9 medium for 3 days, pre-induced with ATC for 5 days, exposed to ATC and INH (10X-MIC) for 2 days, and allowed to recover in fresh 7H9 medium for 7 days. Images were recorded at 3-hour intervals (15 fps). HupB-sfGFP fluorescence (green) and phase-contrast (magenta) channels are merged. Strains, time in hours, ATC, and INH are indicated. Scale bars, 5 μm.
Given the essentiality of HupB for M. tuberculosis intracellular growth (Pandey et al., 2014), we probed whether this was contingent on RNase E. We examined the behavior of our dual fluorescent reporter of RNase E and HupB (wt_rne) and of the si_rne strain during different stages of macrophage infection (Figures 6G, 6H, S6F and S6G). Intracellular bacilli exhibited a very strong positive correlation between RNase E and HupB fluorescence (Pearson r = 0.92; p < 0.0001), consistent with our in vitro findings (Figures 4 and 5). In particular, RNase E and HupB fluorescence decreased at the beginning of infection compared to in vitro-grown bacilli (Figures 6I and 6J), and fluorescence variation doubled for both reporters, conceivably increasing the single-cell growth heterogeneity and diversification potential of M. tuberculosis in the host. Conversely, as the infection progressed, we measured a significant increase of both RNase E and HupB fluorescence, entailing their greater implication during the late stage of macrophage infection (Figures 6I and 6J). Interestingly, rne silencing at the onset of infection not only impaired HupB induction but also significantly decreased the propagation of infection (Figures 6K, S6H, and S6I). We infer that RNase E and HupB are functionally associated also in the host and that a deficit of these two factors limits the spread of infection, presumably due to decreased intracellular growth and virulence. Our results confirm the concerted action of RNase E and HupB in M. tuberculosis, contributing to the maintenance of balanced growth and division in optimal conditions, during drug treatment and infection.
Discussion
The cell can be considered a dynamic and evolving system, whose behavior depends on the interaction of its internal constituents, and on the relationship with external variables (Gonze et al., 2018). The ability of the cell system to functionally diversify is largely driven by fine-tuning gene expression and underlies its selective ability to make choices in response to an increasingly complex and challenging environment (Dhar et al., 2016; Schröter and Dersch, 2019). Gene expression variation is driven by factors, such as promoter toggling, regulatory feedback, and RNA turnover (Kim et al., 2020; Arbel-Goren et al., 2014; Hansen and Weinberger, 2019). Furthermore, cell growth and dilution rates influence the abundance of macromolecules implicated in gene expression (Taniguchi et al., 2010; Shis et al., 2018; Patange et al., 2018), and the cell-cycle phase and relative nucleic acids architecture shape the recruitment of gene expression machineries and regulatory elements (Megaridis et al., 2018; Dame et al., 2020). Although analysis of gene expression variation primarily focused on transcription, a few recent studies have begun to shed light on the crucial contribution of post-transcriptional mechanisms in both prokaryotic and eukaryotic model organisms (Arbel-Goren et al., 2013, 2016; Baudrimont et al., 2019).
In this study, we focus on the highly conserved RNA-degrading enzyme RNase E (Durand et al., 2015), and its implications for the single-cell physiology and functional phenotypic diversification of mycobacteria. We combine live single-cell imaging with mass spectrometry approaches, to probe the cellular dynamics of RNase E and of the NAP HupB, and find that they jointly contribute to M. tuberculosis adaptation and survival when confronted with drug treatment and with the host.
We implement quantitative time-lapse microfluidic imaging of RNase E fluorescent reporters and observe that clonal mycobacterial populations, grown in optimal conditions, exhibit heterogeneous expression of RNase E at the single-cell level. Both fast-growing non-pathogenic and slow-growing pathogenic mycobacteria weakly express RNase E fluorescence in patches inside the cell, and about half of the population (60% in M. smegmatis and 40% in M. tuberculosis) forms brighter localization foci that quench within the cell generation time. Remarkably, RNase E fluorescence positively correlates with an RNA-specific dye, and this correlation is more pronounced in cells forming foci, which disappear inhibiting both RNase E (Durand et al., 2015) and transcription, implying their relevance in RNA metabolism. Consistent with these findings, RNase E co-immunoprecipitated with known constituents of the RNA degradosome and other proteins related to RNA processing and decay (Kovacs et al., 2005; Bandyra et al., 2018; Płociński et al., 2019).
As mycobacterial mRNA turnover was shown to scale with the metabolic activity of the cell (Vargas-Blanco et al., 2019), we can reason that the subpopulation forming RNase E foci might have both faster mRNA degradation rates and faster rRNA maturation (Taverniti et al., 2011), possibly influencing the single-cell growth rate (Manina et al., 2015). Interestingly, the counterintuitive negative correlation between transcript abundance and half-life (Nouaille et al., 2017) is particularly pronounced in mycobacteria (Rustad et al., 2013), and can have two non-mutually-exclusive explanations. On the one hand, transcription and degradation are tightly associated, and RNase E recruitment increases at sites of active transcription. In support of this first explanation, our RNase E pull-down is significantly enriched with RNA polymerase subunits, sigma factors, and transcriptional regulators. On the other hand, fast transcription can prevent ribosome recruitment and foster RNase E degradation (Kovacs et al., 2005; Rustad et al., 2013), as ribosomes and RNase E compete on the ribosome binding site (McAdams and Arkin, 1997; Van Assche et al., 2015). Indeed, upon RNase E induction, we find significant enrichment of the translation initiation factor InfA, of GTPases and of the methionyl-tRNA formyltransferase, implicated in translation initiation, which could be a compensatory gesture by the cell to counteract RNase E catalytic activity. At the single-cell level, RNase E exhibits positive correlations with InfA, Obg, and RpsJ that couple transcription and translation (Kint et al., 2014; Lawson et al., 2018).
During mycobacterial growth, RNase E foci mostly localize between the mid-cell position and the new cell pole, reminiscent of DNA replication dynamics and septum formation in mycobacteria and Caulobacter crescentus (Santi and McKinney, 2015; Logsdon et al., 2017; Laloux and Jacobs-Wagner, 2013). Indeed, we identify several proteins responsible for cell division in our RNase E pull-down, and we also measure a significant decrease of RNase E fluorescence in single cells treated with a specific FtsZ inhibitor (Chiarelli et al., 2020), consistent with RNase E implication in the regulation of septum formation in E. coli (Tamura et al., 2006). Moreover, variations in RNase E cellular levels, due either to chemical and transcriptional inhibition or to increased expression, significantly affect mycobacterial cell size and growth. RNase E induction is also associated with increase of: MazF toxin (Slayden et al., 2018), which causes growth heterogeneity in E. coli (Nikolic et al., 2018); different players of the stringent response (Prusa et al., 2018); and universal stress proteins (O'Toole and Williams, 2003). Overall this is suggestive of a nexus between RNase E levels and mycobacterial cell growth and division.
The question arises whether the localization of RNase E in the mycobacterial cell is related to its function. Cytological profiling of E. coli showed that RNase E associates with the membrane and forms transient foci on the cytoplasmic membrane (Strahl et al., 2015). This peripheral localization, contingent on an amphipathic helix absent in mycobacteria (Moffitt et al., 2016), could sequester RNase E from undue degradation of nascent transcripts (Campos and Jacobs-Wagner, 2013). In contrast, in C. crescentus, RNase E colocalizes with the DNA and forms clusters close to highly transcribed genes and to sites of rRNA synthesis, also reminiscent of the RNA polymerase location (Montero Llopis et al., 2010; Bayas et al., 2018; Weng et al., 2019). Our findings imply that the mycobacterial RNase E exhibits a localization pattern closer to C. crescentus, without apparent interaction with the membrane. The transient RNase E foci we observe, overlaid with RNA spots, are likely to coincide with active transcriptional hubs, possibly aiding the cell to spatially compartmentalize RNase E where needed and to prevent its toxicity (Surovtsev and Jacobs-Wagner, 2018; Al-Husini et al., 2018). Moreover, while RIF treatment results in the disappearance of RNase E foci, the association between RNase E and the nucleoid is preserved, and this can aid the mycobacterial cell to prevent RNase E dissemination and to spatially regulate its degradative potential.
In this study, we show that mycobacterial RNase E likely interacts with the nucleoid, by means of the NAP HupB, and that RNase E and HupB are co-regulated. NAPs are responsible for nucleoid remodeling and drive chromosomal transitions between scattered distribution, which favors accessibility by gene expression machineries, and compaction, which preserves DNA integrity and suspends gene expression during stressful conditions (Hołówka and Zakrzewska-Czerwińska, 2020). In particular, HupB bends DNA, assists the segregation of newly replicated chromosomes (Hołówka et al., 2018), and localizes toward the new cell pole, as does RNase E. We also find that DNA damage causes a significant fluorescence decrease of both proteins without delocalization, compatible with a stalled fork and chromosomal fragmentation (Manina et al., 2019; Burgess Tornaletti and Manina, 2020). In contrast, inhibition of both transcription and translation, causing nucleoid condensation (Scutigliani et al., 2018; Weng et al., 2019), delocalizes both HupB and RNase E toward the mid-cell position, with one fluorescence cluster per cell, entailing that RNase E dynamics are largely DNA-dependent, as in C. crescentus (Montero Llopis et al., 2010; Bayas et al., 2018). Interestingly, treatment with the cell-wall targeting drug INH causes not only a substantial increase in fluorescence and cell-to-cell fluorescence variation of both RNase E and HupB but also an atypical delocalization toward the cell center, with two main HupB clusters largely superposed by RNase E, suggestive of cell-cycle arrest before septation (Hołówka et al., 2018; Santi and McKinney, 2015; Logsdon et al., 2017).
