Abstract
Pederin is a potent polyketide toxin that causes severe skin lesions in humans after contact with insects of genus Paederus. Due to its potent anticancer activities, pederin family compounds have raised the interest of pharmaceutical industry. Despite the extensive studies on the cluster of biosynthetic genes responsible for the production of pederin, it has not yet been possible to isolate and cultivate its bacterial endosymbiont producer. However, the marine bacterium Labrenzia sp. PHM005 was recently reported to produce labrenzin, the closest pederin analog. By cloning a synthetic pedO gene encoding one of the three O-methyltraferase of the pederin cluster into Labrenzia sp. PHM005 we have been able to produce pederin for the first time by fermentation in the new recombinant strain.
Highlights
-
•
Pederin is produced for the first time by fermentation in the marine bacterium Labrenzia sp. PHM005.
-
•
Methyltransferase PedO was heterologously expressed in Labrenzia sp. PHM005.
-
•
PedO is most likely responsible of the C18–OH methylation of labrenzin.
-
•
The new methylated analog was detected by HPLC-MS from the culture extracts.
1. Introduction
Pederin, a natural polyketide, has a huge therapeutic potential as a highly potent anticancer agent (Richter et al., 1997). However up to now the only source of this compound was the insect Paederus fuscipes, where it was found in very low amounts since twenty-five million field-collected insects had to be used to isolate the minimum amount of pure pederin (Pavan and Bo, 1952) to determine its chemical structure (Cardani et al., 1965). Pederin is produced by one of the first trans-AT mixed type polyketide/non-ribosomal peptide synthases (PKS/NRPS) assigned to a natural product that has been identified in an as-yet uncultivated bacterial symbiont of the insect (Piel, 2002; Piel et al., 2004; Piel et al., 2004a, Piel et al., 2004b, Piel et al., 2004c) and thus, its production by bacterial fermentation has not been achieved yet.
Pederin has among other structural peculiarities an O-methyl instead of the conventional ester or carboxylic acid polyketide terminus (Helfrich and Piel, 2010, 2016). In this sense, the cytotoxic activity of pederin-type compounds can be markedly increased by modifying the methylation pattern. The putative pederin biosynthetic gene cluster (ped) encodes three proteins with similarity to O-methyltransferases (MTs): PedA, PedE, and PedO (Piel, 2002; Piel et al., 2004; Piel et al., 2004). Biochemical in vitro experiments conducted with mycalamide as substrate, a pederin family compound with free C18–OH group, demonstrated that PedO was capable of introducing a methyl group at C18 position (Zimmermann et al., 2009).
Recently, the complete genome of the strain Labrenzia sp. PHM005, a free-living and cultivable producer of a pederin analog 18-O-demethyl pederin (Schleissner et al., 2017; Benítez et al., 2021), hereinafter labrenzin, has been sequenced (Kačar et al., 2019). A gene cluster responsible for the synthesis of labrenzin, named lab cluster, has been identified encoding a trans-AT mixed type PKS/NRPS biosynthetic pathway (Kačar et al., 2019). Interestingly, the lab cluster only encodes two MTs and this observation suggested that Labrenzia sp. PHM005 could be a good chassis to biosynthetically produce pederin by engineering the missing MT tailoring gene of ped cluster.
The aim of this work was to demonstrate that pederin could be produced by fermentation in Labrenzia sp. PHM005 by the heterologous expression of a pedO synthetic gene. In addition, further improvements of the pederin production have been investigated by overexpressing other MTs from the lab cluster.
