Summary
Characterizing the mechanical properties of engineered tissue constructs provides powerful insight into the function of engineered tissues for their desired application. Current methods of mechanical characterization of soft hydrogels used in tissue engineering are often destructive and ignore the effect of 3D bioprinting on the overall mechanical properties of a whole tissue construct. This work reports on using a non-destructive method of viscoelastic analysis to demonstrate the influence of bioprinting strategy on mechanical properties of hydrogel tissue scaffolds. Structure-function relationships are developed for common 3D bioprinting parameters such as printed fiber size, printed scaffold pattern, and bioink formulation. Further studies include mechanical properties analysis during degradation, real-time monitoring of crosslinking, mechanical characterization of multi-material scaffolds, and monitoring the effect of encapsulated cell growth on the mechanical strength of 3D bioprinted scaffolds. We envision this method of characterization opening a new wave of understanding and strategy in tissue engineering.
Subject areas: Tissue Engineering, Materials science, Biomaterials
Graphical abstract

Highlights
-
•
3D Printed hydrogel scaffolds can be gently vibrated to evaluate mechanics
-
•
This method can evaluate complex multi-material scaffold mechanics
-
•
The method can evaluate the temporal evolution of cell-laden bioprinted constructs
-
•
Complementary insight to traditional mechanical analysis (rheometry, etc.) is gained
Tissue Engineering; Materials science; Biomaterials
Introduction
Over recent years, biofabrication strategies have become increasingly capable of generating complex architectures that closely mimic those of natural tissues (Pedde et al., 2017). Advancements in central components of the tissue engineering process, such as design approach, material selection, and biofabrication technology, have enabled better control over important properties of engineered tissues such as the mechanical strength of a scaffold, the way the scaffold interacts with surrounding fluid, and how cells attach and grow into a mature tissue. Three-dimensional (3D) bioprinting is one of the most common biofabrication methods that builds constructs by depositing biocompatible materials, cells, and supporting components layer by layer onto a substrate for applications in regenerative medicine, biotechnology, and pharmaceuticals (Murphy and Atala, 2014; Pedde et al., 2017). This technique is advantageous in the field of tissue engineering for its ability to spatially arrange multiple cell types, biomolecules, and biomaterials to engineer a tissue-like construct with similar arrangement and complexity to the native tissue.
Apart from significant medical and economic feasibility, a major deciding factor for an engineered tissue to move on to animal testing and, ideally, progress forward to clinical trials is the success and thoroughness of the in vitro testing. Typical in vitro assessment of scaffolds for tissue engineering addresses criteria based on biocompatibility, biomimicry, biodegradability, mechanical properties, scaffold architecture, and manufacturing technology (O’Brien, 2011). Conveniently, the adjustable parameters in the 3D bioprinting process have direct influence over these criteria, deeming it a formidable tool in tissue engineering (Khademhosseini and Langer, 2016; Kelly et al., 2018). However, an incomplete understanding of how 3D bioprinting parameters have direct influence over these criteria limits the effective optimization of 3D bioprinted constructs for tissue engineering (Zadpoor, 2017; Kelly et al., 2018). For example, 3D bioprinting induces porosity into the engineered tissue, weakening the overall strength of the scaffold. In addition, the pattern of porosity can impact how mechanical stress is distributed in the construct. Understanding how this porosity and its architecture can be used strategically to optimize the mechanical properties, tissue-fluid interactions, and, ultimately, the overall success of the engineered tissue for its desired application will positively impact the success of engineered tissues for their desired applications.
Strategies in characterizing the physical properties of hydrogel constructs include monitoring the swelling and degradation of the materials, and strategies in monitoring the mechanical properties of hydrogels include measuring the bulk material properties with instruments such as a rheometer or a mechanical tester. The effect of 3D bioprinting on scaffold strength has been extensively studied in areas where materials have enough mechanical strength to undergo traditional mechanical properties analysis. A recent comprehensive review by Kelly et al. points out that studies of 3D bioprinting induced porosity and its’ effect on mechanical strength are conducted using compression testing and are mostly limited to the field of bone tissue engineering (Kelly et al., 2018). The extensivity of this research has demonstrated a power law relationship between porosity and elastic modulus and yield stress as noted in a review by Zadpoor, however, both Zadpoor and Kelly et al. point out a deficit within the literature of so called structure-function relationships (Zadpoor, 2017; Kelly et al., 2018). This deficit is in part due to the young age of the field and to the lack of effective characterization methods for soft hydrogel constructs (Jakus et al., 2016). Moreover, an engineered tissue with good biological and chemical compatibility is not of much use if it cannot withstand the mechanical loads it is exposed to in the intended application.
Traditional methods of mechanical analysis often used in characterization of a bulk hydrogel material, such as rheometry, atomic force microscopy, and particle tracing micro-rheology are effective in measuring the intrinsic material and microstructural mechanical properties of bioinks, but are incompatible with measuring the effect of 3D bioprinted macro-architecture on the scaffold mechanical properties (Kloxin et al., 2010; Hadisi et al., 2020). Furthermore, mechanical testing, such as compression and tensile testing, are destructive methods of testing in which the failure modes of the structure being tested may influence the reproducibility of the measurement and fixation of soft materials to the instrument is challenging. In this case, the sample must deform to produce a measurement. Of interest, architectural analyses of 3D bioprinted scaffolds in the literature are primarily on the MPa scale (Kelly et al., 2018). Soft tissues, which are often engineered with hydrogel scaffolds, are on the order of Pa and kPa stiffness (Discher et al., 2009).
A relatively new characterization technique, viscoelastic testing of bilayered materials (VeTBiM), non-destructively characterizes the viscoelastic properties of hydrogels (Ceccaldi et al., 2017). In this approach, the sample is fabricated or placed in a cylindrical sample cup with rigid walls and a flexible bottom. The measurement is conducted by mechanically inducing a low-amplitude vibration to the sample and monitoring its volume and dynamic displacement with an ultrasonic and laser sensor, respectively (ElastoSens Bio I Rheolution - Rheolution Inc., 2021). A non-destructive and contactless measurement introduces practicality into time-dependent studies of the mechanical properties of hydrogels and lends itself as a platform for monitoring the bulk mechanical properties of 3D bioprinted hydrogel scaffolds. By avoiding failure of the sample being tested, reproducible measurements of soft 3D bioprinted hydrogel scaffolds can be achieved and add complementary data to traditional methods.
VeTBiM for monitoring 3D bioprinted scaffold mechanical properties relies on a few assumptions and sample requirements to generate valuable data. The technique assumes that the sample material is homogeneous and isotropic, which is the case for bulk hydrogel samples. However, when a 3D bioprinted scaffold sample is introduced, there is a need to fill the sample’s pores with an aqueous solution. Considering the hydrogel’s density is very close to 1gml−1, the scaffold and fluid combines to produce a composite sample that is incompressible and has an overall density of ∼1gml−1, thus fulfilling the assumptions of the measurement. When the VeTBiM measurement is obtained, the viscoelasticity data generated is a measurement of the composite sample considered as a macroscopically homogeneous material, giving insight into the mechanical properties of the whole tissue construct which can be bi- or multi-phasic.
VeTBiM requires samples that are rigid enough to be fixed to the walls of the instrument sample container and create an incompressible cylinder, imposing requirements on the bioprinted material itself. The use of nano-silicate bioinks with Laponite, a synthetic nano-silicate with a platelet structure and charged surfaces, allows for improved shear thinning behavior as exemplified in many other studies (Gaharwar et al., 2014; Au et al., 2015; Avery et al., 2016; Chimene et al., 2016; Jin et al., 2017; Peak et al., 2018). Similarly, nano-cellulose based bioinks offer this type of behavior and have been employed in a number of bioinks (Pääkko et al., 2007; Martínez Ávila et al., 2016; Wang et al., 2020).This shear-thinning behavior is induced by weak electrostatic interactions that can be reversibly broken when shear stress is applied, thus reducing the viscosity of the bioink. Shear-thinning is important in both the formation of consistent free-standing scaffold structures that do not require crosslinking during the printing process and for the ability to over extrude the bioink into the grooved sample cup walls for successful attachment of the scaffold to the sample cup.
This work reports on the quantification of the mechanical properties of 3D bioprinted soft tissues using a non-destructive VeTBiM approach. We have performed a holistic study on the effect of pertinent design parameters such as bioink formulation, fiber size and spacing, infill density (porosity), and the printed pattern on the overall mechanical properties of 3D bioprinted soft tissue scaffolds (see Figure 1). These studies were complemented with time-dependent studies on degradation, crosslinking, and swelling. To further exemplify the versatility of the characterization method and the controlled influence of 3D bioprinting strategy on scaffold properties, the effect of multi-material bioprinting on the mechanical properties of the final construct was evaluated. Finally, the developed method was used to assess the effect of encapsulated cell growth on the mechanical properties of bioprinted soft tissue scaffolds. To our best knowledge, we are the first to perform such a holistic study on the mechanical properties of 3D bioprinted soft tissue scaffolds.
Figure 1.
Schematic of 3D bioprinting hydrogel scaffolds inside VeTBiM sample cups for mechanical evaluation
The proposed technique can be used to measure and indirectly measure the properties drawn around the central concept that are important factors to be considered in tissue engineering.