These findings prompted us to investigate the role of RNase E and HupB in the context of INH tolerance, as phenotypic variation is being increasingly associated with both spontaneous and stress-induced drug persistence in mycobacteria (Dhar et al., 2016; Logsdon et al., 2017; Sakatos et al., 2018; Manina et al., 2019; Goossens et al., 2020). Furthermore, both RNase E and HupB were shown to affect the expression of the catalase-peroxidase KatG, which converts the pro-drug INH into its active form, and is associated with different forms of INH evasion (Ando et al., 2011; Whiteford et al., 2011; Niki et al., 2012; Wakamoto et al., 2013). Here, we show that cell-to-cell variation in RNase E expression is associated with discrete single-cell responses to INH. In particular, bacilli that experience lysis or survive exhibit higher RNase E levels before INH treatment, and survivors are also more likely to form RNase E foci. Although it may seem counterintuitive that stochastic upregulation of RNase E can lead to two opposite cell fates, this emphasizes the extent to which mycobacterial phenotypic variation can jeopardize the outcome of tuberculosis treatment. We find that RNase E induction causes KatG decrease, endorsing bacilli survival under INH (Wakamoto et al., 2013), and that RNase E is massively induced upon INH exposure prior to HupB. In turn, HupB was shown to downregulate KatG, promoting INH tolerance at the population level (Niki et al., 2012). Additionally, the RNase E-overexpressing pull-down is also enriched with polyphosphate biosynthetic enzymes (Prusa et al., 2018), formerly implicated in INH tolerance (Wang et al., 2018). Conversely, the enrichment of MazF and other toxins upon RNase E induction is consistent with increased mortality, as the accumulation of MazF toxin in M. smegmatis cells subject to INH was conducive to cytolysis (Burgess Tornaletti and Manina, 2020). Furthermore, cells that express significantly lower RNase E levels prior to INH treatment, and that are poor RNase E inducers upon INH exposure, no longer give signs of viability. As rne depletion increases KatG levels, we can speculate that lower RNase E levels are associated with drug activation in individual cells, consistent with increased mortality of M. smegmatis cells experiencing stochastic KatG pulses (Wakamoto et al., 2013). Overall, while too low or too high levels of RNase E and HupB lead to cell death, possibly also due to irreversible DNA compaction (Scutigliani et al., 2018), a dynamic increase followed by the restoration of basal levels of expression is a hallmark of bacilli that better tolerate INH.
To the best of our knowledge, the role of RNase E during infection remains unexplored, albeit it was shown to regulate type III secretion systems of Gram-negative pathogens in vitro (Yang et al., 2008; Lodato et al., 2017; Sharp et al., 2019). In contrast, HupB is required for M. tuberculosis initiation of infection, intracellular iron utilization, and growth (Pandey et al., 2014; Kalra et al., 2018; Yaseen et al., 2018). Thus, we hypothesized that cooperation between RNase E and HupB might benefit M. tuberculosis to cope with the host. Although RNase E and HupB fluorescence levels decrease at the beginning of macrophage infection, they increase as the infection progresses. Furthermore, fluorescence variation markedly increases inside macrophages, consistent with rRNA variation during both macrophage and mouse infection (Manina et al., 2015). Given the role of RNase E in rRNA processing and maturation (Taverniti et al., 2011) and in growth homeostasis we cannot disregard its likely implication in fine-tuning the intracellular growth of individual bacilli.
Remarkably, upon RNase E induction in vitro, several virulence factors are upregulated, including the mammalian cell entry (Mce) proteins (Zhang and Xie, 2011); the stress sensor DevS, belonging to a two-component system critical for virulence (Converse et al., 2009); the VapBC toxin-antitoxin system, critical for long-term survival in macrophages and murine lung (Slayden et al., 2018); and universal stress proteins, relevant for stress endurance in different intracellular pathogens (O'Toole and Williams, 2003). We can infer that RNase E and HupB are crucial for M. tuberculosis regulation of intracellular growth, virulence, and propagation up to the late stage of macrophage infection. It is also tempting to speculate that marked RNase E and HupB intracellular variation account for the diversification of M. tuberculosis intracellular fates and success of infection.
In conclusion, the heterogeneous but synchronized behaviors of RNase E and HupB both in vitro and in macrophages corroborate their functional association in supporting M. tuberculosis homeostasis and persistence under severe conditions. In an era of public health emergencies, when new weapons are increasingly needed to combat refractory infections (Fisher et al., 2017; Schröter and Dersch, 2019), investigating the drivers of phenotypic variation and their functional relationship serves not only to fathom the single-cell physiology of pathogenic microorganisms but also to identify molecular targets responsible for phenotypic diversification, and whose inhibition would radically undermine the ability of pathogens to adapt and persist.
Limitations of study
In this study, we performed a multiscale characterization of the mycobacterial RNase E and showed its association and co-regulation with HupB. Nonetheless, owing to technical limitations, we were unable to clarify whether the RNase E-HupB interaction is mediated by RNA, DNA, or other molecular players. Further investigations will also be required to assess whether there are other true interactors among the proteins identified in the RNase E pull-down, having a functional role. Additionally, to address the single-cell dynamics of RNase E, we could not fluorescently tag the native locus due to its essentiality, but we had to generate a stable merodiploid strain. The same strategy was also applied to analyze HupB. Finally, although we show instances of the implication of RNase E and HupB in the adaptive landscape of M. tuberculosis, their direct role in regulating the intracellular growth rate and the consequences for subpopulation fitness and long-term persistence remain to be assessed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-6xHist tag (H3) | Santa Cruz Biotechnology | Cat# sc-8036; RRID: AB_627727 |
| Mouse monoclonal anti-Ag85 (HYT 27) | Santa Cruz Biotechnology | Cat# sc-57611; RRID: AB_628733 |
| ECL HRP-conjugated anti-mouse m-IgGκ | Santa Cruz Biotechnology | Cat# sc-516102; RRID: AB_2687626 |
| Rabbit monoclonal anti-GFP (3E6) | Thermo Fisher | Cat# A-11120; RRID: AB_221568 |
| Anti·His HRP Conjugate (RGSHHHH) | QIAGEN | Cat# 652503; RRID: AB_2734520 |
| ECL HRP-conjugated anti-rabbit | Amersham | Cat# NA934; RRID: AB_772206 |
| GFP-Trap_A for immunoprecipitation | Chromotek | Cat# gta-20; RRID: AB_2631357 |
| Bacterial strains | ||
| Escherichia coli DH5α | Invitrogen | Cat# 12297016 |
| Mycobacterium smegmatis mc2155 | Lab collection | ATCC 700084 |
| ATCC®700084 - pAG202; inducible strain overexpressing wild type Rne_6X-His-tag | This paper | AGS2 |
| ATCC®700084 – pDV201; attB::rne-linker-mKate2 | This paper | DVS2 |
| ATCC®700084 – pAG212; attB::rne-linker-mKate2 | This paper | AGS13 |
| AGS13 – pAG219, attB::rne-linker-mKate2_attT::deaD-linker-mCitrine | This paper | AGS20 |
| AGS13 – pAG215, attB::rne-linker-mKate2_attT::rraA-linker-mCitrine | This paper | AGS16 |
| AGS13 – pAG218, attB::rne-linker-mKate2_ attT_obg-linker-mCitrine | This paper | AGS19 |
| AGS13 – pAG220, attB::rne-linker-mKate2_ attT::relA-linker-mCitrine | This paper | AGS21 |
| AGS13 – pAG221 attB::rne-linker-mKATE2_attT::ppk-linker-mCitrine | This paper | AGS22 |
| AGS13 – pAG217 attB::rne-linker-mKate2_attT::uvrA-linker-mCitrine | This paper | AGS18 |
| AGS13 – pAG213 attB::rne-linker-mKate2_ attT::lsr2-linker-mCitrine | This paper | AGS23 |
| AGS13 – pAG214 attB::rne-linker-mKate2_attT::infA-linker-mCitrine | This paper | AGS15 |
| AGS13 – pAG216 attB::rne-linker-mKate2_attT::hupB-linker-mCitrine | This paper | AGS17 |
| ATCC®700084 – pGM255; rne (MSMEG_4626) CRISPRi/dCas9 | This paper | mc2155/pGM255 |
| ATCC®700084 – pGM257; scrambled CTsgRNA/dCas9 | This paper | mc2155/pGM257 |
| AGS2 – pAG216; attT::hupB-linker-mCitrine | This paper | AGS33 |
| mc2155/pGM255 - pAG216; attT::hupB-linker-mCitrine | This paper | AGS32 |
| Mycobacterium tuberculosis Erdman | Lab collection | ATCC 35801 |
| ATCC®35801 – gfp::rpsJ (gfp chromosomal integration by two-step homologous recombination using pGM213) | This paper | GMT14 |
| ATCC®35801 – gfp::rpsJ_attB::rne-linker-mCherry | This paper | GMT14/pGM301 |
| ATCC®35801 – pGM301 – pGM305; attB::rne-linker-mCherry_attT::hupB-linker-sfgfp | This paper | GMT25 |
| ATCC®35801 – pGM305 – pGM306; attT::hupB-linker-sfgfp; | This paper | GMT26 |
| ATCC®35801 – pGM305 – pGM309; CRISPRi/dCas9_attT::hupB-linker- sfgfp | This paper | GMT27 |
| Chemicals, peptides, and recombinant proteins | ||
| LB Broth Base | Invitrogen | Cat# 12780052 |
| Agar powder | Invitrogen | Cat# 30391049 |
| Middlebrook 7H9 Broth | BD Difco | Cat# DF0713-17-9 |
| Middlebrook 7H10 Agar | BD Difco | Cat# DF0627-17-4 |
| Middlebrook OADC Growth Supplement | BD Difco | Cat# BD212351 |
| Glycerol | Sigma-Aldrich | Cat# G5516 |
| Tween 80 | Sigma-Aldrich | Cat# P5188 |
| Bovine Serum Albumin (BSA) Fraction V | Sigma-Aldrich | Cat# A9647 |
| Sodium chloride | Sigma-Aldrich | Cat# S9888 |
| Dextrose | Sigma-Aldrich | Cat# G8270 |
| Sucrose | Sigma-Aldrich | Cat# 84100 |
| Hartmans-de Bont minimal medium for mycobacteria | (Smeulders et al., 1999) | N/A |