2. Materials and methods
2.1. Bacterial strains, media and growth conditions
Plasmids and strains are shown in Table S1. Standard overnight Escherichia coli MFDpir (Ferrières et al., 2010) cultures were grown aerobically in Luria-Bertani (LB) broth or LB agar at 37 °C (Bertani, 1951). The medium was supplemented with 1 mM diamine-pymelic acid (DAP) and the corresponding antibiotic, when appropriate. Labrenzia sp. PHM005 wild type and recombinant strains were grown in Marine Broth (MB) Difco 2216 (Sigma-Aldrich) or Marine Agar (MA) Difco 2216 (Sigma-Aldrich), supplemented with antibiotics, when appropriate. All the strains were cultured in 50 mL falcon tubes or 100 mL flasks with 10 and 20 mL of medium, respectively. Culture medium used to study labrenzin and pederin production in Labrenzia sp. PHM005 was modified using marine basal medium supplemented with vitamins (MBM + vit) (Kačar et al., 2019). The culture medium was supplemented with 0.2 mM 3-methyl-benzoate, when the cloned gene was expressed under the control of the inducible Pm promoter. The strains were grown overnight in falcon tubes in MB at 30 °C with shaking at 200 rpm. The overnight culture was washed in 0.85% NaCl solution and diluted to an optical density (OD600) ≈ 0.1 in 20 mL of fresh medium. To determine the production of labrenzin, pederin and analogs by HPLC/MS analyses, the strains were cultured for 72 h.
2.2. Plasmid DNA transformation and clone selection
E. coli MFDpir electro-competent cells were prepared and transformed by electroporation as described (Wirth et al., 1989). To select the transformants, chloramphenicol (34 μg/mL) and 1 mM DAP was added to the LB agar plates. A biparental conjugation was used for transformation of Labrenzia sp. PHM005 using E. coli MFDpir carrying the plasmid of interest as a donor strain. Samples of 1 mL of overnight cultures of Labrenzia and E. coli MFDpir were collected by centrifugation and the pellets were washed with 500 μL of 0.85% NaCl and resuspended into 200 μL of the same solution. Samples of 50 μL of each strain were mixed and deposited into a 0.22 μL filter mating disc placed on the surface of a MA agar plate that was further incubated for 4–6 h at 37 °C. The cells deposited on the filter disc were collected with 1 mL of 0.85% NaCl and vortexed thoroughly to detach the cells from the filter. Afterwards, the appropriate dilutions of cells were plated on MA plates containing chloramphenicol (5 μg/mL).
2.3. Construction of plasmids for expression of MTs
Plasmid pSEVA338 from pSEVA collection (http://seva-plasmids.com/) was used as vector to create an artificial operon containing three genes encoding the Lab6 and Lab16 MTs from Labrenzia and PedO MT from Paederus symbiont. The artificial operon design and sequences are indicated in the supplementary information (Fig. S3), where each gene with the corresponding RBS was flanked by specific restriction sites (blunt cut) in a way that individual genes or the combination of genes could be easily generated by digestion and re-ligation. The operon was synthetized and cloned using SacI and SpeI restriction sites by GenScript yielding plasmid pSEVA338_MTs. Other plasmids derived from pSEVA338_MTs were pSEVA338_lab6, pSEVA338_lab16, pSEVA338_pedO, pSEVA338_lab6_lab16, pSEVA338_pedO_lab6 and pSEVA338_pedO_lab16. The inducible promoter Pm from pSEVA338 was replaced by the strong P14g promoter in plasmid pSEVA227M_P14g (kind gift from Gonzalo Durante) using restriction enzymes PacI and AvrII generating a new construct pSEVA338_P14g_ pedO_lab6.
2.4. Extraction, purification and identification of polyketide compounds
Upon the fermentation of 20 mL of culture medium collected by centrifugation was frozen at −80 °C and subsequently freeze dried. The lyophilized product was then dissolved in 4 mL of distilled water and equal volume of ethyl acetate. After homogenization and centrifugation, the organic phase was collected by pipetting and the extraction was repeated once more. The collected organic phase was dried by vacuum centrifugation and the pellet was dissolved in 150 μL of methanol and filtered for a HPLC/MS analysis.