Results
Developing a method for characterizing mechanical properties of 3D bioprinted hydrogel scaffolds
For this method of characterization to work, the 3D bioprinted scaffolds had to fulfill the same assumptions that VeTBiM makes for measuring the mechanical properties of bulk hydrogel samples. By 3D bioprinting directly into the sample cup, the sample can be effectively attached to the sample cup walls and membrane. Furthermore, filling the pores in the hydrogel scaffold with aqueous solution allows for a sample density of ∼1gmL−1. In theory, the sample fulfills the assumptions that the VeTBiM instrument makes for accurate measurement; however, a curious researcher might presume that the heterogeneity in solid and liquid phases of the sample might generate some questionable behavior when a microscale vertical vibration is applied to the sample (Henni and Schmitt, 2019).
To investigate whether the liquid phase vacates the scaffold micropores during minor vibration, a simple test of turning the sample in the sample cup upside down exemplified that the scaffold filled in with aqueous solution acts as a single unit when exposed to the force of gravity (Video S1). This simple experiment shows two things: The sample was effectively attached to the sample cup and the attractive forces between the filling solution and the scaffold were enough to make the biphasic structure act as one. Moreover, filling the scaffold with aqueous solution mimics the in vivo environment in which an implanted tissue scaffold is engulfed in bodily fluid.
Validating VeTBiM with rheometry
VeTBiM has been previously validated with hydrogels such as chitosan and agar (Ceccaldi et al., 2017). However, rheometry was used to validate the method with the bioinks used in this work. As shown in Figure 2C, no significant difference was seen when measuring the storage (G′) and loss (G″) moduli with both methods. This confirms the accuracy of VeTBiM to measure the mechanical properties of hydrogels with a widely accepted method often used for hydrogels.
Figure 2.
Proposed method of sample preparation for measurement of mechanical properties of 3D bioprinted scaffolds and bioink characterization
(A) Microextrusion bioprinting is employed to print a scaffold in the sample cup with a border around the scaffold to attach the sample to the walls of the sample cup.
(B) The scaffold is filled in with aqueous solution and analyzed with VeTBiM. Ultrasonic and laser sensors are used to monitor dynamic displacement during mechanically induced vibration.
(C) Rheometry and VeTBiM are used to show that both methods obtain the same values for all three bioinks.
(D) All three bioinks exhibit fast recovery of mechanical strength after exposure to high strain condition.
(E) All three bioinks exhibit high cross-section circularity (scale bar = 500 μm).
Data are represented as mean ±SD.
Bioink selection & characterization
Microextrusion bioprinting was chosen for this proof-of-concept for its ability to print higher viscosity inks through long needles, a requirement of printing inside the VeTBiM sample cups. Both adherence to the sample cup silicone membrane and attachment to the sample cup wall were critical in obtaining accurate measurements. Degassing the bioink was an effective method of promoting adherence to the sample cup membrane and fulfilling the assumption of an overall density of 1 g/mL. The shear-thinning behavior of the bioinks used in this study was exemplified using a stress recovery test and measuring the cross-section circularity of printed fibers. All three Laponite-based bioinks exhibited high printed fiber shape-fidelity with cross-section circularities above 0.8 (a value of 1.0 is a perfect circle). This high circularity is a result of fast recovery time after being exposed to shear stress, as shown in Figures 2D and 2E.
Both the printhead speed and needle gauge were useful tools in tuning the size of printed fibers. All three bioinks exhibited fiber diameters that were dependent on the speed that the printhead was moving (see Figure S2). A smaller fiber size was associated with a faster printhead speed showing that the fibers either overextrude at slower speeds or become stretched at higher print speed. The choice of needle gauge exhibited further control over fiber diameter with a wider range of printed fiber sizes and less variability in fiber diameter between samples. The fiber diameter ranges of all three bioinks were in the range of ∼400 μm in diameter up to ∼1 mm. This range in fiber diameter is sufficient for selected studies in varying the printed fiber size.
Considerations in bioink cell adherence and biodegradability
Three cell types were chosen to compare the cell adhering capability of the three bioinks selected for the study. HNDF, a primary human cell line, HaCaT, an immortalized human keratinocyte cell line, and U87, an aggressive glioblastoma cancer cell line, were chosen as a representative group for this proof-of-concept study. Cell adherence was assessed using a PrestoBlue assay after 2 days of growth on the bioinks cast in bulk sheets. All three cell types exemplified preference to the Gel/Lap bioink when compared to the Alg/Lap and PEGDA/Lap bioinks (Figure 3A). This is likely a result of gelatin containing Arginine-Glycine-Aspartate (RGD) cell binding motifs (Panwar and Tan, 2016).
Figure 3.
Bioink cell adherence and biodegradability of Gel/Lap scaffold
(A) PrestoBlue Assay of cells seeded on all three bioinks showed Gel/Lap to have the highest cell adherence.
(B) Schematic of enzymatic degradation of Gel/Lap using collagenase.
(C) Images of the bulk and 3D bioprinted scaffolds in VeTBiM sample cups.
(D) Degradation (weight remaining) of Gel/Lap bulk bioink and Gel/Lap scaffolds.
(E) G′ of degrading Gel/Lap bulk bioink and Gel/Lap scaffolds.
Data are represented as mean ±SD.
An important part of the tissue engineering process is selecting a bioink based on biocompatibility or the ability to act as an appropriate substrate for cell growth, however, this selection still needs to consider the bioink’s ability to act as a scaffold throughout the entire period of tissue maturation (Akalp et al., 2016; Zhou et al., 2018). Although Gel/Lap was exemplified as the best cell adhering substrate bioink from the three cell candidates, gelatin can be easily degraded by collagenases and gelatinases, naturally secreted enzymes by skin fibroblasts, keratinocytes, and glioblastoma (Tandara and Mustoe, 2011; Zhao et al., 2016; Roomi et al., 2017). Furthermore, the act of 3D bioprinting Gel/Lap increases the surface area of the scaffold, resulting in increased exposure and diffusion of both crosslinking and degrading enzymes into the engineered tissue. Both the bulk material and 3D bioprinted scaffolds were crosslinked for 30 min using transglutaminase and subsequently degraded at a fast rate using 200 μg/mL collagenase. As shown in Figure 3D, the weight remaining degradation of the 3D bioprinted scaffold was slightly faster than the bulk material. However, when measuring the mechanical properties throughout degradation over the same time period, the bulk material showed a fast decrease in the mechanical properties over the first 2 h in comparison to the 3D bioprinted scaffold. This proof-of-concept study exemplifies that there are multiple steps in the biofabrication process in which 3D bioprinting can introduce unexpected results. In this case, increased surface area in the 3D bioprinted scaffold provided a more porous structure to speed up the crosslinking process which may have later decreased the rate of degradation when exposed to collagenase. Comparing the bulk sample and the porous, 3D printed sample both in sample cups, the bulk samples had to crosslink by allowing the enzyme to diffuse from the top surface of the 5 mm height cylinder, whereas the porous sample had, in effect, an immediate ubiquitous distribution of enzyme that was able to crosslink throughout the sample and had a maximum distance of less than 0.5 mm to diffuse through hydrogel. Furthermore, the enzyme (transglutaminase) is a large molecule (38 kDa), which enhances the effect of crosslinking at the surface of the scaffolds and the porous samples have a much higher surface area, resulting in a higher amount of crosslinking in a fixed crosslinking period. Finally, the degrading enzyme (collagenase type II) is 68–130 kDa, further enhancing the effect of diffusion throughout the samples on the degradation rate. This experiment functions as a proof of concept in showing the developed characterization method’s capability to measure the effect of degradation on the mechanical properties for both bulk and 3D bioprinted scaffolds. With more experimental control, one could use this capability in monitoring mechanical properties degradation to optimize the scaffold structure with crosslinking and degradation time.
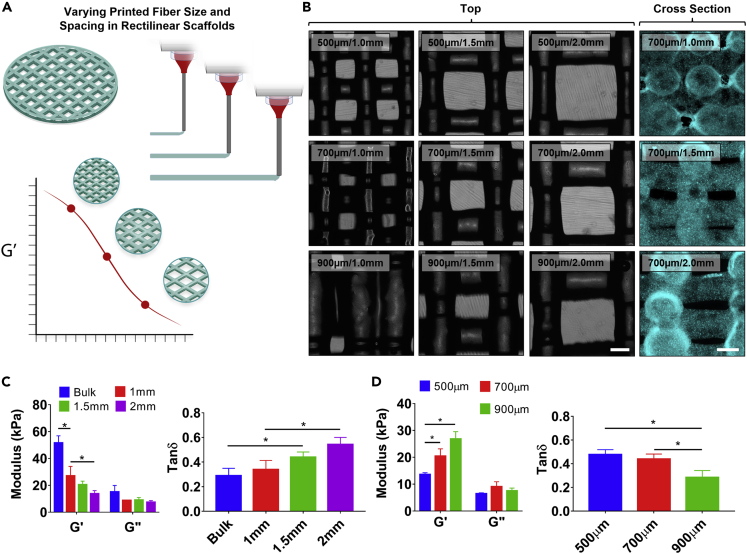
Optimizing rectilinear scaffold architecture
Alg/Lap rectilinear scaffolds of varying fiber size and fiber spacing were 3D bioprinted in ElastoSens sample cups (Figures 4A and 4B. These parameters were chosen to be expanded upon because rectilinear scaffolds are a common scaffold design due to their simple structure which can easily be altered to adjust porosity (Li et al., 2005). Three scaffold conditions of increasing fiber diameter with spacing held constant were printed and analyzed with VeTBiM. As shown in Figure 4D, increasing fiber size resulted in an increasing G′which shows increasing mechanical strength of the scaffold with increasing fiber diameter. The G″ did not display any measurable trend, likely because of there not being a large change in the overall liquid content of the scaffold with change in fiber size. The decreasing trend in the loss tangent (tan(δ)) with increasing fiber diameter further supports the idea of increasing scaffold strength with increasing fiber size by showing predominantly elastic behavior.