| Sodium propionate | Sigma-Aldrich | Cat# P1880 |
| Kanamycin solution | Sigma-Aldrich | Cat# K0254 |
| Hygromycin B solution | Sigma-Aldrich | Cat# H0654 |
| Isoniazid | Sigma-Aldrich | Cat# I3377 |
| Rifampicin | EUROPEAN PHARMACOPOEIA | Cat# R0700000 |
| Mitomycin C | Sigma-Aldrich | Cat#M4287 |
| FtsZ inhibitor: Methyl [(4-nitro-2,1,3-benzothiadiazol-5-yl)thio]acetate | Lab collection (Chiarelli et al., 2020) | C109 |
| Chloramphenicol | Sigma-Aldrich | Cat# C0378 |
| M5 inhibitor: ethyl N-[(1-{[5-chloro-3-(trifluoromethyl)-2-pyridyl]amino}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)carbonyl]carbamate | Maybridge | KM08782 |
| Critical commercial assays | ||
| Q5® Site-Directed Mutagenesis Kit | NEB | E0554S |
| HisPurTM Cobalt Superflow Agarose | Thermo Fisher | 252229 |
| Deposited data | ||
| Whole-cell mass spectrometry-based proteomics of M. smegmatis rne overexpressing (AGS2), silenced (mc2155/pGM255) and control strains | This paper (Table S1), PRIDE repository (Perez-Riverol et al., 2019) | ProteomeXchange ID: PXD025674 |
| Whole-cell mass spectrometry-based proteomics of RNase E-His pull-down from M. smegmatis rne overexpressing (AGS2) and control strain | This paper (Table S2) PRIDE repository (Perez-Riverol et al., 2019) | ProteomeXchange ID: PXD025674 |
| Experimental models: Cell lines | ||
| RAW 264.7 murine macrophages | Lab collection | ATCC TIB-71 |
| Oligonucleotides | ||
| see Table S3 | ||
| Recombinant DNA | ||
| pCR2.1-TOPO TA cloning plasmid, AmpR, KmR | Invitrogen | pCR2.1-TOPO |
| pMYC mycobacterial expression vector, for C-term fusion of 6X-His-tag, HygR | Addgene (Beckham et al., 2020) | # 42192 |
| pJG1100 – Suicide vector expressing KmR, HygR resistance cassettes and sacB marker | Lab collection | pJG1100 |
| pTTP1A-based integrative vector, containing Tweety phage integration site (attT), KmR | Addgene (Pham et al., 2007) | # 91721 |
| mCitrine-N1, containing mEYFP, KmR | Addgene (Griesbeck et al., 2001) | # 54594 |
| pLJR962, tetracycline-inducible dCas9 attB-integrative vector for M. smegmatis gene silencing, KmR | Addgene (Rock et al., 2017) | # 115162 |
| pLJR965, tetracycline-inducible dCas9 attB-integrative vector for M. tuberculosis gene silencing, KmR | Addgene (Rock et al., 2017) | # 115163 |
| pMYC rne (MSMEG_4626), acetamide-inducible Rne-6xHist tag | This paper | pAG202 |
| pMV361-based integrative vector, containing attB phage attachment site, KmR | Lab collection (Peña et al., 1996) | pND200 |
| pND200 expressing mCherrywt from UV15 strong promoter, HygR | Lab collection | pGM218 |
| pND200 expressing Pnative-rne-linker-mKate2, KmR (MSMEG_4626) | This paper | pDV201 |
| pGM218 expressing Pnative-rne-linker-mKate2, HygR (MSMEG_4626) | This paper | pAG212 |
| pTTP1A expressing Pnative-deaD-linker-mCitrine, KmR | This paper | pAG219 |
| pTTP1A expressing Pnative-rraA-linker-mCitrine, KmR | This paper | pAG215 |
| pTTP1A expressing Pnative-obg-linker-mCitrine, KmR | This paper | pAG218 |
| pTTP1A expressing Pnative-relA-linker-mCitrine, KmR | This paper | pAG220 |
| pTTP1A expressing Pnative-ppk-linker-mCitrine, KmR | This paper | pAG221 |
| pTTP1A expressing Pnative-uvrA-linker-mCitrine, KmR | This paper | pAG217 |
| pTTP1A expressing Pnative-lsr2-linker-mCitrine, KmR | This paper | pAG213 |
| pTTP1A expressing Pnative-infA-linker-mCitrine, KmR | This paper | pAG214 |
| pTTP1A expressing Pnative-hupB-linker-mCitrine, KmR | This paper | pAG216 |
| pGM218 expressing Pnative-rne-linker-mCherry, HygR (rv2444c) | This paper | pGM301 |
| pJG1100-based vector for chromosomal knock-in of gfp-linker fused in frame to rpsJ (rv0700) start codon | This paper | pGM213 |
| pTTP1A expressing Pnative-hupB-linker-sfgfp, KmR | This paper | pGM305 |
| pLJR962 modified with HygR cassette | This paper | pGM256 |
| pGM256 carrying sgRNA for rne (MSMEG_4626) silencing, HygR | This paper | pGM255 |
| pGM256 carrying a scrambled CTsgRNA, HygR | This paper | pGM257 |
| pLJR965 modified with HygR cassette | This paper | pGM309 |
| pGM309 carrying sgRNA for rne (rv2444c) silencing, HygR | This paper | pGM306 |
| Software and algorithms | ||
| Mycobrowser | (Kapopoulou et al., 2011) | https://mycobrowser.epfl.ch/ |
| Cytoscape Version 3.8.2 | Cytoscape | https://cytoscape.org/ |
| PANTHER Classification System | (Mi et al., 2013) | http://pantherdb.org |
| ImageJ 1.51s | (Schneider et al., 2012) | https://imagej.net/Fiji |
| Prism Versions 8.4 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| RStudio Version 1.1.423 | RStudio, Inc | https://rstudio.com/pri |
| Python Version 3.8.3 | Python | https://www.python.org/ |
| Other | ||
| Custom R script for single-cell image data post-processing | (Manina et al., 2019) | TL_PostProc_Script_AG-IT.R |
| Custom Python script for single-cell image data post-processing | This paper | post_process_fluorescence.py |
| Custom ImageJ macros for analysis of infected macrophages | This paper | InfectedCells_Analysis_GM.ijm |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Giulia Manina (giulia.manina@pasteur.fr).
Materials availability
Plasmids and bacterial strains generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Bacterial strains and cell line
All DNA cloning and sequencing steps were performed in chemically competent E. coli DH5α, grown in LB medium and selected with the appropriate antibiotics: 100 μg/mL ampicillin; 50 μg/mL kanamycin for E. coli; 100 μg/mL hygromycin; and 25 μg/mL kanamycin with 50 μg/mL hygromycin. Mycobacteria were cultured in Middlebrook 7H9 broth supplemented with 0.5% BSA, 0.2% glucose, 0.085% NaCl, 0.5% glycerol and 0.05% Tween-80. Mycobacterial transformants were selected on 20 μg/mL kanamycin; 50 μg/mL hygromycin; and 15 μg/mL kanamycin with 50 μg/mL hygromycin, according to the selective markers. Hartman’s de Bond (HDB) and modified HDB were used as minimal media (Smeulders et al., 1999) in the absence of Tween-80. Middlebrook 7H10 agar was enriched with 10% OADC and 0.5% glycerol. Bacterial stocks were prepared from exponentially growing cultures derived from single colonies, supplemented with 15% glycerol, and stored at −80°C. Each aliquot was used only once to start primary cultures. Primary cultures of M. smegmatis (ATCC 700084) and M. tuberculosis (ATCC 35801) were grown in complete Middlebrook 7H9 with appropriate antibiotic selection, at 37°C at 150 and 50 RPM, respectively, until reaching mid-log phase (OD600 0.5–0.8). Secondary cultures, used for final assays, were started from dilutions of the primary cultures into the appropriate medium (1:50 for minimal media, and 1:100 for standard 7H9), in the absence of antibiotic selection, except for the expression plasmid pMYC (HygR). Growth curves were measured from secondary cultures that had reached mid-log phase and diluted to OD600 0.025. Induction of RNase E-His in M. smegmatis was achieved with 0.05% acetamide. Transcriptional silencing of rne was achieved in M. smegmatis with 150 ng/mL and in and M. tuberculosis with 100 ng/mL anhydrotetracycline (ATC).
RAW 264.7 murine macrophages (ATCC TIB-71) were cultured in Dulbecco’s Modified Eagle Medium (Gibco, DMEM) supplemented with 10% fetal bovine serum (Gibco) in the absence of antibiotics. For each experiment a fresh stock of about 10ˆ6 cells frozen in 1 mL of DMEM and 10% DMSO was inoculated in a T-175 flask, and propagated at 37°C in humidified 5% CO2 atmosphere until confluence. For microscopy experiments, confluent cells were diluted to a concentration of 10ˆ5 per mL in complete DMEM without antibiotics and without phenol red.
Method details
Strains construction
Oligonucleotides, plasmids and strains are listed in the Key resources table and Table S3. Translational reporter strains were constructed using stable variants of mCherry, mKate2, mCitrine, GFP or sfGFP. Genes encoding proteins of interest and their native regulatory region were PCR amplified and fused in frame with the appropriate fluorescent marker via a linker sequence. Final constructs were electroporated into M. smegmatis or M. tuberculosis, to achieve chromosomal integration into specific phage attachment sites, as formerly described (Manina et al., 2019). M. tuberculosis RpsJ translational reporter was generated by N-terminal fusion of GFP to the native translational start codon, using the pJG1100 suicide vector, by two-step homologous recombination as formerly described (Manina et al., 2015). M. smegmatis expressing inducible rne was generated by cloning the MSMEG_4626 open reading frame into the pMYC vector. The M. smegmatis and M. tuberculosis ATC-inducible rne knock-down strains were generated using the CRISPRi gene silencing system, as formerly described (Rock et al., 2017). Small guide RNA (sgRNA), 20-nucleodide long, were designed complementary to MSMEG_4626 or rv2444c, with a specific protospacer adjacent motif (5′-NNAGAAG-3′). A scrambled sgRNA was also designed as control (Rock et al., 2017). The kanamycin resistance cassette was replaced with hygromycin in both pLJR962 and pLJR965 vectors, to be co-transformed into the HupB fluorescent reporters.
Total RNA extraction
For total RNA extraction, 12 mL of cultures were collected by centrifugation at 4200 x g for 15 min at 4°C. Cell pellets were resuspended in 700 μl of TRIzol and transferred into a 2-mL vials prefilled with glass beads (Precellys). Cells were lysed by bead-beating two times at 4234 x g for 60 sec, with 5 min intervals on ice, and centrifuged at 12000 x g for 10 min. The supernatant was precipitated with 1 volume of isopropanol enriched with 0.5% carrier (Invitrogen), washed in 70% ethanol, and resuspended in 50 μl of nuclease-free water. gDNA was depleted with Turbo DNase (Ambion).