HPLC/MS analysis was carried out using a HPLC/MS system and a separation column previously described (Kačar et al., 2019). The reversed phase separation was performed on a C18 column (ZORBAX Eclipse plus C18, 5 μm, 4.6 × 250 mm, Agilent Technologies, Santa Clara, CA, United States) and MSionization source was electrospray ionization (ESI). The running method was as follows: solvent A was 100% water and solvent B was 100% acetonitrile. The flow rate was 500 μL min−1 using the following gradient: t = 2 min, 100% A; t = 8 min, 95% A; t = 40 min, 55% A; t = 53 min, 0% A; t = 55 min, 0% A; t = 57 min, 100% A; t = 65 min, 100% A.
2.5. Bioinformatic analyses
Clustal Omega service at https://www.ebi.ac.uk/Tools/msa/clustalo/was used to perform multiple protein alignments. Basic Local Alignment Search Tool (Blast) service at https://blast.ncbi.nlm.nih.gov/Blast.cgi was used to compare protein sequences.
3. Results
3.1. Genetic analyses
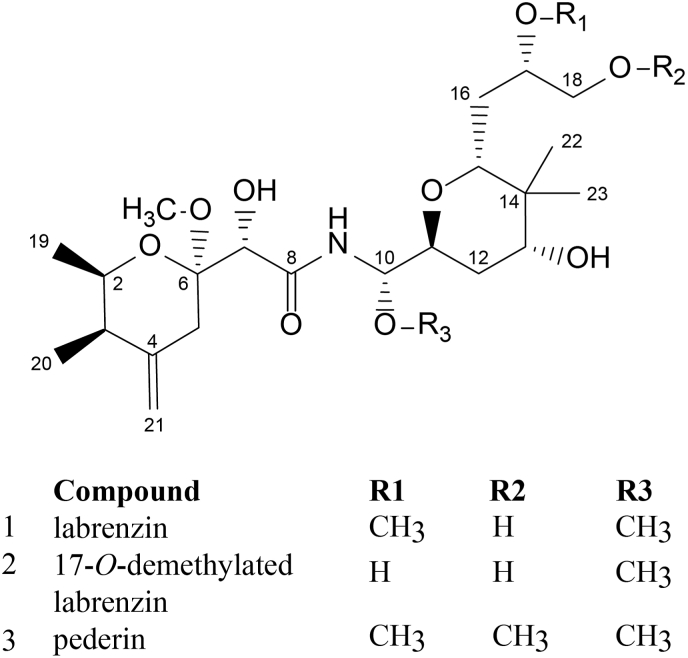
The unique structural difference between pederin and labrenzin is the absence of an O-methylation of C18–OH in labrenzin (Fig. 1). The absence of such methylation can be justified assuming that the lab cluster contains only two MTs, i.e., Lab6 (MT6) and Lab16 (MT16) (Kačar et al., 2019), whereas ped cluster contains three MTs, i.e., PedA, PedE and PedO (Fig. S1). A protein homology comparison between MTs from lab and ped clusters showed that Lab16 is homologous to PedE with a 51% amino acid sequence identity and Lab6 shares 47% sequence identity with PedA. According to Blast analysis the sequence amino acid identity of PedO with Lab16 was very low (28% with a query cover of 63%) but Lab6 and PedO showed a 53% sequence identity. Clustal alignments have also shown that Lab16 is more similar to PedE than to PedO, PedA and Lab6, that showed a high similarity between them (Fig. S2). Only small differences in the length of the three proteins and in their N-terminal regions suggest that PedA and Lab6 could be more closely related. Although the genetic analysis cannot exclude Lab6 from the experimental setting, the finding that pedO is located far from pedA and pedE in the ped cluster together with the observation that in vitro biochemical analysis carried out with PedO using mycalamide A as substrate have demonstrated that PedO is responsible for the methylation of the C18–OH group in pederin (Zimmermann et al., 2009), strongly suggested that PedO could be the missing gene in Labrenzia responsible of the C18–OH methylation in the bacterial symbiont. Nevertheless, considering that PedO methylation activity was only tested in vitro using mycalamide as substrate the hypothesis required an experimental in vivo demonstration. However, these in vivo experiments might not work as expected, as PedO might not methylate labrenzin because precursor concentrations and conditions within Labrenzia cells may be very different from those used for in vitro methylation. Furthermore, different labrenzin intermediates could compete as putative substrates or even function as inhibitors, and although PedO was actively produced in Escherichia coli after optimization of its expression (Zimmermann et al., 2009), the gene and/or enzyme could require some additional optimization. to activate in Labrenzia.