Figure 4.
Rectilinear Alg/Lap scaffold optimization with fiber size and spacing
(A) Schematic displaying the experimental design – changing the fiber size and spacing influences scaffold strength.
(B) Alg/Lap scaffolds with varying spacing and fiber size imaged from above (bright field) and from the cross section (turquoise) (scale bar = 500 μm).
(C) Increased fiber spacing reduces the overall strength and increases the damping capability.
(D) Increased fiber diameter increases the overall strength and reduces the damping capability of rectilinear scaffolds. ∗p < 0.05.Data are represented as mean ±SD.
Increasing the fiber spacing while maintaining constant fiber size yielded similar results (Figure 4C). With increasing fiber spacing, a decrease in G′ and no measurable trend in G″ were observed, resulting in an increasing trend for the loss tangent. Both experiments agree with the idea that increasing the porosity in a 3D bioprinted scaffold, either by decreased fiber sized or increased fiber spacing, results in a weakening of the mechanical strength of the structure and increased damping capability. This agrees with other results in the literature that have been generated for stiffer materials used in bone tissue engineering (Li et al., 2005; Kelly et al., 2018). It should be noted that the bulk material was also measured for this experiment and showed significantly increased mechanical strength in comparison to all 3D bioprinted scaffolds.
To exemplify how this method can be used to optimize a rectilinear scaffold, the porosity of a tissue scaffold could be optimized with using a desired spacing between fibers while adjusting the strength of the scaffold with fiber size.
Tuning diffusion of molecules into tissue scaffolds exemplified with chemical crosslinking
Altering the printed fiber size in scaffolds can influence diffusion of ions and biomolecules into the hydrogel. This effect was indirectly measured by analyzing the effect of fiber diameter on the ionic crosslinking time of Alg/Lap scaffolds. As shown in Figure 5B, reduced crosslinking time for 3D bioprinted scaffolds was shown by a plateau in the normalized G′ around 45 min for 500 μm sized fibers and around 1 h for the 900 μm fiber size. The bulk sample did not exhibit a plateau within the 2-h time interval. These results were corroborated by imaging the diffusion of rhodamine, a fluorescent molecule, into the scaffolds and bulk material over the same time period. Similar plateaus were seen in the plotted cross-sectional fluorescence intensity for the 3D bioprinted samples. An increasing trend was still being observed at the 2-h time point for the bulk sample showing that rhodamine had still not saturated the bulk material. The diffusion of rhodamine into the scaffolds and bulk bioink was linearly correlated with the measured G’. These results suggest that the crosslinking reaction between calcium ions and alginate occurs very quickly after diffusing into the bioink. Both the 3D bioprinted scaffolds and the bulk control samples were treated with deionized water. They exhibited no increase in storage modulus because of the lack of crosslinker in solution. Interestingly, the bulk sample storage modulus decreased over the 2-h period, most likely an effect of material swelling. Swelling did not affect the storage moduli of the 3D bioprinted scaffolds due to their inherent macro-porosity.
Figure 5.
Real-time crosslinking of Alg/Lap rectilinear scaffolds
(A) Changing fiber size influences ion diffusion and the crosslinking time.
(B) Real-time crosslinking measurement of bulk Alg/Lap and 3D bioprinted scaffolds of with 900 and 500 μm fiber size.
(C) Fluorescence images of rhodamine diffusing into samples over 2 h (scale bar = 1 mm).
(D) Fluorescence values measured in the center of the scaffold fibers and the bulk bioink over the 2 h crosslinking period.
(E) The diffusion of rhodamine into the scaffolds and bulk material is linearly correlated with the storage modulus of the scaffold while crosslinking with CaCl2 solution.
Data are represented as mean ±SD.
This type of real-time crosslinking analysis is possible only for the 3D bioprinted scaffolds because of the macro-porosity of the structures allowing for crosslinking solution to be applied during measurement. For the bulk material, the crosslinking solution must be removed from the top of the sample to conduct a measurement at each time point. Furthermore, a real time study such as this would not be possible with traditional methods of mechanical properties characterization because of their destructive nature and the fact that the sample is confined (Oyen, 2013; Roeder, 2013). A similar study of the crosslinking time may be possible with micro-rheology, in which micro-particle flow within a hydrogel is observed to determine rheological properties, however, this technique is only suitable for bulk materials and not 3D bioprinted scaffolds (Xia et al., 2018). Hence, faster crosslinking times for 3D bioprinted structures would not be determined as was in this study. Another interesting result to notice is that the bulk material final G′ value (51.6 ± 5.3 kPa) was ∼4.5 times larger than its initial value, whereas both 3D bioprinted scaffolds were ∼2.5 times larger than their initial values. This effect is likely due to the difference in the overall amount of “crosslinkable” bioink in the samples and the homogeneity of the entire sample.
Altering printed pattern for scaffold strength
For scaffolds which have non-rectilinear patterns, the infill density (a complementary term to porosity) is an adjustable scaffold design parameter that is often altered. PEGDA/Lap scaffolds were 3D bioprinted in a honeycomb pattern. Honeycomb scaffolds have been shown to be beneficial for many tissue types including heart, liver, and bone (George et al., 2006; Tanaka et al., 2006; Engelmayr et al., 2008). The infill density of the honeycomb pattern was 3D bioprinted at 20, 40, and 60% (Figure 6B). Similar to altering the porosity with fiber size and spacing, an increase in infill density, or decrease in porosity, resulted in an increase in the G′ of the honeycomb scaffolds (Figure 6D). No measurable trend was observed in the G” resulting in a decreasing trend of the loss tangent with increasing infill density. This experiment continues to support the notion that decreasing the porosity will result in an increase in mechanical strength and a decrease in damping capability. Similar results have been obtained in engineering intervertebral disks with a honeycomb scaffold, albeit a much stiffer material on the order of MPa (Hu et al., 2018).
Figure 6.
Altering printing pattern, infill density, and bioink formulation all impact the scaffold mechanical properties in PEGDA/Lap scaffolds
(A) Three different patterns were printed with 5% PEGDA/Lap and UV crosslinked.
(B) Honeycomb, rectilinear, and random line patterns printed with PEGDA/Lap at 20% infill density.
(C) The honeycomb pattern exhibited a higher G′ than the rectilinear and random line patterns.
(D) Increasing the infill density increases the mechanical strength of honeycomb scaffolds. (E) Increasing the bioink concentration increases the mechanical strength of honeycomb scaffolds. ∗p < 0.05.Data are represented as mean ±SD.
3D bioprinting enhances scaffold-fluid interactions with surrounding media
Imposing porosity into a tissue scaffold by 3D bioprinting can improve nutrient, waste, and other biomolecule uptake into the scaffold. 3D bioprinted PEGDA/Lap scaffolds were freeze-dried and employed in a swelling study to demonstrate this effect. This study did not require the use of the developed method of characterization; however, it supports the concepts that are being discussed about the usefulness of 3D bioprinting in tissue engineering. As shown in Figure S3B, swelling occurred at a slightly faster rate for 3D bioprinted samples. However, there was no difference between the 3D bioprinted samples because they were printed with the same fiber sizes. After 6 h all samples showed no significant difference in swelling because they were all prepared from the same bioink (Figure S3C). These results demonstrate improved interaction with the surrounding fluid environment in 3D bioprinted samples. This would allow for improved diffusion of nutrients, waste, and other biomolecules throughout the scaffold.
Optimizing bioink formulation for a scaffold design
An important step in the 3D bioprinting process is the determination of an optimal bioink formulation (Murphy and Atala, 2014). Often, the formulation is optimized to fine-tune the bioink to a desirable stiffness to support tissue growth (Kolesky et al., 2016; Zhao et al., 2016). However, the bioink formulation’s effect on scaffold mechanical strength is often ignored. 2.5, 5, and 10% PEGDA with 6% Laponite bioinks were prepared and 3D bioprinted into honeycomb scaffolds with 20% infill density. As shown in Figure 6E, increasing the concentration of the bioink resulted in an increase in G′ or overall mechanical strength of the 3D bioprinted scaffolds. No measurable trend was observed for both the G″ and loss tangent. This observation is different than the previous studies where decreasing the macro-porosity has resulted in a decrease in the loss tangent. This effect is likely due to the scaffold structure remaining the same with only changes in the bioink concentration.
Using printing pattern to optimize scaffold mechanical properties
In scaffold design, the printing pattern can play an important role for some tissue. Honeycomb scaffolds are often used for heart and liver tissues, and have been shown to exhibit higher strength with even distribution of stress throughout the scaffold (Tanaka et al., 2006; Engelmayr et al., 2008; Lu et al., 2015; Soufivand et al., 2020). A study of the effect of pattern on 3D bioprinted scaffolds using PEGDA/Lap was conducted with a rectilinear, honeycomb, and random line pattern (Figures 6A and 6B). Honeycomb and rectilinear patterns were chosen for reasons previously mentioned, and the random line pattern was chosen to determine if large variance in interconnected pore size would have a detrimental effect on the mechanical strength. All patterns were printed with the same bioink formulation (5% PEGDA/Lap) and infill density (20%) so that the only changing parameter was the pattern. As shown in Figure 6C, the rectilinear and line patterns exhibited similar G′ and G″ values, whereas the honeycomb pattern was significantly higher for both G′ and G”. No significant difference was observed in the loss tangents of each pattern. This was likely because of all three of the patterns having the same infill density. The honeycomb scaffold loss tangent was slightly higher than the rectilinear and random line scaffolds, suggesting that the honeycomb scaffold is better able to dissipate mechanical energy. A property which would be beneficial for withstanding repeated deformation in a beating heart. These results agree with another study which compared honeycomb and rectilinear patterned scaffolds for intervertebral disk regeneration using stiffer scaffolds in the order of MPa stiffness (Hu et al., 2018). Considering that the same amount of material and total scaffold volume was achieved, altering the pattern to achieve higher scaffold strength is a valuable tool.