Rapid amplification of 5′ complementary DNA ends (5′ RACE)
5′-RACE was carried out starting from 3 μg of total RNA that was reverse transcribed into cDNA. The reaction was conducted by adding 1 μL of random hexamers (Thermo Fisher) to the total RNA in a final volume of 13 μL, and incubating at 65°C for 5 min. Next, the reaction was incubated for at least 1 min on ice, and then 5X-SSIV Buffer, 100 mM DTT and 200 U/μl of SuperScript IV reverse transcriptase (Invitrogen) were added in a final volume of 20 μl. The reaction was incubated at 23°C for 10 min, then at 55°C for 10 min and lastly at 80°C for 10 min. The cDNA was treated with 4U of RNase H (NEB) at 37°C for 20 min before cDNA purification by QIAquick PCR Purification Kit (Qiagen). Next dATP tails were added to 20 μL of purified cDNA, using 10 mM dATP, 10 U of terminal deoxynucleotidyl transferase (TdT) (NEB), 10X terminal transferase buffer, and 2.5 mM CoCl2 solution, in a final volume of 50 μL. The reaction was incubated at 37°C for 30 min, and TdT was heat-inactivated at 70°C for 10 min. Afterwards, two PCR-amplification rounds were carried out, as follows. First, a PCR mixture was prepared containing 5 μl of dATP-tailed cDNA template, 0.15 mM MgCl2, 0.2 mM dNTPs, 200 nM of gene-specific primer (GSP_4626), QT primer (Scotto-Lavino et al., 2006), 10X DreamTaq DNA polymerase buffer and 1 U of DreamTaq DNA polymerase (Thermo Fisher) in a final volume of 50 μL. Denaturation was carried out at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec, amplification at 72°C for 30 sec. Second, a PCR mixture was prepared with 5 μL of the first PCR product, 200 nM of GSP_4626 and QI primer (Scotto-Lavino et al., 2006). Denaturation was carried out at 95°C for 3 min, followed by 25 cycles of denaturation at 95°C for 30 sec, annealing at 65°C for 30 sec, amplification at 72°C for 30 sec. The final reaction was purified by QIAquick PCR Purification Kit (Qiagen) and analyzed by sequencing using specific primer GSP_4646.
Real-time quantitative PCR
M. smegmatis and M. tuberculosis wild-type and fluorescent reporter strains were cultured in Middlebrook 7H9 broth until OD600 0.25. Samples were split and re-incubated at 37°C, in the absence or presence of a given stress or inducer. At regular intervals, culture volumes corresponding to OD600 3.0 were withdrawn and total RNA was extracted. cDNA was generated starting from 250 ng of total RNA using SuperScrip IV (Invitrogen) and random hexamers (Thermo Fisher), according to manufacturer’s instruction. qRT-PCR was carried out using the SYBR Green PCR Master Mix (Applied Biosystems), 0.3 μM primers, and 1 μL cDNA diluted 1:4. Absolute quantification was run on the LightCycler®480 Instrument (Roche Life Science) as follows: activation step at 50°C for 2 min (ramp rate °C/s 4.8) and 95°C for 10 minutes; 40 amplification steps at 95°C for 15 sec (ramp rate °C/s 4.8), 60°C for 30 sec (ramp rate °C/s 2.5) and 72°C for 30 sec (ramp rate °C/s 4.8); melting curve at 60°C for 15 sec (ramp rate °C/s 2.5) and 95°C for 15 sec (ramp rate °C/s 0.29). Standard curves were generated using serial dilutions of M. smegmatis and M. tuberculosis gDNA, and were used to calculate transcripts copy numbers.
RNase E-His overexpression and purification
Primary M. smegmatis cultures carrying were diluted to OD600 0.03 and incubated at 37°C until they reached OD600 0.5. Secondary cultures were induced at 30°C for 24 hours. Next, 25 mL of culture were collected at 4200 x g for 15 min at 4°C. Pellets were processed under native conditions in 500 μL of protein lysis buffer (10 mM Tris-HCl pH 7.5; 150 mM NaCl; 0.5% glycerol and 1X-Protease Inhibitor Cocktail, Roche). Cell suspensions were sonicated for 5 cycles of 30 sec at 100% amplitude, with 45-sec intervals on ice, and then centrifuged at 10000 x g for 1 hour at 4°C. Pellets were resuspended in 250 μL of protein lysis buffer, quantified by Bradford reagent (Sigma), and the concentration of the protein extracts were adjusted to 1 mg/mL. RNase E pull-down was carried out from 100 μg of protein extract, under native conditions. All purification steps were performed either on ice or at 4°C, to prevent RNase E degradation. 600 μL of HisPurTM Cobalt Superflow Agarose (Thermo Fisher), at a ratio 1:6 v/v to the sample volume, were transferred to a clean 2 mL low-binding tube (Eppendorf) and equilibrated with 1.2 mL of equilibration buffer (20 mM of Na3PO4; 300 mM NaCl; 5 mM imidazole). The resin was sedimented by centrifugation at 700 x g for 2 min at 4°C, and the supernatant discarded. Before adding the cellular suspension to the resin, the samples were equilibrated with 300 μL of equilibration buffer. The equilibrated samples were added to the resin and incubated for 4 hours at 4°C, ensuring gentle but continuous mixture. The sample-containing resin suspension was centrifuged at 700 x g for 2 min at 4°C. The supernatant was carefully removed and stored for downstream analysis. The resin was washed three times for 2 min in the washing buffer (20 mM Na3PO4; 300 mM NaCl; 15 mM imidazole). For each washing step, the supernatant was stored and used for downstream analysis. The bound RNase E-6X-His was eluted two times for 10 min with 0.5 mL of elution buffer (20 mM Na3PO4; 300 mM NaCl; 150 mM imidazole) and precipitated with trichloroacetic acid (TCA)/acetone at 4°C. One volume of cold 40% TCA (Sigma) was added to the eluted sample, followed by 30 min incubation and then centrifuged at 14000 x g for 15 min. Next, the pellet was washed twice with 1 volume of cold acetone (Sigma), incubated for 10 min, and centrifugated at 14000 x g for 10 min. The supernatant was discarded, samples dried and stored at 4°C before Mass Spectrometry.
Immunoprecipitation of HupB-mCitrine and Lsr2-mCitrine
Secondary cultures (30 mL) were collected after 24 hours of RNase E-His induction with 0.05% acetamide, by centrifugation at 4200 x g for 15 min at 4°C. Pellets were washed once and resuspended in 500 μL of protein lysis buffer (10 mM Tris-HCl pH 7.5; 150 mM NaCl; 0.5% glycerol; 0.01% tyloxapol and 1X-Protease Inhibitor Cocktail, Roche). Cell suspensions were lysed with 5 cycles of bead beating for 60 sec at 4234 x g, with 5 min intervals on ice. Cell lysates were centrifuged at 10000 x g for 15 min at 4°C and the soluble fraction was quantified and used for the assay. The NAPs-mCitrine fractions were immunoprecipitated by using the GFP-Trap (Chromotek), as per manufacturer’s instructions. Protein dissociation from agarose beads was carried in 80 μL of 2X Laemmli sample buffer and incubation at 95°C for 5 min. Eluates (35 μL) were run on Novex Tris-Glycine gels 4-20% gels at 200 V for 1 hour, for both Coomassie staining and western blot analysis.
Western blot
Cellular extracts were quantified by Bradford (Sigma), and 15 μg were used for the assay. Proteins were transferred onto a PVDF membrane by wet-transfer performed at 30 V for 1 hour, or by dry-transfer using the iBlot Transfer Apparatus (Invitrogen). The membrane was blocked with 5% non-fat dry milk in 0.01% Tween-20 TBS (TBS-T) for 1 hour, incubated with primary antibody overnight at 4°C, and washed three times with TBS-T for 5 min. The membrane was incubated with the peroxidase-conjugated secondary antibody in TBS-T containing 1% non-fat dry milk for 1 hour at room temperature, and washed three times with TBS-T for 5 min. Proteins were detected using the Pierce ECL Western Blotting Substrate (Thermo Fisher) on a Bio-Rad ChemiDoc MP Imager, or using a chemiluminescence Amersham Hyperfilm MP (Cytiva). Primary and secondary antibodies were diluted in TBS-T and 1% non-fat milk as follows: anti-GFP (1 : 5000), anti-Ag85 (1 : 5000), anti-6X-His (1 : 2000), HRP-conjugated anti-His (1 : 2000); HRP-conjugated anti-rabbit (1 : 10000) and HRP-conjugated anti-mouse (1 : 10000).
Samples preparation for mass spectrometry (MS)
Secondary cultures were diluted to OD600 0.05, induced at 37°C for 24 hours. After induction, 12 mL of culture were centrifuged at 4200 x g for 15 min at 4°C. Pellets were processed in 500 μL of denaturing lysis buffer (100 mM NH4HCO3 and 8 M CH4N2O). Cell suspensions were sonicated for 5 cycles of 30 sec at 100% amplitude, with 45-second intervals on ice. Total cell lysates were quantified by Bradford reagent (Sigma), samples concentration was adjusted to 0.5 mg/mL, and samples were stored at −80°C until use. Bacterial lysate before pull-down (input), the RNase E TCA-precipitated pull-down and whole cell lysate were diluted (1 : 2) using the denaturing lysis buffer. Proteins were reduced with 5 mM Tris(2-carboxyethyl) phosphine hydrochloride solution (TCEP, Sigma) for 30 min and then alkylated with 20 mM iodoacetamide (Sigma) for 1 hour in the dark. Reduced and alkylated proteins were then digested over-night at 37°C with Sequencing Grade Modified Trypsin (Promega) using an enzyme: protein ratio (1 : 50). The digestion was stopped with 4% formic acid (FA). For MS analysis, the resulting peptides from the input, the pull-down and the whole-cell lysate were desalted with stage-tip using C18 Empore disc and eluted with 80% acetonitrile (ACN) and 0.1% FA. All samples were dried in a Speed-Vac, and peptides were resuspended in 2% ACN, 0.1% FA prior to Liquid Chromatography–MS analysis.