Fig. 1.
Chemical structure of pederin family polyketides.
3.2. Production of pederin in Labrenzia sp. PHM005
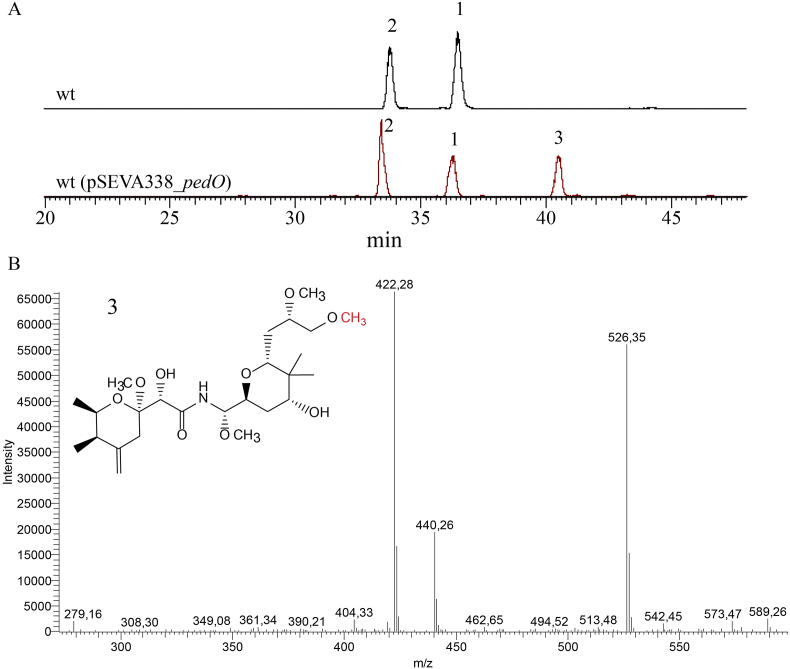
To test our hypothesis, this is, to be able to synthetize pederin for the first time in a cultivable bacterium, a synthetic pedO gene was heterologously expressed in Labrenzia sp. PHM005 previously transformed with the recombinant plasmid pSEVA338_pedO harbouring the pedO synthetic gene under the control of the Pm inducible promoter. As predicted, when a culture extract of Labrenzia sp. PHM005 (pSEVA338_pedO) was analysed by HPLC/MS we observed a new intermediate, more hydrophobic than the previous pederin analogs, i.e., compound (1) (labrenzin) and compound (2) (17-O-demethylated labrenzin) produced by the wild type strain (Figure 2). The MS spectrum revealed a new peak (compound 3) observed at RT = 40.4 min and with ion fragmentation m/z = 526; 440; 422. This fragmentation pattern correlates with both labrenzin and 17-O-demethylated labrenzin (Fig. S4) and indicates an additional methyl group (M+14), as it is in pederin. The scale-up production of pederin was performed in PharmaMar under the industrial fermentation conditions (data not shown) with the strain Labrenzia sp. PHM005 (pSEVA338_pedO) and the new compound (3) with the additional methyl group was also detected (Fig. S5). However, it is worth to mention that under these culture conditions the amount of compound (3) relative to labrenzin was lower than under our flask experiments (Fig. S5A) and insufficient for the proper isolation and structure elucidation by NMR. Nevertheless, the MS analysis allows us to discard that new methylation might have occurred at positions 13-OH or 7-OH. The MS analysis shows that the fragmentation pattern of compound (3) did not change comparing with the compounds (1) and (2) (Fig. S3). In compound (3) all ions contain one additional methyl group (+14). If we take a closer look, compound (1) leaves a “footprint” of the ions 408, 426 and 512, compound (2) shows the ions 394 (408–14), 412 (426–14) and 498 (512–14), while compound (3) marks the ions 422 (408 + 14), 440 (426 + 14) and 526 (512 + 14). This is possible because both methylations occur close to each other at 17-OH and 18-OH. If the additional methylation were performed at 13-OH or 7-OH, the expected fragmentation of compound (3) would be entirely different. Taken together these observations and the in vitro experiments carried out by Zimmermann et al. (2009) we might conclude that compound (3) is more probably pederin.