Multi-material scaffold mechanical assessment
A more recent strategy in scaffold design is to 3D bioprint multi-material scaffolds with the intention of using the different materials or structures for varying functions or supporting multiple cell types with the scaffold (Hong et al., 2015; Rutz et al., 2015; Kolesky et al., 2016; Skylar-Scott et al., 2019; Jahanshahi et al., 2020). In some studies, 3D printed sacrificial structures provide channels for seeding vascular tissue with surrounding densely encapsulated cell tissue (Kolesky et al., 2016; Skylar-Scott et al., 2019). In another example, the ability to spatially organize cells is possible by arranging materials with different properties inside of a scaffold. Rutz et al. guided cell growth in their engineered tissues to be along particular struts of a rectilinear scaffold by using cell-adherent gelatin and non-adherent PEG as the perpendicular struts of a single tissue scaffold (Rutz et al., 2015). This strategy may be beneficial for cell and tissue growth, however, the effects on the mechanical strength of a scaffold are somewhat unpredictable. To mimic the style of multi-material scaffold produced by Rutz et al., PEGDA/Lap and Alg/Lap were employed in 3D bioprinting a multi-material rectilinear scaffold (1:1 ratio of PEGDA/Lap:Alg/Lap) with a two-step crosslinking method (See Figure 7). Both the individual bioink scaffolds with the same structural parameters were also measured. Alg/Lap exhibited a much higher G′ of 13 kPa compared to PEGDA/Lap at 2 kPa. The multi-material scaffold storage modulus was significantly different than both individual bioink samples; however, at 4 kPa it was much closer to the PEGDA/Lap bioink scaffold G’. There are many factors which could have caused this result, for example, a lack of crosslinking between PEGDA/Lap and Alg/Lap layers, damping from the softer material, and reduced clarity in the scaffold impeding UV crosslinking for lower layers of PEGDA/Lap. A systematic study should be conducted to determine an accurate argument for this effect. However, the results are interesting, and the method could prove very useful for engineering complex tissues with spatial arrangement of multiple materials and cell types.
Figure 7.
Evaluating the mechanical properties of multi-component scaffolds with Alg/Lap and PEGDA/Lap bioinks
(A) A dual printhead system was used to bioprint multi-component scaffolds which were then (B) crosslinked sequentially with UV and Ca2+ ions.
(C) The multi-component scaffold G′ and G″ were in between the individual material scaffold values, but closer to the softer material. PEGDA/Lap.
(D) Fluorescently labeled multi-component scaffold imaged with a fluorescent microscope (scale bar = 1 mm).
∗p < 0.05; ∗∗p < 0.01.Data are represented as mean ±SD.
Temporal effect of cell proliferation on mechanical properties of bioprinted rectilinear scaffolds
Optimizing the mechanical performance of engineered tissues for the desired end application should consider the temporal effect of tissue growth on the mechanical strength of a scaffold. With the goal of providing enough support for cells to attach, proliferate, and appropriately functionalize their morphology for the intended tissue structure, a scaffold should be able to degrade at an appropriate rate and allow for cells to multiply and secrete ECM. This dynamic environment in which the cells are altering the microstructure of the scaffold and growing in overall population impacts the overall mechanical strength of the engineered tissue. To demonstrate this effect, acellular and U87 cell-laden rectilinear scaffolds were 3D bioprinted and assessed over 7 days. Commercially available Cellink Bioink was used for this study for its demonstrated printability, biocompatibility, and ability to host encapsulated cells through the bioprinting process (Markstedt et al., 2015; Martínez Ávila et al., 2016; Bojin et al., 2021). At day 0, the acellular and cellular scaffolds had identical mechanical properties, demonstrating that diluting the acellular scaffold ink with the same volume of media as was used to load the cell-laden ink with cells prepared an effective control. A PrestoBlue assay was used to show cell proliferation in the scaffold material over the 7-day period, demonstrating an increase in cell metabolic activity between day 1–4 and maintenance of that metabolic activity through to day 7 (Figure 8B). Acellular and cell-laden scaffold mechanical properties were assessed using VeTBiM and showed that the cell-laden construct maintained a more-robust mechanical strength over the 7-day period when compared to the acellular scaffold. Both the acellular and cell-laden scaffolds G′ decreased from day 0 to day 1, likely an effect of swelling. Throughout day 3–7, the acellular scaffold G′ continued to decrease, suggesting the degradation or dissolution of the scaffold material in the surrounding media. However, the cell-laden scaffold G′ plateaued and became significantly higher than the acellular structure on day 5 and 7. This difference in mechanical strength can be attributed to the proliferation of cells in the 3D bioprinted structure. As shown in Figures 8D and 8E, the cell morphology of encapsulated cells was circular on day 0, demonstrating that the newly encapsulated cells were not attached to the host material. On day 7, live-dead and actin-DAPI stains were used to show the viability and morphology of the encapsulated cells. The cells are highly viable and cellular body extensions are clearly visible, showing that the cells are adhering to the host material. Considering the increased cell proliferation and cell morphology, the difference in mechanical strength is supported by the notion that cell proliferation and growth is enough to impact the mechanical strength of a 3D bioprinted construct and that it can be measured using the developed methodology.
Figure 8.
The effect of cell proliferation and growth on the mechanical properties of 3D bioprinted scaffolds
(A) Acellular and cell-laden scaffolds were 3D printed in rectilinear scaffolds and monitored over 7 days.
(B) Cell proliferation over the 7 days was correlated with (C) maintaining mechanical robustness in the cell-laden scaffold when compared with the acellular scaffold. The G′ of the cell-laden scaffold was significantly higher than the acellular scaffold on days 5 and 7.
(D) Day 0 cell morphology is circular in shape and some cells died in the 3D bioprinting process as shown by the live/dead stain ((1) scale bar = 500 μm and (2) scale bar = 100 μm)).
(E) Day 7 live/dead staining ((1) & (2)) and actin/dapi staining ((3) & (4)) show cell morphology with prolonged extensions demonstrating attachment to the scaffold ((1), (3) scale bar = 500 μm and (2), (4) scale bar = 100 μm).
∗p < 0.05.Data are represented as mean ±SD.
In the case of implantation, 3D bioprinted soft tissues must withstand external forces and loads of the implant site whereas the tissue grows and incorporates itself into the surrounding native tissue. A mismatch of the degradation rate of the scaffold material, growth of encapsulated cells, and invasion of native tissue can expose the engineered tissue to damage if the structure becomes too weak to endure the loads imposed by the surrounding environment. Being able to measure a 3D bioprinted soft tissue structure’s mechanical properties over a relevant timeframe gives strong insight into how the dynamics of tissue growth and scaffold degradation will play out when implanting in an animal or, eventually, a human. Finally, the optimizable parameters of scaffold porosity, printing pattern, and multi-material organization can be used to fine-tune the degradation dynamics irrespective of the material chosen to host the cells.
Discussion
In this work, we report on a non-destructive approach for assessing the mechanical properties of 3D bioprinted soft tissues. The developed method of characterization is capable of measuring the effect of 3D bioprinted architectures on the mechanical properties of scaffolds for tissue engineering applications. As a proof-of-concept, several experiments were performed to demonstrate how the method of characterization would benefit the development of 3D bioprinted hydrogel scaffolds for engineered tissues. We demonstrated that the non-destructive approach used herein can be used to measure the viscoelastic properties of bioinks with accuracies that are comparable to the current gold standard approach (rheometry). In addition, the unique features of the used approach that allows measuring the mechanical properties of scaffolds over a few hours enable dynamic measurement of the mechanical properties of 3D bioprinted soft scaffolds in the presence of tissue remodeling enzymes and cells. Such a feature has significant implications in the field of tissue engineering and drug delivery, where the knowledge about the kinetics of material interaction with the host tissues has utmost importance. Moreover, because the same sample is always used at each time point, the errors associated with batch-to-batch material variations and inconsistencies in sample preparation were eliminated. The ability to print directly into sample holders enabled us to the measure the mechanical properties of constructs with various structural features. Interestingly, the effect of printing pattern was shown to have a significant influence on the mechanical strength. This could prove very useful, for example, when using a set infill density while wanting to maximize mechanical strength. Finally, the method proves useful in measuring multi-material scaffolds.
The developed methodology best operates as a complementary means of analysis to more traditional mechanical analysis that expands the understanding of the structural mechanics of 3D bioprinted tissues, addressing an unmet area of need in validating engineered soft tissues. In particular, rheometry and other traditional methods can be used to assess the intrinsic material properties of bioinks used in tissue engineering, whereas the developed methodology extends the understanding of mechanics to 3D printed multi-phasic composites encased in aqueous solution, much like the in situ environment of the human body. That being said, there are some practical advantages of VeTBiM over traditional methods that are driven by the fact that vibrating the sample does not deform or destroy the sample and a single sample can be tested multiple times. This enabled the demonstrated temporal studies around crosslinking, degradation, and cell proliferation. In addition, the summative properties of a multi-phasic composite can be assessed as a whole structure to gauge the impact of fabrication strategies on the structural mechanics of complex tissues.