Liquid chromatography–MS
For the input: a nanochromatographic system (Proxeon EASY-nLC 1200 - Thermo Fisher Scientific) was coupled on-line to a Q Exactive Plus Mass Spectrometer (Thermo Fisher Scientific) using an integrated column oven (PRSO-V1 - Sonation GmbH). For each sample, peptides were loaded on a 50 cm column (EASY-Spray column, 50cm x 75 μm ID, PepMap C18, 2 μm particles, 100 A pore size - ES803 - Thermo Fisher Scientific) after an equilibration step in 100 % solvent A (H2O, 0.1% FA). Peptides were eluted with a multi-step gradient using 2 to 7% solvent B (80% ACN, 0.1% FA) during 5 min, 7 to 23% during 70 min, 23 to 45% during 30 min and 45 to 95% during 5 min, at a flow rate of 250 nL/min over 132 min. Column temperature was set to 60°C.
For the pull-down: the same procedure was used with an in-house packed 38 cm nano-HPLC column (75-μm inner diameter) with C18 resin (1.9-μm particles, 100 Å pore size, Reprosil-Pur Basic C18-HD resin, Dr. Maisch GmbH).
For whole-cell proteome: the same procedure was used with an in-house packed 50 cm nano-HPLC column (75-μm inner diameter) with C18 resin (1.9-μm particle, 100 Å pore size, Reprosil-Pur Basic C18-HD resin, Dr. Maisch GmbH).
MS data were acquired using Xcalibur software using a data-dependent Top 10 method with a survey scans (300-1700 m/z) at a resolution of 70000 and a MS/MS scans (fixed first mass 100 m/z) at a resolution of 17500. The AGC target and maximum injection time for the survey scans and the MS/MS scans were set to 3E6, 20 ms and 1E6, 60 ms respectively. The isolation window was set to 1.6 m/z and normalized collision energy fixed to 28 for HCD fragmentation. A minimum AGC target of 1E4 was used for an intensity threshold of 1.7E5. Unassigned precursor ion charge states as well as 1, 7, 8 and >8 charged states were rejected and peptide match was disable. Exclude isotopes was enabled and selected ions were dynamically excluded for 45 sec.
Snapshot microscopy
Phase-contrast and fluorescence snapshot imaging were acquired using an inverted DeltaVision Elite Microscope (GE Healthcare) equipped with an UPLFLN100XO2/PH3/1.30 objective (Olympus). All samples were prepared dispensing 0.7 μL of bacteria between two #1.5 coverslips.
RNase E-mKate2 cells were stained with 1 μM RNASelect (Invitrogen) for 1 hour. Exposure conditions: phase-contrast 100% T, 150 msec; mCherry (Ex 555/90, Em 600/675) 100% T, 600 msec; YFP (Ex 513/17, Em 548/22) 32% T, 250 msec. Primary cultures of dual-fluorescent reporters were split and re-incubated at 37°C for 6 hours, either without stress or in HDB-based starvation medium. Exposure conditions: phase-contrast 50% T, 150 msec; mCherry (Ex 575/25, Em 625/45) 100% T, 600 msec; YFP (Ex 513/17, Em 548/22) 100% T, 600 or 250 msec. When appropriate exponential M. smegmatis RNase-HupB dual-fluorescent reporter was treated with 0.2 μg/mL MMC, 20 μg/mL RIF, 64 μg/mL CAP, 16 μg/mL C109 (methyl [(4-nitro-2,1,3-benzothiadiazol-5-yl)thio]acetate), and 20 μg/mL INH at 37°C for 6 hours. Exposure conditions: phase-contrast 50% T, 150 msec; mCherry (Ex 575/25, Em 625/45) 100% T, 600 msec, and YFP (Ex 513/17, Em 548/22) 100% T, 250 msec.
Time-lapse microscopy
Bacterial clumps were eliminated by culture filtration through a 5-μm filter. The single-cell bacterial suspensions were inoculated into our custom-made microfluidic hexa-device, as formerly described (Manina et al., 2019). Growth medium in the absence or presence of INH or ATC was injected into device using a syringe pump at 10 μl/min. The assembled microfluidic system was mounted on the motorized microscope stage of our DeltaVision Elite Microscope (Cytiva), under constant temperature of 37°C. Images were acquired every 30 min for M. smegmatis and every 3 hours for M. tuberculosis, using a UPLFLN100XO2/PH3/1.30 objective (Olympus) and a high-speed sCMOS camera, 2,560 x 2,160 pixels, pixel size 6.5 x 6.5 μm, 15-bit, spectral range of 370–1,100 nm. Exposure conditions for M. smegmatis: phase-contrast 100% T, 150 msec; and mCherry (Ex 575/25, Em 625/45) 100% T, 600 msec. Exposure conditions for M. tuberculosis: phase-contrast 100 % T, 200 msec, FITC (Ex 492/28, Em 523/23) 100% T, 200 msec; TRITC (Ex 556/25, Em 611/47) 100% T, 600 msec. Different reporter strains or independent cultures of the same reporter strain were seeded in separate areas of the hexa-device. For each reporter strain, both technical and biological replicates were analyzed. Plots representing single-cell analysis were generated by pooling data obtained from at least two independent experiments.
Macrophage infection
Confluent Raw 264.7 macrophages (10ˆ5 in 1 mL of complete DMEM) were seeded in a 35-mm μ-Dish (Ibidi), and incubated at 37°C with 5% CO2 for 24 hours before infection. Macrophages were infected with exponentially-growing M. tuberculosis cultures diluted in complete DMEM without phenol red, at OD600 0.005 (MOI 1:5) and OD600 0.01 (MOI 1:10). After 6 hours post infection, cells were washed five times with 1 mL of complete DMEM without phenol red, and ATC 100 ng/mL was added to induce rne silencing. Infected cells were replenished with fresh medium and ATC 100 ng/mL on day 3 post infection. Infected macrophages were imaged using a 100X oil immersion objective, 1.4NA, WD 0.12mm, at day 1, 3 and 6 post infection. Images were sequentially acquired on each field of view, through a z-stack of 6 μm, by acquiring one snapshot every 1-μm. Exposure conditions: bright field 32% T, 50 msec; FITC (Ex 475/28, Em 525/48) 100% T, 200 msec; TRITC (Ex 556/25, Em 611/47) 100% T, 600 msec.
Quantification and statistical analysis
Statistics
Plots and statistical analyses were generated using Prism 8.4 (GraphPad Software), R (v1.3.1093), and Python (V3.8.2). Pearson r correlation coefficients were computed on (x, y) datasets. Two-way ANOVA, followed by correction for multiple comparisons, was performed to evaluate statistical significance between multiple groups. Brown-Forsythe, Welch and one-way ANOVA was computed to compare the variation of a single parameters over multiple groups. Chi square test of independence was used to compare two variables in a contingency table and check their relatedness. Plots merge datasets deriving from at least two independent replicates. Significant p-values, sample size and statistical tests are reported in the legends.
MS data processing, analysis and statistics
Raw data were analyzed using MaxQuant software version 1.5.5.1 (input and IP) or version 1.6.10.43 (whole cell proteome) (Tyanova et al., 2016) using the Andromeda search engine (Cox et al., 2011). The MS/MS spectra were searched against the UniProt M. smegmatis (strain ATCC 700084/mc2155) database containing 6601 proteins, usual known mass spectrometry contaminants and reversed sequences of all entries. Andromeda searches were performed choosing trypsin as specific enzyme with a maximum number of two missed cleavages. Possible modifications included carbamidomethylation (Cys, fixed), oxidation (Met, variable), N-terminal acetylation (variable). The mass tolerance in MS was set to 20 ppm for the first search then 4.5 ppm for the main search and 20 ppm for the MS/MS. Maximum peptide charge was set to seven and seven amino acids were required as minimum peptide length. The “match between runs” feature was applied for samples having the same experimental condition with a maximal retention time window of 0.7 min. One unique peptide to the protein group was required for the protein identification. A false discovery rate (FDR) cutoff of 1 % was applied at the peptide and protein levels. Quantification was performed using the XIC-based LFQ algorithm with the Fast LFQ mode (Cox et al., 2014) and skip normalization was applied only for IP. Unique and razor peptides, included modified peptides, with at least 2 ratio count were accepted for quantification.
For the differential analysis, proteins identified in the reverse and contaminant databases and proteins “only identified by site” were first discarded from the list of identified proteins. Then, only proteins with at least four LFQ values in a condition were kept when comparing M. smegmatis rne overexpressing and silenced strains, and at least two LFQ values in a condition were kept when comparing M. smegmatis Rne pull-down versus control samples, to find potential interactors while ensuring a minimum of replicability of the measured LFQ values. After Log2 transformation, LFQ values were normalized by median centering within conditions, using wrapper normalizeD function of the R package DAPAR (Wieczorek et al., 2017). Remaining proteins without any LFQ value in one of the both conditions have been considered as proteins quantitatively present in a condition and absent in another. They have, therefore, been set aside and considered as differentially abundant proteins. Next, missing values were imputed using the impute.mle function of the R package imp4p (Giai Gianetto et al., 2020). Proteins with a fold-change under 2.0 were considered not significantly different in abundance. Statistical testing of the remaining proteins (having a fold-change over 2.0) was conducted using a limma t-test. An adaptive Benjamini-Hochberg procedure was applied on the resulting p-values thanks to the function adjust.p of R package cp4p (Giai Gianetto et al., 2016). The proteins associated to an adjusted p-value inferior to 1% FDR were considered as significant differentially abundant proteins. Lastly, the proteins of interest are those emerging from this analysis, supplemented by those present in one condition and absent in the comparative condition.
For the analysis of RNase E pull-downs and control samples, the proteins of interest are potential interactors of Rne. Their stoichiometry was studied using intensity-based absolute quantification (iBAQ) values. These values are relevant to rank the absolute abundance of different proteins within a single sample (Schwanhäusser et al., 2011) and to study protein stoichiometry (Smits et al., 2013). The relative stoichiometry ratio (RS) was computed to compare the abundance variation of each protein between the RNase E pull-down and the control samples, to the one of the bait protein (RNase E):
where is the average iBAQ measured in the Rne pull-down samples and in the control samples. If is superior to 1, the potential interactor abundance increases in average more strongly than the one of the bait protein (RNase E) between the pull-down and the control samples. Additionally, a standard deviation of this quantity was computed for each protein.
The protein-protein interaction networks (Figure 2) for the whole-cell proteome analyses were created in Cytoscape (version 3.8.2) using STRING (Szklarczyk et al., 2019). Medium confidence interactions (co-expression, experiments, neighborhood, co-occurrence and database with a score of 0.4) relative to the query proteins were considered. Predicted proteins and protein with an unknown function were excluded from the protein-protein networks.