Fig. 2.
Pederin biosynthesis in Labrenzia sp. PHM005 (pSEVA338_pedO). A) HPLC-MS chromatograms of the supernatant extracts obtained after 72 h of cultivation of Labrenzia sp. PHM005 cultures in MBM + vit medium presenting extracted ions in the range m/z = 498–526. Ion masses include sodium adduct. Intermediates compound (1) (labrenzin), compound (2) (17-O-demethylated labrenzin) and compound (3) (pederin) are indicated. B) ESIMS ion fragmentation of compound (3) (pederin).
3.3. Co-overexpression of lab6, lab16 and PedO methylases
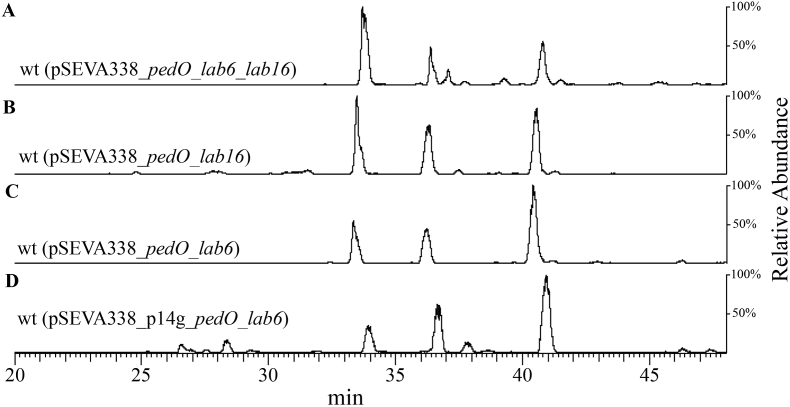
Although pederin was produced in Labrenzia sp. PHM005 (pSEVA338_pedO), it was not the most abundant intermediate in the extract. The amount of labrenzin extracted from the cultures of Labrenzia sp. PHM005 has been estimated to be around 0.5 mg/L (Schleissner et al., 2017). In flasks under our culture conditions we have determined that the wild type strain produces about 5–10 times less labrenzin than under the previous bioreactor conditions and about the same amount of 17-O-demethylated labrenzin, i.e., about 0.08 mg/L each. The amounts of 17-O-demethylated labrenzin, labrenzin, and pederin produced by the new recombinant strain expressing PedO in the same flask conditions are about 0.07 mg/L, 0.04 mg/L, and 0.04 mg/L, respectively. Apparently, labrezin has been partially transformed into pederin by PedO activity. Assuming that the total amount of core polyketide produced by Labrenzia sp. PHM005 will be constant we have trayed to shift the synthesis from 17-O-demethylated labrenzin to pederin. In this sense, we analysed the production in different strains co-expressing the pedO gene in different combinations with lab6 and lab16. To this aim, plasmids pSEVA338_ pedO_lab6, pSEVA338_pedO_lab16, pSEVA338_pedO_lab6_lab16 were constructed and transformed in Labrenzia sp. PHM005. The resulting strains were cultivated in the MBM + vit medium and the production of the labrenzin analogs was analysed. Fig. 3 (A-D) shows HPLC/MS chromatograms of culture extracts of strains harnessing different expression combinations of MTs using pSEVA338 plasmid. Although the relative abundance of the peaks might slightly differ depending of the culture conditions, it appears that the overexpression of pedO and lab6 increases the amount of compound (3) (0.8 mg/L) when compared to compounds (1) (0.6 mg/L) and (2) (0.2 mg/L). Moreover, the large reduction of compound (2) strongly suggests that Lab6 is responsible of the transformation of compound (2) into compound (1) (labrenzin) (paper in preparation).