Although microextrusion bioprinting was selected as the bioprinting method of choice for this proof of concept, this method of characterization has potential for compatibility with other methods of 3D bioprinting. In this scenario, 3D bioprinted scaffolds may have to be printed outside of the sample cup and attached to the walls of the sample cup post-print. Furthermore, rising popularity in techniques that allow for more biocompatible or complex structures, such as bioprinting in suspension baths, microfluidic printing techniques, and multi-component fibers have potential for compatibility with this method (Ashammakhi et al., 2019; McCormack et al., 2020).
Cell-biomaterial interactions and their effects on mechanobiology of engineered tissue is also a rising area of research (Discher et al., 2009; Chen et al., 2014; Vining and Mooney, 2017). This method has potential for more complex time dependent studies of cell-encapsulated hydrogels and their effects on the mechanical evolution of engineered tissue. For example, demonstrating tissue remodeling and correlating it to mechanical changes in the sample can provide insight into how tissues will evolve in an in vivo environment. In addition, 4D bioprinting is an emerging field in which bioprinted constructs are designed to change their properties over time in response to specific stimuli (Gao et al., 2016; Yang et al., 2020). This method can be used to assess the impact of scaffold architecture on mechanical responses to stimuli and, vice versa, the impact of varying stimuli on the mechanical properties of scaffolds.
Future studies employing this method will elucidate trends and changes affected by tissue engineering strategies on the mechanics of complex tissue structures with an empirical method. This approach has potential to impact the forefront of the tissue engineering field in which inhomogeneous structures that effectively mimic complex tissue are being constructed to evolve over time.
Limitations of the study
A limitation worth mentioning with the developed method is that the vibration of samples in VeTBiM is in the vertical direction. Thus, if the scaffold architecture is anisotropic, then the measurement of mechanical properties of 3D bioprinted scaffolds is anisotropic. However, this is not true for bulk materials in which no directional architecture is imposed on the sample. In practice, the 3D bioprinted scaffold pattern could be prepared and rotated 90° about a horizontal axis (before fixing in the sample cup) to have a complete mechanical characterization of the scaffold. Despite this limitation, the characterization method was exemplified to generate valuable data when altering the chemical, physical, and architectural parameters of the 3D bioprinting process. Another limitation includes the use of VeTBiM for characterizing 3D bioprinted scaffolds made of softer, more liquid-like bioinks (on the order of Pascals). With these materials, achieving fixation to the walls of the sample cup may remain a significant challenge; however, the use of these materials in engineering large, structured tissues remains challenging because of weak mechanical properties and reduced print fidelity (Panwar and Tan, 2016). In addition, obtaining reliable results for soft bioinks entails using large quantities of the material (up to 7 mL). This can affect the cell viabilities in bioinks when the mechanical properties of bulk materials are studied.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Laponite | BYK Additives | Product Identifier: XLG |
| Moo Glue TI microbial transglutaminase | The Modernist Pantry | SKU: 1203-50 |
| Poly-ethylene glycol diacrylate | Sigma Aldrich | Cat#437441; CAS: 26570-48-9 |
| 2-Hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (photo-initiator | Sigma Aldrich | Cat#410896; CAS: 1067-53-59 |
| Gelatin from porcine skin (type A, 300 bloom gel strength) | Sigma Aldrich | Cat#G1890; CAS: 9000-70-8 |
| Alginic acid sodium salt | Sigma Aldrich | Cat#180947; CAS: 9005-38-3 |
| Collagenase, Type II | Worthington Biochemical Corporation | Product Code CLS-2; Cat# LS004174 |
| Cellink Bioink | Cellink | SKU: IK1020000303 |
| Critical commercial assays | ||
| Live/dead viability kit | Thermo Fisher (Invitrogen) | Cat# L3224 |
| Alexa Fluor™ 488 Phalloidin | Thermo Fisher (Invitrogen) | Cat# A12379 |
| PrestoBlue Cell Viability Reagent | Thermo Fisher (Invitrogen) | Cat# A13262 |
| Deposited data | ||
| 3D Printing Codes Repository | This paper | Zenodo: https://doi.org/10.5281/zenodo.5759866 |
| Experimental models: Cell lines | ||
| Human glioblastoma cells (U87 MG) | ATCC | ATTC: HTB-14 |
| Human Neonatal Dermal Fibroblasts | ATCC | ATCC: PCS-201-010 |
| HaCaT Cells | Addex Bio | Cat#: T0020001 |
| Software and algorithms | ||
| Heartware (V. 2.4.1) | Cellink | https://cellink.freshdesk.com/support/solutions/articles/35000027693-how-to-install-and-set-up-heartware-bioprinting-assisting-software-on-a-windows-pc |
| Slic3r (V. 1.2.9) | Open source | https://slic3r.org/ |
| ImageJ (V. 1.6.4) | U.S. National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
| Solidworks 2017 | Dassault Systems | https://www.solidworks.com/domain/design-engineering |
| Cura (V. 3.4.1) | Ultimaker | https://ultimaker.com/software/ultimaker-cura |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mohsen Akbari (makbari@uvic.ca).
Materials availability
This study did not generate new unique reagents.
Method details
Materials
Alginic acid sodium salt (Alginate; Catalogue #180947), poly-ethylene glycol diacrylate (PEGDA; Catalogue # 437441), gelatin from porcine skin (type A, 300 bloom gel strength; Catalogue #G1890), calcium chloride, 2-Hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone (photo-initiator; Catalogue # 410896), Triton-X100 (Catalogue #X100), bovine serum albumin (BSA; Catalogue #A3803), and 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI; Catalogue #D9542) were all acquired from Sigma Aldrich, St. Louis, Missouri. Laponite XLG, a synthetic nanosilicate, was acquired from BYK additives, Germany. Moo Gloo TI microbial transglutaminase (TG; Item ID 1203-50) was acquired from The Modernist Pantry, Eliot, Maine. Collagenase, Type II (Code CLS-2) was acquired from Worthington Biochemical Corporation, Lakewood, New Jersey. Fetal bovine serum (FBS; Catalogue #16000069), Dulbecco’s phosphate buffered saline (DPBS; Catalogue #14040133), penicillin/streptomycin (Catalogue #15070063), trypsin/EDTA (Catalogue #15400054), neutral buffered formalin (NBF; Catalogue #22-220682) PrestoBlue Cell Viability Reagent (Catalogue # A13262), and Dulbecco’s modified Eagle’s medium (DMEM; Catalogue #11965092) were obtained from ThermoFisher Scientific (Waltham, Massachusetts, USA). Alexa Fluor™ 488 Phalloidin (Catalogue #A12379) was acquired from Invitrogen (Waltham, Massachusetts, United States). For the purposes of this paper, all solutions prepared are expressed in concentration as w/v% unless otherwise stated.
Preparation of nano-silicate bioinks
0.5% alginate, 6% Laponite (Alg/Lap) was prepared by, first, dissolving alginate in deionized water and cooling the solution to 4°C. Laponite was added to the solution and immediately vortexed for 2-4 minutes to ensure homogenous distribution of laponite in solution. Laponite takes time for water molecules to adsorb to the surface of nanoparticles, delaying the formation of a more viscous gel. While the laponite was not fully hydrated and the mixture exhibited a low viscosity, the solution was degassed in a vacuum chamber at ∼ −90 kPa.
2.5%, 5%, and 10% PEGDA, 6% Laponite, and 1% photo-initiator (PEGDA/Lap) was prepared by, first, dissolving photo-initiator in deionized water, adding the necessary volume of PEGDA, and cooling the solution to 4°C. Laponite was added to the solution and immediately vortexed for 2–4 minutes to ensure homogenous distribution of laponite in solution. While the laponite was not fully hydrated, the solution was degassed in a vacuum chamber at ∼ −90 kPa. Where the concentration of PEGDA is not explicitly labeled, the 5% PEGDA/Lap formulation is used.
5.6% gelatin, 3.4% Laponite (Gel/Lap) was prepared by, first, preparing a solution of 18% gelatin in de-ionized water at 50°C. A solution of 9% Laponite was prepared by adding Laponite to 4°C deionized water and immediately vortexing until a stable, homogenous gel formed. The 9% Laponite solution was heated to 50°C. The gelatin solution was diluted by half with 50°C deionized water without mixing, and enough 9% Laponite was added to dilute to the final concentrations of 5.6% gelatin, 3.4% Laponite. The mixture was immediately vortexed until a homogenous, viscous gel formed. The gel was reheated to 50°C and degassed in a vacuum chamber at ∼ −90 kPa.
All bioinks were stored in the refrigerator and allowed to hydrate overnight before 3D bioprinting. PEGDA/Lap was protected from light to prevent unwanted crosslinking. 2% calcium chloride in deionized water was prepared to crosslink Alg/Lap post bioprinting. 365 nm UV light was used to crosslink PEGDA/Lap post bioprinting. 12 μg/ml TG was used to crosslink Gel/Lap post bioprinting.
3D bioprinting assessment of bioinks
All 3D bioprinting experiments for this work were conducted using a Cellink Inkredible+ (Cellink, Sweden) microextrusion 3D bioprinter with pneumatic pressure to extrude materials. Cellink Heartware (V. 2.4.1) and Slic3r (V. 1.2.9) software were used to manually prepare codes for the printing assessment of each bioink. Both printhead speed and different needle gauges were used to alter the diameter of printed fibers and determine a working size range of printable fibers (Figure S2). The printing air pressure was determined for each individual bioink and optimized to match the printing speed so that circular fibers would be deposited in the scaffold architectures. Gel/Lap was printed with a heated aluminum print cartridge (Cellink) at 50°C. Printed scaffolds and cross-sections were imaged on a Zeiss Axio Observer 5 microscope (Zeiss, Germany) and the cross-section circularity of fibers was determined by analysis with ImageJ (V. 1.6.4) software (NIH) using the following formula:
| (Equation 1) |
The cross-section circularity was used as a measure of print fidelity to show the retention of circular shape after being deposited on the substrate.