Single-cell analysis
ROI Manager Macro of ImageJ 1.52q software was used to perform single-cell segmentation of both snapshots and time-lapse images. The selection freehand tool was used to cover the profile of individual cells. The thickness was set to 7 for M. smegmatis, and 5 for M. tuberculosis. From freehand segmented lines, covering the cell profile, diverse parameters were extracted, namely, cell size and length, the total fluorescence and the intensity fluorescence profile. Both the total fluoresce and the fluorescence profile were expressed as the sum of intensity of the pixels normalized to the thickness of the line drawn over the cell. To homogenize the fluorescence values across experiments and fields of view, the background signal was subtracted at each measurement. The cell position in the lineage and pole age were manually annotated. Two customized scripts, written in R and Python, were used for post-processing of the .csv and .txt data files, deriving from Image J ROI Manager. The image stacks of infected macrophages were projected by SoftWorx maximum-intensity method, and processed using a customized Image J macro, which automatically segments intracellular bacterial foci, capturing their area, total fluorescence, and subtracting the background fluorescence.
Calculation of single-cell parameters and subpopulations
The growth rate was measured by fitting an exponential curve to single-cell size measurements over the generation time of the cell from birth to division. Single-cell localization events (foci) were identified in M. smegmatis if one or more cell segments had RNase E-mKate2 fluorescence values equal or greater than 500 (a.u.) after subtracting the minimum cell fluorescence, and in M. tuberculosis, if one or more cell segments had RNase E-mCherry and HupB-sfGFP fluorescence values respectively equal to or greater than 150 (a.u.) and 120 (a.u.) after subtraction of the minimum cell fluorescence. M. tuberculosis microcolony fluorescence was measured from a region of interest of constant size throughout the whole time-lapse image sequence, and subtracting the background fluorescence. Division and lysis rates (Manina et al., 2019) were analyzed from the total number of cells constituting a microcolony at the time of INH exposure, adding division events and subtracting lysis events over 16 days. Division and lysis events were normalized to the total number of cells at the corresponding time point, resulting in the division and lysis rates at three-hour intervals for the same microcolony. Rates of individual microcolonies were binned over 2-day intervals.
Acknowledgments
We thank Blaise Li (Institut Pasteur Bioinformatics and Biostatistics Hub), for support in writing a python script for sorting image analysis data. We thank Delphine Vergoz for her contribution to constructing the DVS2 strain. This work was funded by grants to G.M. from the French National Research Agency (ANR-17-CE11-0007-01; ANR-10-LABX-62-IBEID). This project also received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 853989. The JU receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA and Global Alliance for TB Drug Development non-profit organization, Bill & Melinda Gates Foundation, University of Dundee. Work in G.M.’s laboratory also received financial support from the Institut Pasteur and a donation from Mr. Lacoste. The content is the sole responsibility of the authors and does not necessarily represent the views of the funders.
Author contributions
Conceptualization, G.M.; Methodology, G.M. and A.G.; Data curation, A.G., T.D., G.M., Validation, A.G., G.M., T.D., Q.G.G., and M.M.; Formal Analysis, A.G., G.M., T.D., and Q.G.G.; Investigation, A.G., and G.M.; Resources, A.G., G.M., T.D., Q.G.G., and M.M.; Writing − Original Draft, G.M.; Writing – Review & Editing, G.M., A.G., T.D., Q.G.G., and M.M.; Visualization, G.M. and A.G.; Supervision, G.M.; Project Administration, G.M.; Funding Acquisition, G.M.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104233.
Supplemental information
Mass spectrometry data and functional analyses of the whole-cell M. smegmatis proteome upon rne induction (oe_rne), silencing (si_rne). and in the control strain (CT). Label-free quantification (LFQ) intensities values are quantified if at least one unique peptide is identified. LFQ intensities are logged (Log2). Reverse (potentially misidentified), potential contaminants, and only identified by site proteins are deleted. Only proteins with at least four LFQ values in a condition are kept. The remaining missing values of the dataset are imputed using the impute.mle function of R package “imp4p” (Giai Gianetto et al., 2020). In each condition, the intensity values are normalized. Exclusive proteins are those present in a condition but absent in another, and are assumed to be differentially abundant between conditions. Differential analysis is performed by LIMMA t-test, and the corresponding p-values are next adjusted using an adaptive Benjamini-Hochberg correction. The proportion of differentially abundant proteins is estimated using the R package “cp4p” (Giai Gianetto et al., 2016). False discovery rate (FDR) is fixed to 1%. Functional analysis reports both exclusive and differentially abundant proteins classified according to their biological functions on Mycobrowser (Kapopoulou et al., 2011). Each functional category is color coded. UniProt and gene identifiers are indicated.
Mass spectrometry data, functional and stoichiometry analyses of Rne-His pull-down, upon rne induction, compared to the CT. LFQ intensities values are quantified if at least one unique peptide is identified. LFQ intensities are logged (Log2). Reverse (potentially misidentified), potential contaminants, and only identified by site proteins are deleted. Only proteins with at least two LFQ values in a condition are kept. The remaining missing values of the dataset are imputed using the impute.mle function of R package “imp4p” (Giai Gianetto et al., 2020). In each condition the intensity values are normalized. Exclusive proteins are those present in a condition but absent in another, and are assumed to be differentially abundant between conditions. Differential analysis is performed by LIMMA t-test, and the corresponding p-values are next adjusted using an adaptive Benjamini-Hochberg correction. The proportion of differentially abundant proteins is estimated using the R package “cp4p” (Giai Gianetto et al., 2016). False discovery rate (FDR) is fixed to 1%. Differentially abundant proteins are separated per condition (Rne > CT and Rne < CT). Functional analysis reports both exclusive and differentially abundant proteins, classified according to their biological functions on Mycobrowser (Kapopoulou et al., 2011). Each functional category is color coded. UniProt and gene identifiers are indicated. Relative stoichiometry and standard deviation for the eluates and the inputs are reported.
The purpose, sequence, and direction of the oligonucleotides are indicated.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE database (Perez-Riverol et al., 2019), with the unique dataset identifier PXD025674. This paper does not report original code. The scripts used in this study are available upon request to the lead contact. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Al-Husini N., Tomares D.T., Bitar O., Childers W.S., Schrader J.M. α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell. 2018;71:1027–1039.e14. doi: 10.1016/j.molcel.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon J., Titgemeyer F., Burkovski A. Common patterns – unique features: nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol. Rev. 2010;34:588–605. doi: 10.1111/j.1574-6976.2010.00216.x. [DOI] [PubMed] [Google Scholar]
- Ando H., Kitao T., Miyoshi-Akiyama T., Kato S., Mori T., Kirikae T. Downregulation of katG expression is associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 2011;79:1615–1628. doi: 10.1111/j.1365-2958.2011.07547.x. [DOI] [PubMed] [Google Scholar]
- Arbel-Goren R., Tal A., Friedlander T., Meshner S., Costantino N., Court D.L., Stavans J. Effects of post-transcriptional regulation on phenotypic noise in Escherichia coli. Nucleic Acids Res. 2013;41:4825–4834. doi: 10.1093/nar/gkt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel-Goren R., Tal A., Parasar B., Dym A., Costantino N., Muñoz-García J., Court D.L., Stavans J. Transcript degradation and noise of small RNA-controlled genes in a switch activated network in Escherichia coli. Nucleic Acids Res. 2016;44:6707–6720. doi: 10.1093/nar/gkw273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel-Goren R., Tal A., Stavans J. Phenotypic noise: effects of post-transcriptional regulatory processes affecting mRNA. Wiley Interdiscip. Rev. RNA. 2014;5:197–207. doi: 10.1002/wrna.1209. [DOI] [PubMed] [Google Scholar]
- Bakkeren E., Diard M., Hardt W.-D. Evolutionary causes and consequences of bacterial antibiotic persistence. Nat. Rev. Microbiol. 2020;18:479–490. doi: 10.1038/s41579-020-0378-z. [DOI] [PubMed] [Google Scholar]
- Bandyra K.J., Luisi B.F., Storz G., Papenfort K. RNase E and the high-fidelity orchestration of RNA metabolism. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.RWR-0008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrimont A., Jaquet V., Wallerich S., Voegeli S., Becskei A. Contribution of RNA degradation to intrinsic and extrinsic noise in gene expression. Cell Rep. 2019;26:3752–3761. doi: 10.1016/j.celrep.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Bayas C.A., Wang J., Lee M.K., Schrader J.M., Shapiro L., Moerner W.E. Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U S A. 2018;115:E3712–E3721. doi: 10.1073/pnas.1721648115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham K.S.H., Staack S., Wilmanns M., Parret A.H.A. The pMy vector series: a versatile cloning platform for the recombinant production of mycobacterial proteins in Mycobacterium smegmatis. Protein Sci. 2020;29:2528–2537. doi: 10.1002/pro.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess Tornaletti L., Manina G. Delving into the functional meaning of phenotypic variation in mycobacterial persistence: who benefits the most from programmed death of individual cells? Microbiol. Insights. 2020;13 doi: 10.1177/1178636120945304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J.E., Cagliero C., Quan S., Squires C.L., Jin D.J. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M., Jacobs-Wagner C. Cellular organization of the transfer of genetic information. Curr. Opin. Microbiol. 2013;16:171–176. doi: 10.1016/j.mib.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarégola S., Jacq A., Laoudj D., McGurk G., Margarson S., Tempête M., Norris V., Holland I.B. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- Castro R.A.D., Borrell S., Gagneux S. The within-host evolution of antimicrobial resistance in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2020:fuaa071. doi: 10.1093/femsre/fuaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarelli L.R., Scoffone V.C., Trespidi G., Barbieri G., Riabova O., Monakhova N., Porta A., Manina G., Riccardi G., Makarov V., et al. Chemical, metabolic, and cellular characterization of a FtsZ inhibitor effective against Burkholderia cenocepacia. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse P.J., Karakousis P.C., Klinkenberg L.G., Kesavan A.K., Ly L.H., Allen S.S., Grosset J.H., Jain S.K., Lamichhane G., Manabe Y.C., et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 2009;77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack R.S., Genereaux J.L., Mackie G.A. RNase E activity is conferred by a single polypeptide: overexpression, purification, and properties of the ams/rne/hmp1 gene product. Proc. Natl. Acad. Sci. U S A. 1993;90:9006–9010. doi: 10.1073/pnas.90.19.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame R.T., Rashid F.-Z.M., Grainger D.C. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 2020;21:227–242. doi: 10.1038/s41576-019-0185-4. [DOI] [PubMed] [Google Scholar]