Fig. 3.
HPLC-MS chromatograms of the culture extracts obtained after 72 h of cultivation of wt recombinant strains transformed with different plasmids in MBM + vit medium. Extracted ions range is m/z = 498–526. Ion masses include sodium adduct. Intermediates 1 (m/z = 512), 2 (m/z = 498) and 3 (m/z = 526) are indicated. A) Labrenzia sp. PHM005 (pSEVA338_lab6_lab16_pedO); B) Labrenzia sp. PHM005 (pSEVA338_lab16_pedO); C) Labrenzia sp. PHM005 (pSEVA338_lab6_pedO); D) Labrenzia sp. PHM005 (pSEVA338_P14g_lab6_pedO).
3.4. Engineering the promoter driven the expression of the lab6_pedO MTs
The further increase the expression of pedO and lab6 genes we replaced the inducible Pm promoter in plasmid pSEVA338_pedO _lab6 with the constitutive strong P14g promoter. Previously, we have tested the P14g promoter strength in Labrenzia sp. PHM005 by constructing a transcriptional fusion with GFP showing that it is functional in Labrenzia and has high expression levels (Figs. S6 and S7). The resulting plasmid named pSEVA338_P14g_pedO_lab6 was transformed in Labrenzia sp. PHM005 and the production of the recombinant strain was analysed. The promoter switch did not seem to alter the peak ratio and increase the production of pederin as observed in the MS chromatograms (Fig. 3D), suggesting that the other bottlenecks of the process should be considered. In this sense, taking into account that the polarity of the intermediates is different, their concomitant excretion to the culture medium as they are produced in the cytoplasm can be a factor that can influence the proportion of more methylated intermediates.
4. Discussion
O-Methylation modulates the pharmacokinetic and pharmacodynamic properties of natural products, affecting their bioavailability, stability, and binding to targets. Tailoring the polyketide structures allows an additional level of functional complexity, and thus, polyketide pathway engineering has generated new-to-nature products through novel glycosylation, acyltransferase, hydroxylation, epoxidation, alkylation, transamination and desaturation reactions acting on naturally occurring products (Cummings et al., 2014). However, as far as we know, very few in vivo experiments have been carried out to expand a polyketide pathway to generate novel polyketides by cloning tailoring O-MTs (Struck et al., 2012; Wang et al., 2020). Fu et al. (1996) expressed the TcmO O-MT of the tetracenomycin biosynthetic pathway of Streptomyces glaucescens in Streptomyces coelicolor CH999 together with the actinorhodin polyketide synthase (PKS) gene cluster, which is responsible for the biosynthesis of 3,8-dihydroxy-methylanthraquinone carboxylic acid (DMAC) and its decarboxylated analog, aloesaponarin. The resulting recombinant strain produced approximately equal quantities of aloesaponarin and a new product but no DMAC. More recently, Wang et al. (2019) have studied the use of two fungal MTs to produce unnatural O-methylated benzenediol lactone polyketides. Other studies have shown that O-methyltransferases can be used to obtain new compounds by modifying the glycosyl moieties of polyketides (Patallo et al., 2001; Han et al., 2018).