Rheometer and VeTBiM measurements with bulk bioinks
Viscoelasticity measurements of the bulk bioinks were conducted with both a rheometer and VeTBiM to validate the accuracy of the VeTBiM measurements. All experiments were conducted using bulk, uncrosslinked bioinks at room temperature. An ElastoSensTM Bio2 made by Rheolution, Inc. (Montreal, QC, Canada) was used for the VeTBiM measurements as it is the only instrument available which uses VeTBiM technology. Samples were prepared by adding 2 mL of bioink to the sample cups quickly after the degassing step during preparation. The sample cup has grooves on its walls to fix the sample to the cup and a silicone membrane for the bottom of the cup to flex with the sample under vibratory conditions (Henni et al., 2010). The rheometer measurements were conducted on an Anton Paar MCR 502 rheometer (Graz, Austria) using 25 mm sandblasted parallel plates with 1 Hz frequency and 0.5% strain. The storage and loss moduli were measured with both methods for comparison. Bioink recovery testing was conducted at 1 Hz by alternating the application of 100% strain and 1% strain for 5-minute intervals on bulk uncrosslinked bioinks.
Using VeTBiM for characterizing 3D bioprinted scaffolds
VeTBiM measurements of 3D bioprinted scaffolds had to fit within a set of assumptions that the instrument makes about the sample. This set of assumptions is associated with the theory behind how the instrument works, which is outlined in the patent for the technology (Henni and Schmitt, 2019). Assumptions that the instrument makes include the following: (1) there is no movement of the sample at the walls of the sample cup; (2) the sample is in contact with the silicone membrane on the bottom of the cup; (3) the sample is in the shape of a disk and; (4) the sample has a density of ∼1 g/ml (which is the case for hydrogels).
Considering the theory behind how the instrument works and the assumptions that are made, the instrument is fit for analysing the overall mechanical properties of 3D bioprinted scaffolds with varying architectural designs provided that the sample in the cup meets all the above-mentioned assumptions.
3D bioprinted scaffold preparation for VeTBiM
In order to print inside of the VeTBiM sample cups, 1″ long stainless-steel dispensing needles were used. For consistent calibration and centring the 3D bioprinter needle in the sample cup, a fixture was designed in SolidWorks 2017 software (Dassault Systems, France) to hold the sample cup in place while printing. The fixture 3D model file was sliced into G-code and 3D printed using Ultimaker Cura (V. 3.4.1) software and an Ultimaker 2 3D printer, respectively (Ultimaker, Netherlands) (see Figure S1). The fixture was made of poly-lactic acid (PLA). Once the sample cup was fixed on the bioprinter sample stage, the printing needle’s coordinate system was calibrated to be centred in the middle of the sample cup with the elastic membrane surface as the Z = 0 coordinate. After calibration, the sample was printed in the sample cup, printing took anywhere from 5–10 minutes depending on the sample being printed. For each layer, the scaffold pattern was printed followed by a border to attach the printed layer of scaffold to the grooved wall of the sample cup (see Figure 2). The scaffold patterns were manually generated using Slic3r’s selection of infill patterns and infill density settings with the appropriate fiber diameter and layer height for all generated G-codes. Scaffolds were either filled in with crosslinking solution or with deionized water after crosslinking. All 3D bioprinted scaffold dimensions were a 22 mm diameter circular cylinder with 5 mm height.
Cell culture
Human Neonatal Dermal Fibroblasts (HNDFs), immortalized human keratinocytes (HaCaTs), and U87 human glioblastoma were cultured in 75cm2 flasks. High glucose DMEM supplemented with 10%v/v fetal bovine serum (FBS), 1%v/v penicillin/streptomycin was used as the cell media for all cell types and was changed every other day. The flasks were stored at 37°C with 95% humidity and 7.5% CO2.
Bioink cell attachment study
Sterile bioink components were prepared by exposing dry gelatin and Laponite to 365 nm UV light for 15 minutes in a closed petri dish and sterile filtering deionized water and solutions of alginate, calcium chloride, and transglutaminase. The components were used to prepare sterile bioinks in a biosafety cabinet and the bioinks were cast into 12 well plates quickly after degassing to form thin sheets. The sheets were kept at 4°C overnight to allow Laponite hydration. 30 minutes before cell seeding, the bioink sheets were treated to 2 ml of culture media and heated in the incubator to 37°C. 250,000 cells per well were seeded and grown for 2 days before analysing. The bioink sheets were rinsed with DPBS to remove dead and unattached cells and Presto Blue Cell Viability reagent (ThermoFisher) was used according to the manufacturer’s protocol to compare cell to bioink adherence between the three bioinks.
Enzymatic degradation of bulk and 3D bioprinted scaffolds
Gel/Lap and TG were used to conduct this study. Bulk samples were prepared as previously described, and 3D bioprinted rectilinear scaffolds were prepared with 1 mm fiber diameter (18G needle) and the 20% infill density setting. The bulk and 3D bioprinted scaffolds were crosslinked with transglutaminase for 30 minutes. Both conditions were measured before treating with 200 μg/ml collagenase solution to achieve fast degradation and then measured every 2 hours for up to 6 hours. All measurements were conducted at 37°C. Degradation by weight was determined using the following formula:
| (Equation 2) |
Effect of fiber diameter and spacing on viscoelastic properties in a rectilinear scaffold
Alg/Lap and 2% CaCl2 were used to bioprint scaffolds for this study. Rectilinear scaffold designs were prepared with three different spacings between printed fibers: 1, 1.5, and 2 mm. The G-codes for bioprinting these scaffolds were manually prepared using Cellink Heartware software so that spacing could be customized. Three different needle gauges, 18, 22, and 25, with inner diameters of 0.84, 0.41, and 0.26 mm, respectively, were used to generate fiber diameters of ∼500, ∼700, and ∼900 μm. Scaffolds were filled in with 2% CaCl2 and allowed to crosslink for 2 hours at room temperature. Scaffolds were imaged on an Axio Observer 5 microscope (Zeiss, Germany). The bioink was mixed with fluorescent microparticles for imaging the cross sections of the scaffolds. Bulk material samples were prepared as previously described. Scaffold mechanical properties were measured at 37°C using VeTBiM with the method previously described.
Effect of 3D bioprinting and fiber diameter on ionic crosslinking time of 3D bioprinting rectilinear scaffolds
3D bioprinted Alg/Lap scaffolds with ∼500 and ∼900 μm fiber size and 1.5 mm spacing were prepared in sample cups as previously stated. For the 3D bioprinted scaffolds, 2% CaCl2 was used to fill in the scaffolds and the samples were immediately placed in the ElastoSensTM Bio2 for real-time assessment of crosslinking over 2 hours. As a control, deionized water was used to fill in the scaffolds. Three drops of silicone oil (Sigma Aldrich, St. Louis, Missouri) were added to the top of the scaffolds to prevent evaporation of the crosslinking solution during the experiment. For the bulk samples, crosslinking solution or deionized water was added on top of the bioink sheets and removed for measurement at 0, 1, and 2 hours. The measured G′ of samples was normalized to the initial value before crosslinking for simple comparison on the same graph. To exemplify the effect of diffusion of Ca2+ ions into the bulk material and 3D bioprinted scaffolds, samples were prepared and immersed in a fluorescent solution of 10μgml−1 rhodamine B (Sigma Aldrich, St. Louis, Missouri). Diffusion into the scaffolds and bulk material was imaged over 2 hours on a fluorescent microscope and the fluorescence intensity in the centre of 3D bioprinted fibers and bulk material were plotted against time. The fluorescence intensity was subsequently plotted against the storage modulus to exemplify the correlation between the diffusion and crosslinking rate.
Effect of scaffold infill density on swelling rate of a honeycomb scaffold
PEGDA/Lap honeycomb scaffolds were prepared at 20, 40, and 60% infill density as previously described. Bulk PEGDA/Lap was also prepared in a 5 mm thick sheet. Samples were sectioned into smaller pieces and freeze-dried. To assess the swelling rate of samples, samples were weighed at their dry weight and then immersed in 0.1 M phosphate buffered saline (Sigma Aldrich, St. Louis, Missouri). Samples were weighed swollen at predetermined time points over 2 hours. Then a final swelling percent was measured at 6 hours. The swelling was calculated using the following formula:
| (Equation 3) |
Effect of bioink formulation on viscoelastic properties of 3D bioprinted honeycomb scaffolds
2.5, 5, and 10% PEGDA/Lap were printed in a 40% infill density honeycomb pattern scaffold and crosslinked for 5 minutes under UV. Samples were filled in with deionized water and analyzed in using VeTBiM at 37°C.
Effect of printing pattern on the viscoelastic properties of 3D bioprinted scaffolds
PEGDA/Lap scaffolds were bioprinted in three different patterns while keeping the infill density and bioink formulation constant at 20% and 5% PEGDA/Lap, respectively. The patterns, rectilinear, honeycomb, and random line, were selected from the patterns available in Slic3r software. Samples were crosslinked for 5 minutes under UV light before VeTBiM measurement at 37°C.