- Deana A., Celesnik H., Belasco J.G. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Dhar N., McKinney J.D., Manina G. Phenotypic heterogeneity in Mycobacterium tuberculosis. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.TBTB2-0021-2016. [DOI] [PubMed] [Google Scholar]
- Durand S., Tomasini A., Braun F., Condon C., Romby P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol. Rev. 2015;39:316–330. doi: 10.1093/femsre/fuv007. [DOI] [PubMed] [Google Scholar]
- Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- Giai Gianetto Q., Combes F., Ramus C., Bruley C., Couté Y., Burger T. Calibration plot for proteomics: a graphical tool to visually check the assumptions underlying FDR control in quantitative experiments. Proteomics. 2016;16:29–32. doi: 10.1002/pmic.201500189. [DOI] [PubMed] [Google Scholar]
- Giai Gianetto Q.G., Wieczorek S., Couté Y., Burger T. A peptide-level multiple imputation strategy accounting for the different natures of missing values in proteomics data. bioRxiv. 2020 doi: 10.1101/2020.05.29.122770. Preprint at. [DOI] [Google Scholar]
- Gonze D., Coyte K.Z., Lahti L., Faust K. Microbial communities as dynamical systems. Curr. Opin. Microbiol. 2018;44:41–49. doi: 10.1016/j.mib.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Goossens S.N., Sampson S.L., Van Rie A. Mechanisms of drug-induced tolerance in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2020;34 doi: 10.1128/CMR.00141-20. e00141−20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O., Baird G.S., Campbell R.E., Zacharias D.A., Tsien R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Hansen M.M.K., Weinberger L.S. Post-transcriptional noise control. Bioessays. 2019;41:1900044. doi: 10.1002/bies.201900044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett E.C., Rubin E.J. Bacterial growth and cell division: a Mycobacterial perspective. Microbiol. Mol. Biol. Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hołówka J., Trojanowski D., Janczak M., Jakimowicz D., Zakrzewska-Czerwińska J., Henkin T.M. The origin of chromosomal replication is asymmetrically positioned on the mycobacterial nucleoid, and the timing of its firing depends on HupB. J. Bacteriol. 2018;200 doi: 10.1128/JB.00044-18. e00044−18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hołówka J., Zakrzewska-Czerwińska J. Nucleoid associated proteins: the small organizers that help to cope with stress. Front. Microbiol. 2020;1 doi: 10.3389/fmicb.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui M.P., Foley P.L., Belasco J.G. Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 2014;48:537–559. doi: 10.1146/annurev-genet-120213-092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sardesai A.A., Basu D., Muniyappa K., Hasnain S.E. DNA clasping by mycobacterial HU: the C-terminal region of HupB mediates increased specificity of DNA binding. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra P., Mishra S.K., Kaur S., Kumar A., Prasad H.K., Sharma T.K., Tyagi J.S. G-quadruplex-forming DNA aptamers inhibit the DNA-binding function of HupB and Mycobacterium tuberculosis entry into host cells. Mol. Ther. Nucleic Acids. 2018;13:99–109. doi: 10.1016/j.omtn.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapopoulou A., Lew J.M., Cole S.T. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb.) 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Garcia-Alcala M., Balleza E., Cluzel P. Stochastic transcriptional pulses orchestrate flagellar biosynthesis in Escherichia coli. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime L., Vincent H.A., Gendoo D.M.A., Jourdan S.S., Fishwick C.W.G., Callaghan A.J., McDowall K.J. The first small-molecule inhibitors of members of the ribonuclease E family. Sci. Rep. 2015;5 doi: 10.1038/srep08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint C., Verstraeten N., Hofkens J., Fauvart M., Michiels J. Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit. Rev. Microbiol. 2014;40:207–224. doi: 10.3109/1040841X.2013.776510. [DOI] [PubMed] [Google Scholar]
- Kołodziej M., Trojanowski D., Bury K., Hołówka J., Matysik W., Kąkolewska H., Feddersen H., Giacomelli G., Konieczny I., Bramkamp M., et al. Lsr2, a nucleoid-associated protein influencing mycobacterial cell cycle. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-82295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs L., Csanadi A., Megyeri K., Kaberdin V.R., Miczak A. Mycobacterial RNase E-associated proteins. Microbiol. Immunol. 2005;49:1003–1007. doi: 10.1111/j.1348-0421.2005.tb03697.x. [DOI] [PubMed] [Google Scholar]
- Kumar V., Tomar A.K., Sahu V., Dey S., Yadav S. Structural insights of Mycobacterium GTPase-Obg and anti-sigma-F factor Usfx interaction. J. Mol. Recognit. 2017;30 doi: 10.1002/jmr.2636. [DOI] [PubMed] [Google Scholar]
- Laloux G., Jacobs-Wagner C. Spatiotemporal control of PopZ localization through cell cycle–coupled multimerization. J. Cell Biol. 2013;201:827–841. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson M.R., Ma W., Bellecourt M.J., Artsimovitch I., Martin A., Landick R., Schulten K., Berger J.M. Mechanism for the regulated control of bacterial transcription termination by a universal adaptor protein. Mol. Cell. 2018;71:911–922. doi: 10.1016/j.molcel.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhan X., Gao J., Qiu J., Feng Y., Meganathan R., Cohen S.N., Georgiou G. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- Lodato P.B., Thuraisamy T., Richards J., Belasco J.G. Effect of RNase E deficiency on translocon protein synthesis in an RNase E-inducible strain of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon M.M., Ho P.-Y., Papavinasasundaram K., Richardson K., Cokol M., Sassetti C.M., Amir A., Aldridge B.B. A parallel adder coordinates mycobacterial cell-cycle progression and cell-size homeostasis in the context of asymmetric growth and organization. Curr. Biol. 2017;27:3367–3374. doi: 10.1016/j.cub.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G.A. RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 2013;11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- Manina G., Dhar N., McKinney J.D. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Manina G., Griego A., Singh L.K., McKinney J.D., Dhar N. Preexisting variation in DNA damage response predicts the fate of single mycobacteria under stress. EMBO J. 2019;38 doi: 10.15252/embj.2019101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams H.H., Arkin A. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. U S A. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megaridis M.R., Lu Y., Tevonian E.N., Junger K.M., Moy J.M., Bohn-Wippert K., Dar R.D. Fine-tuning of noise in gene expression with nucleosome remodeling. APL Bioeng. 2018;2 doi: 10.1063/1.5021183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S., Hoffmann A. Identifying noise sources governing cell-to-cell variability. Curr. Opin. Syst. Biol. 2018;8:39–45. doi: 10.1016/j.coisb.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J.R., Pandey S., Boettiger A.N., Wang S., Zhuang X. Spatial organization shapes the turnover of a bacterial transcriptome. eLife. 2016;5 doi: 10.7554/eLife.13065.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero Llopis P., Jackson A.F., Sliusarenko O., Surovtsev I., Heinritz J., Emonet T., Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki M., Niki M., Tateishi Y., Ozeki Y., Kirikae T., Lewin A., Inoue Y., Matsumoto M., Dahl J.L., Ogura H., et al. A novel mechanism of growth phase-dependent tolerance to isoniazid in mycobacteria. J. Biol. Chem. 2012;287:27743–27752. doi: 10.1074/jbc.M111.333385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic N., Bergmiller T., Vandervelde A., Albanese T.G., Gelens L., Moll I. Autoregulation of mazEF expression underlies growth heterogeneity in bacterial populations. Nucleic Acids Res. 2018;46:2918–2931. doi: 10.1093/nar/gky079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouaille S., Mondeil S., Finoux A.-L., Moulis C., Girbal L., Cocaign-Bousquet M. The stability of an mRNA is influenced by its concentration: a potential physical mechanism to regulate gene expression. Nucleic Acids Res. 2017;45:11711–11724. doi: 10.1093/nar/gkx781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole R., Williams H.D. Universal stress proteins and Mycobacterium tuberculosis. Res. Microbiol. 2003;154:387–392. doi: 10.1016/S0923-2508(03)00081-0. [DOI] [PubMed] [Google Scholar]
- Pandey S.D., Choudhury M., Sritharan M. Transcriptional regulation of Mycobacterium tuberculosis hupB gene expression. Microbiology. 2014;160:1637–1647. doi: 10.1099/mic.0.079640-0. [DOI] [PubMed] [Google Scholar]
- Patange O., Schwall C., Jones M., Villava C., Griffith D.A., Phillips A., Locke J.C.W. Escherichia coli can survive stress by noisy growth modulation. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-07702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C.E., Stoner J.E., Hatfull G.F. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J. Bacteriol. 1996;178:5533–5536. doi: 10.1128/jb.178.18.5533-5536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.T., Jacobs-Sera D., Pedulla M.L., Hendrix R.W., Hatfull G.F. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–2723. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Płociński P., Macios M., Houghton J., Niemiec E., Płocińska R., Brzostek A., Słomka M., Dziadek J., Young D., Dziembowski A. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 2019;47:5892–5905. doi: 10.1093/nar/gkz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic S., Husson R.N., Hatfull G.F., Jacobs W.R., Jr. Mycobacterium tuberculosis serine/threonine protein kinases. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MGM2-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusa J., Zhu D.X., Stallings C.L. The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 2018;76 doi: 10.1093/femspd/fty054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert L., Ollion J., Robert J., Song X., Matic I., Elez M. Mutation dynamics and fitness effects followed in single cells. Science. 2018;359:1283–1286. doi: 10.1126/science.aan0797. [DOI] [PubMed] [Google Scholar]