In this work, we have confirmed the role of PedO MT found in Paederus bacterial symbiont. In addition, we have demonstrated that its heterologous expression in recombinant strains of Labrenzia sp. PHM005 has allowed expanding the labrenzin biosynthetic pathway generating a new compound, pederin, providing labrenzin with an additional methylation on C18–OH. As mentioned above, pederin can be only isolated from beetle extraction so far, and thus, this is the first time pederin is produced by direct fermentation in a cultivable bacterium. Therefore, pederin could be now produced by fermentation at large scale to be tested and used as an antitumoral drug. Nevertheless, although we have developed different pedO expression systems for the production of pederin further improvements should be made for its efficient industrial scale production since the production levels of labrenzin and pederin are still low. In addition to increasing production of the labrenzin polyketide scaffold, we have shown that a fine-tuning of the expression of lab6 and pedO genes should be optimized to increase the pederin production. It is possible that sequential O-methylation tailoring would depend on the substrate preference of MTs resulting the synthesis of the intermediates more favorable than that of pederin. A similar finding was observed when PedO was unable to methylate the C18–OH group of mycalamide A intermediate when the neighboring –OH was methylated, i.e., C18 O-methylation appeared only possible when the C17–OH was non methylated (Zimmermann et al., 2009). On the other hand, the synthetic pedO gene used in this study originates from an uncultured symbiont bacterium (Piel et al., 2005), therefore, its expression might not be very efficient in Labrenzia sp. PHM005. In that sense, for more efficient pederin transformation from labrenzin in the industrial setting, we suggest to carry out codon usage modifications or to approach a directed evolution of PedO enzyme. Finally, it is important to consider that the production of a combination of different sequentially methylated intermediates, all of them secreted to the surrounding medium, can provide an advantage for the producing bacteria, since different products can develop different roles in their surrounding environment. Thus, the simultaneous production and secretion of the intermediates can be detrimental for the industrial overproduction of pederin, since we have determined that labrenzin once secreted to the medium cannot be uptake by the cells (data not shown) and therefore, it cannot be methylated.
Author statement
FC and JG conceived and designed the study. DK performed the experiments and analysed the data. BG, CS and CC participated in the experiment design. LC developed the HPLC-MS method and prepared the standard. PR performed cultivation experiments. DK, BG and JG wrote the manuscript. BG and CS contributed to preparing the final version of the manuscript. All authors have read and approved the final manuscript.
Declaration of compeing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The present study was funded by the Ministry of Economy and Competitiveness of Spain under the program Retos/Colaboración with the project number RTC-2016-4892-1 (DESPOL). The authors would like to thank Ana Valencia for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2022.e00198.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Benítez X., González E.G., García J., Zúñiga P., de la Calle F., Cuevas C. Detection of a pederin-like compound using a dilution-to-extinction-based platform for the isolation of marine bacteria in drug discovery strategies. Microb. Biotechnol. 2021;14:241–250. doi: 10.1111/1751-7915.13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardani C., Ghiringhelli D., Mondelli R., Quilico A. The structure of pederin. Tetrahedron Lett. 1965;6:2537–2545. [Google Scholar]
- Cummings M., Breitling R., Takano E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol. Lett. 2014;351:116–125. doi: 10.1111/1574-6968.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrières L., Hémery G., Nham T., Guérout A.M., Mazel D., Beloin C., Ghigo J.M. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 2010;192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Alvarez M.A., Khosla C., Bailey J.E. Engineered biosynthesis of novel polyketides: regiospecific methylation of an unnatural substrate by the tcmO O-methyltransferase. Biochemistry. 1996;35:6527–6532. doi: 10.1021/bi952957y. [DOI] [PubMed] [Google Scholar]