Effect of bioprinting multiple materials on viscoelastic properties of a rectilinear scaffold
PEGDA/Lap and Alg/Lap were prepared and loaded into the two separate printheads on the Cellink Inkredible+ bioprinter. The G-code for printing the scaffold was prepared by manually editing the code for a rectilinear scaffold of one bioink in Cellink Heartware software. The code was post-processed in the software to make it compatible with the Inkredible+ bioprinter. Alternating layers of each bioink were printed with the dual printhead system. The scaffold struts of each bioink were oriented perpendicularly. The PEGDA/Lap bioink was crosslinked first by exposing the scaffold to 5 minutes of UV light, and the Alg/Lap bioink was crosslinked second by filling in the scaffold with 2% CaCl2 and crosslinking for 2 hours. PEGDA/Lap and Alg/Lap scaffolds with the same scaffold pattern and infill density were prepared and crosslinked with the same conditions for comparison. The samples were then measured with VeTBiM at 37°C. The two bioinks were mixed with different coloured fluorescent microbeads and imaged using a Zeiss Axio Observer 5 fluorescent microscope to visualize the scaffold structure.
Effect of encapsulated cell growth on viscoelastic properties of a rectilinear scaffold
A biocompatible and commercially available bioink comprised of cellulose and alginate, Cellink Bioink (SKU: IK1020000303), was used to investigate the effects of cell encapsulation on the mechanical properties of bioprinted scaffolds. The cell-laden bioink was prepared by resuspending U87 cells in 500 ul of media and combining this cell suspension with 5 mL of the Cellink Bioink to produce a final cell density of 2 × 106 cells/ml. A rectilinear pattern with an infill density of 40% and height of 3.5 mm was bioprinted from an 18G needle using the cell-laden bioink. To provide a baseline of the scaffold’s viscoelastic properties without the influence of cells, 5 ml of the Cellink Bioink was diluted in 500 ul of cell media that did not contain cells and bioprinted in the same geometry. The viscoelastic properties of both the cellular and acellular scaffolds were measured using VeTBiM with the method previously described at days 0, 1, 3, 5, and 7 post-printing.
Evaluation of cell viability and proliferation in bioprinted scaffolds
A live/dead viability kit (Invitrogen, L3224) was used on days 1 and 7 to evaluate the viability of the bioprinting process and cell survival within the scaffold during culture. Briefly, 0.5 μl of the stock calcien AM solution and 2 μl of the stock ethidium homodimer-1 solution were added to 1 ml of sterile PBS. The cell-laden scaffolds were incubated in this solution for 30 minutes at room temperature, washed with PBS, then imaged with a fluorescent microscope.
To visualize cell morphologies within the cell-laden scaffolds, samples were fixed with 10% NBF for 1 hour. These scaffolds were washed three times with PBS and were then permeabilized with 0.3% Triton-X100 in PBS for 15 minutes. A stock solution of Alexa Fluor™ 488 Phalloidin prepared in methanol as per the manufacturers instructions was diluted 1:40 in PBS containing 0.3% Triton-X100 and 1% BSA. Scaffolds were incubated in the phalloidin solution at room temperature while protected from light for 1 hour. Nuclei were counterstained with 5 μg/ml DAPI for 15 minutes and then the scaffolds were washed with PBS and imaged with a fluorescent microscope.
To evaluate cell proliferation within the Cellink Bioink, a long single fiber was bioprinted, then cut into equal 1 cm long segments which were cultured individually in a 24 well plate. At days 1, 4, and 7, a Prestoblue assay (Invitrogen, A13262) was conducted to quantify cellular metabolic activity. The Prestoblue reagent was diluted with media by a ratio of 1:10 and 250 μl of this solution was added to each well containing a segment of cell-laden bioprinted fiber. Following a 30-minute incubation at 37°C, 100 μl from each well was transferred to a 96 well plate and read with a microplate reader.
Quantification and statistical analysis
All experiments were conducted in triplicate and the data were expressed as the mean ±standard deviation. Statistical analysis was conducted using two-way ANOVA on the appropriate experimental groups. P values <0.05 were deemed as statistically different and p values <0.01 were deemed extremely significant.
Acknowledgments
All authors would like to acknowledge the support they received from the Natural Sciences and Engineering Research Council of Canada (NSERC-RGPIN-2016-04024), Michael Smith Foundation for Health Research (MSFHR-18743), the Canadian Foundation for Innovation (CFI-35570), B.C. Knowledge Development Fund (BCKDF), and Rheolution Instruments.
Author contributions
Conceptualization – M. Akbari and B.G.; Methodology – M. Akbari and B.G; Investigation – B.G., E.S., S.S.G., M. Amereh, and Z.M.; Writing – Original Draft, B.G., E.S. and E.P.; Writing, Review & Editing – M. Akbari, B.G., S.A., and E.S.; Funding Acquisition – M.A.; Resources – M. Akbari, B.G., E.S., S.S.G., M. Amereh, Z.M. and E.L.S.; Supervision – M. Akbari.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104251.
Contributor Information
Brent Godau, Email: brentgodau@gmail.com.
Mohsen Akbari, Email: makbari@uvic.ca.
Supplemental information
Data and code availability
Original data reported in this paper will be shared by the lead contact upon request. All original code has been deposited at Zenodo and is publicly available as of the date of publication. The repository DOI is: https://doi.org/10.5281/zenodo.5759866. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akalp U., Bryant S.J., Vernerey F.J. Tuning tissue growth with scaffold degradation in enzyme-sensitive hydrogels: a mathematical model. Soft Matter. 2016;12:7505–7520. doi: 10.1039/c6sm00583g. Royal Society of Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashammakhi N., Ahadian S., Xu C., Montazerian H., Ko H., Nasiri R., Barros N., Khademhosseini A. Bioinks and bioprinting technologies to make heterogeneous and biomimetic tissue constructs. Mater. Today Bio. 2019;1:100008. doi: 10.1016/j.mtbio.2019.100008. Elsevier BV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au P.I., Hassan S., Liu J., Leong Y.K. Behaviour of laponite gels: Rheology, ageing, pH effect and phase state in the presence of dispersant. Chem. Eng. Res. Des. 2015;101:65–73. doi: 10.1016/j.cherd.2015.07.023. Institution of Chemical Engineers. [DOI] [Google Scholar]
- Avery R.K., Albadawi H., Akbari M., Zhang Y.S., Duggan M.J., Sahani D.V., Olsen B.D., Khademhosseini A., Oklu R. An injectable shear-thinning biomaterial for endovascular embolization. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aah5533. [DOI] [PubMed] [Google Scholar]
- Bojin F., Robu A., Bejenariu M.I., Ordodi V., Olteanu E., Cean A., Popescu R., Neagu M., Gavriliuc O., et al. 3D bioprinting of model tissues that mimic the tumor microenvironment. Micromachines. 2021;12:535. doi: 10.3390/MI12050535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi C., Strandman S., Hui E., Montagnon E., Schmitt C., Hadj Henni A., Lerouge S. Validation and application of a nondestructive and contactless method for rheological evaluation of biomaterials. J. Biomed. Mater. Res. B Appl. Biomater. 2017;105:2565–2573. doi: 10.1002/jbm.b.33797. [DOI] [PubMed] [Google Scholar]
- Chen W., Shao Y., Li X., Zhao G., Fu J. Nanotopographical surfaces for stem cell fate control: engineering mechanobiology from the bottom. Nano Today. 2014;9:759–784. doi: 10.1016/j.nantod.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimene D., Lennox K.K., Kaunas R.R., Gaharwar A.K. Advanced bioinks for 3D printing: a materials science perspective. Ann. Biomed. Eng. 2016;44:2090–2102. doi: 10.1007/s10439-016-1638-y. [DOI] [PubMed] [Google Scholar]
- Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science (New York, N.Y.) 2009;324:1673–1677. doi: 10.1126/science.1171643. American Association for the Advancement of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElastoSens Bio I Rheolution - Rheolution Inc 2021. https://www.rheolution.com/elastosens-bio2.html
- Engelmayr G.C., Jr., Cheng M., Bettinger C.J., Borenstein J.T., Langer R., Freed L.E. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat. Mater. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaharwar A.K., Avery R.K., Assmann A., Paul A., McKinley G.H., Khademhosseini A., Olsen B.D. Shear-thinning nanocomposite hydrogels for the treatment of hemorrhage. ACS Nano. Am. Chem. Soc. 2014;8:9833–9842. doi: 10.1021/nn503719n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Yang Q., Zhao X., Jin G., Ma Y., Xu F. 4D bioprinting for biomedical applications. Trends Biotechnol. 2016;34:746–756. doi: 10.1016/J.TIBTECH.2016.03.004. Elsevier Current Trends. [DOI] [PubMed] [Google Scholar]
- George J., Kuboki Y., Miyata T. Differentiation of mesenchymal stem cells into osteoblasts on honeycomb collagen scaffolds. Biotechnol. Bioeng. 2006;95:404–411. doi: 10.1002/bit.20939. John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
- Hadisi Z., Walsh T., Hossein Dabiri S.M., Seyfoori A., Godeau B., Charest G., Fortin D., Akbari M. 3D printing for the future of medicine. J. 3D Printing Med. 2020 doi: 10.2217/3dp-2019-0010. Future Medicine Ltd London, UK. [DOI] [Google Scholar]