- Rock J.M., Hopkins F.F., Chavez A., Diallo M., Chase M.R., Gerrick E.R., Pritchard J.R., Church G.M., Rubin E.J., Sassetti C.M., et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad T.R., Minch K.J., Brabant W., Winkler J.K., Reiss D.J., Baliga N.S., Sherman D.R. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res. 2013;41:509–517. doi: 10.1093/nar/gks1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi I., McKinney J.D. Chromosome organization and replisome dynamics in Mycobacterium smegmatis. mBio. 2015;6 doi: 10.1128/mBio.01999-14. e01999−14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatos A., Babunovic G.H., Chase M.R., Dills A., Leszyk J., Rosebrock T., Bryson B., Fortune S.M. Posttranslational modification of a histone-like protein regulates phenotypic resistance to isoniazid in mycobacteria. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aao1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter L., Dersch P. Phenotypic diversification of microbial pathogens—cooperating and preparing for the future. J. Mol. Biol. 2019;431:4645–4655. doi: 10.1016/j.jmb.2019.06.024. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M.A. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- Scutigliani E.M., Scholl E.R., Grootemaat A.E., Khanal S., Kochan J.A., Krawczyk P.M., Reits E.A., Garzan A., Ngo H.X., Green K.D., et al. Interfering with DNA decondensation as a strategy against mycobacteria. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J.S., Rietsch A., Dove S.L., DiRita V.J. RNase E promotes expression of type III secretion system genes in Pseudomonas aeruginosa. J. Bacteriol. 2019;201 doi: 10.1128/JB.00336-19. e00336−19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shis D.L., Bennett M.R., Igoshin O.A. Dynamics of bacterial gene regulatory networks. Annu. Rev. Biophys. 2018;47:447–467. doi: 10.1146/annurev-biophys-070317-032947. [DOI] [PubMed] [Google Scholar]
- Slayden R.A., Dawson C.C., Cummings J.E. Toxin–antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 2018;76 doi: 10.1093/femspd/fty039. [DOI] [PubMed] [Google Scholar]
- Smeulders M.J., Keer J., Speight R.A., Williams H.D. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits A.H., Jansen P.W., Poser I., Hyman A.A., Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H., Turlan C., Khalid S., Bond P.J., Kebalo J.-M., Peyron P., Poljak L., Bouvier M., Hamoen L., Luisi B.F., et al. Membrane recognition and dynamics of the RNA degradosome. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtsev I.V., Jacobs-Wagner C. Subcellular organization: a critical feature of bacterial cell replication. Cell. 2018;172:1271–1293. doi: 10.1016/j.cell.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain P.S. Efficient attenuation of stochasticity in gene expression through post-transcriptional control. J. Mol. Biol. 2004;344:965–976. doi: 10.1016/j.jmb.2004.09.073. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M., Lee K., Miller C.A., Moore C.J., Shirako Y., Kobayashi M., Cohen S.N. RNase E maintenance of proper FtsZ/FtsA ratio required for nonfilamentous growth of Escherichia coli cells but not for colony-forming ability. J. Bacteriol. 2006;188:5145–5152. doi: 10.1128/JB.00367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y., Choi P.J., Li G.W., Chen H., Babu M., Hearn J., Emili A., Xie X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti V., Forti F., Ghisotti D., Putzer H. Mycobacterium smegmatis RNase J is a 5′-3′ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol. Microbiol. 2011;82:1260–1276. doi: 10.1111/j.1365-2958.2011.07888.x. [DOI] [PubMed] [Google Scholar]
- Tejada-Arranz A., de Crécy-Lagard V., de Reuse H. Bacterial RNA degradosomes: molecular machines under tight control. Trends Biochem. Sci. 2020;45:42–57. doi: 10.1016/j.tibs.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Van Assche E., Van Puyvelde S., Vanderleyden J., Steenackers H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Blanco D.A., Zhou Y., Zamalloa L.G., Antonelli T., Shell S.S. mRNA degradation rates are coupled to metabolic status in Mycobacterium smegmatis. mBio. 2019;10 doi: 10.1128/mBio.00957-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y., Dhar N., Chait R., Schneider K., Signorino-Gelo F., Leibler S., McKinney J.D. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cumming B.M., Mao C., Zhu Y., Lu P., Steyn A.J.C., Chen S., Hu Y. RbpA and σB association regulates polyphosphate levels to modulate mycobacterial isoniazid-tolerance. Mol. Microbiol. 2018;108:627–640. doi: 10.1111/mmi.13952. [DOI] [PubMed] [Google Scholar]
- Warner D.F., Tønjum T., Mizrahi V. DNA metabolism in mycobacterial pathogenesis. Curr. Top. Microbiol. Immunol. 2013;374:27–51. doi: 10.1007/82_2013_328. [DOI] [PubMed] [Google Scholar]
- Weng X., Bohrer C.H., Bettridge K., Lagda A.C., Cagliero C., Jin D.J., Xiao J. Spatial organization of RNA polymerase and its relationship with transcription in Escherichia coli. PNAS. 2019;116:20115–20123. doi: 10.1073/pnas.1903968116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford D.C., Klingelhoets J.J., Bambenek M.H., Dahl J.L. Deletion of the histone-like protein (Hlp) from Mycobacterium smegmatis results in increased sensitivity to UV exposure, freezing and isoniazid. Microbiology. 2011;157:327–335. doi: 10.1099/mic.0.045518-0. [DOI] [PubMed] [Google Scholar]
- Wieczorek S., Combes F., Lazar C., Giai Gianetto Q., Gatto L., Dorffer A., Hesse A.-M., Couté Y., Ferro M., Bruley C., et al. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics. 2017;33:135–136. doi: 10.1093/bioinformatics/btw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Jain C., Schesser K. RNase E regulates the Yersinia type 3 secretion system. J. Bacteriol. 2008;190:3774–3778. doi: 10.1128/JB.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen I., Choudhury M., Sritharan M., Khosla S. Histone methyltransferase SUV 39H1 participates in host defense by methylating mycobacterial histone-like protein HupB. EMBO J. 2018;37:183–200. doi: 10.15252/embj.201796918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller M.E., Csanadi A., Miczak A., Rose T., Bizebard T., Kaberdin V.R. Quaternary structure and biochemical properties of mycobacterial RNase E/G. Biochem. J. 2007;403:207–215. doi: 10.1042/BJ20061530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Xie J.-P. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol. Cell Biochem. 2011;352:1–10. doi: 10.1007/s11010-011-0733-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative video of exponential-phase bacteria seeded into our customized microfluidic device (Manina et al., 2019), and grown under constant flow of fresh 7H9 medium. Images were recorded at 30-minute intervals (15 fps). RNase E-mKate2 fluorescence (magenta) and phase-contrast (blue) channels are merged. Time is indicated in hours. Scale bar, 10 μm.
Representative video of exponential-phase bacteria, grown in fresh medium for 18 hours, and stressed with the putative M5 RNase E inhibitor for 14 hours. Images were recorded at 30-minute intervals (15 fps). RNase E-mKate2 fluorescence (magenta) and phase-contrast (blue) channels are merged. Time in hours and the presence of the inhibitor are indicated. Scale bar, 10 μm.
Representative video of exponential-phase bacilli seeded into our customized microfluidic device (Manina et al., 2019), grown in fresh 7H9 medium for 4 days, exposed to INH (10X-MIC) for 2 days, allowed to recover in fresh 7H9 medium for 10 days, and exposed a second time to INH (10X-MIC) for 7 days. Images were recorded at 3-hour intervals (15 fps). RNase E-mCherry (magenta) and HupB-sfGFP (green) fluorescence and phase-contrast (gray) channels are merged. Time in hours and INH are indicated. Scale bar, 5 μm.
Representative videos of exponential-phase bacilli grown in fresh 7H9 medium for 3 days, pre-induced with ATC for 5 days, exposed to ATC and INH (10X-MIC) for 2 days, and allowed to recover in fresh 7H9 medium for 7 days. Images were recorded at 3-hour intervals (15 fps). HupB-sfGFP fluorescence (green) and phase-contrast (magenta) channels are merged. Strains, time in hours, ATC, and INH are indicated. Scale bars, 5 μm.
Mass spectrometry data and functional analyses of the whole-cell M. smegmatis proteome upon rne induction (oe_rne), silencing (si_rne). and in the control strain (CT). Label-free quantification (LFQ) intensities values are quantified if at least one unique peptide is identified. LFQ intensities are logged (Log2). Reverse (potentially misidentified), potential contaminants, and only identified by site proteins are deleted. Only proteins with at least four LFQ values in a condition are kept. The remaining missing values of the dataset are imputed using the impute.mle function of R package “imp4p” (Giai Gianetto et al., 2020). In each condition, the intensity values are normalized. Exclusive proteins are those present in a condition but absent in another, and are assumed to be differentially abundant between conditions. Differential analysis is performed by LIMMA t-test, and the corresponding p-values are next adjusted using an adaptive Benjamini-Hochberg correction. The proportion of differentially abundant proteins is estimated using the R package “cp4p” (Giai Gianetto et al., 2016). False discovery rate (FDR) is fixed to 1%. Functional analysis reports both exclusive and differentially abundant proteins classified according to their biological functions on Mycobrowser (Kapopoulou et al., 2011). Each functional category is color coded. UniProt and gene identifiers are indicated.
Mass spectrometry data, functional and stoichiometry analyses of Rne-His pull-down, upon rne induction, compared to the CT. LFQ intensities values are quantified if at least one unique peptide is identified. LFQ intensities are logged (Log2). Reverse (potentially misidentified), potential contaminants, and only identified by site proteins are deleted. Only proteins with at least two LFQ values in a condition are kept. The remaining missing values of the dataset are imputed using the impute.mle function of R package “imp4p” (Giai Gianetto et al., 2020). In each condition the intensity values are normalized. Exclusive proteins are those present in a condition but absent in another, and are assumed to be differentially abundant between conditions. Differential analysis is performed by LIMMA t-test, and the corresponding p-values are next adjusted using an adaptive Benjamini-Hochberg correction. The proportion of differentially abundant proteins is estimated using the R package “cp4p” (Giai Gianetto et al., 2016). False discovery rate (FDR) is fixed to 1%. Differentially abundant proteins are separated per condition (Rne > CT and Rne < CT). Functional analysis reports both exclusive and differentially abundant proteins, classified according to their biological functions on Mycobrowser (Kapopoulou et al., 2011). Each functional category is color coded. UniProt and gene identifiers are indicated. Relative stoichiometry and standard deviation for the eluates and the inputs are reported.
The purpose, sequence, and direction of the oligonucleotides are indicated.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE database (Perez-Riverol et al., 2019), with the unique dataset identifier PXD025674. This paper does not report original code. The scripts used in this study are available upon request to the lead contact. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.