- Han J.W., Ng B.G., Sohng J.K., Yoon Y.J., Choi G.J., Kim B.S. Functional characterization of O-methyltransferases used to catalyse site-specific methylation in the post-tailoring steps of pradimicin biosynthesis. J. Appl. Microbiol. 2018;124:144–154. doi: 10.1111/jam.13619. [DOI] [PubMed] [Google Scholar]
- Helfrich E.J.N., Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- Helfrich E.J.N., Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016;33:231–316. doi: 10.1039/c5np00125k. [DOI] [PubMed] [Google Scholar]
- Kačar D., Schleissner C., Cañedo L.M., Rodríguez P., Calle F. De, Galán B., García J.L. Genome of Labrenzia sp. PHM005 reveals a complete and active trans-AT PKS gene cluster for the biosynthesis of labrenzin. Front. Microbiol. 2019;10:1–14. doi: 10.3389/fmicb.2019.02561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patallo E.P., Blanco G., Fischer C., Brana A.F., Rohr J., Méndez C., Salas J.A. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus. Characterization of three methyltransferase genes. J. Biol. Chem. 2001;276:18765–18774. doi: 10.1074/jbc.M101225200. [DOI] [PubMed] [Google Scholar]
- Pavan M., Bo G. Ricerche sulla differenziabilita, natura e attivita del principio tossico di Paederus fuscipes Curt.(Col. Staph.) Mem. Soc. Ent It. 1952:67–82. [Google Scholar]
- Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J., Butzke D., Fusetani N., Hui D., Platzer M., Wen G., Matsunaga S. Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the Pederin family. J. Nat. Prod. 2005;68:472–479. doi: 10.1021/np049612d. [DOI] [PubMed] [Google Scholar]
- Piel J., Höfer I., Hui D. vol. 186. 2004. pp. 1280–1286. (Evidence for a Symbiosis Island Involved in Horizontal Acquisition of Pederin Biosynthetic Capabilities by the Bacterial Symbiont of Paederus Fuscipes Beetles). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J., Hui D., Fusetani N., Matsunaga S. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ. Microbiol. 2004;6:921–927. doi: 10.1111/j.1462-2920.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- Piel J., Wen G., Platzer M., Hui D. Unprecedented diversity of catalytic domains in the first four modules of the putative pederin polyketide synthase. Chembiochem. 2004;5:93–98. doi: 10.1002/cbic.200300782. [DOI] [PubMed] [Google Scholar]
- Richter A., Kocienski P., Raubo P., Davies D. The in vitro biological activities of synthetic 18-O-methyl mycalamide B, 10-epi-18-O-methyl mycalamide B and pederin. Anti Cancer Drug Des. 1997;12(11):217–227. [PubMed] [Google Scholar]
- Schleissner C., Cañedo L.M., Rodríguez P., Crespo C., Zúñiga P., Peñalver A., de la Calle F., Cuevas C. Bacterial production of a pederin analogue by a free-living marine Alphaproteobacterium. J. Nat. Prod. 2017;80:2170–2173. doi: 10.1021/acs.jnatprod.7b00408. [DOI] [PubMed] [Google Scholar]
- Struck A.W., Thompson M.L., Wong L.S., Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem. 2012;13:2642. doi: 10.1002/cbic.201200556. 1655. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang R., Chen X., Sun X., Yan Y., Shen X., Yuan Q. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb. Cell Factories. 2020;19:110. doi: 10.1186/s12934-020-01367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang C., Duan L., Zhang L., Liu H., Xu Y.M., Liu Q., Mao T., Zhang W., Chen M., Lin M., Gunatilaka A.A.L., Xu Y., Molnár I. Rational reprogramming of O-methylation regioselectivity for combinatorial biosynthetic tailoring of benzenediol lactone scaffolds. J. Am. Chem. Soc. 2019;141:4355–4364. doi: 10.1021/jacs.8b12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., Friesenegger A., Fiedler S. Transformation of various species of gram-negative bacteria belonging to 11 different genera by electroporation. MGG Mol. Gen. Genet. 1989;216:175–177. doi: 10.1007/BF00332248. [DOI] [PubMed] [Google Scholar]
- Zimmermann K., Engeser M., Blunt J.W., Munro M.H.G., Piel J. Pederin-type pathways of uncultivated bacterial symbionts: analysis of O-methyltransferases and generation of a biosynthetic hybrid. J. Am. Chem. Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.