- Henni A.H., Schmitt C. 2019. System and Method for Measurements of Viscoelastic Parameters in Soft Materials. U.S. Patent288.541. [Google Scholar]
- Henni A.H., Schmitt C., Cloutier G. Shear wave induced resonance elastography of soft heterogeneous media. J. Biomech. 2010;43:1488–1493. doi: 10.1016/j.jbiomech.2010.01.045. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- Hong S., Sycks D., Chan H.F., Lin S., Lopez G.P., Guilak F., Leong K.W., Zhao X. 3D printing of highly stretchable and tough hydrogels into complex, cellularized structures. Adv. Mater. 2015;27:4035–4040. doi: 10.1002/adma.201501099. John Wiley & Sons, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Wu D., Huang L., Jiao Y., Li L., Lu L., Zhou C. 3D bioprinting of cell-laden scaffolds for intervertebral disc regeneration. Mater. Lett. 2018;223:219–222. doi: 10.1016/J.MATLET.2018.03.204. North-Holland. [DOI] [Google Scholar]
- Jahanshahi M., Hamdi D., Godau B., Samiei E., Sanchez-Lafuente C., Neale K., Hadisi Z., Dabiri S., Pagan E., et al. An engineered infected epidermis model for in vitro study of the skin’s pro-inflammatory response. Micromachines 2020. 2020;11:227. doi: 10.3390/MI11020227. Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakus A.E., Rutz A.L., Shah R.N. Advancing the field of 3D biomaterial printing. Biomed. Mater. 2016;11:014102. doi: 10.1088/1748-6041/11/1/014102. IOP Publishing. [DOI] [PubMed] [Google Scholar]
- Jin Y., Liu C., Chai W., Compaan A., Huang Y. Self-supporting nanoclay as internal scaffold material for direct printing of soft hydrogel composite structures in air. ACS Appl. Mater. Inter. 2017;9:17456–17465. doi: 10.1021/acsami.7b03613. [DOI] [PubMed] [Google Scholar]
- Kelly C.N., Miller A.T., Hollister S.J., Guldberg R.E., Gall K. Design and structure–function characterization of 3D printed synthetic porous biomaterials for tissue engineering. Adv. Healthc. Mater. 2018;7:1–16. doi: 10.1002/adhm.201701095. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A., Langer R. A decade of progress in tissue engineering. Nat. Protoc. 2016;11:1775–1781. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- Kloxin A.M., Kloxin C.J., Bowman C.N., Anseth K.S. Mechanical properties of cellularly responsive hydrogels and their experimental determination. Adv. Mater. 2010;22:3484–3494. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky D.B., Homan K.A., Skylar-Scott M.A., Lewis J.A. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. 2016;113:3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.P., De Wijn J.R., Van Blitterswijk C.A. Porous Ti6Al4V scaffolds directly fabricated by 3D fibre deposition technique: effect of nozzle diameter. J. Mater. Sci. Mater. Med. 2005;16:1159–1163. doi: 10.1007/s10856-005-4723-6. Kluwer Academic Publishers. [DOI] [PubMed] [Google Scholar]
- Lu C., Zhao M., Jie L., Wang J., Gao Y., Cui X., Chen P. Stress distribution on composite honeycomb sandwich structure suffered from bending load. Proced. Eng. 2015;99:405–412. doi: 10.1016/J.PROENG.2014.12.554. Elsevier. [DOI] [Google Scholar]
- Markstedt K., Mantas A., Tournier I., Martinez Avila H., Hagg D., Gatenholm P. 3D bioprinting human chondrocytes with nanocellulose–alginate bioink for cartilage tissue engineering applications. Biomacromolecules. 2015;16:1489–1496. doi: 10.1021/ACS.BIOMAC.5B00188. Am. Chem. Soc. [DOI] [PubMed] [Google Scholar]
- Martínez Ávila H., Schwarz S., Rotter N., Gatenholm P. 3D bioprinting of human chondrocyte-laden nanocellulose hydrogels for patient-specific auricular cartilage regeneration. Bioprinting. 2016;1–2:22–35. doi: 10.1016/j.bprint.2016.08.003. Elsevier. [DOI] [Google Scholar]
- McCormack A., Highley C.B., Leslie N.R., Melchels F.P. 3D printing in suspension baths: keeping the promises of bioprinting afloat. Trends Biotechnol. 2020;38:584–593. doi: 10.1016/j.tibtech.2019.12.020. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- Murphy S.V., Atala A. 3D bioprinting of tissues and organs. Nature Biotechnol. 2014;32:773–785. doi: 10.1038/nbt.2958. Nat. Publishing Group. [DOI] [PubMed] [Google Scholar]
- O’Brien F.J. Biomaterials & scaffolds for tissue engineering. Mater.Today. 2011;14:88–95. doi: 10.1016/S1369-7021(11)70058-X. Elsevier. [DOI] [Google Scholar]
- Oyen M.L. Mechanical characterisation of hydrogel materials. Int. Mater. Rev. 2013;59:44–59. doi: 10.1179/1743280413y.0000000022. [DOI] [Google Scholar]
- Pääkko M., Ankerfors M., Kosonen H., Nykanen A., Ahola S., Osterberg M., Ruokolainen J., Laine J., Larsson P.T., Ikkala O., et al. Enzymatic hydrolysis combined with mechanical shearing and high-pressure homogenization for nanoscale cellulose fibrils and strong gels. Biomacromolecules. 2007;8:1934–1941. doi: 10.1021/BM061215P. [DOI] [PubMed] [Google Scholar]
- Panwar A., Tan L.P. Current status of bioinks for micro-extrusion-based 3D bioprinting. Molecules. 2016;21:685. doi: 10.3390/molecules21060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peak C.W., Stein J., Gold K.A., Gaharwar A.K. Nanoengineered colloidal inks for 3D bioprinting. Langmuir. 2018;34:917–925. doi: 10.1021/acs.langmuir.7b02540. [DOI] [PubMed] [Google Scholar]
- Pedde R.D., Mirani B., Navaei A., Styan T., Wong S., Mehrali M., Thakur A., Mohtaram N.K., Bayati A., Dolatshahi-Pirouz A., et al. Emerging biofabrication strategies for engineering complex tissue constructs. Adv. Mater. 2017;29:1–27. doi: 10.1002/adma.201606061. [DOI] [PubMed] [Google Scholar]
- Roeder R.K. Mechanical characterization of biomaterials. Character Biomater. 2013:49–104. doi: 10.1016/B978-0-12-415800-9.00003-6. Academic Press. [DOI] [Google Scholar]
- Roomi M.W., Kalinovsky T., Rath M., Niedzwiecki A. Modulation of MMP-2 and MMP-9 secretion by cytokines, inducers and inhibitors in human glioblastoma T-98G cells. Oncol. Rep. 2017;37:1907–1913. doi: 10.3892/or.2017.5391. Spandidos Publications. [DOI] [PubMed] [Google Scholar]
- Rutz A.L., Hyland K.E., Jakus A.E., Burghardt W.R., Shah R.N. A multimaterial bioink method for 3D printing tunable, cell-compatible hydrogels. Adv. Mater. 2015;27:1607–1614. doi: 10.1002/adma.201405076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar-Scott M.A., Uzel S.G.M., Nam L.L., Ahrens J.H., Truby R.L., Damaraju S., Lewis J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Science Advances. Am. Assoc. Adv. Sci. 2019;5 doi: 10.1126/SCIADV.AAW2459/SUPPL_FILE/AAW2459_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufivand A.A., Abolfathi N., Hashemi S.A., Lee S.J. Prediction of mechanical behavior of 3D bioprinted tissue-engineered scaffolds using finite element method (FEM) analysis. Addit. Manuf. 2020;33:101181. doi: 10.1016/j.addma.2020.101181. Elsevier B.V. [DOI] [Google Scholar]
- Tanaka M., Nishikawa K., Okubo H., Kamachi H., Kawai T., Matsushita M., Todo S., Shimomura M. Control of hepatocyte adhesion and function on self-organized honeycomb-patterned polymer film. Colloids Surf. A: Physicochem. Eng. Aspects. 2006;284–285:464–469. doi: 10.1016/J.COLSURFA.2005.11.098. Elsevier. [DOI] [Google Scholar]
- Tandara A.A., Mustoe T.A. MMP- and TIMP-secretion by human cutaneous keratinocytes and fibroblasts - impact of coculture and hydration. J. Plast.Reconstr. Aesthet. Surg. 2011;64:108–116. doi: 10.1016/j.bjps.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining K.H., Mooney D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Q., Xu C. Nanocellulose-based inks for 3D bioprinting: Key aspects in research development and challenging perspectives in applications—a mini review. Bioengineering. 2020;7:40. doi: 10.3390/BIOENGINEERING7020040. Multidisciplinary Digital Publishing Institute. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., Xiao H., Pan Y., Wang L. Microrheology, advances in methods and insights. Adv. Colloid Interf. Sci. 2018;257:71–85. doi: 10.1016/j.cis.2018.04.008. Elsevier B.V. [DOI] [PubMed] [Google Scholar]
- Yang Q., Gao B., Xu F. Recent advances in 4D bioprinting. biotechnology Journal. 2020;15:1900086. doi: 10.1002/BIOT.201900086. John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
- Zadpoor A.A. Mechanics of additively manufactured biomaterials. J. Mech. Behav. Biomed.Mater. 2017;70:1–6. doi: 10.1016/j.jmbbm.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Zhao X., Lang Q., Yildirimer L., Lin Z.Y., Cui W., Annabi N., Ng K.W., Dokmeci M.R., Ghaemmaghami A.M., Khademhosseini A. Photocrosslinkable gelatin hydrogel for epidermal tissue engineering. Adv. Healthc. Mater. 2016;5:108–118. doi: 10.1002/adhm.201500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Stukel J., AlNiemi A., Willits R.K. Novel microgel-based scaffolds to study the effect of degradability on human dermal fibroblasts. Biomed. Mater. 2018;13:055007. doi: 10.1088/1748-605X/aaca57. Inst. Phys. Publishing. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data reported in this paper will be shared by the lead contact upon request. All original code has been deposited at Zenodo and is publicly available as of the date of publication. The repository DOI is: https://doi.org/10.5281/zenodo.5759866. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.